Abstract
Purpose
CR increases fatty acid oxidation to decrease tissue lipid content. The Nuclear factor E2-related factor 2 (Nrf2)-Kelch like ECH associated Protein 1 (Keap1) pathway is an antioxidant gene regulatory pathway that has been previously investigated in weight gain. However, limited interaction of Nrf2/Keap1 and CR exists. The purpose of this study was to determine how Keap1 knockdown (Keap1-KD), which is known to increase Nrf2 activity, affects the CR response, such as weight loss, hepatic lipid decrease, and induction of fatty acid oxidation gene expression.
Methods
C57BL/6 and Keap1-KD mice were maintained on 40% CR or fed ad libitum for 6 weeks. Hepatic lipid content, lipid metabolic gene, and miRNA expression was quantified.
Results
CR lowered hepatic lipid content, and induced fatty acid oxidation gene expression to a greater degree in Keap1-KD compared to C57BL/6 mice. CR differentially altered miRNA 34a, 370, let-7b* in livers of Keap1-KD compared to C57BL/6 mice.
Conclusions
CR induced induction of fatty acid oxidation gene expression was augmented with Keap1 knockdown, which was associated with differential expression of several miRNAs implicated in fatty acid oxidation and lipid accumulation.
Keywords: caloric restriction, gene expression, liver, Nfe2l2, nuclear receptor
INTRODUCTION
Nuclear Factor E2-Related Factor 2 (Nrf2) belongs to the basic leucine zipper family of transcription factors, tethered into the cytoplasm via a complex with Kelch like ECH-associated Protein 1 (Keap1), which upon activation, translocates to the nucleus and activates gene transcription (1,2). Nrf2 is best described for its role in xenobiotic metabolism and oxidative stress response (3,4) Nrf2 regulates expression of genes important in metabolism and oxidative stress response such as NAD(P)H:quinone oxidoreductase (Nqo1), Glutamate cysteine ligase catalytic subunit (Gclc), and Glutathione S-Transferase (Gst) (5) Nrf2 has been described to inhibit Lxrα dependent increase in Srebp1 expression, resulting in decreased lipogenesis (6). Shin et al. 2009 and Tanaka et al. 2008 demonstrated increased hepatic lipid accumulation in absence of Nrf2 in mice fed a standard diet or upon high fat diet feeding (7,8).
A single study has demonstrated an anti-carcinogenic, protective activity of Nrf2 in caloric restriction (9). However, no studies present an account for whether increased Nrf2 activity can affect the response to CR or modulate fatty acid oxidation enzyme expression, although a relationship between Sirtuin 1 (Sirt1) activation and regulation of Nrf2 binding has been described in vitro (10).
Caloric restriction (CR) induces many beneficial processes in the body and is has been shown to protect against diseases of metabolic etiology, such as insulin resistance, diabetes, and NAFLD (11,12). Beneficial effects of CR are attributed to activation of Sirt1, which activates gluconeogenic and fatty acid oxidation gene expression via Peroxisome proliferator-activated receptor gamma coactivator 1-alpha (Pgc-1α) co-activation of gene transcription (13,14). Sirt1 activates the nuclear receptor Peroxisome Proliferator activated receptor α (Pparα) via deacetylation of Pparα coactivator (Pgc-1α) and upregulation of fatty acid oxidation genes, such as Cpt1a (15,16). Concurrently, CR activation of Sirt1/Pgc-1α cascade inhibits expression of lipogenic factors and regulators such as Sterol regulatory element binding protein 1c (Srebp1c), Liver x receptor (Lxr) and respective target genes Fatty acid synthase (Fas), Acetyl-CoA carboxylase (Acc1), Acyl-CoA thioesterase 1 (Acot1), Fatty acid binding protein 4 (Fabp4), Stearoyl-CoA desaturase (Scd1).
Small non-coding RNAs, microRNAs have been recently demonstrated to provide an additional layer of regulatory control in development, homeostasis and pathology (17). microRNAs, 19–22 nucleotides long, act as gene silencers with the RNA induced silencing complex (RISC) and modestly regulate gene expression (18). Mammalian microRNAs do not necessarily exhibit complete homology with the target mRNA and along with classically known cleavage of target mRNA, they can also affect mRNA stability (17). Due to partial homology, a single miRNA is shown to regulate expression of multiple target mRNAs (19). miRNA expression and activity is altered in various disease conditions including metabolic syndrome (20). miRNA expression and regulation are being explored in detail as potential therapeutic targets and biomarkers in metabolic syndrome and cancer (21,22). Limited information exists regarding the effect of caloric restriction on regulation of miRNAs, but certainly implicates them as regulators of Sirt1. For example, miR-34a, miR-132, and miR-199a target Sirt1 to regulate biological functions, such as hepatic lipid metabolism, islet β-cell exocytosis, cell apoptosis, stress-induced chemokine production, and hypoxia preconditioning (23,24). More studies have focused miRNA regulation and fatty liver disease, but can be useful to understanding mechanisms regarding CR. For example, miR-122 has been shown to be involved in regulation of cholesterol and triglyceride synthesis, silencing miR-122 drastically decreased cholesterol synthesis, decreased fatty acid levels and triglyceride load on the liver in mice fed a high fat diet (25,26). Patients with Non-alcoholic fatty liver disease (NAFLD) demonstrated increased circulating miR-122 levels along with miR-34a (27). miR-34a is another miRNA that has been shown to regulate important aspects of fatty acid oxidation via silencing Sirtuin1 (Sirt1), a master regulator of energy homeostasis in the cell (28). Sirt1 is known to inhibit miR-34a, while positively upregulating Farnesoid X receptor (FXR) expression via histone deacetylation (29). miR-33a has been shown to be important for its regulation of Srebp expression and hence steatotic development (30,31). miR221 has been demonstrated to be important in development of liver fibrosis and hepatocellular carcinoma (32). Let-7 miRNA increases insulin resistance via modulating insulin receptor (InsR) expression along with Glut4 expression in skeletal muscle (33,34). How CR regulates miRNAs in liver are largely undescribed.
In the present study, we hypothesized that Keap1-KD, which has been associated with down regulation of lipid synthesis gene expression (35), will augment caloric restriction effects. We demonstrated that Keap1-KD increased induction of fatty acid oxidation gene expression, which was associated with novel changes in the miRNA regulatory circuit.
MATERIALS AND METHODS
Animals and Treatments
Keap1-KD and Nrf2-null breeders congenic to a C57BL/6 background as previously described (36,37) were obtained from Dr. Curtis Klaassen (Kansas University Medical Center, Kansas). Age-matched male C57BL/6 mice were purchased from Harlan laboratories (Indianapolis, IN, USA) at 13 weeks old and were allowed to acclimate for 3 weeks before the study commenced. The colonies were amplified and maintained at the Fogarty Hall Animal Care Facility, University of Rhode Island. The average caloric consumption was calculated for each mouse on a purified diet (AIN93-G) obtained from Testdiet, IN, USA over a period of 10 days. This semi-purified diet was selected to provide enhanced nutrition. Male C57BL/6 and Keap-1KD mice (16 weeks age) were each divided into two groups of n=5 and n=6. One group of each genotype (n=5) was maintained as a control group which was fed ad libitum (AL) over the length of the experiment. The other group of each phenotype (n=6) was placed on a 40% reduced caloric diet (CR) for a period of 7 weeks with access to water ad libitum. Body weight and food consumption was monitored at least weekly throughout the study. At week three, blood was collected by cheek pouch puncture for serum chemistry. The study was terminated when weight loss plateau was reached (After 7 weeks CR) and blood and livers were collected. Mice were not evaluated for markers of malnutrition. Livers were snap frozen in liquid nitrogen and stored in −80°C until further analysis. Serum glucose (Cayman Chemicals, Ann Arbor, MI), serum and liver triglyceride (Pointe scientific, MI, USA) and free fatty acid concentrations (Wako diagnostics, VA) were determined by spectrophometeric assay kits. Male C57BL/6 and Nrf2-null mice (n=5 and 6 respectively, 24 week old) that were bred in-house were euthanized and livers collected for basal miRNA quantification. However, no malnutrition end points were measured. The study herein was reviewed and approved by the University of Rhode Island Institutional Animal Care and Use Committee and the number of mice used was based upon required power analysis.
RNA Isolation and mRNA Quantification
Total RNA was isolated from liver by phenol-chloroform extraction with RNAzol B reagent (Tel-Test Inc., Friends-wood, TX) according to the manufacturer’s instructions. RNA concentration was determined by measuring UV absorbance at 260 nm using NanoDrop™ and the integrity were confirmed by formaldehyde gel electrophoresis. mRNA expression was quantified by RT2-PCR, with expression normalized to 18S rRNA expression.
miRNA Quantification and Array Analysis
Total RNA enriched for small nucleotide fraction was isolated from livers obtained from AL and CR C57BL/6 and Keap1-KD mice using the miRNA easy mini kit (Qiagen Inc, MD, USA). 1 μg total RNA was used for miRNA cDNA preparation using RT2 miRNA first strand kit (Qiagen Inc, MD, USA) and mature miR-34a, miR-370, miR144, let-7b*, miR-221, miR-146b, miR-692, miR-205 and miR485 expression was measured by RT2-PCR. The miRNA expression was normalized to mean expression level of Snord65, Snord66, and Snord85. PCR arrays (mouse genome V2.0) (SABiosciences, Frederick, MD, USA) were used to determine expression of miRNA in pooled liver RNA samples isolated using miRNA easy kit (Invitrogen Corp, MD, USA) as per the manufacturer’s instructions. Data analysis was performed with the web-based software package for the miRNA PCR array system (http://www.sabiosciences.com/pcr/arrayanalysis.php).
Statistics
Groups were analyzed by a one-way ANOVA followed by a Duncan’s Multiple Range post hoc test. A planned comparison between C57BL/6 and Keap1-KD groups was performed among CR groups after performing the one-way ANOVA. Groups without a common letter are considered significantly different from each other (p ≤0.05).
RESULTS
Effect of Keap1-KD on CR-Induced Weight Loss and Liver Lipid Content
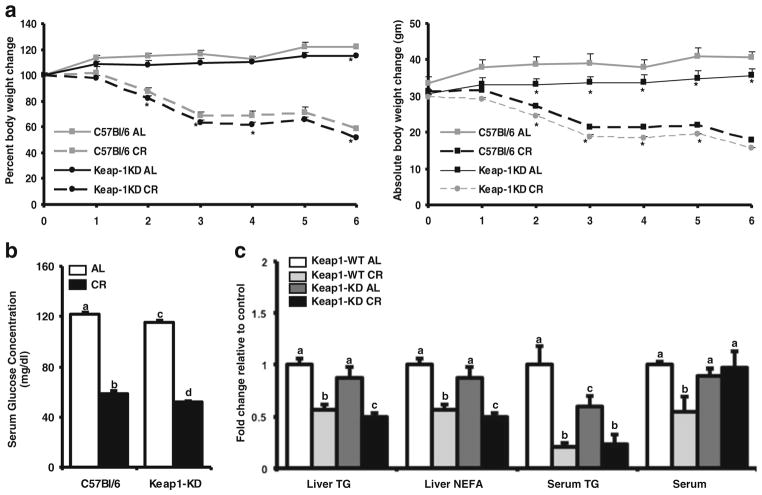
To determine whether Keap1 knockdown changed susceptibility to weight loss and fat mobilization, C57BL/6 and Keap1-KD mice were fed ad libitum or placed on a 40% CR diet for 6 weeks. As depicted in Fig. 1a, Keap1-KD mice fed ad libitum gained less weight over time than C57BL/6 counterparts. While C57BL/6 mice fed ad libitum gained approximately 15% of their initial weight at the end of the study, Keap1 KD mice gained a lesser percentage (8%) of their initial weight at the starting point (week 0) (Fig. 1a). Upon CR, both C57BL/6 and Keap1-KD mice lost approximately 43% of their initial weight (Fig. 1a). At the age of 23 weeks, the Keap1KD mice did not show significantly different liver weights (Supplementary Fig. 1). However, they had a slightly higher liver-to-body weight ratio, which is in agreement with previously published studies using the Keap1KD mouse model (38). After CR, there was no difference in liver-to-body weight ratios between the two genotypes indicating a slightly higher susceptibility of the Keap1KD mice to CR induced changes in the liver.
Fig. 1.
Keap1-KD mice gained less weight and had lower hepatic lipid content after CR compared to C57BL/6 mice. (a) Body weight and percent body weight in C57BL/6 and Keap1-KD mice during CR. During 6 weeks of 40% reduced caloric diet (CR), body weight of ad libitum (AL) fed (n=5) or CR (n=5) mice (both C57BL/6 and Keap1-KD) was determined weekly. The percent body weight changes were calculated with the body weight at initial time point considered as 100% and each mouse being its own control. The data is represented as average ± SEM (n=5) body weight or percent body weight change over the weeks of CR. (b) Serum glucose, was quantified from C57BL/6 and Keap1-KD mice, at the end of the study. The data is represented as average ± SEM (n=5) mg/dl glucose. P<0.05 was considered statistically significant. Letters different from each other represent a significant difference between groups (p<0.05). (c) Serum and liver triglyceride and free fatty acid levels were quantified from C57BL/6 and Keap1-KD mouse livers upon CR at the end of 6 weeks. The data is represented as average ± SEM (n=5) fold change in serum biochemical factors over the ad libitum fed controls. Groups without a common letter are considered significantly different from each other (p≤0.05).
After 3 weeks of CR, serum glucose concentrations decreased by ~20% in the C57BL/6, but decreased twice as much (~40%) in the Keap1-KD mice (Table I). Keap1-KD AL mice had higher basal serum glucose concentrations compared to C57BL/6 mice when measured after 3 weeks of starting the study (Table I). At the end of the study however; after 6 weeks of CR, the basal glucose level in Keap1-KD AL mice was lower than C57BL/6 mice and upon CR, Keap-1KD mice had lower glucose levels than C57BL/6 mice (Fig. 1b). After 3 weeks of CR, Keap1-KD mice had significantly lower serum TG (Table I). While C57BL/6 mice demonstrated ~80% decrease in serum TG content over the AL fed mice, Keap1-KD mice did not show any significant changes in serum TG levels, potentially due to low basal TG values. At the end of the study, serum TG values remained lower in Keap-1KD mice fed AL, without notable difference between C57BL/6 and Keap1-KD mice that were subjected to CR (Fig. 1c). Liver TG levels in C57BL/6 and Keap1-KD mice fed AL were similar. CR decreased the hepatic TG levels by ~43% in the C57BL/6 mice, but more so in the Keap-1 KD mice (~ 50%) (Fig. 1c). Serum NEFA remained unchanged in these mice at week three (Table I). However, at the end of the study, C57BL/6 mice had a ~35% decrease in serum NEFA levels, whereas Keap-1 KD mice showing no difference upon CR at all. Keap1-KD mice had slightly lower hepatic NEFA levels compared to C57BL/6 mice after CR (Fig. 1c).
Table I.
Serum Biochemical Measurements after 3 week CR
| Group | Glucose (mg/dL) | Triglycerides (mg/dL) | NEFA (mg/dL) |
|---|---|---|---|
| C57BL/6 AL | 149±15.6a | 117±29.6a | 0.006±0.0006a |
| C57BL/6 CR | 119±4.72b | 18±0.6b | 0.005±0.0012a |
| Keap1-KD AL | 185±13.2c | 53±7.5c | 0.007±0.0016a |
| Keap1-KD CR | 112±11.4b | 38±8.53d | 0.005±0.0008a |
Serum glucose, triglyceride, and free fatty acid levels were quantified from C57BL/6 and Keap1-KD mice fed ad libitum (AL) or placed on 40% caloric restriction (CR) for 3 weeks. Blood was collected by a cheek pouch bleed (submandibular vein puncture), serum was isolated and used for quantification. The data is represented as average ± SEM (n=5) per group with p<0.05 considered statistically significant. Groups without a common letter are considered significantly different from each other (p≤0.05)
CR-Induced Fatty Acid Oxidation Gene Expression is Higher in Keap1-KD than C57BL/6 Mice
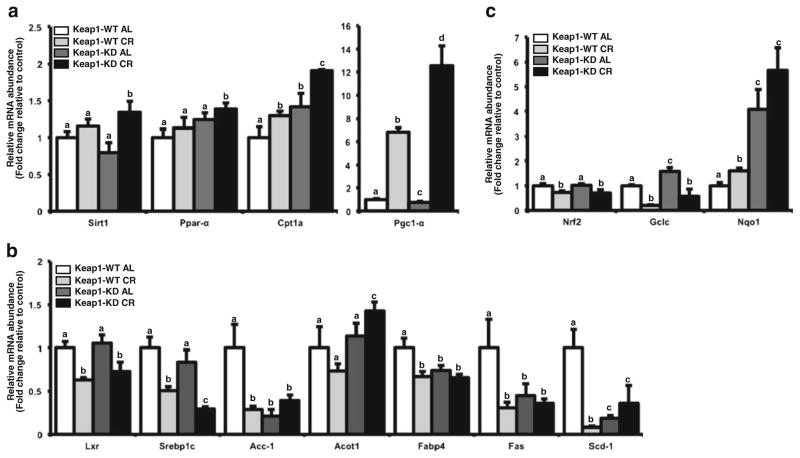
To identify whether key genes related to fatty acid oxidation are activated upon CR, mRNA expression of Sirt1, Pgc1α, and Ppar α was quantified along with Cpt1 α. In livers from AL fed mice, Cpt1 α and Ppar α expression was increased in Keap1KD mice compared to C57BL/6 mice (Fig. 2a). As observed in Fig. 2a, CR restriction induced the expression of Sirt1, Pgc-1α, Pparα in livers of both C57BL/6 mice, which is consistent with previous reports (39). Pgc1α, Pparα induction was highest in livers of Keap1-KD mice that were calorically restricted (Fig. 2a), presenting a novel observation for Keap1 knockdown and induction of the Sirt1/Pgc1α cascade.
Fig. 2.
Keap1 knockdown enhances CR-related induction of fatty acid oxidation genes and inhibition of lipogenic genes. Total RNA was isolated from livers of male C57BL/6 and Keap1-KD mice fed ad libitum (AL) or placed on 40% reduced caloric diet (CR) for 6 weeks. (a) Effect of CR on fatty acid oxidation gene expression in C57BL/6 and Keap1-KD mouse liver: Sirt1, Pparα, Cpt1a and Pgc-1α mRNA expression was quantified by RT2-PCR using SYBR green chemistry. (b) Effect of CR on Nrf2 target gene expression in C57BL/6 and Keap1-KD mouse liver. mRNA expression of Nqo1 and Gclc was quantified by RT2-PCR. (c) Effect of CR on lipogenic gene expression in C57BL/6 and Keap1-KD mouse liver: mRNA expression of Srebp1c, Lxr, Fas, Acc1, Acot-1, Fabp4 and Scd1 was quantified by RT2-PCR. All data is represented as average ± SEM (n=5) fold changes in expression over the controls. Groups without a common letter are considered significantly different from each other (p≤0.05).
Keap1-KD Increased CR-Induced Down Regulation of Gene Expression for Some Lipogenic Genes in Mouse Liver
CR is known to decrease the expression of gene involved in biosynthesis of fats (39). Hence, the mRNA expression of lipogenic transcription factors, such as Srebp1c and Lxr, and downstream target genes such as Fas, Acc1, Fabp4 and Scd1 was measured. In C57BL/6 mice, CR decreased mRNA expression of lipogenic gene expression regulators, Srebp1c and Lxr, to ~50% of the ad libitum fed controls (Fig. 2b). Correspondingly, CR decreased the mRNA expression of target genes regulated by these transcription factors (e.g. Fas, Fabp4, Acc1) compared to AL mice (Fig. 2b). The mRNA expression of some lipogenic genes, such as Acc1, Fabp4, Fas and Scd1, was lower in the Keap1-KD mice as compared to ad libitum fed C57BL/6 mice (Fig. 2b). Upon CR, the expression of these lipogenic genes was not altered in Keap1-KD mice, although the gene down regulation of the regulatory Srebp1c was significantly lower in the Keap1-KD mouse livers over the calorically restricted C57BL/6 mice (Fig. 2b). In assessing similarities and differences in gene expression, Lxr and Srbpc expression are consistent with some level of Sirt1 regulation, whereas Acot differed. CR based Srebp1c downregulation in the liver could potentially be via inhibition of LXR (Srebp1c being a target gene for LXR in the liver), whereas Acot1 is also regulated by Ppar alpha pathway (40) along with LXR (41). Perhaps the effect observed in Acot1 expression during CR is a combination of or a crosstalk between both these pathways.
Keap1-KD Enhanced the Effect of CR on Nrf2 Target Gene Expression
To determine whether CR has an effect of Nrf2 target gene expression, we quantified the mRNA expression of Nrf2 and its target genes, Nqo1 and Gclc, in C57BL/6 and Keap1-KD mice on CR. As depicted in Fig. 2c, CR induced Nqo1 (~1.6 fold) mRNA expression in liver and downregulated the expression of Gclc (~30% and 79% respectively) in C57BL/6 mice, which is consistent with a previous observation (42). The basal expression of Nqo1 and Gclc, Nrf2 target genes, was significantly higher in Keap1-KD mouse livers, which is also consistent with a previous publication (43). However, the observed decrease in Gclc mRNA expression by CR was lesser in Keap1-KD mice (80% in C57BL/6 CR versus 60% in Keap1-KD CR mouse livers) and Nqo1 induction via CR was augmented in Keap1-KD mice (~1.6 fold in C57BL/6 CR versus 1.4 fold in Keap1-KD CR mouse livers) (Fig. 2c).
Caloric Restriction Significantly Altered Expression of miRNA Implicated in Regulating Fatty Acid Oxidation and Lipogenic Gene Expression
To determine whether the induction of fatty acid oxidation genes upon CR is corresponds to changes in the miRNA regulatory circuit, miRNA expression of various miRNAs found in the mouse genome were quantified in C57BL/6 CR and Keap1-KD CR liver samples that were pooled. CR regulated miRNA differently in C57BL/6 and Keap1-KD mice (Tables II).
Table II.
Differential Increase (A) and Decrease (B) in miRNA Expression in Livers of C57BL/6 and Keap1-KD mice After CR
| Identity | Fold Change |
|---|---|
| A | |
| mmu-miR-193 | 3.36 |
| mmu-miR-149 | 3.20 |
| mmu-miR-434-5p | 3.73 |
| mmu-miR-34b-5p | 8.33 |
| mmu-miR-205 | 10.61 |
| mmu-miR-455 | 3.60 |
| mmu-miR-343 | 3.20 |
| mmu-miR-692 | 102.54 |
| B | |
| mmu-miR-184 | −4.63 |
| mmu-miR-135a | −4.18 |
| mmu-miR-146b | −14.55 |
| mmu-miR-802 | −6.07 |
| mmu-miR-665 | −4.32 |
| mmu-miR-383 | −4.24 |
| mmu-miR-200b* | −4.21 |
| mmu-miR-449b | −4.57 |
| mmu-miR-667 | −4.63 |
| mmu-miR-150* | −5.11 |
| mmu-miR-411* | −5.91 |
| mmu-miR-466j | −6.42 |
| mmu-miR-485* | −11.26 |
| mmu-miR-92a* | −6.69 |
| mmu-miR-1898 | −5.06 |
| mmu-miR-1899 | −4.06 |
RNA fraction enriched for small RNAs was isolated from livers of C57BL/6 and Keap1-KD mice that were placed on caloric restriction and miRNA expression was quantified from pooled RNA samples by RT2 -PCR using a Mouse Genome V2.0 miRNA array from SABiosciences as per manufacturer’s instructions. Data analysis was performed with the web-based software package for the miRNA PCR array system. (http://www.sabiosciences.com/pcr/arrayanalysis.php). Data is presented as fold increase or decrease in miRNA expression of Keap-1KD CR relative to C57BL/6 CR
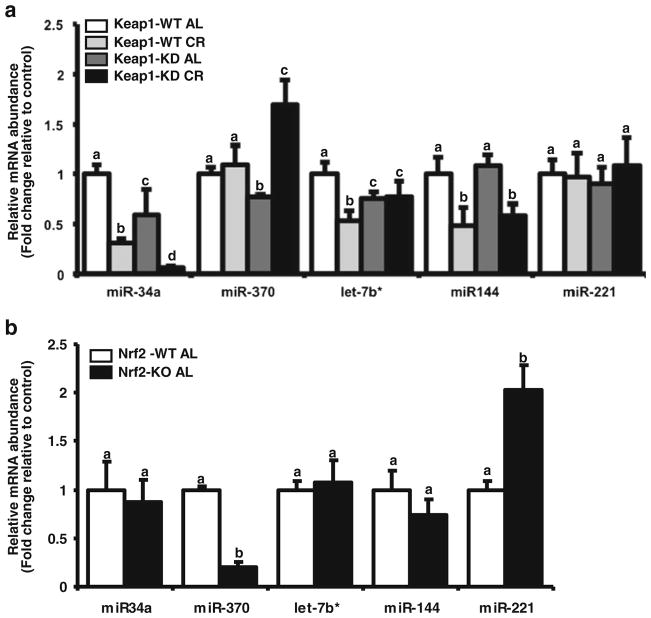
Based on this observation and a reported relationship to steatosis or fatty acid oxidation gene expression, miR-34a, miR-370, let-7b*, miR-144 was quantified. Keap1-KD significantly decreased basal expression of miR-34a, miR-370 and let-7b* (Fig. 3a). This observation is consistent with the higher basal expression of Pgc-1α and Pparα (Fig. 2b), as these miRNAs have been described to regulate the latter transcripts (44,45). CR significantly decreased miR-34a, let7-b* and miR-144 expression in C57BL/6 mouse livers (Fig. 3a). Keap1-KD did not significantly affect the decrease in miR-144 expression upon CR, whereas it enhanced the decrease in expression of miR-34a (Fig. 3a). In contrast, CR decreased miR-370 in Keap1-KD mouse livers, but did not in C57BL/6 mice. Conversely, CR decreased let7b* expression in C57BL/6 mice but did not in Keap1-KD mice (Fig. 3a). These observations are consistent with the mRNA expression levels, in that Keap-1KD mice on CR have the highest expression of fatty acid oxidation genes (Fig. 2b).
Fig. 3.
Effect of Keap1 knockdown and targeted Nrf2 deletion on miRNA expression in mouse livers after CR. The RNA fraction enriched for small RNAs was isolated from livers of male C57BL/6 and Keap1-KD mice fed ad libitum (AL) or placed on 40% CR for 6 weeks. (a) Effect of CR on miRNA expression in livers of C57BL/6 and Keap1-KD mice. miR-34a, miR-370, Let-7b, miR-144 expression was quantified by RT2-PCR and normalized to mean Snord66, Snord 68 and Snord 85 expression. (b) Effect of CR on miRNA expression in livers of C57BL/6 and Nrf2-null mice fed ad libitum. miRNA expression of miR34a, miR370, Let-7b, miR144 was quantified by RT2-PCR and normalized to mean Snord66, Snord 68 and Snord 85 expression. All data is represented as average ± SEM (n=4) fold changes in expression over the controls. Groups without a common letter are considered significantly different from each other (p≤0.05).
Targeted Nrf2 Deletion also Affects miRNA Expression Pattern in Mouse Liver
To determine whether basal miRNA expression changes were due to a change in Nrf2 expression in the Keap1-KD mouse model, the miRNA expression was also quantified in livers of adult Nrf2-null male mice (Fig. 3b). miR-34a, let-7b*, and miR-144 levels were similar between C57BL/6 and Nrf2-null mice. However, miR-370 expression in livers of Nrf2-null mice was reduced to about 25% of that in C57BL/6 livers. Four other miRNAs; miR-146b, miR-485, miR-205, miR-692 were also quantified due to tremendous changes observed in the preliminary miRNA array (Tables I and II) with the pooled samples. However, due to large variability in expression the observations with the individual samples were not appreciable. The results are presented in Supplementary Fig. 2.
CR Decreases Expression of some miRNA Processing and RISC Complex Genes
Next, the effect of CR and Keap1-KD on Drosha and some components of the RISC complex were examined to determine whether Keap1-KD affects the expression miRNA synthesis machinery. Dicer mRNA expression was similar between all groups. Dicer, Exportin 5, and Drosha mRNA expression was similar between C57BL/6 and Keap1-KD mice fed ad libitum. CR decreased Exportin5 and Drosha mRNA expression in C57BL/6 mice, but this was not observed in Keap1-KD mice (Fig. 4).
Fig. 4.
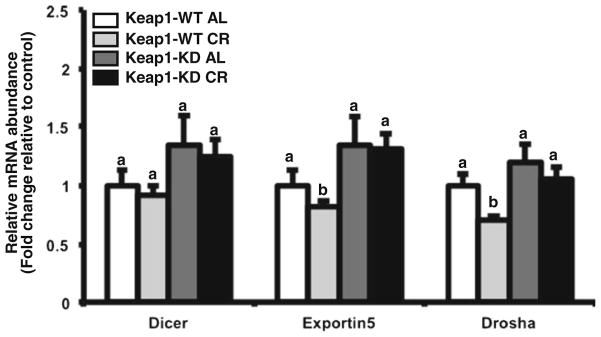
CR and Keap1-KD affects expression of some RISC complex components in liver. Total RNA was isolated from livers of C57BL/6 and Keap1-KD mice fed ad libitum (AL) and placed on 40% CR for 6 weeks. mRNA expression of Dicer, Exportin 5 and Drosha was quantified by RT2-PCR and normalized to 18 s rRNA housekeeping gene. All data is represented as average ± SEM (n=5) fold changes in expression over the controls. Groups without a common letter are considered significantly different from each other (p≤0.05).
DISCUSSION
The purpose of this study was to identify whether knockdown of Keap1, which is associated with increased Nrf2, alters the effects of CR on expression of fatty acid oxidation and lipogenic genes, as well as, identifying changes expression of miRNAs that regulate lipid metabolism and biosynthetic gene expression. The present study puts forth undescribed data illustrating that Keap1 and Nrf2 alter miRNA expression corresponding with induction of fatty acid oxidation gene expression in liver. Data from this study should be interpreted carefully, as caloric restriction was performed in lean mice instead of obese mice (as Keap1 knockdown has been previously described to have effects on diet-induced weight gain and liver lipid content (38,46). Therefore the results herein might not accurately represent the effect of CR in obese mice with Keap1 knockdown. Thus, we consider this study to examine a role for Keap1 knockdown in some aspects of the caloric restriction response.
In the present study, it was observed that Keap1 knockdown slightly increased CR effects. This was observed by an increased percent weight loss after CR, greater liver lipid decrease, increased induction of fatty acid oxidation genes (e.g. Sirt1, Pgc-1α, Pparα, and Cpt1α), and greater decreased expression of lipogenic regulatory genes (Srebp-1c). This observation is similar to some previous publications in lean mice, which have indicated that there is a dose related effect of Nrf2 on suppression of lipid synthetic pathways at the mRNA and protein level in liver (47) – with lack of Nrf2 resulting in increased expression of expression of genes that are associated with lipid synthetic pathways. Also, it is consistent with a previous study that used the triterpenoid Nrf2 activator, CDDO-IM, which increased exercise-induced weight loss (7).
These data differ from other findings in obese mice, which indicate that targeted Nrf2 deletion prevents hepatic steatosis in mice fed a high fat diet and Keap1 knockdown increases hepatic lipid accumulation in leptin-deficient obese mice (7,38). Careful interpretation of the role of Nrf2 and Keap1 role in fatty acid oxidationand the regulation of the genes that dictate the process, is necessary. First, the processes that dictate fat utilization and mobilization in liver under caloric restriction versus lipid accumulation during diet-induced hepatic steatosis are not exactly opposite – with induction of different signaling pathways, coactivator/repressor recruitment, and epigenetic modifications for each condition. Second, current studies indicate that regulation the Nrf2-Keap1 pathway might be different depending upon the metabolic phenotype (lean versus obese, standard versus dietary fat content), length of high fat diet challenge, and presence of leptin. More specifically, regulation of the Nrf2-Keap1 pathway by CR in lean mice might differ with how regulation occurs in obese mice with steatosis. The current study was performed in lean mice and induction of Nqo1 mRNA was detected in liver, which was also reported previously (9). How CR regulates Nrf2 activity and target gene expression in obese or steatotic mice is not described.
Epigenetic regulation has emerged as an important contributor to downstream effects of CR. Preliminary studies have demonstrated that CR intervention causes DNA methylation changes at loci of gene important in insulin signaling and weight control in human adipose tissue (48) and these changes are being considered for biomarker studies during weight loss (49). For example, CR has also been shown to change histone acetylation (39) via changes in expression of histone acetyl transferases and histone deacetylases, specifically Sirt1 models in vitro and in vivo. Scant but convincing data demonstrates the importance of miRNA regulation in effects of CR, with studies addressing effects in brain and adipose tissue. For example, in brain, CR decreased miR-34a expression in brains of aged mice, which is consistent with our observation that CR decreased miR-34a in liver. Herein, we describe the effects of CR on the expression of several miRNAs, which have been identified to have a role in either steatosis or lipid oxidation pathways.
The effect of miRNA expression in the current study can be divided into three parts: miRNA expression altered at the level of basal expression by both Keap1 and Nrf2 knockdown (miR-34a, miR-370, let-7b*), miRNAs altered only by CR (miR-144), and miRNAs that were differentially expressed between C57BL/6 and Keap1-KD mice under CR (miR-34a, miR-370, and let-7b*). In the present study, CR affected miR-34a, miR-370, let7-b* expression in mouse livers. miR-34a, which is known to regulate and be regulated by Sirt1(29), was decreased after CR in C57BL/6 mouse livers. This decrease corresponded to the observed increase in Sirt1 and Pgc-1α mRNA expression. miR-34a expression was lower in Keap-1KD livers in AL mice (Fig. 3a), and decreased further upon CR which corresponded to a highest levels of Sirt1 and Pgc-1α mRNA expression in Keap1-KD mice after CR. In contrast, CR increased miR-34a expression in Nrf2 null mouse livers, indicating and important effect of Nrf2 on regulating miR-34a expression upon activation of CR pathways, such as the AMP-Kinase pathway (50). These results indicate an important, novel effect of Keap1 knockdown, as well as, absence of Nrf2 on the regulation of miR-34a expression and hence, potentially Sirt1 and Pgc-1α expression. As Sirt1-Pgc-1α mediated transcriptional regulation has implications for nutrient metabolism, aging, metabolic disease, response to dietary micronutrients and polyphenolic compound, and cancer (51), a role for the Nrf2-Keap1 pathway in the regulation of Pgc-1α has significant implications.
Similarly, Keap1 knockdown basally decreased expression of miR-370 and let-7b*. miR-370 is known to target the 3′UTR of Pgc-1α (52) and decrease fatty acid oxidation with triglyceride accumulation. In the same study, a dominant negative form of c-jun increased miR-370 expression along with Srebp1c mRNA expression while decreasing Pgc-1α mRNA expression, a pattern which is similar to the observations made in the Keap1 knockdown livers in this study. This potentially indicates towards a positive correlation between c-Jun and Keap1. Nrf2 null mouse livers however, did not have increased miR-370 expression, which potentially indicates towards the importance of Keap1, and not Nrf2 in miR-370 regulation. It has been indicated that Keap1 and c-Jun act as negative regulators of Nrf2 activity (53), thus indicating that Keap1 and c-Jun directly or indirectly via Nrf2 regulate the expression of miR-370 and by association, miR122 (52). The let-7 family of miRNAs affects the expression of genes insulin pathway such as insulin receptor and Insulin Receptor substrate (IRS) (54). The let-7 family also enhances the expression of hemoxygenase-1 (HO-1), an important Nrf2-Keap1 target gene important in anti-oxidant response by inhibiting Bach1 repressor in vitro (55). In this study, let-7b* expression is decreased livers of Keap1-KD mice, whereas it was unaffected in Nrf2 null mice fed AL. These preliminary observations imply a potential negative feedback regulation of let-7b expression by Nrf2, or Keap1 regulation independent of Nrf2. CR did not alter the expression of let-7b in Keap1-KD, as well as, Nrf2 null mice indicating that CR effects of let-7b that are independent of Keap1, and perhaps Nrf2.
CR decreased miR-144, which has not been described previously. Increased miR-144 is associated with oxidative stress in red blood cells and decreased Nrf2 expression and activity (56). However, Keap1KD or Nrf2KO did not alter the basal level expression of hepatic miR-144 in this study, indicating that the miR-144 regulation of Nrf2 expression basally could be via alternate mechanisms. However, CR decreased miR-144 expression with could be correlated to a decreased Nrf2 mRNA expression, thus indicating a potential relationship between Nrf2 and miR-144 expression in CR. These observations are in line with the expression of Gclc, a target gene of Nrf2 which is also significantly downregulated in the CR livers.
The outcome of Keap-1 KD CR mediated effects primarily indicates towards a potential higher efficiency of Keap-1KD mice in weight loss and fatty acid oxidation compared to C57BL/6 mice. Future studies will need to address whether this is a function of Keap1 regulation of transcriptional pathways that are a result of increased Nrf2 activation and/or independent of Nrf2. The miRNA regulatory circuit was differentially expressed in Keap1-KD or Nrf2-null mice, along with a differential effect of CR in Keap1-KDs. Along with other recent studies that document a role for Nrf2 in weight loss or regulation of Srebp-1c (9,57), the present study indicates an important regulatory role of Keap1-Nrf2 pathway in development and progression of steatosis, obesity-diabetes and metabolic syndrome associated adverse outcomes.
The study herein presents novel observations regarding the regulation of miRNAs after CR, the effect of CR on the miRNA processing complex and upstream processors of miRNA processing, including the RISC complex. Basal expression of RISC components measured was not affected by Keap1-KD. CR decreased the expression of Drosha and Exportin5 in AL C57BL/6 mice, but not in Keap1-KD mice. One study has published a decrease in dicer expression upon aging (58), making mice more susceptible to oxidative stress.
Our findings indicate that in lean mice, knockdown of Keap1, and potentially Nrf2 activation, increases the beneficial effects of CR via induction of the genes that promote fatty acid oxidation and suppress lipogenesis. This effect was accompanied by differential expression of some miRNAs known to regulate these processes. Overall, Keap1 knockdown may have beneficial effects in conjunction with CR.
Supplementary Material
ABBREVIATIONS
- Acc1
Acetyl-CoA carboxylase
- Acot1
Acyl-CoA thioesterase 1
- AL
Ad libitum
- Cpt1a
Carnitine palmitoyltransferase 1A
- CR
Caloric Restriction
- Fabp4
Fatty acid binding protein 4
- Fas
Fatty acid synthase
- FXR
Farnesoid X receptor
- GCLC
Glutamate cysteine ligase catalytic subunit
- GST
Glutathione S-Transferase
- InsR
insulin receptor
- Keap1
Kelch like ECH-associated Protein 1
- Lxr
Liver x receptor
- NAFLD
Non-alcoholic fatty liver disease
- NQO1
NAD(P) H: quinone oxidoreductase
- Nrf2
Nuclear factor E2-Related factor 2
- Pgc-1α
Peroxisome proliferator-activated receptor gamma coactivator 1-alpha
- Pparα
Peroxisome Proliferator activated receptor α
- RISC
RNA induced silencing complex
- Scd1
Stearoyl-CoA desaturase
- Sirt1
Sirtuin1
- Srebp1c
Sterol regulatory element binding protein 1c
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s11095-013-1138-9) contains supplementary material, which is available to authorized users.
References
- 1.Itoh K, Igarashi K, Hayashi N, Nishizawa M, Yamamoto M. Cloning and characterization of a novel erythroid cell-derived CNC family transcription factor heterodimerizing with the small Maf family proteins. Mol Cell Biol. 1995;15:4184–93. doi: 10.1128/mcb.15.8.4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moi P, Chan K, Asunis I, Cao A, Kan YW. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc Natl Acad Sci U S A. 1994;91:9926–30. doi: 10.1073/pnas.91.21.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maher JM, Cheng X, Slitt AL, Dieter MZ, Klaassen CD. Induction of the multidrug resistance-associated protein family of transporters by chemical activators of receptor-mediated pathways in mouse liver. Drug Metab Dispos. 2005;33:956–62. doi: 10.1124/dmd.105.003798. [DOI] [PubMed] [Google Scholar]
- 4.Maher JM, Dieter MZ, Aleksunes LM, Slitt AL, Guo G, Tanaka Y, et al. Oxidative and electrophilic stress induces multidrug resistance-associated protein transporters via the nuclear factor-E2-related factor-2 transcriptional pathway. Hepatology. 2007;46:1597–610. doi: 10.1002/hep.21831. [DOI] [PubMed] [Google Scholar]
- 5.McWalter GK, Higgins LG, McLellan LI, Henderson CJ, Song L, Thornalley PJ, et al. Transcription factor Nrf2 is essential for induction of NAD(P)H:quinone oxidoreductase 1, glutathione S-transferases, and glutamate cysteine ligase by broccoli seeds and isothiocyanates. J Nutr. 2004;134:3499S–506S. doi: 10.1093/jn/134.12.3499S. [DOI] [PubMed] [Google Scholar]
- 6.Kay HY, Kim WD, Hwang SJ, Choi HS, Gilroy RK, Wan YJ, et al. Nrf2 inhibits LXRalpha-dependent hepatic lipogenesis by competing with FXR for acetylase binding. Antioxid Redox Signal. 2011;15:2135–2146. doi: 10.1089/ars.2010.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shin S, Wakabayashi J, Yates MS, Wakabayashi N, Dolan PM, Aja S, et al. Role of Nrf2 in prevention of high-fat diet-induced obesity by synthetic triterpenoid CDDO-imidazolide. Eur J Pharmacol. 2009;620:138–44. doi: 10.1016/j.ejphar.2009.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanaka Y, Aleksunes LM, Yeager RL, Gyamfi MA, Esterly N, Guo GL, et al. NF-E2-related factor 2 inhibits lipid accumulation and oxidative stress in mice fed a high-fat diet. J Pharmacol Exp Ther. 2008;325:655–64. doi: 10.1124/jpet.107.135822. [DOI] [PubMed] [Google Scholar]
- 9.Pearson KJ, Lewis KN, Price NL, Chang JW, Perez E, Cascajo MV, et al. Nrf2 mediates cancer protection but not prolongevity induced by caloric restriction. Proc Natl Acad Sci U S A. 2008;105:2325–30. doi: 10.1073/pnas.0712162105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kawai Y, Garduno L, Theodore M, Yang J, Arinze IJ. Acetylation-deacetylation of the transcription factor Nrf2 (nuclear factor erythroid 2-related factor 2) regulates its transcriptional activity and nucleocytoplasmic localization. J Biol Chem. 2011;286:7629–40. doi: 10.1074/jbc.M110.208173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chalasani N, Younossi Z, Lavine JE, Diehl AM, Brunt EM, Cusi K, et al. The diagnosis and management of non-alcoholic fatty liver disease: practice guideline by the American Gastroenterological Association, American Association for the Study of Liver Diseases, and American College of Gastroenterology. Gastroenterology. 2012;142:1592–609. doi: 10.1053/j.gastro.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 12.Chalasani N, Younossi Z, Lavine JE, Diehl AM, Brunt EM, Cusi K, et al. The diagnosis and management of non-alcoholic fatty liver disease: practice guideline by the American Association for the Study of Liver Diseases, American College of Gastroenterology, and the American Gastroenterological Association. Hepatology. 2012;55:2005–23. doi: 10.1002/hep.25762. [DOI] [PubMed] [Google Scholar]
- 13.Cantoand C, Auwerx J. PGC-1alpha, SIRT1 and AMPK, an energy sensing network that controls energy expenditure. Curr Opin Lipidol. 2009;20:98–105. doi: 10.1097/MOL.0b013e328328d0a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeninga EH, Schoonjans K, Auwerx J. Reversible acetylation of PGC-1: connecting energy sensors and effectors to guarantee metabolic flexibility. Oncogene. 2010;29:4617–24. doi: 10.1038/onc.2010.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodgersand JT, Puigserver P. Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc Natl Acad Sci U S A. 2007;104:12861–6. doi: 10.1073/pnas.0702509104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nemoto S, Fergusson MM, Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1{alpha} J Biol Chem. 2005;280:16456–60. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- 17.Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107:823–6. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- 18.Pasquinelliand AE, Ruvkun G. Control of developmental timing by micrornas and their targets. Annu Rev Cell Dev Biol. 2002;18:495–513. doi: 10.1146/annurev.cellbio.18.012502.105832. [DOI] [PubMed] [Google Scholar]
- 19.Millarand AA, Waterhouse PM. Plant and animal microRNAs: similarities and differences. Funct Integr Genomics. 2005;5:129–35. doi: 10.1007/s10142-005-0145-2. [DOI] [PubMed] [Google Scholar]
- 20.Ebertand MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–24. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ajit SK. Circulating microRNAs as biomarkers, therapeutic targets, and signaling molecules. Sensors (Basel) 2012;12:3359–69. doi: 10.3390/s120303359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rottiersand V, Naar AM. MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol. 2012;13:239–50. doi: 10.1038/nrm3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang XW, Heegaard NH, Orum H. MicroRNAs in liver disease. Gastroenterology. 2012;142:1431–43. doi: 10.1053/j.gastro.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, et al. miR-122 regulation of lipid metabolism revealed by in vivo anti-sense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 26.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–9. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 27.Cermelli S, Ruggieri A, Marrero JA, Ioannou GN, Beretta L. Circulating microRNAs in patients with chronic hepatitis C and non-alcoholic fatty liver disease. PLoS One. 2011;6:e23937. doi: 10.1371/journal.pone.0023937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci U S A. 2008;105:13421–6. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leeand J, Kemper JK. Controlling SIRT1 expression by microRNAs in health and metabolic disease. Aging (Albany NY) 2010;2:527–34. doi: 10.18632/aging.100184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davalos A, Goedeke L, Smibert P, Ramirez CM, Warrier NP, Andreo U, et al. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc Natl Acad Sci U S A. 2011;108:9232–7. doi: 10.1073/pnas.1102281108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rayner KJ, Suarez Y, Davalos A, Parathath S, Fitzgerald ML, Tamehiro N, et al. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328:1570–3. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogawa T, Enomoto M, Fujii H, Sekiya Y, Yoshizato K, Ikeda K, et al. MicroRNA-221/222 upregulation indicates the activation of stellate cells and the progression of liver fibrosis. Gut. 2012;61:1600–9. doi: 10.1136/gutjnl-2011-300717. [DOI] [PubMed] [Google Scholar]
- 33.Sun T, Fu M, Bookout AL, Kliewer SA, Mangelsdorf DJ. MicroRNA let-7 regulates 3T3-L1 adipogenesis. Mol Endocrinol. 2009;23:925–31. doi: 10.1210/me.2008-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu H, Shyh-Chang N, Segre AV, Shinoda G, Shah SP, Einhorn WS, et al. The Lin28/let-7 axis regulates glucose metabolism. Cell. 2011;147:81–94. doi: 10.1016/j.cell.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu KC, Cui JY, Klaassen CD. Beneficial role of Nrf2 in regulating NADPH generation and consumption. Toxicol Sci. 2011;123:590–600. doi: 10.1093/toxsci/kfr183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reisman SA, Yeager RL, Yamamoto M, Klaassen CD. Increased Nrf2 activation in livers from Keap1-knockdown mice increases expression of cytoprotective genes that detoxify electrophiles more than those that detoxify reactive oxygen species. Toxicol Sci. 2009;108:35–47. doi: 10.1093/toxsci/kfn267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun. 1997;236:313–22. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 38.Xu J, Kulkarni SR, Donepudi AC, More VR, Slitt AL. Enhanced Nrf2 activity worsens insulin resistance, impairs lipid accumulation in adipose tissue, and increases hepatic steatosis in leptin-deficient mice. Diabetes. 2012;61:3208–18. doi: 10.2337/db11-1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen D, Bruno J, Easlon E, Lin SJ, Cheng HL, Alt FW, et al. Tissue-specific regulation of SIRT1 by calorie restriction. Genes Dev. 2008;22:1753–7. doi: 10.1101/gad.1650608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dongol B, Shah Y, Kim I, Gonzalez FJ, Hunt MC. The acyl-CoA thioesterase I is regulated by PPARalpha and HNF4alpha via a distal response element in the promoter. J Lipid Res. 2007;48:1781–91. doi: 10.1194/jlr.M700119-JLR200. [DOI] [PubMed] [Google Scholar]
- 41.Stulnig TM, Steffensen KR, Gao H, Reimers M, Dahlman-Wright K, Schuster GU, et al. Novel roles of liver X receptors exposed by gene expression profiling in liver and adipose tissue. Mol Pharmacol. 2002;62:1299–305. doi: 10.1124/mol.62.6.1299. [DOI] [PubMed] [Google Scholar]
- 42.Zhang YK, Saupe KW, Klaassen CD. Energy restriction does not compensate for the reduced expression of hepatic drug-processing genes in mice with aging. Drug Metab Dispos. 2010;38:1122–31. doi: 10.1124/dmd.110.032599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reisman SA, Csanaky IL, Aleksunes LM, Klaassen CD. Altered disposition of acetaminophen in Nrf2-null and Keap1-knockdown mice. Toxicol Sci. 2009;109:31–40. doi: 10.1093/toxsci/kfp047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee AP, Sharma A, Song G, Miao J, Mo YY, Wang L, et al. A pathway involving farnesoid X receptor and small heterodimer partner positively regulates hepatic sirtuin 1 levels via microRNA-34a inhibition. The J Biol Chem. 2010;285:12604–12611. doi: 10.1074/jbc.M109.094524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Castro RE, Ferreira DM, Afonso MB, Borralho PM, Machado MV, Cortez-Pinto H, et al. miR-34a/SIRT1/p53 is suppressed by ursodeoxycholic acid in the rat liver and activated by disease severity in human non-alcoholic fatty liver disease. J Hepatol. 2013;58(1):119–25. doi: 10.1016/j.jhep.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 46.More VR, Xu J, Shimpi PC, Belgrave C, Luyendyk JP, Yamamoto M, et al. Keap1 knockdown increases markers of metabolic syndrome after long-term fat diet feeding. Free Radic Biol Med. 2013 doi: 10.1016/j.freeradbiomed.2013.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang YK, Yeager RL, Tanaka Y, Klaassen CD. Enhanced expression of Nrf2 in mice attenuates the fatty liver produced by a methionine- and choline-deficient diet. Toxicol Appl Pharmacol. 2010;245:326–34. doi: 10.1016/j.taap.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bouchard L, Rabasa-Lhoret R, Faraj M, Lavoie ME, Mill J, Perusse L, et al. Differential epigenomic and transcriptomic responses in subcutaneous adipose tissue between low and high responders to caloric restriction. Am J Clin Nutr. 2010;91:309–20. doi: 10.3945/ajcn.2009.28085. [DOI] [PubMed] [Google Scholar]
- 49.Milagro FI, Campion J, Cordero P, Goyenechea E, Gomez-Uriz AM, Abete I, et al. A dual epigenomic approach for the search of obesity biomarkers: DNA methylation in relation to diet-induced weight loss. FASEB J. 2011;25:1378–89. doi: 10.1096/fj.10-170365. [DOI] [PubMed] [Google Scholar]
- 50.Chaudharyand N, Pfluger PT. Metabolic benefits from Sirt1 and Sirt1 activators. Curr Opin Clin Nutr Metab Care. 2009;12:431–7. doi: 10.1097/MCO.0b013e32832cdaae. [DOI] [PubMed] [Google Scholar]
- 51.Yamakuchi M, Lowenstein CJ. MiR-34, SIRT1 and p53: the feedback loop. Cell Cycle Georgetown, Tex. 2009;8:712–5. doi: 10.4161/cc.8.5.7753. [DOI] [PubMed] [Google Scholar]
- 52.Iliopoulos D, Drosatos K, Hiyama Y, Goldberg IJ, Zannis VI. MicroRNA-370 controls the expression of microRNA-122 and Cpt1alpha and affects lipid metabolism. J Lipid Res. 2010;51:1513–23. doi: 10.1194/jlr.M004812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jaiswal AK. Nrf2 signaling in coordinated activation of antioxidant gene expression. Free Radic Biol Med. 2004;36:1199–207. doi: 10.1016/j.freeradbiomed.2004.02.074. [DOI] [PubMed] [Google Scholar]
- 54.Frostand RJ, Olson EN. Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proc Natl Acad Sci USA. 2011;108:21075–21080. doi: 10.1073/pnas.1118922109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hou W, Tian Q, Steuerwald NM, Schrum LW, Bonkovsky HL. The let-7 microRNA enhances heme oxygenase-1 by suppressing Bach1 and attenuates oxidant injury in human hepatocytes. Biochim Biophys Acta. 2012;1819:1113–22. doi: 10.1016/j.bbagrm.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sangokoya C, Telen MJ, Chi JT. microRNA miR-144 modulates oxidative stress tolerance and associates with anemia severity in sickle cell disease. Blood. 2010;116:4338–48. doi: 10.1182/blood-2009-04-214817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu KC, Liu J, Klaassen CD. Role of Nrf2 in preventing ethanol-induced oxidative stress and lipid accumulation. Toxicol Appl Pharmacol. 2012;262:321–329. doi: 10.1016/j.taap.2012.05.010. [DOI] [PubMed] [Google Scholar]
- 58.Mori MA, Raghavan P, Thomou T, Boucher J, Robida-Stubbs S, Macotela Y, et al. Role of microRNA processing in adipose tissue in stress defense and longevity. Cell Metab. 2012;16:336–47. doi: 10.1016/j.cmet.2012.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.