Abstract
Pantherine felids (‘big cats’) include the largest living cats, apex predators in their respective ecosystems. They are also the earliest diverging living cat lineage, and thus are important for understanding the evolution of all subsequent felid groups. Although the oldest pantherine fossils occur in Africa, molecular phylogenies point to Asia as their region of origin. This paradox cannot be reconciled using current knowledge, mainly because early big cat fossils are exceedingly rare and fragmentary. Here, we report the discovery of a fossil pantherine from the Tibetan Himalaya, with an age of Late Miocene–Early Pliocene, replacing African records as the oldest pantherine. A ‘total evidence’ phylogenetic analysis of pantherines indicates that the new cat is closely related to the snow leopard and exhibits intermediate characteristics on the evolutionary line to the largest cats. Historical biogeographic models provide robust support for the Asian origin of pantherines. The combined analyses indicate that 75% of the divergence events in the pantherine lineage extended back to the Miocene, up to 7 Myr earlier than previously estimated. The deeper evolutionary origin of big cats revealed by the new fossils and analyses indicate a close association between Tibetan Plateau uplift and diversification of the earliest living cats.
Keywords: first appearance, Himalaya, Pantherinae, Felidae, Miocene, Asia
1. Introduction
At top of the food chain, living big cats, or the pantherines (including clouded leopard, Sunda clouded leopard, snow leopard, tiger, jaguar, leopard and lion), are apex predators in each of the continents/regions where they reside. Despite their important roles in modern ecosystems and the critically endangered status of several species, our knowledge of their evolutionary history is extremely poor, and based almost exclusively on molecular phylogenies. Accumulation of new fossil records is painstakingly slow, and when such records do become available, they often contradict relationships suggested by molecular phylogenies that indicate long ghost lineages or long ‘fuses’ leading to recent explosive diversification [1,2]. The elusive fossil records, in turn, cause much confusion regarding the geographical centre of origins for the big cats and their subsequent intercontinental emigrations.
The great uncertainty in understanding the context of pantherine evolution is partly exacerbated by exclusion of fossil taxa in phylogenetic reconstruction [1,2]. In particular, a 40–50% difference in divergence times of big cat species can be created by an approximately 3.8 Myr old, phylogenetically uncertain fossil calibration for the origin of Panthera, based on fragmentary remains in Africa [2–4]. There is potential for fossil pantherines to inform phylogenetic and divergence time analyses, but a combined approach has never been attempted. Furthermore, putative early Panthera fossils from Pliocene deposits of Asia are either too fragmentary to be useful in phylogenetic analyses [5], or were collected from uncertain localities and thus have no reliable age estimates associated with them [6,7]. All these issues compounded to the existing approximately 4 m.y. time gap between adequately described Panthera species being restricted exclusively to the interval from the Pleistocene to Present on the one hand, and estimated molecular divergence time of pantherines in the Miocene on the other hand. This current state of knowledge has not changed with increased molecular sampling [1,2], thus the resolution of incongruent results hinges on improvement in the pantherine fossil record.
In this report, we describe new fossils of a pantherine cat from the Zanda Basin of the northwestern Himalaya Range, extending the pantherine record back by 2 Myr. We perform a phylogenetic analysis of four fossil and six living pantherine species using the first combined morphological and molecular dataset for this group. Our analysis includes DNA sequences previously used in molecular studies of felid evolution [1,2], plus ancient mitochondrial DNA (mtDNA) previously published for extinct ‘cave lions’ Panthera spelaea and ‘American lions’ Panthera atrox [8]. Time-calibrated trees of 10 pantherines and two out-group feline species are generated from Bayesian and parsimony analyses of the combined dataset, treating fossil pantherine species as terminal taxa. A new set of divergence time estimates is produced based on this complete dataset, and character transitions mapped. We then conduct a historical biogeographic analysis using a maximum-likelihood framework to reconstruct the likely ancestral range of the hypothetical pantherine ancestor, with consideration of multiple scenarios of origination.
2. Systematic palaeontology
The new fossil pantherine is described as follows: Carnivora Bowdich, 1821; Felidae Fischer, 1817; Pantherinae Pocock, 1917; Panthera Oken, 1816. Panthera blytheae sp. nov. Holotype. A partial cranium preserving the first left incisor, canines, and the third and fourth premolars, representing a full adult individual, Institute of Vertebrate Paleontology and Paleoanthropology (IVPP), Beijing, China, specimen number V18788.1 (figure 1; electronic supplementary material, S3 and table S1); discovered by J.L. on 7 August 2010 and excavated by a field team under the direction of G.T.T. Referred material. IVPP V18788.2, ramus fragment (IVPP locality ZD1001). IVPP V18788.3, partial premaxilla–maxilla fragment with left canine (IVPP locality ZD1001). IVPP V18789.1–3, isolated fourth premolar, partial maxillary and dentary fragments (IVPP locality ZD1208). IVPP V18790, partial right dentary with third and fourth premolars and the first molar (IVPP locality ZD1223) (see electronic supplementary material, figures S3 and S4). Etymology. The specific name, blytheae, is made in honour of the daughter of avid supporters of the Natural History Museum of Los Angeles County, Paul and Heather Haaga. Type locality and horizon. IVPP locality ZD1001 (31°39'58″ N, 79°44'57″ E, elevation 4114 m) is adjacent to the Zanda Canyon trail approximately 15 km north of the Zanda county seat, Ngari District, Tibet Autonomous Region, China [9] (see electronic supplementary material, figures S1 and S2). IVPP V18788.1–3 were collected from a small bone bed in the middle part of the Zanda Formation, within a lens of greenish, coarse-grained sandstone (see the electronic supplementary material for details). Age. The type locality IVPP ZD1001 is stratigraphically correlated to chron C3n.1r with an estimated age of 4.42 Ma [9]. The stratigraphic range of occurrence of the species based on all available material is from 5.95 Ma (IVPP locality ZD1223, correlated to chron C3r) to 4.10 Ma (IVPP locality ZD1208, correlated to chron C2Ar), or from the end of the Late Miocene to the Early Pliocene [9] (figure 3).
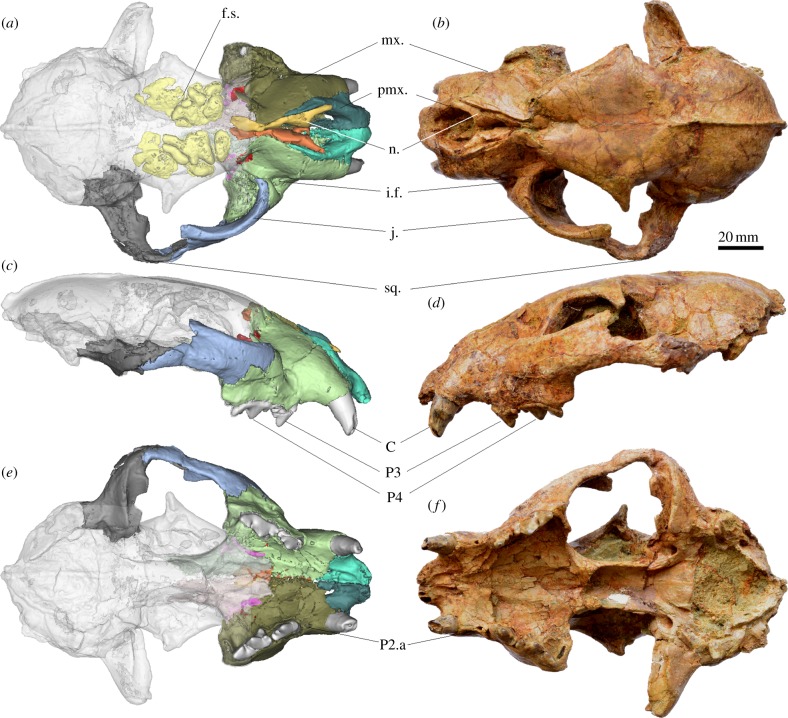
Figure 1.
Holotype cranium of P. blytheae, IVPP V18788.1. (a) Three-dimensional reconstruction of cranium, dorsal view. (b) Cranium dorsal view. (c) Three-dimensional reconstruction of cranium, left lateral view. (d) Cranium left lateral view. (e) Three-dimensional reconstruction of cranium, ventral view. (f) Cranium ventral view. f.s., frontal sinus; mx., maxilla; pmx., premaxilla; n., nasal; i.f., infraorbital foramen; j., jugal; sq., squamosal; C, upper canine; P3, upper third premolar; P4, upper fourth premolar (carnassial), P2.a, alveolus of upper second premolar. (Online version in colour.)
Figure 3.
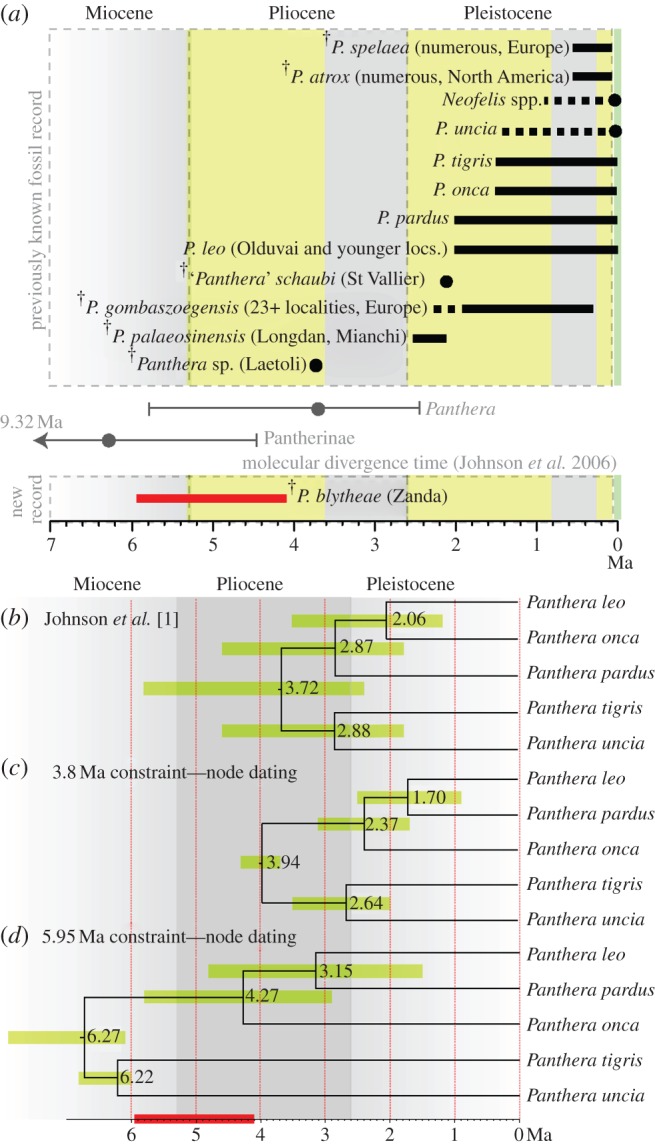
Pantherine fossil record and fossil calibration points. (a) Stratigraphic ranges of previously known fossil record of Pantherinae (Panthera + Neofelis), compared with the age of P. blytheae and previous divergence time estimates based on molecular data and nodal calibration points. Geologic age data taken from [4,5,8,9,13–16]. (b) Divergence time estimates for extant Panthera based on node dating using Bayesian relaxed-clock analysis of nDNA sequences. Estimates from Johnson et al. [1]. (c) Estimates using 3.8 Ma as the minimum age of Panthera. (d) Estimates using 5.95 Ma, lower age limit of P. blytheae, as the minimum age of Panthera. Shaded bars indicate 95% HPD, node values are means. Dark bar on the timescale indicates the stratigraphic range of P. blytheae. (Online version in colour.)
3. Diagnosis
Panthera blytheae possesses characteristics shared with other species of the genus, including frontoparietal suture located at the postorbital constriction, the absence of an anterior bulge overhanging the infraorbital canal, truncated and tapered dorsal maxilla, tip of parasagittal crest perpendicular to the sagittal crest, and an angular connection between the maxillary flange and the palatal bone at the posterior edge of the palate [10,11] (see electronic supplementary material). Panthera blytheae shares with the snow leopard Panthera uncia in having an almost round canine cross section, a weakly inclined mandibular symphysis, a smooth transition between mandibular ramus and symphysis, the presence of fronto-nasal depression, narrow distance between anterior edge of bulla and glenoid ridge, a sharp-turning ventral premaxilla–maxilla border at the canines, and straight and symmetrical p4 cusp alignment (see electronic supplementary material, figure S3 and S4). Panthera blytheae is unique in possessing a small labial cusp on the posterior cingulum of the upper third premolars, and the presence of converging ridges on the labial surface of the upper fourth premolars (see electronic supplementary material for additional discussion).
4. Description and comparison
The holotype specimen is diagenetically compressed in the dorsoventral direction; therefore, the morphology of each bone in the face was reconstructed using high-resolution X-ray computer tomography (figure 1 and electronic supplementary material). The cranium of P. blytheae is around the size of a clouded leopard (Neofelis nebulosa), about 10% smaller than the living snow leopard (figure 2). The digital reconstruction shows a well-developed frontal sinus area in the posterior postorbital region, a shared feature of the living pantherines [10,11]. The incisors and canines are heavily worn, and in comparison the premolars are sharp and unworn. The presence of highly reduced second upper premolars and relatively large first upper molars is evident from their alveoli, and their relative sizes are more similar to the condition in the clouded leopard than to other pantherines. Lower premolars are similar in size to those in small felines such as the ocelot, but the lower first molar is relatively enlarged. The width of the muzzle relative to the rest of the cranium is intermediate between the narrow morphology seen in the clouded leopard, and the widened appearance present in all other pantherines. Mandible depth, P3 parastyle and lingual expansion, and frontal sinus expansion in P. blytheae are more similar to the large extant pantherines than to the snow leopard. Undulating Hunter–Schreger Band enamel microstructure is present throughout the cheek teeth of P. blytheae.
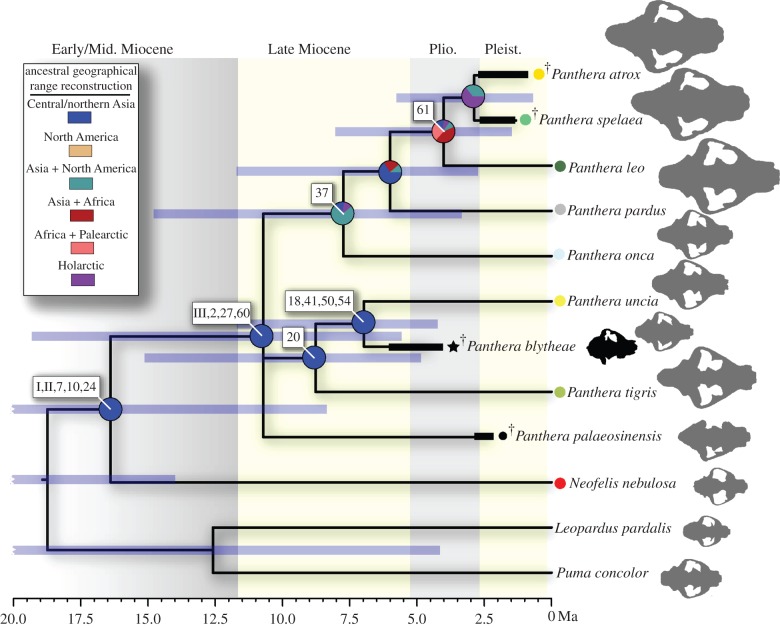
Figure 2.
Time-calibrated phylogeny of the Pantherinae based on a partial scaffold of the combined dataset, using an IGR relaxed-clock Bayesian analysis. 95% HPD intervals are shown in shaded bars; ranges older than 20.0 Ma are trimmed for sake of illustration (see table 1 for full range of data). Reconstructed ancestral geographical ranges using DEC model M3 (Central Asian pantherine ancestor; see electronic supplementary material, table S8) are shown as pie graphs over ancestral nodes, indicating probabilities of single versus more inclusive regions of ancestral distribution. Modern and historical geographical ranges and fossil localities are indicated for terminal nodes as in figure 4. Stratigraphic ranges of fossil species used in the analyses are indicated by thick black bars. White boxes indicate genetic and morphological character transitions: I. SRY3(+4), II. Numt insert 2, III. SRY3(–7), based on [1]; other numbers refer to morphological characters, see data matrix in MorphoBank. Panthera atrox, ‘American lion’; P. spelaea, ‘cave lion’; Panthera leo, lion; Panthera pardus, leopard; Panthera onca, jaguar; Panthera uncia, snow leopard; Neofelis nebulosa, clouded leopard; Leopardus pardalis, ocelot; Puma concolor, cougar. †extinct species. (Online version in colour.)
5. Phylogeny
We conducted a series of phylogenetic analyses based on a combined dataset of 81 soft tissue, behavioural and skeletal morphological characters and nearly 43 000 base pairs (bp) of nDNA and mtDNA sequences (see electronic supplementary material, tables S2 and S3; matrix available in MorphoBank). Six living pantherine species (excluding Neofelis diardi), four fossil pantherine species and two living feline out-group species were included in our analyses. Parsimony analysis of the combined dataset returned four most parsimonious trees (MPT). The strict consensus MPT is largely consistent with previous results based only on the molecular dataset [1,2] and shows two major clades within Pantherinae: at the base are the clouded leopard N. nebulosa and the fossil Panthera palaeosinensis, this is followed by a clade that includes the living tiger P. tigris and snow leopard P. uncia, plus P. blytheae. The final clade includes the living jaguar, leopard, lion and two fossil species P. atrox and P. spelaea (see electronic supplementary material, figure S5). The monophyly of Pantherinae received high support in all parsimony analyses of data partitions (Bremer support = 4+, jackknife support = 89–100%, bootstrap support = 85–100%; see electronic supplementary material), but within this clade only the grouping of P. uncia and P. blytheae received robust support (Bremer = 2–4, jackknife = 61–85%, bootstrap = 58–87%). Relationships among other groups of pantherines received moderate to low support in the parsimony analyses, but the topology among extant species are recovered with high support in Bayesian analyses of molecular and morphological data in both combined and partitioned analyses [1,2,10] (see electronic supplementary material, table S4).
6. Divergence times
We used the topology of living pantherines recovered in both parsimony and Bayesian analyses as a partial scaffold in Bayesian estimations of divergence times based on the combined dataset, using all four fossil species as terminal nodes. Different relaxed molecular clock models (IGR, CPP, TK02) were tested [12] (see electronic supplementary material, table S5). The resulting divergence time estimates are consistently older than previous estimates done using molecular datasets with fossil ages as calibrated nodal constraints [1,2] (figures 2 and 3; table 1; see electronic supplementary material, S5 and S6). In our estimates, a maximum of six out of eight pantherine divergence events occurred within the Miocene, with the two remaining events (i) divergence of the African lion and fossil ‘lions’ and (ii) divergence between the Eurasian and American ‘lions’, both occurring in the Pliocene (figures 2 and 3; electronic supplementary material, S6). The 95% highest posterior density (HPD) is wide and overlapping for all divergence time estimates, indicating high levels of uncertainty tha is at least partially caused by the rapid diversification of modern felids during Mio-Pliocene times [1] (table 1).
Table 1.
Divergence time estimates for nodes in the pantherine clade. Mean age and 95% HPD were calculated from Bayesian total evidence dating analysis of the strict consensus MPT with fossils as terminal species using the IGR relaxed-clock model. Support values were calculated from the strict consensus MPT in TNT and PAUP* using 1000 replicates for Bootstrap (bs) and Jackknife (jk) resampling. For results based on other relaxed-clock models, see electronic supplementary material.
| clade | mean age (Ma) | 95% HPD age (Ma) | Bremer | bs | jk |
|---|---|---|---|---|---|
| Pantherinae | 16.40 | 8.37–27.68 | 6 | 94 | 94 |
| Panthera | 10.72 | 5.57–19.33 | — | — | — |
| P. tigris–uncia–blytheae | 8.78 | 4.86–15.13 | 1 | 72 | 56 |
| P. uncia–blytheae | 6.97 | 4.23–11.69 | 4 | 81 | 82 |
| P. onca–pardus–leo–atrox–spelaea | 7.74 | 3.34–14.79 | 1 | — | 57 |
| P. pardus–leo–atrox–spelaea | 6.00 | 2.73–11.70 | 1 | — | — |
| P. leo–atrox–spelaea | 4.01 | 1.48–8.04 | 1 | — | — |
| P. atrox–spelaea | 2.87 | 0.69–5.76 | 1 | — | — |
7. Historical biogeography
We used the time-calibrated tree from the IGR relaxed-clock model in maximum-likelihood analyses of historical biogeography under the dispersal–extinction–cladogenesis (DEC) model [17], and compared the results with ancestral range reconstructions obtained from parsimony and Bayesian approaches [18] (see electronic supplementary material, tables S6 and S7). In addition, we tested different models of pantherine ancestral geographical range by limiting the ancestral distribution to only one of seven possible geographical regions (figure 4; see electronic supplementary material, table S8). DEC analyses permitting multiple area reconstructions indicate the mostly likely geographical origin of pantherines is in the Central/northern Asia or the Holarctic region. This ancestral distribution was followed by separate dispersals during the Miocene in Southeast Asia by the clouded leopard and the tiger–snow leopard lineages, respectively [1]. This is then followed by later Miocene dispersal of the lion–leopard–jaguar lineage and then Pliocene dispersal of the fossil ‘lions’. According to the DEC models, the last two dispersals probably occurred from Africa-Palearctic or Holarctic ancestral geographical ranges. Out of all single-area origin models, Central-northern Asia had the highest likelihood of being the ancestral geographical region of pantherines (–ln likelihood = 40.56, compared with 41.01–49.03 for all other models; figure 2 and electronic supplementary material, table S8), corroborating interpretations based solely on molecular data [1].
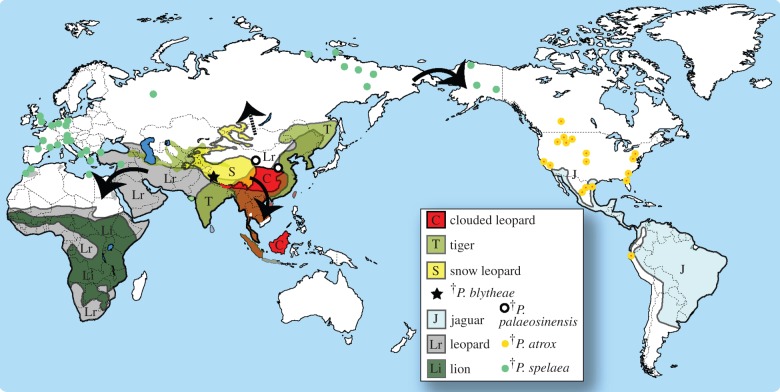
Figure 4.
Biogeographic evolution of big cats. Recent geographical ranges are shaded, and fossil localities indicated by points and star. Recent and historical ranges are based on [19–24], fossil localities were extracted from [8] and downloaded from the Paleobiology Database on 29 April 2013, searched using the group name ‘Panthera leo’. Dark arrows show likely dispersal events suggested by the DEC analysis. (Online version in colour.)
8. Implications
The region within which P. blytheae was found was both a tectonically active and faunally diverse area during the later Cenozoic [25,26]. The presence of both widespread and endemic fossil mammal species points to the southwestern Tibetan Plateau as an important area for understanding biotic connections to the uplift of the Himalaya Range. Unlike previously described fossil mammals from the Zanda Basin [25,26], which represent elements of extinct faunas, the close relationship between P. blytheae and the living snow leopard based on our data indicates the existence of an endemic Central Asian pantherine lineage that is intimately associated with the Himalayan region and the mountains of Central Asia in general (figure 4) [13]. Combined with fossil evidence of horses Equus, pikas Ochotona, foxes Alopex and blue sheep Pseudois from the Zanda Basin, a picture of persistent faunal components that accompanied the last 6 Myr of uplift on the ‘Roof of the World’ is emerging. The ancient predator–prey association between Tibetan pantherines (P. blytheae and then P. uncia), blue sheep, and Tibetan antelopes and a paleoenvironment dominated by open, arid areas accentuated with steep cliffs formed by protruding basement rocks like those observed there today [25,27], both indicate that the present day ecology of cold-adapted snow leopards and their prey has roots that can be traced back for several million years. This evidence indicates that Tibet was not only a ‘training ground’ for Ice Age megaherbivores [26], but perhaps also a refugium for other mammalian lineages that have maintained a faunal association in this part of the world from the Mio-Pliocene to the present.
9. Conclusion
The new pantherine fossils from the Zanda Basin fill in the approximately 4 m.y. time gap between previously known species records and estimated molecular divergence time for pantherines (figure 3). The new specimens permit a robust time-calibrated cladistic analysis of the most comprehensive combined morphological and molecular dataset for this lineage to date (figures 2 and 3). The exact timing of divergence events within the big cat lineage awaits additional data to provide narrower confidence intervals, especially for the early cladogenetic timing in the Middle and Late Miocene (figure 2). However, the evolutionary and biogeographic scenarios offered by concurrent examination of morphological and molecular data, in addition to robust age constraints from our new fossil discovery, partially reconcile traditionally dichotomized paleontological/morphological and molecular frameworks of big cat evolution. The incorporation of historical biogeographic analysis tools with the combined dataset indicates that molecular and fossil evidence are now in agreement regarding the Asian origin of big cats, and further pinpointing a geographical region still occupied today by the earliest branching species in the lineage (figure 4). Evidence of similar environmental conditions and long-term associations between cold-adapted predator and prey species found in the Zanda Basin suggests that future studies of its fossil fauna will provide important data not only on the evolutionary stability of endemic lineages, but also how their eventual dispersal outside the plateau is linked to tectonic, climatic and biotic events. The new fossils also lend support to the prediction that much remains to be found in the geologic record of Central and Southeast Asia. Finally, the ancient age of the big cat lineage and their origin in Central Asia indicate that the dramatic tectonic events on and around the Tibetan Plateau set a backdrop, if not an impetus, to the rise of some of the most dominant predators in today's ecosystems.
Acknowledgements
The authors thank H. Thomas for preparing, and G. Kung for CT scanning, the holotype specimen. S. Goldberg assisted with MorphoBank; W. Johnson provided the concatenated mtDNA sequence file; J. Flynn provided research equipment and space for Z.J.T.; J. Chen provided discussion of historical biogeographic methods; Z. Luo, L. Werdelin and an anonymous reviewer provided constructive comments that improved the manuscript. E. Westig and N. Duncan provided access to the AMNH mammal collection; J.-O. Ebbestad and V. Berg-Madsen provided access to the PMU paleontology collection.
Data accessibility
Phylogenetic data: MorphoBank (http://morphobank.org/permalink/?P898). Final supermatrix assembly uploaded to MorphoBank database (http://morphobank.org/permalink/?P898).
Funding statement
Funding was provided by National Basic Research Program of China (973 program; 2012CB821904) and Chinese Academy of Sciences (XDB03020104); NSF GRF, DDIG and AMNH Frick Post-Doctoral Fellowship to Z.J.T., NSF EAR-0446699, 0444073, 0958704, 1227212 to X.W., NMNH Peter Buck Post-Doctoral Fellowship to G.J.S., and an NGS Waitt Grant (W22-08) to Q.L.
References
- 1.Johnson WE, et al. 2006. The Late Miocene radiation of modern felidae: a genetic assessment. Science 311, 73–77 (doi:10.1126/science.1122277) [DOI] [PubMed] [Google Scholar]
- 2.Davis BW, Li G, Murphy WJ. 2010. Supermatrix and species tree methods resolve phylogenetic relationships within the big cats, Panthera (Carnivora: Felidae). Mol. Phylogenet. Evol. 56, 64–76 (doi:10.1016/j.ympev.2010.01.036) [DOI] [PubMed] [Google Scholar]
- 3.Werdelin L, Yamaguchi N, Johnson WE, O'Brien SJ. 2010. Phylogeny and evolution of cats (Felidae). In Biology and conservation of wild felids (eds Macdonald DW, Loveridge AJ.), pp. 59–82 Oxford, UK: Oxford University Press [Google Scholar]
- 4.Werdelin L, Peigne S. 2010. Chapter 32. Carnivora. In Cenozoic mammals of Africa (eds Werdelin L, Sanders WJ.), pp. 603–657 Berkeley, CA: University of California Press [Google Scholar]
- 5.Qiu Z, Deng T, Wang B. 2004. Early Pleistocene mammalian fauna from Longdan, Dongxiang, Gansu, China. Palaeontol. Sin. C 191, 1–198 [with 139 plates. Chinese with English Summary] [Google Scholar]
- 6.Zdansky O. 1924. Jungtertiäre carnivoren Chinas [Late Tertiary Carnivora of China]. Palaeontol. Sin. C 2, 1–149 (German). [Google Scholar]
- 7.Mazak JH, Christiansen P, Kitchener AC. 2011. Oldest known pantherine skull and evolution of the tiger. PLoS ONE 6, e25483 (doi:10.21371/journal.pone.0025483) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barnett R, et al. 2009. Phylogeography of lions (Panthera leo ssp.) reveals three distinct taxa and a late Pleistocene reduction in genetic diversity. Mol. Ecol. 18, 1668–1677 (doi:10.1111/j.1365-294X.2009.04134) [DOI] [PubMed] [Google Scholar]
- 9.Wang X, et al. 2013. Mio-Pleistocene Zanda Basin biostratigraphy and geochronology, pre-Ice Age fauna, and mammalian evolution in western Himalaya. Palaeogeogr. Palaeoclimatol. Palaeoecol. 374, 81–95 (doi:10.1016/j.palaeo.2013.01.007) [Google Scholar]
- 10.Christiansen P. 2008. Phylogeny of the great cats (Felidae: Pantherinae), and the influence of fossil taxa and missing characters. Cladistics 24, 977–992 (doi:10.1111/j.1096-0031.2008.00226.x) [DOI] [PubMed] [Google Scholar]
- 11.Salles LO. 1992. Felid phylogenetics: extant taxa and skull morphology (Felidae, Aeluroidea). Ame. Mus. Novitates 3047, 1–67 (http://hdl.handle.net/2246/5011) [Google Scholar]
- 12.Ronquist F, Klopfstein S, Vilhelmsen L, Schulmeister S, Murray DL, Rasnitsyn AP. 2012. A total-evidence approach to dating with fossils, applied to the early radiation of the Hymenoptera. Syst. Biol. 61, 973–999 (doi:10.1093/sysbio/sys058) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hemmer H. 1972. Uncia uncia. Mamm. Species 20, 1–5 (doi:10.2307/3503882) [Google Scholar]
- 14.Turner A, Antón M. 1997. The big cats and their fossil relatives, p. 234 New York, NY: Columbia University Press [Google Scholar]
- 15.Burger J, et al. 2004. Molecular phylogeny of the extinct cave lion Panthera leo spelaea. Mol. Phylogenet. Evol. 30, 841–849 (doi:10.1016/j.ympev.2003.07.020) [DOI] [PubMed] [Google Scholar]
- 16.O'Regan HJ. 2002. A phylogenetic and palaeoecological review of the Pleistocene felid Panthera gombaszoegensis. Doctoral dissertation, p. 349 Liverpool John Moores University, Liverpool, UK [Google Scholar]
- 17.Ree RH, Smith SA. 2008. Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Syst. Biol. 57, 4–14 (doi:10.1080/10635150701883881) [DOI] [PubMed] [Google Scholar]
- 18.Yu Y, Harris AJ, He X. 2010. S-DIVA (Statistical Dispersal-Vicariance Analysis): a tool for inferring biogeographic histories. Mol. Phylogenet. Evol. 56, 848–850 (doi:10.1016/j.ympev.2010.04.011) [DOI] [PubMed] [Google Scholar]
- 19.Haas SK, Hayssen V, Krausman PR. 2005. Panthera leo. Mamm. Species 762, 1–11 (doi:10.1644/1545-1410(2005)762[0001:PL]2.0.CO;2) [Google Scholar]
- 20.Uphyrkina O, Johnson WE, Quigley H, Miquelle D, Marker L, Bush M, O'Brien SJ. 2001. Phylogenetics, genome diversity and origin of modern leopard, Panthera pardus. Mol. Ecol. 10, 2617–2633 (doi:10.1046/j.0962-1083.2001.01350.x) [DOI] [PubMed] [Google Scholar]
- 21.Kitchener AC, Beaumont MA, Richardson D. 2006. Geographical variation in the clouded leopard, Neofelis nebulosa, reveals two species. Curr. Biol. 16, 2377–2383 (doi:10.1016/j.cub.2006.10.066) [DOI] [PubMed] [Google Scholar]
- 22.Mazak V. 1981. Panthera tigris. Mammalian Species 152, 1–8 (doi:10.2307/3504004) [Google Scholar]
- 23.McCarthy TM, Chapron G. 2003. Snow Leopard survival strategy, p. 105 Seattle, WA: ISLT and SLN [Google Scholar]
- 24.Seymour KL. 1989. Panthera onca. Mamm. Species 340, 1–9 (doi:10.2307/3504096) [Google Scholar]
- 25.Deng T, Li Q, Tseng ZJ, Takeuchi GT, Wang Y, Xie G, Wang S, Hou S, Wang X. 2012. Locomotive implication of a Pliocene three-toed horse skeleton from Tibet and its paleo-altimetry significance. Proc. Natl Acad. Sci. USA 109, 7374–7378 (doi:10.1073/pnas.1201052109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deng T, et al. 2011. Out of Tibet: Pliocene woolly rhino suggests high-plateau origin of Ice Age megaherbivores. Science 333, 1285–1288 (doi:10.1126/science.1206594) [DOI] [PubMed] [Google Scholar]
- 27.Wang Y, et al. 2013. Diet and environment of a mid-Pliocene fauna from southwestern Himalaya: Paleo-elevation implications. Earth Planet. Sci. Lett. 376, 43–53 (doi:10.1016/j.epsl.2013.06.014) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Phylogenetic data: MorphoBank (http://morphobank.org/permalink/?P898). Final supermatrix assembly uploaded to MorphoBank database (http://morphobank.org/permalink/?P898).