Abstract
Objective(s): Klebsiella infections are caused mainly by K. pneumoniae and K. oxytoca. In the last two decades, a new type of invasive Klebsiella pneumoniae which contains mucoviscosity-associated gene (magA) has emerged. The aim of this study was to investigate the prevalence of magA gene and to detect antimicrobial susceptibility patterns of Klebsiella spp. isolated from clinical samples.
Materials and Methods: Klebsiella isolates were collected from patients admitted to referral hospitals of Hamadan, Iran, during a 12-month period from 2007 to 2008. The samples were analyzed by conventional microbiological methods and polymerase chain reaction (PCR). The hypermucoviscosity (HV) phenotype of Klebsiella isolates was characterized by formation of viscous strings >5 mm as a positive test. The susceptibility of isolates to routine antibiotics was assessed by agar disk diffusion method.
Results: Out of 105 Klebsiella isolates, 96.2% was identified as K. pneumoniae and 3.8% as K. oxytoca by PCR. magA gene was detected in 4 (3.8%) isolates of K. pneumoniae. The isolates of K. oxytoca contained no magA gene. From 4 isolates with positive magA gene, two of them were HV+ and two were HV- phenotype. Overall, sixty-four isolates (60.95%) of K. pneumoniae showed an HV positive phenotype and all isolates of K. oxytoca were HV- phenotype. The most effective antibiotics against the isolates were tobramycin (79.05%), ceftazidime (79.05%), ceftizoxime (78.09%), ciprofloxacin (76.19%), ceftriaxone (76.24%) and amikacin (74.29%).
Conclusion: The results suggest that there is also magA associated serotype of the K. pneumoniae in this region. In addition, the presence of HV+ phenotype may not be associated with magA.
Key Words: Drug resistance, Iran, Klebsiella, Bacterial Proteins
Introduction
Klebsiella infections are caused mainly by K. pneumoniae and K. oxytoca. They are opportunistic bacterial pathogens associated with nosocomial infections such as urinary tract infection (UTI), pneumonia and septicaemia (1, 2). For the first time in 1998, a new type of invasive K. pneumoniae emerged in Taiwan, which was typically presented as a community-acquired primary liver abscess (3-5). Several reports, especially from the Asia Pacific region and the United States, have also shown that this pathogen has become the predominant cause of liver abscess (6-8). A new virulence gene which is mucoviscosity-associated gene A (magA), has recently been identified in this pathogen (9). magA is detected in a vast majority of K. pneumoniae liver abscess isolates and is associated with hypermucoviscosity (HV) and resistance to killing by human serum and phagocytosis (10).
Based on many recent reports all indicating the emergence of multi-drug resistance K. pneumoniae (11-14). It seems that the determinantion of antimicrobial susceptibility patterns are essential for appropriate therapy (12). Antibiotic susceptibility pattern of isolates has not been previously determined in our region. Therefore, the aim of this study was to investigate the prevalence of magA gene in Klebsiella spp. isolated from clinical samples and to detect their antimicrobial susceptibility in Hamadan, Iran.
Materials and Methods
A cross-sectional study was conducted on 105 Klebsiella isolates collected from patients. The patients were admitted to referral hospitals in Hamadan, Iran, during a 12-month period from 2007 to 2008. They had no history of antibiotic therapy before sampling and informed consents were obtained from them.The samples were transferred to bacteriology laboratory, plated on MacConkey agar, eosin methylen blue agar, blood agar, incubated for 18–24 hr at 37°C and identified by conventional microbiological methods (15).
DNA extraction
A single colony was taken from each eosin methylen blue agar which had been incubated overnight and emulsified into 100 μl of phosphate buffer salt. After incubation for 10 min at 95°C, 50 μl of proteinase K (100 mg/l) and 150 μl of TE (1 mM EDTA/10 mM Tris, pH 7.5) were added to the suspension and incubated for a further 20 min at 37°C (16).
For PCR detection, the bacterial DNAs were extracted and amplified using primer pairs targeting specific sequences (Table 1). To identify K. pneumoniae isolates, 40 pair primers were designed by Oligo software using urease-D gene. These primers were tested by Blast software in the web and the best pair was chosen. For detecting K.oxytoca and magA, specific primers of phenylalanine X (PehX) and magA genes were chosen from literature (9, 17-19).
Table 1.
Details of specific oligonucleotides which were used as primers to amplify particular sequences of Klebsiela pneumonia, K. oxytoca and magA gene
| Gene | primers | Size of amplicon | GenBank Accession no. | Ref. |
|---|---|---|---|---|
| Ure-D | 5'-CCC GTT TTA CCC GGA AGA AG-3' | 243 bp | L07039 | This study |
| 5'-GGA AAG AAG ATG GCA TCC TGC-3' | ||||
| PehX | 5'-GAT ACG GAG TAT GCC TTT ACG GTG-3' | 344 bp | AYO65648 | (19) |
| 5-TAG CCT TTA TCA AGC GGA TAC TGG-3 | ||||
| magA | 5'-CGC CGC AAA TAC GAG AAG TG-3' | 540 bp | AB085741 | (9) |
| 5'-GCA ATC GAA GTG AAG AGT GC -3' |
The 20 μl final volume of the PCR mixture contained 2 µl 10x buffer (500 mM-KCl, 100 mM Tris-HCl,pH 8.4, 15 mM MgCl2), 0.4, μl of deoxynucleotide mixture (dGTP, dTTP, dATP, and dCTP; 10 mM each), 0.8 μl of MgCl2 (50mM), 1 μl of each primer (10 mM), 0.1 μl Taq polymerase (5 units), 1 μl of template DNA and 13,7 µl distilled water.For amplifying condition, the initial denaturation step of 2 min at 94°C was followed by 35 cycles of 45 s at 94°C, 45 s at 60°C for ure-D, 59°C for PehX and 52°C for magA, 45 s at 72°C and the extension step of 5 min at 72°C. PCR products were detected by electrophoresis on 1% agarose gel. Finally, for magA, the related bond was prepared by DNA extraction kit from gel and was sequenced (Milegen, France) (20).
The hypermucoviscosity (HV) phenotype of Klebsiella isolates was also characterized and formation of viscous strings >5 mm in length showed a positive string test or a mucoviscose shape when a loop was passed through a colony (17).
In order to detect susceptibility of isolates to routine antibiotics, all isolates of K. pneumoniae and K. oxytoca were assessed by an agar disk diffusion method recommended by Clinical and Laboratory Standard Institute (21). Ten antibiotics including tobramycin (10 µg), amikacin (30 µg), gentamicin (10 µg), ceftriaxone (30 µg), ceftizoxime (30 µg), cefalotine (30 µg), cefatazidime (30 µg), cefazolin (30 µg), nalidixic acid (30 µg), and ciprofloxacin (5 µg) were used for the antibiogram. K. pneumoniae ATCC 1290, K.oxytoca ATCC 1402 and E. coli ATCC 11303 were used as reference strains (20).
Results
Out of 105 Klebsiella isolates, 38.1% were isolated from urine, 30.4% from stool, 11.4% from liver abscess, 4.8% from blood, 3.8% from wound, 3.8% from sinus, 2.9% from sputum and 4.8% from unknown samples. From 105 Klebsiella isolates, 92 strains (87.6%) were identified as K.pneumoniae, 5 strains (4.7%) as K. oxytoca and 8 strains (7.6%) as Klebsiella spp. by conventional microbiological tests, compared to 101 strains (96.2%) as K. pneumoniae and 4 strains (3.8%) as K. oxytoca by PCR.
MagA gene was detected in 4 isolates (3.8%) of K. pneumoniae, but none of the isolates of K. oxytoca contained magA gene. From the 4 isolates, three (75%) were obtained from blood samples and one (25%) from an abscess sample. Sixty-four isolates (60.95%) of K. pneumoniae showed an HV positive phenotype. From 4 isolates with positive magA gene, two of them were HV+ and two were HV- phenotype. The 4 isolates (100%) of K. oxytoca were HV- phenotype.
The results of the susceptibility of 105 isolates of K. pneumoniae and K. oxytoca to ten routine antibiotics are shown in the Table 2.
Table 2.
Susceptibility of 105 isolates of Klebsiela pneumoniae and K. oxytoca to ten routine antibiotics
| Antibiotic | Sensitive No (%) | Resistant No (%) | Antibiotic | Sensitive (%) | Resistant (%) |
|---|---|---|---|---|---|
| Tobramycin | 83 (79.05) | 22 (20.95) | Amikacin | 78 (74.290 | 27 (25.71) |
| Ceftazidime | 83 (79.05) | 22 (20.95) | Gentamicin | 75 (71.430) | 30 (28.57) |
| Ceftizoxime | 82 (78.09) | 23 (21.90) | Nalidixic acid | 75 (71.430) | 30 (28.57) |
| Ciprofloxacin | 80 (76.19) | 25 (23.81) | Cefalotine | 65 (61.91) | 40 (38.09) |
| Ceftriaxone | 79 (75.24) | 26 (24.76) | Cefazolin | 64 (60.95) | 41 (39.05) |
Tobramycin (10 µg), Ceftazidime (30 µg), Ceftizoxime (30 µg), Ciprofloxacin (5 µg), Ceftriaxone (30 µg), Amikacin (30 µg), Gentamicin (10 µg), Nalidixic acid (30 µg), Cefalotine (30 µg) and Cefazolin (30 µg
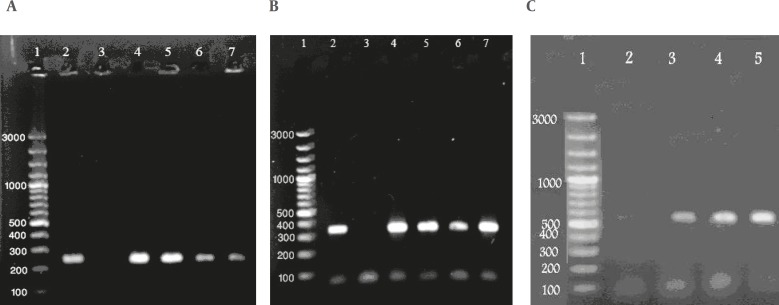
Figure 1.
Agarose gel electrophoresis analysis for the Ure-D gene in Klebsiela pneumoniae (A), the PehX gene in K. oxytoca (B) and magA gene in K. pneumonia (C) strains isolates
Lane 1, molecular size marker, expressed in base pairs. Lanes 2A, K. pneumoniae (ATCC 1290), 243 bp and 2B, K. oxytocae (ATCC 1402), 344 bp as positive controls. Lanes 3A, 3B and 2C, Escherichia coli (ATCC 11303) as negative controls and the other wells as positive isolates
Discussion
The purpose of the study was to investigate the presence of magA gene in K. pneumoniae and K. oxytoca, which were isolated from clinical samples. In Fang’s study, 52 out of 53 (98 %) K. pneumoniae isolated from liver abscess carried this specific virulence gene and the presence of one magA-negative isolate was thought to be due to patient’s underlying disease of liver cirrhosis and hepatic failure. Thus, Fang et al. concluded that magA is an essential virulence gene for K. pneumoniae strains causing liver abscess and could be used as a diagnostic tool. Fang believed that magA gene is exclusivly limited to liver abscess and HV positive phenotype (9, 10, 22). With extension of global research in other countries such as North American countries, Singapore and Korea, on Klebsiella isolates, they showed magA gene isolated from other cases like acquired bacteraemia, sepsis, meningitis and endophthalmitis (5, 6, 8, 17, 23, 24). In contrast to Fang’s studies, these samples included HV+ and HV- phenotypes. Therefore, based on the results of the present as well as other studies, containing HV+ phenotype is not a certain reason for the presence of magA gene since the HV- phenotype may have magA gene, too (17, 25). In our study, we found 4 positive magA gene isolates while two of them contained HV+ and two were HV- phenotype. Based on the data of this project, magA gene can specially belong to K. pneumoniae because we examined the K. oxytoca isolates for the presence of magA gene but none of them carry this gene.
The survey of antibiotic susceptibility of K. pneumoniae and K. oxytoca showed different percentages of susceptibility to the tested antibiotics. In this study resistance to nalidixic acid, cefalotine and cefazolin was relatively high in contrast to other studies (13, 14, 26). In our study, tobramycin was mostly active against strains of Klebsiella followed by ceftazidime and ceftizoxime.
Conclusion
The K. pneumoniae magA positive strains exist in our area and are isolated from various samples. In addition, the presence of HV+ phenotype is not associated with magA gene.
Acknowledgment
This study was funded by the Research Deputy of Hamadan University of Medical Sciences and approved by its ethics committee. The authors have no conflict of interest.
References
- 1.Podschun R, Ullmann U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev . 1998;11:589–603. doi: 10.1128/cmr.11.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kang CI, Kim SH, Bang JW, Kim HB, Kim NJ, Kim EC, et al. Community-acquired versus nosocomial Klebsiellapneumoniae bacteremia: clinical features, treatment outcomes, and clinical implication of antimicrobial resistance. J Korean Med Sci . 2006;21:816–822. doi: 10.3346/jkms.2006.21.5.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu WL, Ko WC, Cheng KC, Lee CC, Lai CC, Chuang YC. Comparison of prevalence of virulence factors for Klebsiellapneumoniae liver abscesses between isolates with capsular K1/K2 and non-K1/K2 serotypes. Diagn Microbiol Infect Dis . 2008;62:1–6. doi: 10.1016/j.diagmicrobio.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 4.Wang JH, Liu YC, Lee SS, Yen MY, Chen YS, Wang JH, et al. Primary liver abscess due to Klebsiellapneumoniae in Taiwan. Clin Infect Dis . 1998;26:1434–1438. doi: 10.1086/516369. [DOI] [PubMed] [Google Scholar]
- 5.Liu YC, Cheng DL, Lin CL. Klebsiellapneumoniae liver abscess associated with septic endophthalmitis. Arch Int Med. 1986;146:1913–1916. [PubMed] [Google Scholar]
- 6.Chung DR, Lee SS, Lee HR, Kim HB, Choi HJ, Eom JS, et al. Emerging invasive liver abscess caused by K1 serotype Klebsiellapneumoniae in Korea. J Infect . 2007;54:578–583. doi: 10.1016/j.jinf.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 7.Lederman ER, Crum NF. Pyogenic liver abscess with a focus on Klebsiellapneumoniae as a primary pathogen: an emerging disease with unique clinical characteristics. Am J Gastroenterol . 2005;100:322–331. doi: 10.1111/j.1572-0241.2005.40310.x. [DOI] [PubMed] [Google Scholar]
- 8.Ohmori S, Shiraki K, Ito K, Inoue H, Ito T, Sakai T, et al. Septic endophthalmitis and meningitis associated with Klebsiellapneumoniae liver abscess. Hepatol Res . 2002;22:307–312. doi: 10.1016/s1386-6346(01)00153-x. [DOI] [PubMed] [Google Scholar]
- 9.Fang CT, Chuang YP, Shun CT, Chang SC, Wang JT. A novel virulence gene in Klebsiellapneumoniae strains causing primary liver abscess and septic metastatic complications. J Exp Med . 2004;199:697–705. doi: 10.1084/jem.20030857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee HC, Chuang YC, Yu WL, Lee NY, Chang CM, Ko NY, et al. Clinical implications of hypermucoviscosity phenotype in Klebsiellapneumoniae isolates: association with invasive syndrome in patients with community-acquired bacteraemia. J Int Med . 2006;259:606–614. doi: 10.1111/j.1365-2796.2006.01641.x. [DOI] [PubMed] [Google Scholar]
- 11.Blahova J, Kralikova K, Krcmery V Sr, Babalova M, Menkyna R, Glosova L, et al. Four years of monitoring antibiotic resistance in microorganisms from bacteremic patients. J Chemother (Florence, Italy) 2007;19:665–669. doi: 10.1179/joc.2007.19.6.665. [DOI] [PubMed] [Google Scholar]
- 12.Biedenbach DJ, Moet GJ, Jones RN. Occurrence and antimicrobial resistance pattern comparisons among bloodstream infection isolates from the SENTRY Antimicrobial Surveillance Program (1997-2002) Diagn Microbiol Infect Dis . 2004;50:59–69. doi: 10.1016/j.diagmicrobio.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 13.Ivanov DV, Egorov AM. [Spreading and mechanisms of antibiotic resistance of microorganisms, producing beta-lactamases. Molecular mechanisms of resistance to beta-lactams of Klebsiella spp. strains, isolated in cases of nosocomial infections] Biomed khim . 2008;54:104–113. [PubMed] [Google Scholar]
- 14.Lagamayo EN. Antimicrobial resistance in major pathogens of hospital-acquired pneumonia in Asian countries. Am J Infect Control . 2008;36:S101–108. doi: 10.1016/j.ajic.2007.10.020. [DOI] [PubMed] [Google Scholar]
- 15.Forbes BA, Sahm DF, Weissfeld AS. Bailey & Scott’s Diagnostic Microbiology. UK: Mosby; 2007. [Google Scholar]
- 16.Zamani A, Sadeghian S, Ghaderkhani J, Alikhani MY, Najafimosleh M, Goodarzi MT, et al. Detection of methicillin-resistance (mec-A) gene in Staphylococcus aureus strains by PCR and determination of antibiotic susceptibility. Ann Microbiol . 2007;57:273–276. [Google Scholar]
- 17.Fang FC, Sandler N, Libby SJ. Liver abscess caused by magA+ Klebsiellapneumoniae in North America. J Clin Microbiol . 2005;43:991–992. doi: 10.1128/JCM.43.2.991-992.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurupati P, Chow C, Kumarasinghe G, Poh CL. Rapid detection of Klebsiellapneumoniae from blood culture bottles by real-time PCR. J Clin Microbiol . 2004;42:1337–1340. doi: 10.1128/JCM.42.3.1337-1340.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kovtunovych G, Lytvynenko T, Negrutska V, Lar O, Brisse S, Kozyrovska N. Identification of Klebsiellaoxytoca using a specific PCR assay targeting the polygalacturonase pehX gene. Res Microbiol . 2003;154:587–592. doi: 10.1016/S0923-2508(03)00148-7. [DOI] [PubMed] [Google Scholar]
- 20.Mashouf RY, Zamani A, Farahani HS. Diagnostic multiplex polymerase chain reaction assay for the identification of Pseudomonas aeruginosa from the skin biopsy specimens in burn wound infections and detection of antibiotic susceptibility. Saudi Med J . 2008;29:1109–1114. [PubMed] [Google Scholar]
- 21.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing, 17th informational supplement. M100-S17. Wayne: PA: Clinical and Laboratory Standards Institute; 2007. [Google Scholar]
- 22.Struve C, Krogfelt KA. Role of capsule in Klebsiellapneumoniae virulence: lack of correlation between in vitro and in vivo studies. FEMS Microbiol lett . 2003;218:149–154. doi: 10.1111/j.1574-6968.2003.tb11511.x. [DOI] [PubMed] [Google Scholar]
- 23.Yeh KM, Kurup A, Siu LK, Koh YL, Fung CP, Lin JC, et al. Capsular serotype K1 or K2, rather than magA and rmpA, is a major virulence determinant for Klebsiellapneumoniae liver abscess in Singapore and Taiwan. J Clin Microbiol . 2007;45:466–471. doi: 10.1128/JCM.01150-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fung CP, Chang FY, Lee SC, Hu BS, Kuo BI, Liu CY, et al. A global emerging disease of Klebsiellapneumoniae liver abscess: is serotype K1 an important factor for complicated endophthalmitis. Gut . 2002;50:420–424. doi: 10.1136/gut.50.3.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Struve C, Bojer M, Nielsen EM, Hansen DS, Krogfelt KA. Investigation of the putative virulence gene magA in a worldwide collection of 495 Klebsiella isolates: magA is restricted to the gene cluster of Klebsiellapneumoniae capsule serotype K1. J Med Microbiol . 2005;54:1111–1113. doi: 10.1099/jmm.0.46165-0. [DOI] [PubMed] [Google Scholar]
- 26.Kuo LC, Yu CJ, Kuo ML, Chen WN, Chang CK, Lin HI, et al. Antimicrobial resistance of bacterial isolates from respiratory care wards in Taiwan: a horizontal surveillance study. Int J Antimicrobial Agents . 2008;31:420–426. doi: 10.1016/j.ijantimicag.2008.01.010. [DOI] [PubMed] [Google Scholar]