Abstract
Cancer therapeutics are developed through extensive screening; however, many therapeutics evaluated with 2D in vitro cultures during pre-clinical trials suffer from lower efficacy in patients. Replicating the in vivo tumor microenvironment in vitro with three-dimensional (3D) porous scaffolds offers the possibility of generating more predictive pre-clinical models to enhance cancer treatment efficacy. We developed a chitosan and hyaluronic acid (HA) polyelectrolyte complex 3D porous scaffold and evaluated its physical properties. Chitosan-HA (C-HA) scaffolds had a highly porous network. C-HA scaffolds were compared to 2D surfaces for in vitro culture of U-118 MG human glioblastoma (GBM) cells. C-HA scaffold cultures promoted tumor spheroid formation and increased stem-like properties of GBM cells as evidenced by the upregulation of CD44, Nestin, Musashi-1, GFAP, and HIF-1α as compared with 2D cultures. Additionally, the invasiveness of GBM cells cultured in C-HA scaffolds was significantly enhanced compared to those grown in 2D cultures. C-HA scaffold cultures were also more resistant to chemotherapy drugs, which corresponded to the increased expression of ABCG2 drug efflux transporter. These findings suggest that C-HA scaffolds offer promise as an in vitro GBM platform for study and screening of novel cancer therapeutics.
Keywords: spheroid, cancer stem cell, multidrug resistance, invasion, glioma
1. Introduction
Glioblastoma multiforme (GBM) is the most aggressive and deadly malignant primary brain tumor in humans [1]. Despite aggressive surgical resection combined with radiochemotherapy, the prognosis for GBM patients remains dismal with a median survival of approximately 14 months after diagnosis [2, 3]. Novel therapeutic agents undergo significant testing and evaluation before they reach patients: during pre-clinical development they are initially screened with in vitro studies, which are valuable as they allow for high throughput and cost efficient exploration. However, traditional 2D in vitro cultures often fail to simulate the in vivo tumor microenvironment, which has significant effect on cell phenotype, malignancy, and treatment efficacy [4-7]. Therefore, researchers have investigated the use of 3D material scaffolds to create an artificial structure that can mimic the in vivo tumor microenvironment, which could be used as a platform for more representative in vitro study and screening of therapeutics [4-7]. Indeed, cancer cells cultured in 3D structures such as spheroids or porous scaffolds are more malignant than the same cells cultured in 2D, have greater resistance to anti-cancer treatments, and more closely resemble human tumors [4-15]. These 3D structures provide a superior tumor model for in vitro trials due to the arrangement of cancer cells in a 3D structure with increased cell-cell and cell-extracellular matrix (ECM) signaling. The preparation of 3D porous scaffolds with chemical composition resembling native tumor microenvironment ECM could further enhance the malignancy of cultured cancer cells and provide a more predictive analysis of drug efficacy, thus improving the decision on lead compounds.
Porous scaffolds prepared from natural polysaccharides are promising for mimicking the in vivo tumor ECM since they resemble glycosaminoglycans (GAGs), which are essential components of the ECM [16]. Hyaluronic acid (HA) is a natural anionic polymer found in synovial fluid, skin, and cartilage, and is one of the major GAG components in brain ECM [17]. HA is widely used for biomedical applications because of its biocompatibility and water adsorbability [18, 19]. As a result of its remarkable hydrodynamic characteristics, particularly in terms of its viscosity and ability to retain water, HA plays a significant role in the assembly of extracellular and pericellular matrices by regulating porosity and malleability [17, 20]. HA has been known to be associated with GBM tumor growth and invasion, characterized by increased expression in brain tumor stroma and within the advancing edge [17, 21-23]. HA creates a microenvironment that facilitates migration, proliferation, survival, and invasion of GBM cells [17, 21, 22]. Additionally, HA is associated with the malignant state of GBM as it interacts with cell surface receptors, mainly CD44 and RHAMM (receptor for hyaluronic-acid-mediated motility), which activate a wide range of intracellular signaling pathways including those promoting migration and invasion [17, 21, 22, 24].
Despite the significant biophysical properties of HA, its negative charge hinders cell adhesion; therefore, it is blended with other biomaterials to promote cell attachment [25]. Chitosan is a widely used natural cationic polymer derived from crustacean shells that resembles GAGs, and has broad tissue engineering applications in view of its biocompatibility, biodegradability, and hydrophilicity, and is inexpensive and readily available [19]. The cationic nature of chitosan allows it to interact with negatively charged polymers to form a polyelectrolyte complex (PEC) through ionic bonding [26]. The two molecules are prone to swelling individually and therefore do not generate stable scaffolds, but the PEC makes the molecules insoluble [27]. A PEC comprised of chitosan and HA would enhance the properties of the individual polymers, increasing scaffold stability, improving cell adhesion, and increasing the mechanical strength of the material. In fact, chitosan-HA hydrogels have been used for tissue engineering applications [28, 29] and chitosan-HA porous 3D scaffolds for dental pulp regeneration and for cartilage tissue engineering [30, 31]. Chitosan and HA have been used individually for 3D culture of cancer cells [14, 32-34]. Here we demonstrate the use of a C-HA scaffold for mimicking the GBM tumor microenvironment ECM.
In this study, we present the synthesis, materials characterization, and in vitro trials of C-HA 3D porous scaffolds for mimicking the human GBM tumor microenvironment ECM. C-HA scaffolds were prepared by forming PECs between chitosan and HA in solution, then lyophilizing the solution. Our hypothesis was that human GBM cells cultured on C-HA scaffolds would show enhanced malignancy and invasiveness as compared to 2D tissue culture surfaces. The material properties of C-HA scaffolds were characterized with SEM imaging, mechanical testing, mercury porosimetry, FTIR, and swelling behavior. The C-HA scaffolds were seeded with U-118 MG human GBM cells and compared with 2D surfaces to reveal differences in behavior of cultured cells. The in vitro samples were characterized with Alamar blue for proliferation, SEM for cell morphology, immunofluorescence for cell differentiation, qRT-PCR for gene expression, invasion assays, and drug trials.
2. Materials and methods
2.1 Materials
All chemicals were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise indicated. Chitosan (practical grade, > 75% deacetylated, MW = 190,000 – 375,000) and hyaluronic acid (hyaluronic acid sodium salt, from Streptococcus equi) were used as received. Dulbecco’s Modified Eagle Medium (DMEM), antibiotic-antimycotic (AA), Dulbecco’s phosphate buffered saline (D-PBS), Versene, and AlamarBlue reagent were purchased from Invitrogen (Carlsbad, CA). Fetal bovine serum (FBS) was purchased from Atlanta Biologicals (Atlanta, GA). The human GBM cell line (U-118 MG) was purchased from American Type Culture Collection (ATCC, Manassas, VA). Human GBM cells were maintained according to manufacturer’s instruction in fully supplemented DMEM with 10% FBS and 1% AA at 37°C and 5% CO2 in a fully humidified incubator.
2.2 Scaffold synthesis
C-HA scaffolds were prepared by dissolving chitosan (4 wt%) and hyaluronic acid (1 wt%) separately into a 1 wt% acetic acid solution and the solutions were mixed in a Thinky mixer (ARM-300, Thinky USA, Laguna Hills, CA) at 2000 rpm for 3 min twice. The solutions were allowed to completely dissolve by aging at room temperature overnight. The solutions were stirred together, then mixed with the Thinky mixer at 2000 rpm for 5 min twice. The solution was cast in tissue culture plates of 24 wells, refrigerated at 4°C for 12 h, frozen at −20°C overnight, and lyophilized for 24 h. The scaffolds were sectioned into 2 mm thick discs, neutralized with 50 v% ammonium hydroxide for 1 h under vacuum, and washed 4 times with excess DI water to remove any remaining base. The neutralized C-HA scaffolds were frozen at −20°C overnight, lyophilized for over 24 h, and sterilized with ethylene oxide gas for in vitro trials.
2.3. Compressive mechanical testing
C-HA scaffold samples were cut to a size of 4 mm × 4 mm × 4 mm and tested according to the ASTM D5024-07 standard. Dry and wet samples were compressed at a rate of 0.4 mm/min using a micromechanical tester [35] with a 1-kg load cell. Young’s modulus was determined from the sample compressive plots (n = 5 per condition).
2.4. Swelling properties
The swelling behavior of the C-HA scaffolds was evaluated in D-PBS and fully supplemented DMEM. The diameter of dry scaffolds was measured with a micrometer and the measurement axis was marked. The scaffolds were immersed in solution (D-PBS or fully supplemented DMEM) and incubated at 37°C and 5% CO2 in a fully humidified incubator. The samples were measured with a micrometer at regular intervals over a 1-week period. All measurements of the scaffolds were conducted along the marked measurement axis to reduce variation.
2.5. FTIR analysis
The interaction between chitosan and HA molecules in forming polyelectrolyte complexes was characterized with infrared spectroscopy and compared to single polymer samples. Films of the polymer samples were prepared by spin-coating the polymer solution onto 35 mm diameter plastic Petri dishes. The films were allowed to dry overnight in a fume hood. The film was placed in a magnetic holder. Polarized Fourier transformed infrared (FTIR) spectra of 2000 scans at 8 cm−1 were obtained using a Nicolet 5DX spectrometer with a DTGS detector. The sample compartment of the FTIR machine was purged with dry air for 1 hour to remove moisture before characterization.
2.6. Porosity analysis
The porosity, average pore size, and pore size distribution of the scaffolds were measured by an AutoPore IV 9500 mercury porosimeter (Micromeritics Instrument Co., Nacross, GA). The Washburn equation was used to calculate the pore diameter, porosity (%), total pore volume (mL/g), and total pore area (m2/g), and the pore distribution of the scaffolds were determined by measuring the volume of mercury infused. For each measurement, cylindrical scaffolds of 3 mm in diameter and 3 mm in length were placed in a 10 mL penetrometer, subjected to a vacuum of 50 mm Hg, and infused with mercury. Sample weights were measured before and after the mercury infusion.
2.7. Cell seeding on scaffolds
Human GBM cells (U-118 MG) were seeded onto 24-well plate wells and dry 3D porous C-HA scaffolds in 24-well plates at 50,000 cells per sample in 50 μL fully supplemented media. The samples were incubated at 37°C and 5% CO2 in a fully humidified incubator to allow cell adhesion to the substrate for 1 h before 1 mL fully supplemented media was added to each well. The samples were cultured for 15 days with regular media changes.
2.8. Cell proliferation analysis
Proliferation of human glioblastoma cells cultured on 2D wells and 3D porous C-HA scaffolds was assessed with the Alamar blue assay following the manufacturer’s protocol. Briefly, 1 mL of Alamar blue solution (10% Alamar blue reagent in fully supplemented DMEM) was added to each well. The samples were incubated at 37°C for 2 h, then the Alamar blue solution was transferred to a black-bottom 96-well plate to obtain fluorescence values using a SpectraMax M5 microplate reader (Molecular Devices, Union City, CA) at excitation wavelength of 570 nm and fluorescence emission read at 585 nm. The cell number was calculated based on standard curves created for each cell line grown as monolayers. Excess Alamar blue solution was aspirated and fresh fully supplemented media was added to each well.
2.9. Scanning electron microscopy analysis
Cell cultured samples for scanning electron microscopy (SEM) analysis were fixed with 2.5% Karnovsky’s fixative overnight at 4°C. The samples were dehydrated in a series of ethanol washes (0%, 30%, 50%, 75%, 95%, 100%), with each wash performed twice. The samples were critical point dried, sectioned, sputter coated with platinum, and then imaged with a JSM-7000F SEM (JEOL, Tokyo, Japan).
2.10. Immunofluorescence imaging
Immunofluorescence analysis was performed on the 15 day scaffold samples to assess protein expression within the cultured cells. Samples were embedded in OCT and sectioned into 10 μm thick sections on a cryotome and mounted on slides. The sections were fixed in chilled acetone for 10 min, and then washed thrice with D-PBS. The sections were blocked with 10% FBS in D-PBS for 30 min at room temperature. The sections were stained with mouse monoclonal Nestin primary antibody (Millipore, Billerica, MA) and rat monoclonal CD44 primary antibody conjugated with FITC (Abcam, Cambridge, MA) at 1:500 dilution in 10% FBS in D-PBS overnight at 4°C. The slides were washed thrice with D-PBS before incubation with Texas Red conjugated rabbit anti-mouse IgG secondary antibody (Abcam, Cambridge, MA) for 1 h at room temperature (1:500 dilution in 10% FBS in D-PBS). Cell nuclei were counterstained with DAPI (300 nM in D-PBS, Invitrogen) for 10 min and mounted with Prolong Gold (Invitrogen). Images were obtained on an inverted fluorescent microscope (Nikon Instruments, Melville, NY) with the appropriate filters using a Nikon Ri1 Color Cooled Camera System (Nikon Instruments, Melville, NY) and 60× Oil Objective Lens (Nikon Instruments, Melville, NY).
2.11. Quantitative RT-PCR
RNA was extracted from cells harvested from scaffolds and 2D surfaces using the Qiagen RNeasy kit (Qiagen, Valencia, CA). cDNA was prepared using a the Qiagen RNeasy kit (Qiagen, Valencia, CA) following the manufacturer’s protocol. qRT-PCR was used to evaluate the expression of genes of interest. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as the control. SYBR Green PCR Master mix (Bio-Rad, Hercules, CA) was used for template amplification with a primer for each of the transcripts in a Bio-Rad CFX96 real-time PCR detection system. Quantitative amplification was monitored by the level of fluorescence reflecting the cycle number at the detection threshold (crossing point) using a standard curve. Thermocycling for all targets was carried out in a 25 μl solution containing 0.2 μM primers (Integrated DNA Technologies, Coralville, IA) and 4 pg cDNA from the reverse transcription reaction under following conditions: 95°C for 15 min, 45 cycles of denaturation (15 s, 94°C), annealing (30 s, 55°C), and extension (30 s, 72°C).
2.12. Boyden chamber invasion assay
Boyden chamber invasion assays were performed using a method adapted from the literature [36]. Cells were harvested from C-HA scaffolds and 2D surfaces at day 10 by incubating in Versene at 37°C for 15 min. The cells were washed off of the scaffolds or surfaces by pipetting the Versene solution, collected in a 15 mL tube, centrifuged, and washed several times with D-PBS. Cells were resuspended at 1,000,000 cells/mL in serum-free DMEM media and stained with 700 nM DRAQ5 in the dark for 25 min at 4°C with gentle shaking. Following staining, cells were washed twice with serum-free DMEM. Invasion was measured in a 96-well chemotaxis chamber (Model AA96, Neuroprobe, Gaithersburg, MD) using filters with a pore diameter of 10 μm. The filter was coated with collagen by incubating a collagen solution (714 μL rat tail collagen, type I (BD Biosciences, Bedford, MA), 57 μL glacial acetic acid (J. T. Baker, Center Valley, PA), and 49.229 mL deionized water) with the filter in a Ziploc bag at 37°C for 15 min. The upper wells of the chamber were loaded with 50,000 cells in 390 μL DMEM with 1% FBS, while the lower wells contained DMEM with 20% FBS. The chamber was left on the bench top for 10 min before being moved to an incubator for 24 h at 37°C and 5% CO2.
Following incubation, cells that did not migrate were removed from the top side of the filter by wiping with a Kimwipe wetted with D-PBS, followed by a thorough D-PBS rinse to remove all cell debris. Invasion was measured by scanning the filter with an Odyssey Infrared Imaging System (LICOR, Lincoln, NE) to detect DRAQ5-labeled cells that had invaded through the pores of the filter. The filter was scanned with the Odyssey at 680 nm and 700 nm, focus offset was set at 1 mm, resolution = 169 μm, quality = low, and intensity = 4. Background fluorescence was measured in wells that contained medium only and subtracted from each fluorescence reading. A square grid that covered individual wells was set manually and fluorescence within each square was quantified and processed as an individual event.
2.13. Dose-response experiments
U-118 MG cells grown on 2D surfaces and C-HA scaffolds for 10 days were treated with doxorubicin-HCl (DOX) and temozolomide (TMZ) for 4 h before being replaced with fresh medium. Drug concentrations used for the trial were 0.001, 0.01, 0.1, 1, 10 and 100 mg/mL for DOX and 100, 200, 400, 800, 1000 and 2000 μM for TMZ, with n = 3 samples for each trial. Cell viability was examined three days after treatment with the Alamar blue assay as described above. Cell viability was reported as percent of viable cells relative to an untreated control.
2.14. Statistical analysis
All of the data were statistically analyzed to express the mean ± standard deviation (SD) of the mean. Statistical significance was set at p < 0.05 and tested with Student’s t-test.
3. Results and discussion
3.1. C-HA scaffold properties
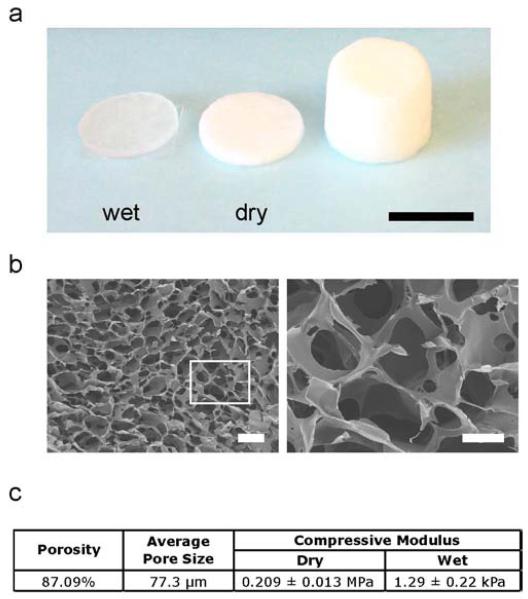
C-HA 3D porous scaffolds were prepared to simulate the native GBM tumor microenvironment ECM. The C-HA scaffolds were prepared at a concentration of 4 wt% chitosan and 1 wt% HA. Figure 1a shows the C-HA scaffolds as produced and cut into discs of 12 mm in diameter and 2 mm in thickness in both dry and wet states. The C-HA scaffolds become nearly transparent when wet, providing a scaffold platform that causes less scattering for microscopy applications than opaque scaffolds. The C-HA scaffolds are highly porous with an interconnected pore network produced by solvent freezing and lyophilizing, as shown in the SEM images in Figure 1b. The scaffold porosity was produced during freezing when the aqueous solvent froze, resulting in phase separation between the solvent and the PEC. The frozen solvent was removed by lyophilizing and left the porous polymer scaffold. Mercury porosimetry indicated that the C-HA scaffolds had an average pore size of 77.31 μm with 87.09% porosity (Figure 1c). The C-HA scaffolds were tested in compression to determine the compressive Young’s modulus, which was 0.209 ± 0.013 MPa and 1.29 ± 0.22 kPa for C-HA, in the dry and wet states, respectively (Figure 1c). The Young’s modulus of brain tissue is in the range of 0.5–1.0 kPa [37], so the C-HA scaffolds have a Young’s modulus in the wet state that more closely matches the stiffness of native brain tissue than tissue culture polystyrene (Young’s modulus of ~106 kPa) [38].
Figure 1.
Physical properties of C-HA scaffold. (a) Photograph of C-HA scaffolds as produced and cut into scaffold discs, wet (left) and dry (right), scale bar is 10 mm. (b) Pore morphology of C-HA scaffolds imaged at low (left) and high magnifications, scale bars are 250 μm (left) and 100 μm (right). (c) C-HA scaffold porosity, average pore size, and compressive mechanical properties in dry and wet conditions.
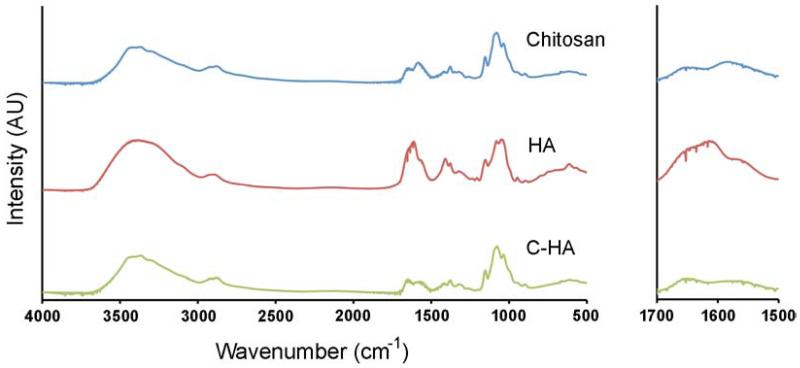
The C-HA scaffolds were examined with FTIR to verify that the chitosan and HA components formed a PEC during mixing. Figure 2 compares the FTIR spectra for 4 wt% chitosan, 1 wt% HA, and 4 wt% – 1 wt% C-HA, which indicates that peaks in the C-HA spectra have shifted from the pure material spectra. The chitosan spectra showed the amide I peak at 1650 cm−1 and the peak at 1580 cm−1 which is due to the overlapping of the amide II peak and the N-H bending vibration of the amine groups present in the deacetylated units [30]. The HA spectra exhibited several highly overlapped peaks in the carbonyl stretching vibration region between 1500–1800 cm−1 [30]. The highest peak in this region (~1612 cm−1) was assigned to the antisymmetrical stretching vibration in the carbonyl group of the carboxylate (COO−) [30]. The amide I and amide II peaks of the acetamide group overlapped with this peak: on the left side of the (COO−) peak as a discrete shoulder (~1647 cm−1) was the amide I peak and on the right side of the peak as a larger shoulder (~1559 cm−1) was the amide II peak [30]. In the C-HA spectra, the amide II peak shifted from 1580 cm−1 to 1566 cm−1 and had a lower intensity peak and the amide I peak at 1650 cm−1 in C-HA spectra was stronger than in chitosan spectra. The C-HA spectra also showed a ratio change in the peaks at 1035 cm−1 and 1080 cm−1, compared to the chitosan spectra. Other HA phase contributions in the C-HA spectra could be seen in minor changes in intensity compared to the chitosan spectra, between 1300 cm−1 and 1410 cm−1, where the C-HA dip at 1339 cm−1 matched HA and the HA peak at 1409 cm−1 increased in the C-HA peak at 1402 cm−1. The influence of HA in the C-HA spectra was muted due to the difference in concentration between 4 wt% chitosan and 1 wt% HA. These changes in the spectra suggest the formation of a chitosan-HA PEC due to the ionic interaction between the negatively charged carboxyl group (−COOH) of HA and the positively charged amino group (−NH2) of chitosan.
Figure 2.
FTIR spectra for 4 wt% chitosan, 1 wt% HA, and 4 wt% – 1 wt% C-HA scaffolds. The right panel highlights the area with the greatest changes in spectra.
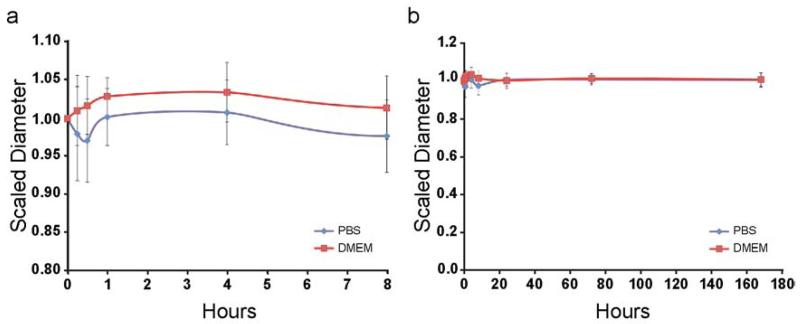
The C-HA scaffolds maintained their shape and showed limited swelling during one-week incubation at 37°C (Figure 3). The scaffolds were stable for one week and did not show any visible signs of degradation during the trial. The scaffolds used for in vitro cultures did not show any signs of degradation during the 15-day cultures. Scaffold neutralization with 50 v% ammonium hydroxide makes the chitosan molecules in the PEC with HA less reactive by deprotonating the unbound amino groups (NH +3 to NH2), promoting scaffold stability. The C-HA scaffold stability is further proof that a PEC between chitosan and HA was formed, since the scaffold would likely have poor stability if a PEC was not formed.
Figure 3.
C-HA scaffold swelling behavior in D-PBS and DMEM. Scaled diameter of C-HA scaffolds as a function of time in incubation (a) in the first 8 hours and (b) over a one-week period.
3.2. GBM cell growth on C-HA scaffolds
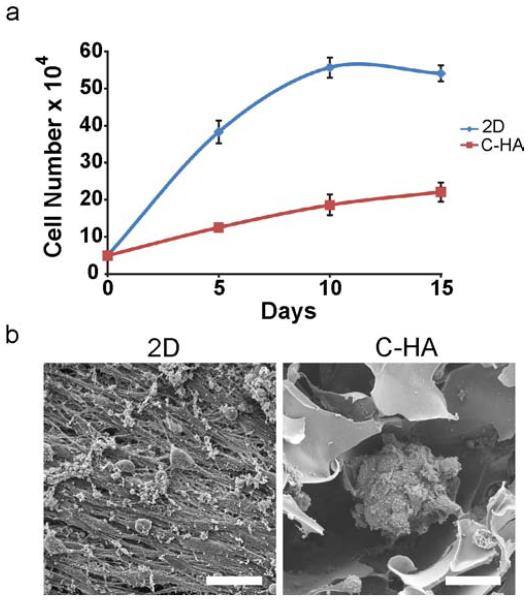
After the C-HA scaffold materials properties were measured, the C-HA scaffolds were evaluated for in vitro culture of U-118 MG human GBM cells for mimicking the in vivo tumor microenvironment ECM. The C-HA scaffolds and 2D controls were seeded with cells and cultured in vitro for 15 days. The U-118 MG cells proliferated on the scaffolds, although at a slower rate than on the 2D controls (Figure 4a). The slower proliferation rate with 3D porous scaffolds is common as the cells adjust to the porous 3D network and due to 3D environmental factors, such as limited diffusion of nutrients and wastes and greater surface area to colonize [5, 6]. Additionally, researchers have shown that human GBM cells proliferate in a stiffness-dependent manner [39], with greater proliferation on stiffer surfaces, partially explaining the difference in proliferation between U-118 MG cells on 2D tissue culture polystyrene and in 3D C-HA scaffolds that has a significantly lower Young’s modulus. The SEM images in Figure 4b revealed the differences in cell morphology between the C-HA scaffolds and 2D controls at day 15. The cells grew as flat monolayers in the 2D controls, while they formed tumor spheroids in C-HA scaffolds, which better resemble the structure of in vivo GBM tumors. The C-HA scaffold culture allowed the cells to form greater numbers of cell-cell and cell-ECM contacts. These results again highlighted the significance of using 3D scaffolds to better mimic in vivo tumor growth and structure.
Figure 4.
C-HA scaffold in vitro performance with U-118 MG human GBM cells. (a) Alamar blue proliferation assay showing U-118 MG proliferation on 2D substrates and C-HA scaffolds over 15 days. (b) SEM images of in vitro U-118 MG cultures at day 15 on 2D substrates and C-HA scaffolds, scale bars are 40 μm.
3.3. Evaluation of HA receptor expression and stem-like properties
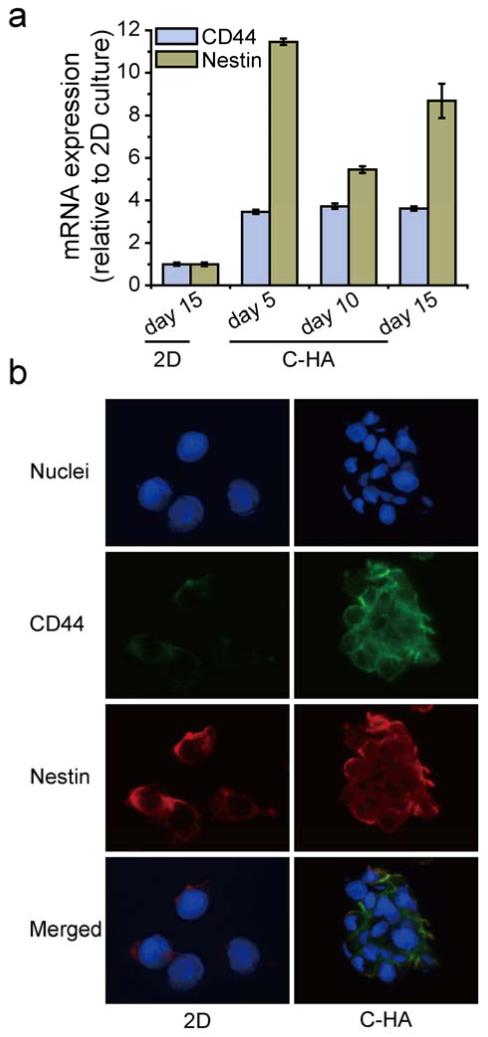
CD44 is the principal HA receptor, and the interaction between CD44 and HA plays a central role in GBM adhesion, proliferation, and invasion through activating downstream cell signaling [17, 40]. U-118 MG human GBM cells cultured on C-HA scaffolds showed consistently increased CD44 expression over 15 days as a result of the increased HA interaction. As shown in Figure 5a, CD44 mRNA expression in U-118 MG cells cultured on C-HA scaffolds for 5 days increased by 3.5 ± 0.1 fold as compared with 2D controls and the expression remained at similar level during the 15-day experiment. In addition to the qRT-PCR results, upregulation of CD44 was also assessed by immunofluorescence at day 15, where C-HA scaffold cultured cells had greater CD44 fluorescence intensity (green, Figure 5b). The continued upregulation of CD44 demonstrates the persistent activated state of GBM cells induced by the C-HA scaffold. Furthermore, CD44 has been used as a GBM stem cell marker [41], suggesting that C-HA scaffold culture may have increased the stemness of GBM cells.
Figure 5.
Expression of CD44 and Nestin by cells cultured for 15 days, measured by (a) qRT-PCR and (b) immunofluorescence. The expression values are relative to the values from 2D cultures at day 15. In immunofluorescence images, CD44 and Nestin are stained green and red, respectively; and nuclei are counterstained blue.
Growing evidence shows that solid brain tumors contain a small subpopulation of slowly cycling cells that undergo self-renewal, termed cancer stem cells or tumor-initiating cells and are believed to be responsible for the relapse of GBM and its resistance to radiochemotherapy [42]. To further test the stem-like properties of GBM cells in C-HA scaffold cultures, qRT-PCR was conducted on additional stem cell markers, including Nestin, Musashi-1, GFAP, and HIF-1α. Nestin is the intermediate filament proteins expressed mainly in neurons where it is implicated in the radial growth of the axon [43]. Nestin expression was also identified in primary brain tumors and has been extensively used as a marker for GBM stem cells and prognostic marker for GBM malignancy [44, 45]. Significantly elevated Nestin expression was demonstrated by qRT-PCR, where C-HA scaffold cultures had 11.5 ± 0.2, 5.5 ± 0.2, and 8.7 ± 0.8 fold higher expression compared to 2D controls at days 5, 10, and 15, respectively (Figure 5a). Nestin upregulation was also indicated by the greater fluorescence intensity compared to 2D controls from immunofluorescence as shown in Figure 5b. Enhanced Nestin expression revealed the GBM stem-like properties promoted by C-HA scaffold cultures. It may also suggest enhanced cell motility, invasive potential, and increased malignancy of GBM cells [46].
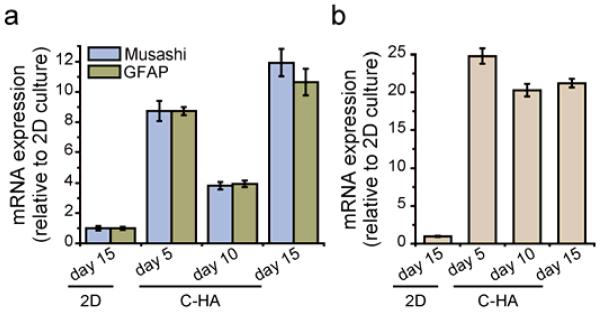
Two other stem cell markers that have been extensively expressed in patient-derived GBM stem cells, Musashi-1 and GFAP, were also investigated. The Musashi family is an evolutionarily conserved group of neural RNA-binding proteins [47]. Musashi-1 protein expression is greater in high-grade tumors than in low-grade tumors, and therefore its expression correlates with the grade of malignancy and proliferative activity in human GBM [48]. GFAP is the astrocyte differentiation marker for neural stem cells that is highly expressed in tumor stem cells [49, 50]. The expression patterns of both gene markers was similar with Nestin, where increased expression in C-HA scaffold cultures was approximately 9, 4, and 11 fold greater than 2D controls at days 5, 10, and 15, respectively (Figure 6a). The expression of these two GBM stem cell markers along with Nestin, all of which are highly expressed in patient-derived GBM stem cells, suggest that C-HA scaffolds improve the tumorigenic properties of GBM cells and facilitate GBM stem-like properties, which therefore makes C-HA scaffold cultured GBM cells more phenotypically similar to the primary GBM tumors. It also demonstrates the application of C-HA scaffolds as a GBM tumor microenvironment-mimetic ECM platform.
Figure 6.
Expression of (a) Musashi and GFAP, and (b) HIF-1α measured by qRT-PCR. Cells were grown on 2D for 15 days.
Hypoxia is a well-recognized tumor microenvironmental condition linked to poor patient outcome and resistance to therapies [51]. Hypoxia-inducible factors (HIFs) are transcription factors upregulated at low oxygen levels and they mediate the cellular hypoxia response influencing angiogenesis, cell survival, invasion, and chemotherapy resistance [52, 53]. Among these factors, HIF-1α is well known as a regulatory factor for stem cell biology and its expression has been identified in GBM stem cells and promotes GBM stemness [54, 55]. In addition, previous studies have shown that hypoxia in tumors can induce resistance to chemotherapeutic agents due to the upregulation of anti-apoptotic proteins [56]. In our studies, significantly increased HIF-1α mRNA expression was observed in C-HA scaffold cultures, with approximately 20-fold greater upregulation compared to 2D cultures throughout the 15-day experiment (Figure 6b). As with previous studies [56, 57], HIF-1α expression in human GBM cells was dramatically elevated in response to 3D culture. This indicates that the C-HA scaffolds may create a local hypoxic environment during in vitro culture, especially in the centers of the spheroids, which could potentially promote the stem-like properties of GBM [55, 57].
3.4. Characterization of invasiveness of GBM cells
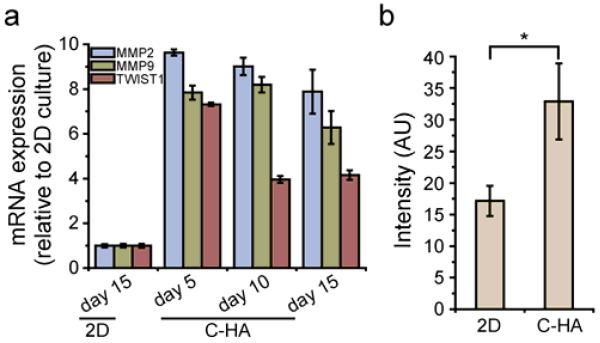
Human GBM is characterized by diffuse invasion into normal brain structures; therefore, markers for cell invasion including MMP-2, MMP-9, and TWIST1 were analyzed with qRT-PCR, and the cultures were evaluated with a Boyden chamber invasion assay. Matrix metalloproteinases (MMPs) have been well recognized as invasive markers for GBM owing to their capability to facilitate primary brain tumor invasion in vivo and migration in vitro by effective degradation of ECM [58, 59]. TWIST1 is an essential regulator of mesodermal development [60], and increased TWIST1 mRNA expression is associated with the transition from low grade to high grade tumor in vivo, indicating a role of TWIST1 in promoting malignant progression and invasion of GBM [61]. As shown in Figure 7a, MMP-2 and MMP-9 expression was elevated for C-HA scaffold cultures at all three time points with an average 8.8 and 7.4 fold increase, respectively, as compared to 2D culture. TWIST1 expression at day 5 was significantly elevated and dropped to a constant, lower level at days 10 and 15. The Boyden chamber invasion assay results correlated with the qRT-PCR results and showed a two-fold increase in invading GBM cells after C-HA scaffold culture. The upregulation of MMP-2, MMP-9 and TWIST1 expression along with the greater cell invasion in the Boyden chamber assay indicates that the C-HA scaffolds promoted the invasiveness in human GBM cells. This could lead to establishment of an in vitro invasion model, which would better replicate the growth of primary tumors in vivo and feature a chemical composition that would promote an invasive phenotype.
Figure 7.
C-HA scaffolds promote invasive phenotype. (a) Expression of MMP2, MMP9, and TWIST1 by cells cultured on C-HA scaffolds over a 15-day period as related to cells cultured on 2D at day 15, as measured by qRT-PCR. (b) Modified Boyden chamber assay results, where 10-day C-HA scaffold cultured cells have a significant difference (p < 0.05) compared with 10-day 2D cultured cells.
3.5. Characterization of drug resistance of GBM cells
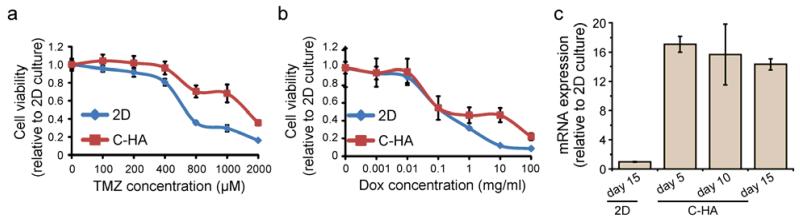
Drug trials were conducted to examine if C-HA scaffolds promoted drug resistance to common cancer chemotherapies, including DOX and TMZ. The C-HA scaffolds cultures showed drug resistance compared to 2D cultures at high doses (Figure 8a, b). The DOX treated cultures showed similar LD50 values for 2D and C-HA scaffolds (0.098 mg/mL), while cultures on C-HA scaffolds had a much greater LD50 values for TMZ, 1171.42 μM and 565.37μM for C-HA scaffolds and 2D controls, respectively. This may due to the spheroid structure of GBM cells on C-HA scaffolds that require the penetration of DOX to the center of the tumor sphere, while cells were equally exposed to the drug on 2D culture. In addition, it was reported that GBM stem cells are mainly localized in the inner core of the tumor spheroid so that cells distributed at the outside layer are less resistant to chemo drugs [62].
Figure 8.
C-HA scaffolds promote drug resistance at high drug concentrations. GBM response to (a) TMZ and (b) Dox. (c) ABCG2 transporter expression measured by qRT-PCR. Characterizations were performed after 10 days of cell culture for (a) and (b).
ATP binding cassette (ABC) transporters superfamily, in particular ABCG2, functions as outward pumps for chemotherapeutic drugs including TMZ and DOX [63, 64]. ABCG2 plays a vital role in GBM survival from chemotherapy and repopulating the tumor after treatment. Moreover, high expression levels of ABC drug transporters have been identified in GBM stem cells and are thought to be responsible for chemoresistance and tumor recurrence [65, 66]. ABCG2 expression was determined using qRT-PCR to investigate if ABCG2 expression increased drug resistance in C-HA scaffold cultures as compared to 2D cultures (Figure 8c). ABCG2 expression was significantly increased on C-HA scaffold cultures compared to 2D cultures, showing increases of 17.1 ± 1.09, 15.6 ± 4.2 and 14.3 ± 0.7 fold on days 5, 10, and 15, respectively, as compared to 2D cultures. Our results revealed the upregulation of this transporter that hampered the cytotoxicity of TMZ and DOX, which conferred drug resistance of C-HA scaffold cultures as compared to 2D cultures. It is possible that other ABC transporters, such as ABCB1, could be involved in the enhanced drug resistance, but ABCG2 is the main stem cell-associated ABC transporter [63], and thus was chosen for this study. This provides further evidence that C-HA scaffolds provide a suitable representation of in vivo tumor ECM.
4. Conclusions
We have demonstrated the production and materials characterization of a 3D porous C-HA scaffold. We showed that C-HA scaffolds provide an in vitro platform for U-118 MG human GBM cells that better mimics the in vivo GBM tumor microenvironment ECM compared to 2D surfaces due to their 3D structure and material composition resembling the native ECM. The cell-ECM contacts, particularly with HA, initiated signaling pathways that produced more malignant cultures than 2D cultures. The C-HA scaffold cultures formed tumor spheroids, had a greater invasive response, and promoted drug resistance compared to 2D monolayers. The C-HA scaffold cultures promoted the overexpression of stem cell markers potentially indicating stem-like properties, but these findings need to be confirmed by functional assays. The C-HA scaffolds demonstrated that they are a viable in vitro platform for GBM culture and evaluation, such as screening of cancer therapeutics, and potentially for other biomedical applications.
Acknowledgements
This work was supported in part by Washington Research Foundation and Kyocera Professorship Endowment to MZ. SJF acknowledges support from the Egtvedt Fellowship. FK acknowledges support from an NIH training grant (T32CA138312). We thank Susan Fung for her assistance with the Boyden Chamber invasion assay. We acknowledge the use of the SEM at the Dept of Materials Science and Engineering at the University of Washington.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Louis D, Ohgaki H, Wiestler O, Cavenee W, Burger P, Jouvet A, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Holland EC. Glioblastoma multiforme: the terminator. Proc Natl Acad Sci U S A. 2000;97:6242–4. doi: 10.1073/pnas.97.12.6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphorn MJB, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–96. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- [4].Bissell MJ, Hines WC. Why don’t we get more cancer? a proposed role of the microenvironment in restraining cancer progression. Nat Med. 2011;17:320–9. doi: 10.1038/nm.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hutmacher DW. Biomaterials offer cancer research the third dimension. Nat Mater. 2010;9:90–3. doi: 10.1038/nmat2619. [DOI] [PubMed] [Google Scholar]
- [6].Hutmacher DW, Loessner D, Rizzi S, Kaplan DL, Mooney DJ, Clements JA. Can tissue engineering concepts advance tumor biology research? Trends Biotechnol. 2010;28:125–33. doi: 10.1016/j.tibtech.2009.12.001. [DOI] [PubMed] [Google Scholar]
- [7].Kimlin LC, Casagrande G, Virador VM. In vitro three-dimensional (3D) models in cancer research: an update. Mol Carcinog. 2013;52:167–82. doi: 10.1002/mc.21844. [DOI] [PubMed] [Google Scholar]
- [8].Benton G, George J, Kleinman HK, Arnaoutova IP. Advancing science and technology via 3D culture on basement membrane matrix. J Cell Physiol. 2009;221:18–25. doi: 10.1002/jcp.21832. [DOI] [PubMed] [Google Scholar]
- [9].Benton G, Kleinman HK, George J, Arnaoutova I. Multiple uses of basement membrane-like matrix (BME/Matrigel) in vitro and in vivo with cancer cells. Int J Cancer. 2011;128:1751–7. doi: 10.1002/ijc.25781. [DOI] [PubMed] [Google Scholar]
- [10].Feder-Mengus C, Ghosh S, Reschner A, Martin I, Spagnoli GC. New dimensions in tumor immunology: what does 3D culture reveal? Trends Mol Med. 2008;14:333–40. doi: 10.1016/j.molmed.2008.06.001. [DOI] [PubMed] [Google Scholar]
- [11].Fischbach C, Chen R, Matsumoto T, Schmelzle T, Brugge JS, Polverini PJ, et al. Engineering tumors with 3D scaffolds. Nat Methods. 2007;4:855–60. doi: 10.1038/nmeth1085. [DOI] [PubMed] [Google Scholar]
- [12].Fischbach C, Kong HJ, Hsiong SX, Evangelista MB, Yuen W, Mooney DJ. Cancer cell angiogenic capability is regulated by 3D culture and integrin engagement. Proc Natl Acad Sci U S A. 2009;106:399–404. doi: 10.1073/pnas.0808932106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hirschhaeuser F, Menne H, Dittfeld C, West J, Mueller-Klieser W, Kunz-Schughart LA. Multicellular tumor spheroids: an underestimated tool is catching up again. J Biotechnol. 2010;148:3–15. doi: 10.1016/j.jbiotec.2010.01.012. [DOI] [PubMed] [Google Scholar]
- [14].Kievit FM, Florczyk SJ, Veiseh O, Park JO, Disis ML, Zhang M. Chitosan-alginate 3D scaffolds as a mimic of the glioma tumor microenvironment. Biomaterials. 2010;31:5903–10. doi: 10.1016/j.biomaterials.2010.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kleinman HK, Martin GR. Matrigel: basement membrane matrix with biological activity. Semin Cancer Biol. 2005;15:378–86. doi: 10.1016/j.semcancer.2005.05.004. [DOI] [PubMed] [Google Scholar]
- [16].Scott JE. Supramolecular organization of extracellular matrix glycosaminoglycans, in vitro and in the tissues. FASEB J. 1992;6:2639–45. [PubMed] [Google Scholar]
- [17].Toole BP. Hyaluronan: from extracellular glue to pericellular cue. Nat Rev Cancer. 2004;4:528–39. doi: 10.1038/nrc1391. [DOI] [PubMed] [Google Scholar]
- [18].Rinaudo M. Main properties and current applications of some polysaccharides as biomaterials. Polym Int. 2008;57:397–430. [Google Scholar]
- [19].Muzzarelli RAA. Chitosan composites with inorganics, morphogenetic proteins and stem cells, for bone regeneration. Carbohydr Polym. 2011;83:1433–45. [Google Scholar]
- [20].Schanté CE, Zuber G, Herlin C, Vandamme TF. Chemical modifications of hyaluronic acid for the synthesis of derivatives for a broad range of biomedical applications. Carbohydr Polym. 2011;85:469–89. [Google Scholar]
- [21].Sironen RK, Tammi M, Tammi R, Auvinen PK, Anttila M, Kosma V-M. Hyaluronan in human malignancies. Exp Cell Res. 2011;317:383–91. doi: 10.1016/j.yexcr.2010.11.017. [DOI] [PubMed] [Google Scholar]
- [22].Toole BP. Hyaluronan-CD44 interactions in cancer: paradoxes and possibilities. Clin Cancer Res. 2009;15:7462–8. doi: 10.1158/1078-0432.CCR-09-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Veiseh M, Turley EA. Hyaluronan metabolism in remodeling extracellular matrix: probes for imaging and therapy of breast cancer. Integr Biol. 2011;3:304–15. doi: 10.1039/c0ib00096e. [DOI] [PubMed] [Google Scholar]
- [24].Naor D, Wallach-Dayan SB, Zahalka MA, Sionov RV. Involvement of CD44, a molecule with a thousand faces, in cancer dissemination. Semin Cancer Biol. 2008;18:260–7. doi: 10.1016/j.semcancer.2008.03.015. [DOI] [PubMed] [Google Scholar]
- [25].Wang XM, He J, Wang Y, Cui FZ. Hyaluronic acid-based scaffold for central neural tissue engineering. Interface Focus. 2012;2:278–91. doi: 10.1098/rsfs.2012.0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Berger J, Reist M, Mayer JM, Felt O, Gurny R. Structure and interactions in chitosan hydrogels formed by complexation or aggregation for biomedical applications. Eur J Pharm Biopharm. 2004;57:35–52. doi: 10.1016/s0939-6411(03)00160-7. [DOI] [PubMed] [Google Scholar]
- [27].Rinaudo M. Properties and degradation of selected polysaccharides: hyaluronan and chitosan. Corros Eng, Sci Technol. 2007;42:324–34. [Google Scholar]
- [28].Tan H, Chu CR, Payne KA, Marra KG. Injectable in situ forming biodegradable chitosan-hyaluronic acid based hydrogels for cartilage tissue engineering. Biomaterials. 2009;30:2499–506. doi: 10.1016/j.biomaterials.2008.12.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Tan H, Rubin JP, Marra KG. Injectable in situ forming biodegradable chitosan-hyaluronic acid based hydrogels for adipose tissue regeneration. Organogenesis. 2010;6:173–80. doi: 10.4161/org.6.3.12037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Coimbra P, Alves P, Valente TAM, Santos R, Correia IJ, Ferreira P. Sodium hyaluronate/chitosan polyelectrolyte complex scaffolds for dental pulp regeneration: synthesis and characterization. Int J Biol Macromol. 2011;49:573–9. doi: 10.1016/j.ijbiomac.2011.06.011. [DOI] [PubMed] [Google Scholar]
- [31].Correia CR, Moreira-Teixeira LS, Moroni L, Reis RL, van Blitterswijk CA, Karperien M, et al. Chitosan scaffolds containing hyaluronic acid for cartilage tissue engineering. Tissue Eng Part C Methods. 2011;17:717–30. doi: 10.1089/ten.tec.2010.0467. [DOI] [PubMed] [Google Scholar]
- [32].Campbell JJ, Davidenko N, Caffarel MM, Cameron RE, Watson CJ. A multifunctional 3D co-culture system for studies of mammary tissue morphogenesis and stem cell biology. PLoS One. 2011;6:e25661. doi: 10.1371/journal.pone.0025661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Leung M, Kievit FM, Florczyk SJ, Veiseh O, Wu J, Park JO, et al. Chitosan-alginate scaffold culture system for hepatocellular carcinoma increases malignancy and drug resistance. Pharm Res. 2010;27:1939–48. doi: 10.1007/s11095-010-0198-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Florczyk SJ, Liu G, Kievit FM, Lewis AM, Wu JD, Zhang M. 3D porous chitosan-alginate scaffolds: a new matrix for studying prostate cancer cell-lymphocyte interactions in vitro. Adv Healthc Mater. 2012;1:590–9. doi: 10.1002/adhm.201100054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Edmondson D, Bhattarai N, Jana S, Kim A, Zhang M. Design and evaluation of a nano-scale differential tensile test device for nanofibers. Appl Phys Lett. 2009;94:103101. [Google Scholar]
- [36].Miller AM, Stella N. Microglial cell migration stimulated by ATP and C5a involve distinct molecular mechanisms: quantification of migration by a novel near-infrared method. Glia. 2009;57:875–83. doi: 10.1002/glia.20813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Pettikiriarachchi JTS, Parish CL, Shoichet MS, Forsythe JS, Nisbet DR. Biomaterials for brain tissue engineering. Aust J Chem. 2010;63:1143–54. [Google Scholar]
- [38].Gilbert PM, Havenstrite KL, Magnusson KEG, Sacco A, Leonardi NA, Kraft P, et al. Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Science. 2010;329:1078–81. doi: 10.1126/science.1191035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Ananthanarayanan B, Kim Y, Kumar S. Elucidating the mechanobiology of malignant brain tumors using a brain matrix-mimetic hyaluronic acid hydrogel platform. Biomaterials. 2011;32:7913–23. doi: 10.1016/j.biomaterials.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Merzak A, Koocheckpour S, Pilkington GJ. CD44 mediates human glioma cell adhesion and invasion in vitro. Cancer Res. 1994;54:3988–92. [PubMed] [Google Scholar]
- [41].Anido J, Saez-Borderias A, Gonzalez-Junca A, Rodon L, Folch G, Carmona MA, et al. TGF-beta receptor inhibitors target the CD44(high)/Id1(high) glioma-initiating cell population in human glioblastoma. Cancer Cell. 2010;18:655–68. doi: 10.1016/j.ccr.2010.10.023. [DOI] [PubMed] [Google Scholar]
- [42].Stiles CD, Rowitch DH. Glioma stem cells: a midterm exam. Neuron. 2008;58:832–46. doi: 10.1016/j.neuron.2008.05.031. [DOI] [PubMed] [Google Scholar]
- [43].Reynolds BA, Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255:1707–10. doi: 10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- [44].Salmaggi A, Boiardi A, Gelati M, Russo A, Calatozzolo C, Ciusani E, et al. Glioblastoma-derived tumorospheres identify a population of tumor stem-like cells with angiogenic potential and enhanced multidrug resistance phenotype. Glia. 2006;54:850–60. doi: 10.1002/glia.20414. [DOI] [PubMed] [Google Scholar]
- [45].Strojnik T, Rosland GV, Sakariassen PO, Kavalar R, Lah T. Neural stem cell markers, nestin and musashi proteins, in the progression of human glioma: correlation of nestin with prognosis of patient survival. Surg Neurol. 2007;68:133–44. doi: 10.1016/j.surneu.2006.10.050. [DOI] [PubMed] [Google Scholar]
- [46].Wan F, Herold-Mende C, Campos B, Centner F-S, Dictus C, Becker N, et al. Association of stem cell-related markers and survival in astrocytic gliomas. Biomarkers. 2011;16:136–43. doi: 10.3109/1354750X.2010.536256. [DOI] [PubMed] [Google Scholar]
- [47].Okano H, Imai T, Okabe M. Musashi: a translational regulator of cell fate. J Cell Sci. 2002;115:1355–9. doi: 10.1242/jcs.115.7.1355. [DOI] [PubMed] [Google Scholar]
- [48].Toda M, Iizuka Y, Yu W, Imai T, Ikeda E, Yoshida K, et al. Expression of the neural RNA-binding protein musashi1 in human gliomas. Glia. 2001;34:1–7. doi: 10.1002/glia.1034. [DOI] [PubMed] [Google Scholar]
- [49].Gürsel DB, Shin BJ, Burkhardt J-K, Kesavabhotla K, Schlaff CD, Boockvar JA. Glioblastoma stem-like cells—biology and therapeutic implications. Cancers. 2011;3:2655–66. doi: 10.3390/cancers3022655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Zhang QB, Ji XY, Huang Q, Dong J, De Zhu Y, Lan Q. Differentiation profile of brain tumor stem cells: a comparative study with neural stem cells. Cell Res. 2006;16:909–15. doi: 10.1038/sj.cr.7310104. [DOI] [PubMed] [Google Scholar]
- [51].Kaur B, Khwaja FW, Severson EA, Matheny SL, Brat DJ, Van Meir EG. Hypoxia and the hypoxia-inducible-factor pathway in glioma growth and angiogenesis. Neuro Oncol. 2005;7:134–53. doi: 10.1215/S1152851704001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Bar EE. Glioblastoma, cancer stem cells and hypoxia. Brain Pathol. 2011;21:119–29. doi: 10.1111/j.1750-3639.2010.00460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Zagzag D, Zhong H, Scalzitti JM, Laughner E, Simons JW, Semenza GL. Expression of hypoxia-inducible factor 1 alpha in brain tumors - association with angiogenesis, invasion, and progression. Cancer. 2000;88:2606–18. [PubMed] [Google Scholar]
- [54].Li Z, Bao S, Wu Q, Wang H, Eyler C, Sathornsumetee S, et al. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell. 2009;15:501–13. doi: 10.1016/j.ccr.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Soeda A, Park M, Lee D, Mintz A, Androutsellis-Theotokis A, McKay RD, et al. Hypoxia promotes expansion of the CD133-positive glioma stem cells through activation of HIF-1 alpha. Oncogene. 2009;28:3949–59. doi: 10.1038/onc.2009.252. [DOI] [PubMed] [Google Scholar]
- [56].Kim JW, Ho WJ, Wu BM. The role of the 3D environment in hypoxia-induced drug and apoptosis resistance. Anticancer Res. 2011;31:3237–45. [PubMed] [Google Scholar]
- [57].Bar EE, Lin A, Mahairaki V, Matsui W, Eberhart CG. Hypoxia increases the expression of stem-cell markers and promotes clonogenicity in glioblastoma neurospheres. Am J Pathol. 2010;177:1491–502. doi: 10.2353/ajpath.2010.091021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Lakka SS, Rajan M, Gondi C, Yanamandra N, Chandrasekar N, Jasti SL, et al. Adenovirus-mediated expression of antisense MMP-9 in glioma cells inhibits tumor growth and invasion. Oncogene. 2002;21:8011–9. doi: 10.1038/sj.onc.1205894. [DOI] [PubMed] [Google Scholar]
- [59].Deryugina EI, Bourdon MA, Luo GX, Reisfeld RA, Strongin A. Matrix metalloproteinase-2 activation modulates glioma cell migration. J Cell Sci. 1997;110:2473–82. doi: 10.1242/jcs.110.19.2473. [DOI] [PubMed] [Google Scholar]
- [60].Mikheeva SA, Mikheev AM, Petit A, Beyer R, Oxford RG, Khorasani L, et al. TWIST1 promotes invasion through mesenchymal change in human glioblastoma. Mol Cancer. 2010:9. doi: 10.1186/1476-4598-9-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Elias MC, Tozer KR, Silber JR, Mikheeva S, Deng M, Morrison RS, et al. TWIST is expressed in human gliomas and promotes invasion. Neoplasia. 2005;7:824–37. doi: 10.1593/neo.04352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Pistollato F, Abbadi S, Rampazzo E, Persano L, Della Puppa A, Frasson C, et al. Intratumoral hypoxic gradient drives stem cells distribution and MGMT expression in glioblastoma. Stem Cells. 2010;28:851–62. doi: 10.1002/stem.415. [DOI] [PubMed] [Google Scholar]
- [63].Bleau A-M, Huse JT, Holland EC. The ABCG2 resistance network of glioblastoma. Cell Cycle. 2009;8:2937–45. [PubMed] [Google Scholar]
- [64].Martin V, Sanchez-Sanchez AM, Herrera F, Gomez-Manzano C, Fueyo J, Alvarez-Vega MA, et al. Melatonin-induced methylation of the ABCG2/BCRP promoter as a novel mechanism to overcome multidrug resistance in brain tumour stem cells. Br J Cancer. 2013;108:2005–12. doi: 10.1038/bjc.2013.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Chua C, Zaiden N, Chong K-H, See S-J, Wong M-C, Ang B-T, et al. Characterization of a side population of astrocytoma cells in response to temozolomide. J Neurosurg. 2008;109:856–66. doi: 10.3171/JNS/2008/109/11/0856. [DOI] [PubMed] [Google Scholar]
- [66].Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5:275–84. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]