Abstract
In this work, we consider the problem of estimating the probability for a specific random genetic mutation to be present in a tumor of a given size. Previous mathematical models have been based on stochastic methods where the tumor was assumed to be homogeneous and, on average, growing exponentially. In contrast, we are able to obtain analytical results for cases where the exponential growth of cancer has been replaced by other, arguably more realistic types of growth of a heterogeneous tumor cell population. Our main result is that the probability that a given random mutation will be present by the time a tumor reaches a certain size, is independent of the type of curve assumed for the average growth of the tumor, at least for a general class of growth curves. The same is true for the related estimate of the expected number of mutants present in a tumor of a given size, if mutants are indeed present.
Keywords: Tumor growth, Genetic mutations, Drug resistance, Stem cells, Ordinary differential equations, Branching processes
1 Introduction
Mathematical modeling and simulation of tumor growth is a growing field in applied mathematics. Among its main goals is the development of a framework which may allow to better understand the large amount of experimental data available today and possibly provide a description of cancer evolution. While mathematical methods are been developed, especially at the multiscale level, to better capture the complexity of cancer (Bellomo et al. 2008), exponential growth is still one of the most commonly used curves in the mathematical modeling of cancer. For describing the initial phase of a tumor’s growth, the exponential curve may indeed be a reasonable approximation. However, this does not appear to be true in general. For example, due to limits in space and nutrients, tumors seem to slow down their growth after having attained a certain size. Exponential growth models cannot reflect this critical saturation. Thus, to account for the more advanced phases of the tumor, the logistic and Gompertz growth models have been successfully introduced, as in the celebrated work on the modeling of human breast cancer by Moolgavkar (1986). For a more recent example, we refer to Nakasu et al. (2010), where various growth curves are compared by the authors in the case of meningiomas in order to determine which one provides the best fit to the data. Nonlinear regression analyses were performed against power, exponential, logistic, and Gompertzian curves. Their conclusion was that Gompertzian and logistic growth curves provided the best fit.
It is not the goal of this paper to consider the various types of tumor growth that have been proposed in the fast-growing literature on tumor modeling. However, it is clear that tumor growth will depend on a very large number of factors such as, for example, the type of tumor, its location, and patient-specific characteristics. Thus, it is surprising that models of tumor growth arguably more realistic than the classical exponential curve have not been fully integrated into existing mathematical models on the development of genetic mutations in cancer. For example, in the mathematical modeling literature on drug resistance, there are some works in which the possibility of considering nonexponential tumor growth types is mentioned, as in Komarova (2006). However, to the best of our knowledge, for the case where a non-exponential tumor growth is assumed, there are no estimates of the probability that, by the time a tumor reaches detection size, drug resistant cancer cells will be already present. One possible reason is given by the fact that to replace the exponential curve with any other growth model is mathematically a more complex undertaking, especially when dealing with stochastic methods. Moreover, the Markovian assumptions, which are usually inherent to the stochastic models, require exponentially distributed times, thus making it very natural to use the corresponding exponential growth.
In contrast, due to the simplicity of our approach, in this paper we are able to obtain analytical results for a general class of tumor growth curves. Our main result is that the probability which a given random mutation will be present by the time a tumor reaches a certain size, is independent of the type of curve assumed for the average growth of the tumor (at least for a large class of curves). The same is true for the related estimate of the expected number of mutants present in a tumor of a given size, if mutants are indeed present.
Due to the fact that the existing literature has been mainly modeling random genetic point mutations which cause cancer cells to become drug resistant, we will also describe the problem using this terminology. Point mutations are random genetic changes that occur during cell division. These mutations cause the replacement of a single base nucleotide or pair, with another nucleotide or pair in the DNA or RNA. This is a random event with a very small (yet nonzero) probability that modifies the cellular phenotype. These random genetic mutations are the main biological mechanism causing the development of drug resistance (Teicher 2006), together with genetic amplifications and kinetic resistance (Kimmel and Axelrod 1990; Harnevo and Agur 1991, 1993, Murray 1997; Panetta and Adam 1995; Gaffney 2005). Kinetic resistance refers to the reduction in effectiveness of a drug which is caused by the cell division cycle. Such resistance is generally only temporary. Many drugs (such as methotrexate, vincristine, and cytosine arabinoside, to name a few) are mainly effective during only one specific phase of the cell cycle, e.g., during the S phase, when the DNA is synthesized. Thus, the cell will be substantially invulnerable if it is out of the cell division cycle, i.e., in the G0 state. Gene amplification is the consequence of an overproduction of a particular gene or genes. This means that a limited portion of the genome is reproduced to a much greater extent than the replication of DNA composing the remainder of the genome. Such a defect amplifies the phenotype that the gene confers on the cell, which in turn, induces resistance by essentially providing the cells with more copies of a particular gene than the drug is able to cope with. Also this type of resistance has been proven to be typically temporary. Thus, given that point random mutations are permanent we will focus only them (also to simplify, make more tractable the model).
The structure of the paper is as follows: In Sect. 2, we briefly review some of the main results found in the existing literature on the mathematical modeling of drug resistance development, where an average exponential tumor growth is assumed, and refer interested readers to the references therein. For expository reasons, in Sect. 3, we first present our model and its results for the case where the average tumor growth is assumed to follow a logistic curve. After a brief treatment of the Gompertzian case in Sect. 4, we present the general result in Sect. 5. Concluding remarks are provided in Sect. 6.
2 Previous Work: The Exponential Case
The modeling of drug resistance due to random point mutations was motivated by the experimental findings of Luria and Delbrück (1943), on the development of resistance to antibiotics in bacteria due to mutations. Using fluctuation analysis, the authors concluded that the development of resistance is primarily a random phenomenon rather than a drug-induced, directed one. In their mathematical model, Luria and Delbrück modeled the occurrence of mutations by a random process, specifically a Poisson process with an intensity function. Importantly, both normal and mutant cells were assumed to grow deterministically in an exponential fashion. This Nobel Prize winning work has been followed by a large literature on the study of the distribution of the number of mutants in a population which grows exponentially, known as the Luria and Delbrück distribution (Zheng 1999).
The first model of resistance to chemotherapy due to point mutations in cancer has been the celebrated model by Goldie and Coldman and its extensions (see Coldman and Goldie 1985, 1986; Goldie and Coldman 1979, 1983a, 1983b, 1998; Goldie et al. 1982). In Goldie and Coldman (1979), the growth of the drug sensitive cancer cell population is approximated by using a deterministic exponential curve. At each division, there is a small positive probability that a cancer drug sensitive cell may give rise to one drug resistant cancer cell daughter because of a random point mutation. Such a mutant will generate a clone that grows according to a birth process. The number of mutations occurring in the drug sensitive population up to a given time is instead approximated by a Poisson distribution. It is also assumed that back mutations cannot occur. The number of mutants present in the cancer cell population is then given by a filtered Poisson process. The probability of having no resistant cells present in a tumor is then calculated, where the nonexistence of resistant cells is assumed to be the condition for being cured. We note that in the model by Goldie and Coldman, the drug-sensitive population is modeled deterministically as in Luria and Delbrück (1943), but the drug resistant population is modeled stochastically rather than deterministically. The main results of Goldie and Coldman (1979) are that the probability of having no drug resistance present in a tumor is inversely related to the tumor size and that more frequent dosage repetitions are more successful in minimizing the risk of drug resistance development than less frequent doses administered for a longer period of time. Interestingly, Goldie and Coldman extended their mathematical model to consider the development of drug resistance when the cancer stem cell hypothesis is considered (Goldie and Coldman 1983b, 1998). Unfortunately, they assumed that a stem cell can either renew symmetrically, producing two daughter stem cells, or differentiate symmetrically, producing two differentiated (not stem cells) daughters. In this way, the two division modes are simply equivalent to either a stem cell birth or death, leaving out from the stem cell dynamics the fundamental case of an asymmetric division, where a stem cell and a differentiated daughter cell are produced. The model then reduces to the usual birth and death process of a growing population. Notwithstanding this limitation their model is the first model on drug resistance that somehow considers the cancer stem cell dynamics.
A more recent study on point mutations is by Komarova and Wodarz (2005, 2009), Komarova (2006), Komarova et al. (2009). For example, in Komarova and Wodarz (2005), Komarova (2006), a model which is based on stochastic birth and death processes on a combinatorial mutation network is used to describe the development of resistance to multidrug treatments. Thus, probabilistic methods and a hyperbolic PDE are used to show how the pretreatment phase is more significant in the development of drug resistance than the treatment phase. This is a very natural, intuitive result given that the treatment will, in general, drastically reduce the cancer population, and consequently also reduce the number of possible cell divisions and mutations. Moreover, the main result obtained in Komarova (2006), Komarova and Wodarz (2005) is the following: in the case of a single drug treatment, the probability to have resistant mutants generated before the beginning of the treatment and present, including their progeny, at some given time afterward, does not depend on the cancer turnover rate. A consequence of such result is that also the probability of treatment success does not depend on such a rate. This result was later proved not to hold at finite times in Tomasetti and Levy (2010a, 2010b).
In a recent work, Iwasa et al. (2006) have shown that the probability that resistance to a drug develops by the detection time is given by
| (1) |
Here, M is the total number of cancer cells found at detection; u the probability of mutation per cell division; and L and D are the birth and death rates, respectively. This result was obtained using Markov chains and continuous time branching processes.
It is important to emphasize that, in all works we just mentioned, it is always being assumed that cancer cells are a homogeneous population (aside from being or not drug resistant). This is the reason why, in (1), M is taken to be the total number of cells. Yet, recent experimental evidence suggests that tumors should not be thought of as homogeneous. Indeed, it appears that tissues are maintained by a small subset of slowly replicating cells. These so-called “stem cells” have the capacity of both self-renewal and differentiation into more mature cells. Stem cells are very long lived, while mature, fully differentiated cells have a variable life span, which depending on the tissue of origin, can typically range from a few days to several months. From the point of view of drug resistance, the heterogeneity hypothesis implies that only the cells that have the capacity for self-renewal can propagate drug resistance. Therefore, these cells should be taken into account in any model of drug resistance in cancer. In fact, these are the only cells that should be taken into account. Thus, we will focus on the dynamics of a growing cancer stem cell population. It has been experimentally observed (see Morrison and Kimble 2006; Wu et al. 2007; McKenzie et al. 2006; Yatabe et al. 2001) that a stem cell may divide in the following three ways:
asymmetric division: a stem cell divides into one progenitor cell and one stem cell (with probability a),
symmetric differentiation: a stem cell divides into two progenitor cells (with probability b),
symmetric renewal: a stem cell divides into two stem cells (with probability c = 1 − a − b).
Generally, these three modes of division coexist in the stem cell population. Clearly, the mode of division chosen by the cancer stem cells will dramatically affect the dynamics of the tumor’s growth.
It is important to realize that the specific division mode chosen by an individual stem cellmay depend on multiple factors, some of which yet unknown. Certainly both intrinsic, developmental as well as extrinsic, environmental signals may be involved. A well-known example of asymmetric division controlled by an intrinsic mechanism (specifically, the asymmetric partitioning, localization of PAR proteins) is given by the C. Elegans zygote, while an example of an extrinsic mechanism for asymmetric division is given by the Drosophila germline stem cell, where the orientation among daughter cells causes one daughter to stay in the stem-cell niche thus retaining a stem-cell identity (see, Morrison and Kimble 2006, for a very detailed review on this topic). Interestingly, there appears to be a link between symmetric self-renewal and an inherent risk of cancer. Due to the above, it is likely that the probabilities of asymmetric and symmetric divisions will be functions of time. While here, for simplicity, we are assigning average values to these probabilities, in Sect. 5 we will be able to model also the time-dependent case.
Denote the total number of wild-type, i.e., drug-sensitive cancer stem cells at time t by S(t). Assume that this population grows exponentially. In Tomasetti and Levy (2010c), it was shown that the dynamics for the averaged behavior of this stem cell population can be described using the following equation:
| (2) |
Furthermore, the “expected number” of mutations occurring when there are exactly x wild-type cancer stem cells, mx, was estimated to be
| (3) |
Note that if the cancer stem cell population is growing, L(1 − a − 2b) − D must be greater than zero, and hence the expression in (3) is well defined.
The next result found in Tomasetti and Levy (2010c) is the calculation of the probability, PR, that at the time of detection, there are cancer stem cells that developed resistance to the drug. This probability is given by
| (4) |
where and M is the number of cancer stem cells found at detection. Note that this M is different than the M that was used in Iwasa et al. (2006), where it denoted the total number of cancer cells found at detection. We note that (4) is an extension of (1) (which can be recovered by setting a = b = 0, and setting M as the total population size).
Given (4), it is possible to calculate the expected value of the number of resistant cells that are found at detection, assuming that resistance has indeed developed by the time of detection. This conditional expectation of resistant cells is shown to be
| (5) |
In this paper, we extend these previous results by replacing the exponential cancer growth model by more realistic models. The simplicity of the methodology allows us to obtain an estimate for PR for the case of logistic, Gompertzian, and a large class of other types of tumor growth. In this sense, this paper represents an extension and a generalization of previous results. For example, the exponential growth model could be seen as a logistic growth model where the carrying capacity is infinite. Please note that it is not the purpose of this work to write down a model that explicitly indicates the source of the time dependent, nonlinear growth. There are too many possible formulations depending on which type of tumor we are considering and (even for a given tumor) depending on which specific variables we choose to model, as the large literature on mathematical modeling of cancer shows. The goal of this work, and its main result, is only to show that the obtained estimates are independent of the specific type of curve assumed for the average growth of the tumor, as long as it satisfies the rather general form given by the equations in the paper. Logistic and Gompertzian growth will be used here simply as examples, given their popularity in a variety of cancer models, without a specific formulation, justification.
3 The Logistic Case
As in Tomasetti and Levy (2010c), we adopt a mixed technique and derive the mathematical model using ordinary differential equations (ODEs) for the wild-type cancer population and branching processes for the mutant cells.
3.1 Ordinary Differential Equations
Recall that the cell division rate is denoted by L, the death rate by D, and the mutation probability per cell division by u. Furthermore, a denotes the probability of an asymmetric division, b the probability of a symmetric differentiation, and c = 1 − a − b the probability of a faithful symmetric renewal. When a tumor is detected, the number of cancer stem cells is assumed to be M. We denote the total number of wild-type, i.e., drug-sensitive cancer stem cells at time t by S(t). The second group of cells, the mutated cancer stem cells that developed resistance to the drug, are denoted by R(t).
Importantly, these populations are now assumed to grow logistically on average. The dynamics of the averaged behavior of these cancer stem cell populations can then be described using the following system:
| (6) |
In (6), the size of the wild-type cell population can increase only as a result of a symmetric renewal, were no mutation occurs. The probability of such an event is (1 − u)(1 − a − b). Multiplying this probability by the division rate L provides the birth rate of S(t). On the other hand, a decrease in the wild-type population will occur in the following cases: a cell death (D), a symmetric differentiation into two progenitors (bL), and if there is an asymmetric division (aL) in which the stem cell daughter is the mutant. The stem cell daughter will be the mutant with probability u/2, since u is the probability of mutations, and in this scenario only one of the two daughter cells is a stem cell. For the second equation in (6), the size of the mutant population can once again increase only due to a symmetric renewal. Only that this time, we assume that mutated cells can no longer mutate back to the wild-type state (due the rarity of this event for point mutations). A decrease in the mutant population will be the result of cell death and of a symmetric differentiation into two progenitors. The last term of the equation are the wild-type cells that mutated. They either come from a symmetric renewal: uL(1 − a − b), or from an asymmetric division in which a mutation hits the daughter stem cell: uLa/2. The sum of these two terms is uL(1 − a/2 − b).
Since we are interested in modeling situations where the resistant cells are a small proportion of the total or they are not present at all (otherwise we would not wonder about the probability of their presence), we will disregard the effect on resource limitations given by the resistant cells, that is, we will consider the following simplified system:
| (7) |
This is also due to the fact that we will be modeling the resistant cell stochastically and, therefore, we will not have a deterministic value for their numbers at time t to add to the number of wild-type stem cells. Given that the mutation rate u is very small, system (7) can be further reduced to the following:
| (8) |
where the initial conditions are S(0) = 1 and R(0) = 0.
We note that the only difference with (2) is the inclusion of the term , which accounts for the carrying capacity K of the logistic growth, where S(t) must be less than K for all t, and also M < K.
3.2 The Expected Number of Mutations When the Size of the Wild-Type Population Is x
From (8), we estimate the expected number of mutations, mx, that occur once the wild-type population is of size x. The solution of (8) for S(t) is given by
| (9) |
This means that the “average” time at which the wild-type cancer stem cell population reaches size x, defined as t(x), where S(t(x)) = x, is given by
| (10) |
Thus, the “average” length of time for which the population will consist of exactly x stem cells is
| (11) |
Hence, the “expected number” of mutations occurring when there are exactly x wild-type cancer stem cells, mx, is given by
| (12) |
This expression is obtained by multiplying the number of wild-type stem cells present at that time, x, by the mutation rate , and by the length of time for which the total stem cell population equals x.
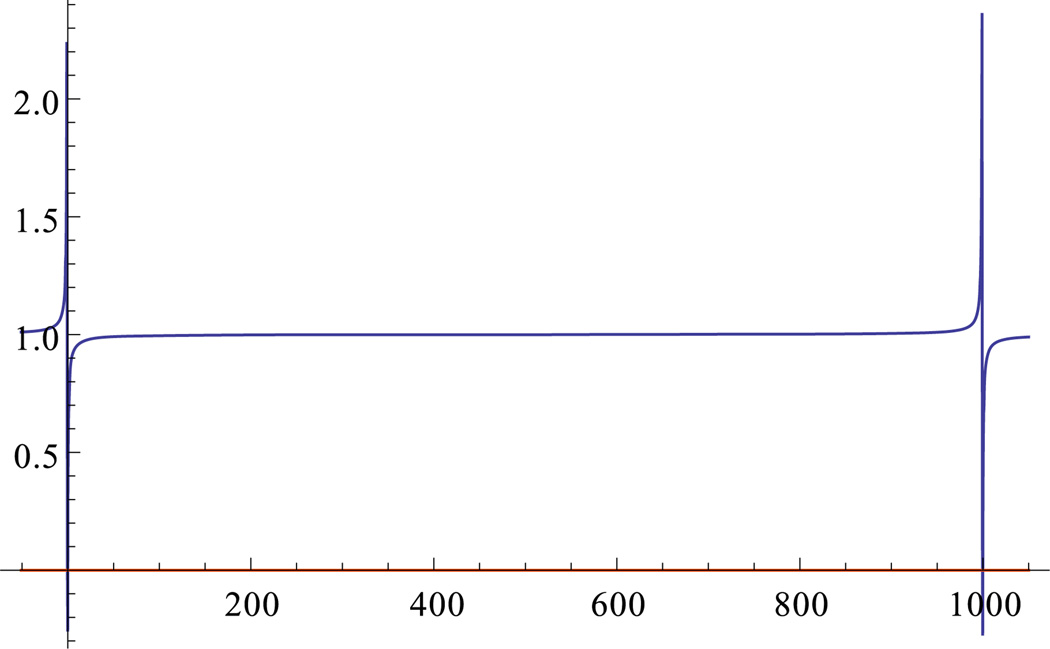
Finally, note that ; see Fig. 1. Thus, (12) can be reduced to
| (13) |
Interestingly, this is exactly the same expression found for the exponential growth case, (3). There is an intuitive way to understand why the expected number of mutations occurring when there are exactly x wild-type cancer stem cells turns out to be the same expression when assuming an exponential model or a logistic model for the cancer growth: the closer the cancer stem cell population is to the carrying capacity K the smaller the division rate but also the larger the time spent by the tumor in the state S(t) = x. The two effects turn out to balance each other.
Fig. 1.
Graph of , for K = 1000
3.3 Branching Processes
Assume now that the tumor population grows logistically starting from one wild-type stem cell. Let rx be the actual number of mutations produced when there are x = 1, 2,…, M − 1 wild-type cancer stem cells. Assume that all random variables, rx, follow a Poisson distribution with mean mx, given by (13), and that they are independent.
Consider the clone initiated by a mutant cancer stem cell which originated when there were x wild-type cancer stem cells. Biologically, we would like the division and death rates to balance each other near the carrying capacity to approach a constant population. Mathematically, we can do this by assuming that the population size of such a clone follows a continuous-time branching process where in each time step of length Δt, a cancer stem cell divides with probability and dies with probability . This means that births and deaths occur according to an exponential distribution with parameters and , respectively. Note that the difference with the exponential growth case is again the presence of the extra factor . By (9),
| (14) |
where r := (L(1 − a − 2b) − D). Thus, for a clone of mutants generated when the wild-type population was equal to S(t) = x, we have that births and deaths in such a clone occur according to an exponential distribution with parameters
| (15) |
and
| (16) |
respectively. Here, time is measured starting from the generation of the clone.
Let gx(ξ) be the generating function of such a clone for which the original mutation happened when S(t) = x. Denote by R(t) the number of mutated cancer stem cells forming the clone. Then, by the Kolmogorov backward equation we have, as for the exponential case (see Tomasetti and Levy 2010c),
| (17) |
since E[ξ R(t) | R(0) = 2] = E[ξ R(t) | R(0) = 1]2, by independence. The time t = 0 in (17) is the time of occurrence of the original mutation that generates the clone, i.e., when the wild-type population is of size x. The time t in (17) measures time starting from this t = 0.
Let gx(ξ, t) = E[ξ R(t) | R(0) = 1]. As Δt →0, we get
| (18) |
again a Riccati equation as for the exponential growth case (see (15) in Tomasetti and Levy 2010c), but this time with variable coefficients rather than constant,
| (19) |
Solving (19) with the initial condition gx(ξ, 0) = ξ gives
| (20) |
Since , where tM−x is the time it takes for the cancer stem cells to go from x to M, by letting into (20), we obtain the generating function
| (21) |
This implies that
| (22) |
3.4 The Probability of Having Resistant Cancer Stem Cells at the Time of Detection
We denote by T the total number of drug resistant cancer stem cells that are present when the cancer is detected, i.e., when the total stem cell population is M. Finally, we let GT (ξ) be its generating function and recall that, by (18) in Tomasetti and Levy (2010c),
| (23) |
By (13), (22), and (23), we have that the probability of having resistant cancer stem cells at the time of detection is
| (24) |
By replacing the summation with an integral, and with a change of variable, we obtain
| (25) |
It is immediate to see that such expression is equivalent to
| (26) |
where .
It is important to note that (26) is precisely (4), obtained for the exponential growth case.
Finally, given (26) and noting that , it follows that the expected value of the number of resistant cells that are found at detection, assuming that resistance has indeed developed by the time of detection, is given by
| (27) |
as in the exponential growth case.
In summary, we have calculated the probability, PR, that by the time a tumor reaches detection size, there are drug resistant cancer stem cells in a logistically growing cancer population. We have found that this probability, given by (26), is the same as for the exponential growth case.
Note that this formulas are estimates for any given size M of the tumor stem cell population and, therefore, are independent of time. Also, they are valid both for the case when this “detection size” M is close to the carrying capacity (as typically in an avascular tumor) as well as for a vascularized tumor (in which case, however, the logistic growth is not a good model).
This result may appear somewhat surprising. However, it can be explained by making the following observation. We note that the closer the cancer stem cell population is to the carrying capacity, K, the smaller the time-dependent division probability (which in the exponential growth case is constant) but also more time is spent by the cancer stem cells in the state S(t) = x, with the two effects balancing each other.
4 The Gompertzian Case
We would like to briefly consider now the case of a tumor population that follows, on average, a Gompertzian growth. This type of growth is arguably the most popular curve which has been used to describe tumor growth (but see Retsky 2011).
Using the Gompertzian equation, the dynamics of the averaged behavior of the drug-sensitive cancer stem cell population can then be described by the following (reduced) equation:
| (28) |
with S(0) = 1. Here, r := L(1 − a − 2b) − D, and .
We note that the only difference with (2) is the inclusion of the term Q exp(−rt), where S(t) must be less than K := S(∞) for all t.
Given that the solution of (28) is given by
| (29) |
it is easy to see that the “expected number” of mutations occurring when there are exactly x wild-type cancer stem cells, mx, is again given by (3), as for the logistic and exponential cases.
For a clone of mutants generated when the wild-type population was equal to S(t) = x, we then let births and deaths in such a clone occur according to an exponential distribution with parameters L̃(t) = L(1 − a − b) ln(K/x)e−rt and D̃(t) = (Lb + D)ln(K/x)e−rt respectively. Note that the difference with the exponential growth case is the presence of the extra factor ln(K/x)e−rt.
Letting g(ξ, t) = E[ξ R(t) | R(0) = 1], we obtain again a Riccati equation with variable coefficients,
| (30) |
Solving (30) with the initial condition gx(ξ, 0) = ξ gives
| (31) |
Since M ≈ x exp[ln(K/x)(1 − e−rtM−x)], by letting into (31), we obtain the generating function
| (32) |
This is exactly the same generating function we obtained for the logistic cases. Thus, the expressions for PR and E(T | resistance) will be again the same as for the exponential and logistic cases.
5 On General Types of Tumor Growth
While exponential, logistic and Gompertzian curves have been used successfully to model tumor growth, there are many other possible types of growth that can be considered. Thus, it is natural to ask whether our results are independent of the average type of growth that the cancer stem cell population follows. The answer is in the affirmative, at least for a very large class of growth curves.
Theorem 1 Let S′ (t) = [L(1 − a − 2b) − D]f (t)S(t), where f (t) is any strictly positive continuous real function. Then PR is given by (4), and E(T | resistance) by (5).
Proof We have that
| (33) |
Thus, if we let ΔS(t) = 1, that is, starting at time t, S goes from x to x + 1 in time Δt, then
| (34) |
Hence, the “expected number” of mutations occurring when there are exactly x wild-type cancer stem cells, mx, is given by
| (35) |
which is the same expression we have found for all types of growth we considered previously; see, for example, (3).
Given that births and deaths in a clone will occur according to an exponential distribution with parameters L̃(t) = L(1 − a − b)f (t) and D̃(t) = (Lb + D)f (t), respectively, we obtain again a Riccati equation for the p.g.f. g(ξ, t),
| (36) |
where g(ξ, 0) = ξ. Since g(t) = 1 is a solution, the solution of (36) is given by , where z(ξ, t) satisfies
| (37) |
and . Let tx and tM be the times when the cancer stem cell population reaches size x and M, respectively. The solution of (37) is then given by
| (38) |
Here, time is now measured from the moment the clone has been generated, i.e., when S = x. However, . Thus, gx(ξ) is exactly the same probability generating function we obtained previously (see (21) and (32)). Thus, the expressions for PR and E(T | resistance) will be identical to (4), and (5), respectively
Note that the same expression for mx is obtained in the even more general case where S′ (t) = [L(1 − a − 2b) − D]f (t). Thus, if S′ (t) = [L(1 − a − 2b) − D]f (t) and if D = 0 and b = 0, then the probability, PR, that by the time a tumor reaches detection size there are drug resistant stem cells in the cancer population will then be given by
| (39) |
since every time a mutation occurs the resulting clone will never become extinct. This is the same expression given by (4), with D = 0 and b = 0. Interestingly, experimental evidence seems to indicate that b is very small (see Booth and Potten 2000; Giebel et al. 2006) and also D is small compared to L (Abkowitz et al. 1995; Hanahan and Weinberg 2000).
6 Conclusions
In this paper, we have extended and generalized previous results on the probability of random point mutations present by the time a tumor is detected. We have showed that the estimate for such probability, and the related estimate for the expected number of mutants found at detection, if mutants are present at detection, are independent of the type of curve assumed in the model for the average tumor growth (at least for a large class of growth curves). While such result may now appear to be intuitive, it was not so both in the modeling and in what we may have guessed initially.
In this model, we have assumed that both the wild-type and the resistant stem cells have the same division and death rates and the same division events probabilities a, b. This assumption was made in order to simplify the presentation, and can be easily modified to model situations where, for example, the mutant cancer cells R(t) have a relative constant or time-dependent fitness advantage or disadvantage with respect to the wild-type cancer cells S(t) (see an example in Iwasa et al. 2006). However, even in those cases, we have here shown that the results would be independent of the type of curve chosen to model the average tumor growth. The inclusion of a state dependent fitness for the resistant cells would represent instead a more complex undertaking.
In conclusion, our results seem to be a good approximation for a rather general model of cancer growth with random point mutations. The simplicity of our methodology allowed us to obtain an estimate for PR and E(T | resistance) for the case where the average cancer stem cell population growth can be modeled by the equation S′ (t) = [L(1 − a − 2b) − D]f (t)S(t). We would like to note that arguably our modeling methodology may be seen as more general than previous works where fully stochastic approaches were taken, in the sense that at least for the wild-type population we only made assumptions on their averaged behavior. We did not assume, for example, that the time to division is exponentially distributed or that the Markov property holds.
Acknowledgements
The author wishes to thank Professor Dmitry Dolgopyat for his helpful discussions and the reviewers for their valuable comments. The work of CT was supported in part by the National Institute of Health under Grant T32 CA009337, by the joint National Science Foundation/National Institute of General Medical Sciences program under Grant DMS-0758374 and by the National Cancer Institute under Grant R01CA130817.
References
- Abkowitz JL, Persik MT, Shelton GH, Ott RL, Kiklevich JV, Catlin SN, Guttorp P. Behavior of hematopoietic stem cells in a large animal. Proc. Natl. Acad. Sci. USA. 1995;92:2031–2035. doi: 10.1073/pnas.92.6.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellomo N, Li NK, Maini PK. On the foundations of cancer modeling: selected topics, speculations, and perspectives. Math. Models Methods Appl. Sci. 2008;18:593–646. [Google Scholar]
- Booth C, Potten CS. Gut instincts: Thoughts on intestinal epithelial stem cells. J. Clin. Invest. 2000;105:1493–1499. doi: 10.1172/JCI10229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coldman AJ, Goldie JH. Role of mathematical modeling in protocol formulation in cancer chemotherapy. Cancer Treat. Rep. 1985;69(10):1041–1048. [PubMed] [Google Scholar]
- Coldman AJ, Goldie JH. A stochastic model for the origin and treatment of tumors containing drug-resistant cells. Bull. Math. Biol. 1986;48(3–4):279–292. doi: 10.1007/BF02459682. [DOI] [PubMed] [Google Scholar]
- Gaffney EA. The mathematical modelling of adjuvant chemotherapy scheduling: incorporating the effects of protocol rest phases and pharmacokinetics. Bull. Math. Biol. 2005;67(3):563–611. doi: 10.1016/j.bulm.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Giebel B, Zhang T, Beckmann J, Spanholtz J, Wernet P, Ho AD, Punzel M. Primitive human hematopoietic cells give rise to differentially specified daughter cells upon their initial cell division. Blood. 2006;107:2146–2152. doi: 10.1182/blood-2005-08-3139. [DOI] [PubMed] [Google Scholar]
- Goldie JH, Coldman AJ. A mathematical model for relating the drug sensitivity of tumors to their spontaneous mutation rate. Cancer Treat. Rep. 1979;63(11–12):1727–1733. [PubMed] [Google Scholar]
- Goldie JH, Coldman AJ. A model for resistance of tumor cells to cancer chemotherapeutic agents. Math. Biosci. 1983a;65:291–307. [Google Scholar]
- Goldie JH, Coldman AJ. Quantitative model for multiple levels of drug resistance in clinical tumors. Cancer Treat. Rep. 1983b;67(10):923–931. [PubMed] [Google Scholar]
- Goldie JH, Coldman AJ. Drug resistance in cancer: mechanisms and models. Cambridge: Cambridge University Press; 1998. [Google Scholar]
- Goldie JH, Coldman AJ, Gudaskas GA. Rationale for the use of alternating non-cross resistant chemotherapy. Cancer Treat. Rep. 1982;66(3):439–449. [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Harnevo LE, Agur Z. The dynamics of gene amplification described as a multitype compartmental model and as a branching process. Math. Biosci. 1991;103(1):115–138. doi: 10.1016/0025-5564(91)90094-y. [DOI] [PubMed] [Google Scholar]
- Harnevo LE, Agur Z. Use of mathematical models for understanding the dynamics of gene amplification. Mutat. Res. 1993;292(1):17–24. doi: 10.1016/0165-1161(93)90004-j. [DOI] [PubMed] [Google Scholar]
- Iwasa Y, Nowak MA, Michor F. Evolution of resistance during clonal expansion. Genetics. 2006;172:2557–2566. doi: 10.1534/genetics.105.049791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel M, Axelrod DE. Mathematical models of gene amplification with applications to cellular drug resistance and tumorigenicity. Genetics. 1990;125(3):633–644. doi: 10.1093/genetics/125.3.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarova N. Stochastic modeling of drug resistance in cancer. J. Theor. Biol. 2006;239(3):351–366. doi: 10.1016/j.jtbi.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Komarova N, Wodarz D. Drug resistance in cancer: principles of emergence and prevention. Proc. Natl. Acad. Sci. U.S.A. 2005;102(27):9714–9719. doi: 10.1073/pnas.0501870102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarova N, Wodarz D. Combination therapies against chronic myeloid leukemia: short-term versus long-term strategies. Cancer Res. 2009;69(11):4904–4910. doi: 10.1158/0008-5472.CAN-08-1959. [DOI] [PubMed] [Google Scholar]
- Komarova N, Katouli AA, Wodarz D. Combination of two but not three current targeted drugs can improve therapy of chronic myeloid leukemia. PLoS ONE. 2009;4(2):e4423. doi: 10.1371/journal.pone.0004423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luria SE, Delbruck M. Mutation of bacteria from virus sensitivity to virus resistance. Genetics. 1943;28:491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie JL, Gan OI, Doedens M, Wang JC, Dick JE. Individual stem cells with highly variable proliferation and self-renewal properties comprise the human hematopoietic stem cell compartment. Nat. Immunol. 2006;7:1225–1233. doi: 10.1038/ni1393. [DOI] [PubMed] [Google Scholar]
- Moolgavkar SH. Carcinogenesis modeling: from molecular biology to epidemiology. Annu. Rev. Public Health. 1986;7:151–169. doi: 10.1146/annurev.pu.07.050186.001055. [DOI] [PubMed] [Google Scholar]
- Morrison SJ, Kimble J. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature. 2006;441:1068–1074. doi: 10.1038/nature04956. [DOI] [PubMed] [Google Scholar]
- Murray JM. The optimal scheduling of two drugs with simple resistance for a problem in cancer chemotherapy. IMA J. Math. Appl. Med. Biol. 1997;14:283–303. [PubMed] [Google Scholar]
- Nakasu S, Nakasu Y, Fukami T, Jito J, Nozaki K. Growth curve analysis of asymptomatic and symptomatic meningiomas. J. Neurooncology. 2010 doi: 10.1007/s11060-010-0319-1. [DOI] [PubMed] [Google Scholar]
- Panetta JC, Adam J. A mathematical model of cycle-specific chemotherapy. Math. Comput. Model. 1995;22:67–82. [Google Scholar]
- Retsky J. Metronomic chemotherapy was originally designed and first used in 1994 for early stage cancer—why s it taking so long to proceed? Bioequiv. Availab. 2011;3(4) [Google Scholar]
- Teicher BA. Cancer drug resistance. Totowa: Humana Press; 2006. [Google Scholar]
- Tomasetti C, Levy D. An elementary approach to modeling drug resistance in cancer. Math. Biosci. Eng. 2010a;7(4):905–918. doi: 10.3934/mbe.2010.7.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasetti C, Levy D. Drug resistance always depends on the turnover rate. In: Herold KE, Bentley WE, editors. IFMBE proceedings. Vol. 32. New York: Springer; 2010b. [Google Scholar]
- Tomasetti C, Levy D. Role of symmetric and asymmetric division of stem cells in developing drug resistance. Proc. Natl. Acad. Sci. USA. 2010c;107(39):16766–16771. doi: 10.1073/pnas.1007726107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Kwon HY, Rattis F, Blum J, Zhao C, Ashkenazi R, Jackson TL, Gaiano N, Oliver T, Reya T. Imaging hematopoietic precursor division in real time. Cell Stem Cell. 2007;1:541–554. doi: 10.1016/j.stem.2007.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatabe Y, Tavare S, Shibata D. Investigating stem cells in human colon by using methylation patterns. Proc. Natl. Acad. Sci. USA. 2001;98:10839–10844. doi: 10.1073/pnas.191225998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q. Progress of a half century in the study of the Luria–Delbrück distribution. Math. Biosci. 1999;162:1–32. doi: 10.1016/s0025-5564(99)00045-0. [DOI] [PubMed] [Google Scholar]