Summary
Although mycoplasmas have a paucity of glycosyltransferases and nucleotidyltransferases recognizable by bioinformatics, these bacteria are known to produce polysaccharides and glycolipids. We show here that mycoplasmas also produce glycoproteins and hence have glycomes more complex than previously realized. Proteins from several species of Mycoplasma reacted with a glycoprotein stain, and the murine pathogen Mycoplasma arthritidis was chosen for further study. The presence of M. arthritidis glycoproteins was confirmed by high-resolution mass spectrometry. O-linked glycosylation was clearly identified at both serine and threonine residues. No consensus amino acid sequence was evident for the glycosylation sites of the glycoproteins. A single hexose was identified as the O-linked modification, and glucose was inferred by 13C labeling to be the hexose at several of the glycosylation sites. This is the first study to conclusively identify sites of protein glycosylation in any of the mollicutes.
Keywords: FT-MS, general glycosylation, glycosyltransferase, lipoprotein, mollicutes
Introduction
Mycoplasmas (Class Mollicutes) are noted for causing chronic diseases of the respiratory and genital tracts and joints in many animals including humans. Factors that contribute to disease chronicity are largely unknown but include polysaccharides that protect the mycoplasma from host defenses (Bolland et al., 2012, Shaw et al., 2013). Mycoplasmas are related to Gram-positive bacteria but lack a cell wall and have a streamlined genome, usually containing 600–800 protein-coding regions. All species of Mycoplasma require cholesterol for growth and hence are found in nature only in an animal host. The fastidious nature of the mycoplasmas is usually overcome in the laboratory by the use of rich culture medium containing undefined components such as animal serum, which introduces an enormous variety of glycoconjugates that can adsorb to the surface of the mycoplasma and confound interpretation of carbohydrate assays. We have adapted the serum-free medium described by Yus et al. for growth of several species of mycoplasma to facilitate studies on glycobiology (Yus et al., 2009).
As the technology for glycoprotein identification has become more refined, it is becoming clear that these molecules are not as rare in bacteria as once thought (Nothaft and Szymanski, 2010, Iwashkiw et al., 2013). A few previous reports suggested the possibility of mycoplasmal glycoproteins, but in most cases it was not determined whether the glycoproteins were produced by the mycoplasma or were components of the serum-containing culture medium that had adhered to the mycoplasma surface (Kobayashi and Watanabe, 1991, Kahane and Brunner, 1977). Two putative glycoproteins were identified in Mycoplasma gallisepticum by staining with Pro-Q Emerald, but the glycan and glycosylation sites were not determined (Demina et al., 2009). The data presented here conclusively demonstrate that numerous proteins in M. arthritidis and probably other species of mycoplasma are glycosylated. The identified glycosylation sites were O-linked at serine or threonine residues. The finding of what appears to be general protein glycosylation in organisms with genomes as minimal as mycoplasmas suggests that glycosylation is fundamentally important and possibly exists in most organisms.
Results
Glycoprotein staining
Proteins from several species of Mycoplasma were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and stained with Pro-Q Emerald 300. Significant levels of staining were apparent near the top of the gels and generalized staining throughout the gels approached a smear (Fig. 1, panel A). M. arthritidis was chosen for further study because numerous proteins appeared to react with the glycostain. We reasoned that if glycoproteins were produced by mycoplasmas, some of these proteins would be at the cell surface. Many surface proteins in M. arthritidis and other species of mycoplasma are lipoproteins (Dybvig et al., 2008). Lipoproteins partition into the detergent phase when extracted with Triton X-114 (TX-114), whereas most other proteins partition into the aqueous phase (Bordier, 1981). Although an analysis of TX-114-extracted protein would not identify the complete set of glycoproteins of M. arthritidis, it was highly likely that at least some of the glycoproteins would be present in this fraction.
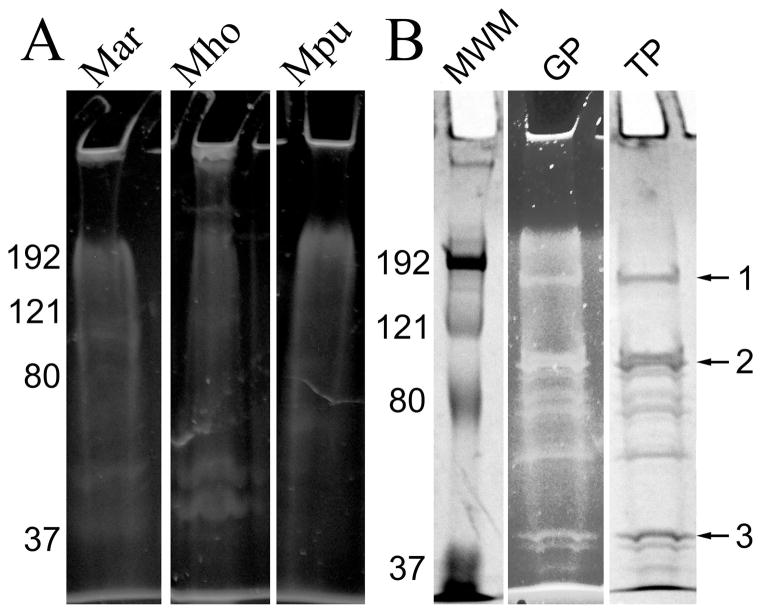
Fig. 1.
Glycoprotein staining of SDS-PAGE gels. Panel A, total proteins from M. arthritidis (Mar), M. hominis (Mho), and M. pulmonis (Mpu), stained for glycoproteins with Pro-Q Emerald 300. Panel B: lane MWM, molecular weight markers (Bio-Rad Kaleidoscope); lane GP, glycoproteins extracted from M. arthritidis with TX-114 and stained with Pro-Q Emerald 300; lane TP, total proteins identified by subsequent staining with Coomassie of lane GP. Numbers on the left of the gels refer to molecular weight standards in kDa. Arrows on the right refer to bands excised for FT-MS analysis.
TX-114-extracted material was analyzed on SDS-PAGE gels, stained for glycoproteins with Pro-Q Emerald 300, and then stained again with Coomassie (Fig. 1B). Multiple bands reacted with the glycoprotein stain, suggesting that glycoproteins were abundant in the lipoprotein fraction. When compared to whole cell lysates, the lipoprotein fraction concentrated the glycoprotein staining into distinct bands. Three bands (identified by arrows in Fig. 1B) that had the same mobility as the bands that reacted with the glycoprotein stain were excised from Coomassie-stained gels and digested with trypsin. The peptides were analyzed by matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF). The results confirmed that all proteins in the bands originated from M. arthritidis. The predominant proteins in bands 1–3 were MARTH_455, MARTH_403, and MARTH_819, respectively. Band 2 also contained a significant amount of the protein MARTH_665. All of these are annotated as lipoproteins of unknown function (GenBank accession number CP001047).
Monosaccharide analysis of TX-114 extracted material and sugar transport
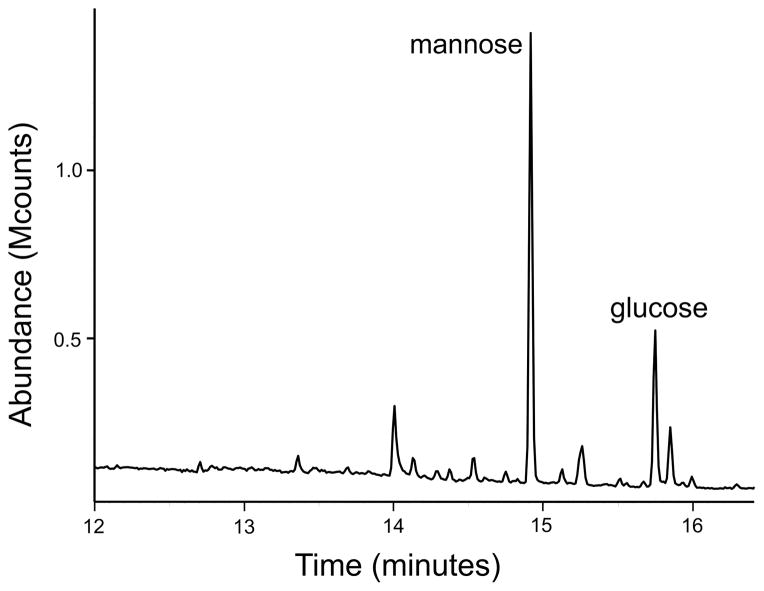
Gas chromatography showed glucose and mannose to be the major sugars present in the lipoprotein fraction of M. arthritidis (Fig. 2). Import of glucose and mannose into the non-glycolytic M. arthritidis was investigated in an effort to clarify which of these sugars was potentially associated with glycoproteins. When incubated with 14C glucose or 14C mannose for periods of time ranging from 0 to 48 hours, no radiolabeling of the mycoplasmas occurred. Using the culture conditions employed in this study, M. arthritidis does not import either monosaccharide.
Fig. 2.
GC/MS of TX-114 extracted material from M. arthritidis.
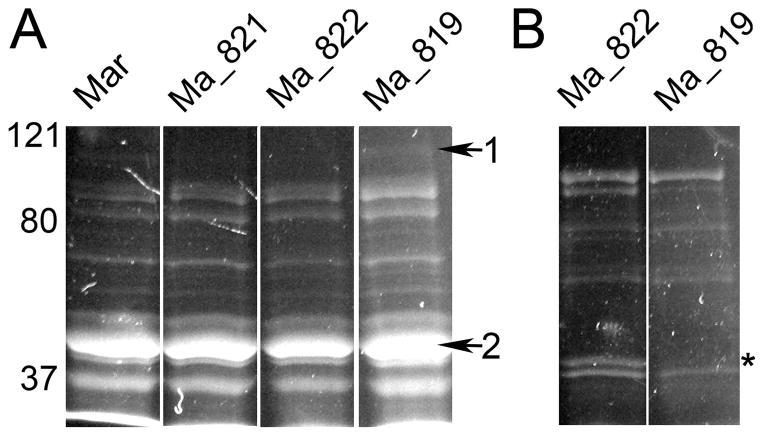
Studies are ongoing that investigate oligosaccharide import in mycoplasmas. M. arthritidis has only one putative operon, containing MARTH_orf 819, -821, -822 and -823, annotated as possibly coding for a sugar import system. The annotation of this operon is uncertain because only the predicted MARTH_821 protein bears strong resemblance to a sugar importer. A transposon library of M. arthritidis has been previously described and contains mutants with disruptions in these genes (Dybvig et al., 2008). The aqueous and detergent phases of TX-114 extracts of these mutants were examined on SDS-PAGE gels stained for glycoproteins. All of the mutants had essentially the same profile with multiple proteins stained from both the aqueous and TX-114 fractions (Fig. 3). The proteins in the bands labeled in Fig. 3A as 1 and 2 were analyzed by mass spectrometry. The predominant protein in band 1 was the MARTH_orf627 product, the LonA protease. The major proteins of band 2 were the products of MARTH_orf411, -530, -812, and -873, coding for the RNA polymerase subunit RpoA, phosphoglycerate kinase, a predicted aminopeptidase, and arginine deiminase, respectively. MARTH_819 is a predicted lipoprotein of 45 kDa, and a stained protein of about this size is missing in the TX-114 detergent phase in the MARTH_orf 819 mutant (Fig. 3B, lane 2), suggesting that this protein is glycosylated as confirmed below.
Fig. 3.
Glycoprotein staining of SDS-PAGE gels of proteins from potential sugar import mutants. Mycoplasmas were grown in MB, and proteins were extracted with TX-114. Panel A, proteins found in the aqueous phase. Lanes labeled as Ma-821, -822, and -819 have transposon disruptions in MARTH_orf821, -822, and -819, respectively. The lane labeled Mar is the wild-type TnCtrl strain. Arrows refer to bands that were excised and analyzed by mass spectrometry for protein identification. Panel B, proteins that partitioned into the TX-114 phase. A protein of about 45 kDa that reacted with the glycostain is missing in the MARTH_orf 819 mutant and is indicated by an asterisk.
FT-MS
Peptides were analyzed by liquid chromatography coupled to a hybrid linear quadrupole ion trap Orbitrap Velos Fourier transform mass spectrometer (Orbitrap MS) for glycosylation analysis. Specific glycopeptides that contained a hexose (162.0795 Da) were identified from the Orbitrap MS scans. Candidate glycopeptides were initially identified from the tandem mass spectra of trypsinized proteins fragmented by collision-induced dissociation (CID) or electron transfer dissociation (ETD). Because the primary sugars found in the lipoprotein fraction were glucose and mannose, the CID and ETD spectra were searched for peptides that had the addition of one or more hexoses. The candidate glycopeptides were individually analyzed to determine whether the fragment ions could be matched with the ions predicted from the amino acid sequence of the proteins identified by MALDI-TOF.
The MS-1 scans of the peptides identified in the initial screening were analyzed for mass shifts with the addition of a glycan. Both the glycosylated and nonglycosylated peptides were identified in multiple charge states for 12 of the glycopeptides (summarized in Table 1), confirming protein glycosylation. In each case a single hexose was attached to serine or threonine. Two pairs of the 12 glycopeptides were overlapping the same glycosylation site. Hence, a total of 10 distinct glycosylation sites were identified. Six of these sites were in the same protein, MARTH_403. Single sites of glycosylation were identified in MARTH_455 and MARTH_665. Two glycosylation sites were found in MARTH_819, consistent with the glycostaining observed in Fig. 3B, lane Ma_819.
Table 1.
Protein glycosylation.
| Glycosylated peptide sequence | Protein designationa | Positionb | Verificationsc |
|---|---|---|---|
| LELAKQVILTLDDGTVKd | MARTH_403 | 117/918 | MS-1, CID, 13C, OP See Figs. S1–S3 |
| RLELAKQVILTLDDGTVK | MARTH_403 | 117/918 | MS-1, CID, 13C, OP See Figs. S4–S6 |
| SINSKQFLEDLKK | MARTH_403 | 164/918 | MS-1, CID, 13C See Figs. S7–S9 |
| KEGATADFENLINK | MARTH_403 | 295/918 | MS-1, ETD, CID, 13C See Figs. 3–5 |
| YQQRPQEKEIFSTR | MARTH_403 | 427/918 | MS-1, CID See Figs. S10 –S11 |
| TWNLYKQGQLSSIPFSTLTQAQQQEAIR | MARTH_403 | 430/918 | MS-1, CID See Figs. S12–S13 |
| LSELAEDLAKYEESHK | MARTH_403 | 779/918 | MS-1, CID, 13C, OP See Figs. S14–S17 |
| YEESHKIIGSIPFGDFDK | MARTH_403 | 779/918 | MS-1, CID, OP See Figs. S18–S19 |
| DILENKDDSLSTQGK | MARTH_455 | 795/1949 | MS-1, CID See Figs. S20–S21 |
| YINKLEALDENDLTPDSLAWAR | MARTH_665 | 168/881 | MS-1, CID, 13C Figs. S22–S23 |
| TAVITDGGDINDISFNQSAWEGVLNFMEQVKAPIQK | MARTH_819 | 79/431 | MS-1, CID See Figs. S24–S26 |
| VIDGTFQEAYKR | MARTH_819 | 107/431 | MS-1, CID, 13C See Figs. S27–S29 |
GenBank accession number CP001047. Each of the proteins identified in the table have unknown function but possess a signal peptide sequence indicative of a lipoprotein.
Amino acid position of the glycan/protein total length
MS-1, LC-MS mass shift corresponding to hexose; CID, tandem mass spectrometry fragments generated by collision induced dissociation; ETD, tandem mass spectrometry fragments generated by electron transfer dissociation; 13C, LC-MS ions of glycosylated peptide in 13C-labeled and unlabeled forms; OP, overlapping peptides with same modification
Underlined amino acid is glycosylated
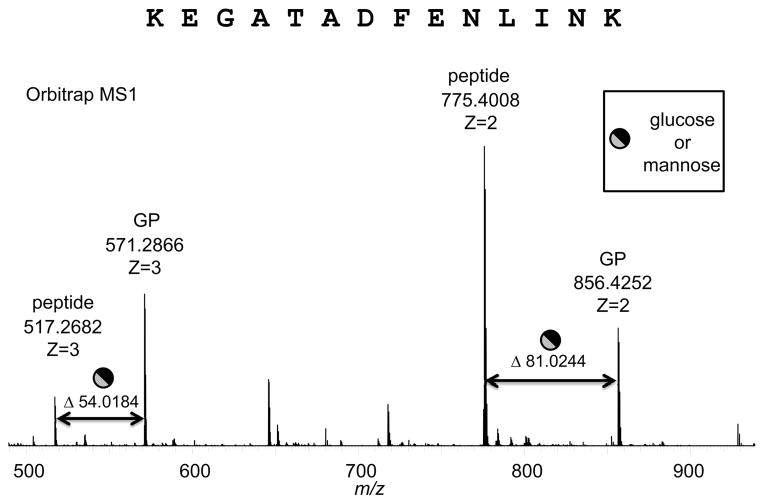
One of the glycosylation sites in MARTH_403 is at Thr295, within the identified peptide sequence of KEGATADFENLINK. The charged species obtained from the Orbitrap MS of the peptide generated two sets of fragments, the doubly-charged species at m/z 856.4252 (glycosylated) and m/z 775.8640 (nonglycosylated) and the triply-charged species at m/z 571.2866 (glycosylated) and m/z 517.2682 (nonglycosylated) (Fig. 4). The mass shift between these two sets is determined to be 162.05 Da, the average calculated mass of a hexose linked through an O-linkage. The H2O (18.0153 Da) lost during the glycosidic bond formation is deducted from the mass of monoisotopic hexose (180.0634 Da) for these calculations, not the amino acid. The mass accuracy associated with these techniques is exquisite and strongly indicates that the glycan mass and glycosylation sites are correctly identified.
Fig. 4.
Glycosylation of Thr295 in the peptide KEGATADFENLINK of MARTH_403. Orbitrap MS1 showing mass shift of the triply- and doubly-charged ions. The 54.0184 shift for z = 3 between the non-glycosylated peptide at m/z 517.2682 and the glycosylated peptide (GP) at m/z 571.2866 equates to a mass shift of 162.0552 Da, which corresponds to hexose glycosylation with a mass accuracy of 0.007 Da. The 81.0244 shift for z = 2 between the non-glycosylated peptide and the glycosylated peptide equates to a mass shift of 162.0488 Da, which corresponds to a hexose with a mass accuracy of 0.0006 Da.
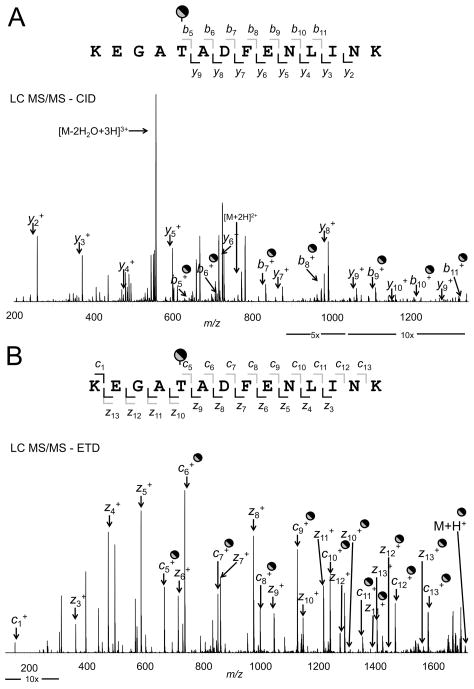
The CID and ETD spectra for KEGAT295ADFENLINK are shown in Fig. 5. LC MS/MS-CID was conducted on the peptide at m/z 571.2866 (z = 3) captured by the Orbitrap MS. The b and y ions corresponded to the predicted pattern from ProteinProspector with the b series showing the addition of a hexose (Fig. 5A). The LC MS/MS-ETD showing the c and z ions for this glycosylated peptide fragment is shown in (Fig. 5B). The fragments at z10, z11, z12, and z13 were identified in both the glycosylated and nonglycosylated forms adding to the support for glycosylation of this protein.]
Fig. 5.
Tandem MS of the glycosylation of the peptide KEGAT295ADFENLINK of MARTH_403. Panel A, LC MS/MS-CID of the glycosylated peptide showing the assigned b and y ions. Panel B, LC MS/MS-ETD of the glycosylated peptide showing the assigned c and z ions. Ion dividers above and below the peptide sequence are gray for glycosylated fragments and black for nonglycosylated fragments.
13C labeling of glycans
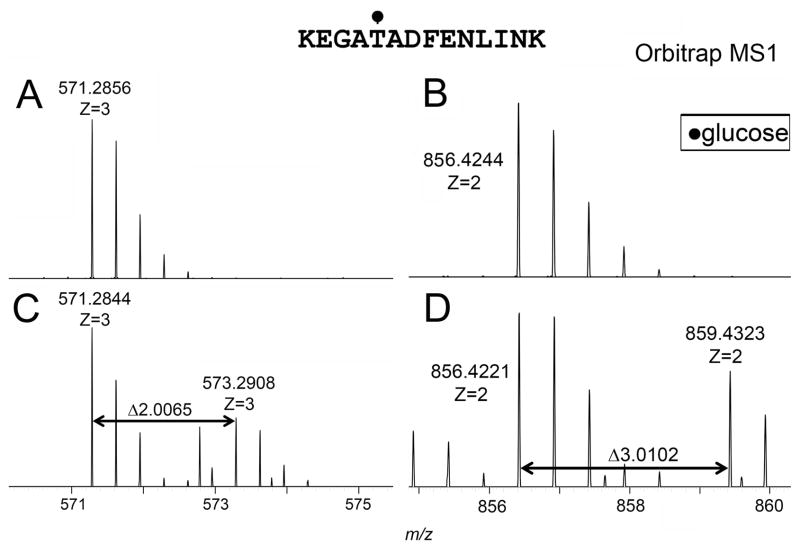
We have shown that [U-13C]starch isotopically labels glucose, but not mannose, residues of glycoconjugates of M. arthritidis (Jordan et al., 2013). The mycoplasma was grown in the presence of a 1:1 mixture of 13C-labeled and unlabeled (12C) starch. Any shift in the mass spectra of peptide ions that corresponded to a 13C-labeled hexose would suggest that the glycan attached by M. arthritidis to the peptide was glucose and not mannose. Fig. 6 shows that a doublet of the ions from KEGAT295ADFENLINK were found in doubly and triply charged states that correspond to the mass of unlabeled hexose and 13C-labeled glucose. Of the twelve glycopeptides identified in Table 1, six were shown to be glucose based on 13C labeling.
Fig. 6.
Ions of glycosylated peptide KEGAT295ADFENLINK of MARTH_403. (A) triply-charged species grown in serum-free medium, (B) doubly-charged species grown in serum-free medium, (C) triply-charged species grown in MB supplemented with 13C starch, and (D) doubly-charged species grown in MB supplemented with 13C starch.
Supplementary information
Shown above are the MS-1, tandem MS, and 13C-labeling for the glycopeptide KEGAT295ADFENLINK. For each of the other glycopeptides identified in this study, the verification data are shown in supplementary information as described in Table 1.
Discussion
Glycosylation is the most common posttranslational modification to proteins (Nothaft and Szymanski, 2010). It has been estimated that about two-thirds of eukaryotic proteins are glycosylated (Brooks, 2009, Apweiler et al., 1999). Examples of bacterial glycoproteins include the well-characterized general N-glycosylation of Campylobacter jejuni proteins (Linton et al., 2005, Wacker et al., 2002) and O-glycosylation of bacterial flagella and pili (Logan, 2006, Ng et al., 2006). Most of the characterized glycoproteins of pathogens are virulence factors (Zhou and Wu, 2009). The function(s) of protein glycosylation in host-dependent organisms such as mycoplasmas could be to inhibit cleavage by proteases, increase protein stability, disguise the bacterium from host defenses, and contribute to host colonization (Yuki, 2007, Ang et al., 2002, Kitazawa et al., 1998, Kusunoki et al., 2001, Asakura et al., 2010, Fletcher et al., 2009, Howard et al., 2009, Szymanski and Wren, 2005, Peng et al., 2008, Wu et al., 2007).
General protein O-glycosylation may be widespread in bacteria. Described in this study are the first protein glycosylation sites identified in any species of mycoplasma. Protein glycosylation in M. arthritidis occurs at serine and threonine residues at no obvious consensus amino acid sequence. Ten glycosylation sites have been identified thus far, and ongoing studies suggest that dozens of proteins are glycosylated. The glycostaining shown in Fig. 1 suggests that multiple proteins are glycosylated not just in M. arthritidis but also in Mycoplasma hominis and Mycoplasma pulmonis. The glycostaining shown in Fig. 3A suggests that glycosylation is not limited to lipoproteins. A single hexose would be expected to react weakly with the glycostain but many glycosylation sites can be present on the same protein, enhancing the fluorescent signal. For example, MARTH_403 has seven glycosylation sites found in this study and probably has additional sites not yet identified. A total of 140 of 918 (15%) of this protein’s amino acids are serine or threonine residues. The methods employed in the current study were not quantitative and the frequency of glycosylation at any given site cannot be estimated.
The 13C-labeling shown in Figs. 5, S6, S9, S16, S17, and S29 confirm that the mycoplasmas have the enzymatic machinery to break down starch and attach glucose to their proteins. As 13C starch labels the glucose but not the mannose residues of mycoplasma glycoconjugates (Jordan et al., 2013), glucose is definitely O-linked to six of the glycopeptides identified in Table 1. The glycans may be variable. Those that did not label with 13C might be mannose, the only hexose identified by GC/MS in the lipoprotein fraction other than glucose.
Mycoplasmas produce a variety of glycomoieites. In addition to glycoproteins, M. arthritidis is known to produce a glucolipid (Li et al., 1997). This species also synthesizes rhamnose and incorporates it into a glycoconjugate of unknown structure (Jordan et al., 2013). Similarly, M. pulmonis produces glycolipid, a glycoconjugate containing rhamnose, the EPS-I polysaccharide, and possibly multiple glycoproteins as suggested by Fig. 1. The single glycosyltransferase gene annotated for M. arthritidis (MARTH_orf849) and for M. pulmonis (MYPU_7700) would be insufficient for synthesis of all the glycomoieties that are produced. Additional glycosyltransferases must exist that are not readily recognizable through bioinformatics. An example might be the MYPU_7410 and 7420 gene products that are required for EPS-I production in M. pulmonis (Daubenspeck et al., 2009). The glycosyltransferase genes MARTH_orf849 and MYPU_7700 have not been disrupted in transposon libraries and are thought to be essential as are the glycosyltransferase genes of Mycoplasma genitalium, underscoring the importance of glycoconjugate synthesis in mycoplasmas (French et al., 2008, Glass et al., 2006, Dybvig et al., 2008).
The inability of M. arthritidis to import glucose and mannose as a monosaccharide suggests that oligosaccharides might be required for glycoconjugate synthesis as described for bifidobacteria (Yamamoto, 2012). The putative oligosaccharide importer is not apparent through bioinformatic analysis, and the results shown in Fig. 3 indicate that it is independent of the MARTH_orf819 operon. The oligosaccharide importer might be essential for viability as synthesis of glycolipids, glycoproteins and polysaccharides are thought to all be dependent on its activity (Jordan et al., 2013).
It is possible that the glycan identified in this study was a single hexose because of the “simple” medium that was used. A culture medium that contained a more complex mixture of oligosaccharides might permit more diversity in the glycans that are attached to the proteins. M. arthritidis is a non-glycolytic species that causes dramatic joint disease in murine animals (Luo et al., 2008). The concentration of the monosaccharide glucose in the murine joint is low and the major available source for precursors for glycan synthesis would be glycosaminoglycans and heavily glycosylated proteins such as collagen. Thus, the host environment would favor a pathway that uses oligosaccharides instead of monosaccharides for bacterial glycoconjugate synthesis. The glycans of M. arthritidis in vivo might differ significantly from what has been identified in vitro.
Experimental procedures
Strains and culture conditions
M. arthritidis strain TnCtrl is a previously described member of a transposon library that has mini-Tn4001 inserted at an intergenic location in strain 158 and serves as a wild-type control (Dybvig et al., 2008, Luo et al., 2008). Mutants of 158 with disruptions in the MARTH_819 operon have the mini-transposon located at nucleotide positions 754593, 755852, and 759050 of the M. arthritidis genome (GenBank accession number CP001047), truncating MARTH_819, -821, and -822 to 32%, 20%, and 85% of the full-length protein, respectively. Also used in this study were M. hominis strain PG21 (ATCC 23114) and M. pulmonis strain CTG (Daubenspeck et al., 2009). TnCtrl, M. hominis and M. pulmonis were grown in serum-free medium as described (Jordan et al., 2013) unless indicated otherwise. Mutants in the MARTH_orf819 operon were grown in mycoplasma broth (MB), prepared as described elsewhere (Dybvig et al., 2008).
Glycoprotein staining of SDS-PAGE gels
Cells harvested from cultures in stationary growth phase were washed three times with phosphate-buffered saline (PBS). The amount of protein in the samples was determined by using a BCA protein assay kit (Pierce). Lipoproteins were extracted from cells containing 2-mg protein as previously described (Bordier, 1981, Wise et al., 1995). Total protein or TX-114-extracted protein were suspended in 20 μl SDS loading buffer and boiled for 10 minutes. The proteins (150 μg) were electrophoresed on 7.5% SDS-PAGE gels (Bio-Rad). Glycoproteins were stained using the Pro-Q Emerald 300 Glycoprotein Gel and Blot Stain Kit (Invitrogen) as described in the package insert protocol and imaged with ultraviolet light using an AlphaImager EC system. After staining for glycoproteins, the same gel or in some cases parallel gels were stained with Coomassie blue to visualize total proteins in the preparation.
GC/MS
TX-114 was removed from lipoprotein-extracted samples with Pierce® Detergent Removal Spin Columns, dessicated, treated with 0.4 ml acidic methanol at 80°C for at least 16 hours and dried again. The resulting material was analyzed by GC/MS as described (Jordan et al., 2013).
Labeling of mycoplasma cell cultures
For 14C labeling, ten μCi of either [U-14C]glucose or [1-14C]mannose was added to 1 ml overnight cultures. At times 0, 2, 24 and 48 hours, 100 μl of culture was centrifuged at 16,000 x g for 10 minutes, washed three times with PBS and suspended in 100-μl water. The suspensions were added to 4 ml ScintiVerse™ BD Cocktail (Fisher-Scientific) and radioactive counts assessed with a Wallac 1410 liquid scintillation counter.
For 13C labeling, [U-13C]starch was obtained from Cambridge Isotopes Laboratories, Inc. Medium was supplemented with a 1:1 ratio of 12C and 13C starch that had been acid solubilized as described (Jordan et al., 2013). For the experiments using starch, the culture medium was MB. 33-mg starch in 3-ml water was added to 30-ml cultures.
FT-MS
Individual protein bands were excised from the gel with a razor blade and subjected to in-gel tryptic digestion (with reduction by 10 mM dithiothreitol and alkylation with 50 mM iodoacetamide). Tryptic digests were loaded onto a liquid chromatography-mass spectrometry system composed of a Micro AS autosampler, LC nanopump (Eksigent), and a linear ion trap-Orbitrap Velos hybrid mass spectrometer (Orbitrap Velos, Thermo Fisher Scientific). The analytical column was a 100-μm diameter, 11-cm column pulled tip packed with Jupiter 5-μm C18 reversed-phase beads (Phenomenex). An acetonitrile gradient from 1 to 25% in 0.1% formic acid was run over 50 min at 650 nl/min. Orbitrap parameters were set to normal mass range (MS1, 300 < m/z < 1800) with a 50,000 resolution scan followed by five data dependent tandem mass spectrometry (MS/MS) scans per cycle in profile mode. Dynamic exclusion was set to exclude ions for 2 min after a repeat count of three within a 45-sec duration. M. arthritidis peptides were identified by use of TurboSEQUEST v.27 (rev.12, Thermo Fisher Scientific), Byonic (Protein Metrics), algorithms with tryptic cleavage and a parent ion mass accuracy of 10.0 ppm with the M. arthritidis database. SEQUEST results were further refined through Scaffold 3.0 (Proteome software) at a 95% confidence at the peptide level and 99% confidence at the protein level (Nesvizhskii et al., 2003). In the algorithm searches, serine and threonine residues were searched for the presence of hexose (162.0528 Da) and deoxyhexose (146.0579 Da) combinations. Sites of glycosylation attachment identified by the search algorithms or identified in the MS1 spectra were confirmed by manual interpretation of CID or ETD MS/MS compared to theoretical fragmentation patterns calculated in ProteinProspector MS product tool (UCSF Mass Spectrometry Facility) and by the observance of nonglycosylated and glycosylated precursor ion species in the MS1 spectra with observed mass differences of hexose residues in different charge states (Renfrow et al., 2007).
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (AI63909, AR44252 and AI93750). We thank Portia Caldwell for technical assistance.
References
- Ang CW, Tio-Gillen AP, Groen J, Herbrink P, Jacobs BC, Van Koningsveld R, et al. Cross-reactive anti-galactocerebroside antibodies and Mycoplasma pneumoniae infections in Guillain–Barré syndrome. J Neuroimmunol. 2002;130:179–183. doi: 10.1016/s0165-5728(02)00209-6. [DOI] [PubMed] [Google Scholar]
- Apweiler R, Hermjakob H, Sharon N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim Biophys Acta. 1999;1473:4–8. doi: 10.1016/s0304-4165(99)00165-8. [DOI] [PubMed] [Google Scholar]
- Asakura H, Churin Y, Bauer B, Boettcher JP, Bartfeld S, Hashii N, et al. Helicobacter pylori HP0518 affects flagellin glycosylation to alter bacterial motility. Mol Microbiol. 2010;78:1130–1144. doi: 10.1111/j.1365-2958.2010.07393.x. [DOI] [PubMed] [Google Scholar]
- Bolland JR, Simmons WL, Daubenspeck JM, Dybvig K. Mycoplasma polysaccharide protects against complement. Microbiology. 2012;158:1867–1873. doi: 10.1099/mic.0.058222-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordier C. Phase separation of integral membrane proteins in Triton X-114 solution. J Biol Chem. 1981;256:1604–1607. [PubMed] [Google Scholar]
- Brooks SA. Strategies for analysis of the glycosylation of proteins: current status and future perspectives. Mol Biotechnol. 2009;43:76–88. doi: 10.1007/s12033-009-9184-6. [DOI] [PubMed] [Google Scholar]
- Daubenspeck JM, Bolland JR, Luo W, Simmons WL, Dybvig K. Identification of exopolysaccharide-deficient mutants of Mycoplasma pulmonis. Mol Microbiol. 2009;72:1235–1245. doi: 10.1111/j.1365-2958.2009.06720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demina IA, Serebryakova MV, Ladygina VG, Rogova MA, Zgoda VG, Korzhenevskyi DA, et al. Proteome of the bacterium Mycoplasma gallisepticum. Biochem (Moscow) 2009;74:165–174. doi: 10.1134/s0006297909020072. [DOI] [PubMed] [Google Scholar]
- Dybvig K, Zuhua C, Lao P, Jordan DS, French CT, Tu AHT, et al. Genome of Mycoplasma arthritidis. Infect Immun. 2008;76:4000–4008. doi: 10.1128/IAI.00516-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher CM, Coyne MJ, Villa OF, Chatzidaki-Livanis M, Comstock LE. A general O-glycosylation system important to the physiology of a major human intestinal symbiont. Cell. 2009;137:321–331. doi: 10.1016/j.cell.2009.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French CT, Lao P, Loraine AE, Matthews BT, Yu H, Dybvig K. Large-scale transposon mutagenesis of Mycoplasma pulmonis. Mol Microbiol. 2008;69:67–76. doi: 10.1111/j.1365-2958.2008.06262.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass JI, Assad-Garcia N, Alperovich N, Yooseph S, Lewis MR, Maruf M, et al. Essential genes of a minimal bacterium. Proc Nat Acad Sci USA. 2006;103:425–430. doi: 10.1073/pnas.0510013103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard SL, Jagannathan A, Soo EC, Hui JP, Aubry AJ, Ahmed I, et al. Campylobacter jejuni glycosylation island important in cell charge, legionaminic acid biosynthesis, and colonization of chickens. Infect Immun. 2009;77:2544–2556. doi: 10.1128/IAI.01425-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwashkiw JA, Vozza NF, Kinsella RL, Feldman MF. Pour some sugar on it: the expanding world of bacterial protein O-linked glycosylation. Mol Microbiol. 2013;89:14–28. doi: 10.1111/mmi.12265. [DOI] [PubMed] [Google Scholar]
- Jordan DS, Daubenspeck JM, Dybvig K. Rhamnose biosynthesis in mycoplasmas requires precursor glycans larger than monosaccharide. Mol Microbiol. 2013;89:918–928. doi: 10.1111/mmi.12320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahane I, Brunner H. Isolation of a glycoprotein from Mycoplasma pneumoniae membranes. Infect Immun. 1977;18:273–277. doi: 10.1128/iai.18.2.273-277.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitazawa K, Tagawa Y, Honda A, Yuki N. Guillain–Barré syndrome associated with IgG anti-GM1b antibody subsequent to Mycoplasma pneumoniae infection. J Neurol Sci. 1998;156:99–101. doi: 10.1016/s0022-510x(98)00020-3. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Watanabe T. Comparison of glycoproteins from ten human Mycoplasma species. Microbios. 1991;67:23–33. [PubMed] [Google Scholar]
- Kusunoki S, Shiina M, Kanazawa I. Anti-Gal-C antibodies in GBS subsequent to mycoplasma infection: Evidence of molecular mimicry. Neurology. 2001;57:736–738. doi: 10.1212/wnl.57.4.736. [DOI] [PubMed] [Google Scholar]
- Li JL, Matsuda K, Takagi M, Yamamoto N. Detection of serum antibodies against phosphocholine-containing aminoglycoglycerolipid specific to Mycoplasma fermentans in HIV-1 infected individuals. J Immunol Methods. 1997;208:103–113. doi: 10.1016/s0022-1759(97)00135-x. [DOI] [PubMed] [Google Scholar]
- Linton D, Dorrell N, Hitchen PG, Amber S, Karlyshev AV, Morris HR, et al. Functional analysis of the Campylobacter jejuni N-linked protein glycosylation pathway. Mol Microbiol. 2005;55:1695–1703. doi: 10.1111/j.1365-2958.2005.04519.x. [DOI] [PubMed] [Google Scholar]
- Logan SM. Flagellar glycosylation - a new component of the motility repertoire? Microbiology. 2006;152:1249–1262. doi: 10.1099/mic.0.28735-0. [DOI] [PubMed] [Google Scholar]
- Luo W, Yu H, Cao Z, Schoeb TR, Marron M, Dybvig K. Association of Mycoplasma arthritidis mitogen with lethal toxicity but not with arthritis in mice. Infect Immun. 2008;76:4989–4998. doi: 10.1128/IAI.00667-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- Ng SY, Chaban B, Jarrell KF. Archaeal flagella, bacterial flagella and type IV pili: a comparison of genes and posttranslational modifications. J Mol Microbiol Biotechnol. 2006;11:167–191. doi: 10.1159/000094053. [DOI] [PubMed] [Google Scholar]
- Nothaft H, Szymanski CM. Protein glycosylation in bacteria: sweeter than ever. Nat Rev Microbiol. 2010;8:765–778. doi: 10.1038/nrmicro2383. [DOI] [PubMed] [Google Scholar]
- Peng Z, Wu H, Ruiz T, Chen Q, Zhou M, Sun B, et al. Role of gap3 in Fap1 glycosylation, stability, in vitro adhesion, and fimbrial and biofilm formation of Streptococcus parasanguinis. Oral Microbiol Immunol. 2008;23:70–78. doi: 10.1111/j.1399-302X.2007.00401.x. [DOI] [PubMed] [Google Scholar]
- Renfrow MB, Mackay CL, Chalmers MJ, Julian BA, Mestecky J, Kilian M, et al. Analysis of O-glycan heterogeneity in IgA1 myeloma proteins by Fourier transform ion cyclotron resonance mass spectrometry: implications for IgA nephropathy. Anal Bioanal Chem. 2007;389:1397–1407. doi: 10.1007/s00216-007-1500-z. [DOI] [PubMed] [Google Scholar]
- Shaw BM, Daubenspeck JM, Simmons WL, Dybvig K. EPS-I polysaccharide protects Mycoplasma pulmonis from phagocytosis. FEMS Microbiol Lett. 2013;338:155–160. doi: 10.1111/1574-6968.12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski CM, Wren BW. Protein glycosylation in bacterial mucosal pathogens. Nat Rev Microbiol. 2005;3:225–237. doi: 10.1038/nrmicro1100. [DOI] [PubMed] [Google Scholar]
- Wacker M, Linton D, Hitchen PG, Nita-Lazar M, Haslam SM, North SJ, et al. N-linked glycosylation in Campylobacter jejuni and its functional transfer into E. coli. Science. 2002;298:1790–1793. doi: 10.1126/science.298.5599.1790. [DOI] [PubMed] [Google Scholar]
- Wise KS, Kim MF, Watson-McKown R. Variant membrane proteins. In: Razin S, Tully JG, editors. Molecular and Diagnostic Procedures in Mycoplasmology. San Diego: Academic Press; 1995. pp. 227–249. [Google Scholar]
- Wu H, Zeng M, Fives-Taylor P. The glycan moieties and the N-terminal polypeptide backbone of a fimbria-associated adhesin, Fap1, play distinct roles in the biofilm development of Streptococcus parasanguinis. Infect Immun. 2007;75:2181–2188. doi: 10.1128/IAI.01544-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K. Biological analysis of the microbial metabolism of hetero-oligosaccharides in application to glycotechnology. Biosci Biotechnol Biochem. 2012;76:1815–1827. doi: 10.1271/bbb.120401. [DOI] [PubMed] [Google Scholar]
- Yuki N. Ganglioside mimicry and peripheral nerve disease. Muscle Nerve. 2007;35:691–711. doi: 10.1002/mus.20762. [DOI] [PubMed] [Google Scholar]
- Yus E, Maier T, Michalodimitrakis K, van Noort V, Yamada T, Chen WH, et al. Impact of genome reduction on bacterial metabolism and its regulation. Science. 2009;326:1263–1268. doi: 10.1126/science.1177263. [DOI] [PubMed] [Google Scholar]
- Zhou M, Wu H. Glycosylation and biogenesis of a family of serine-rich bacterial adhesins. Microbiology. 2009;155:317–327. doi: 10.1099/mic.0.025221-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.