Abstract
Although hormones and downstream transcription factors are considered main drivers directing mammary gland development and oncogenic transformation, an emerging body of evidence suggests these processes are modulated by dynamic histone methylation landscapes. The methyltransferase EZH2 catalyzes the formation of trimethyl groups on lysine 27 of histone 3 (H3K27me3) and loss- and gain-off-unction studies have provided insight into its role in normal mammary development and oncogenic transformation. EZH2 controls the homeostasis of mouse mammary stem cells, and mammary epithelium devoid of EZH2 does not undergo functional development during pregnancy, possibly due to a paucity of stem cells. EZH2 levels are frequently elevated in breast cancer suggesting a link between H3K27me3 and cell proliferation. In addition to its role as epigenetic regulator, recent studies have placed EZH2 into the category of transcriptional co-activators and thus opened the possibility of non-canonical signaling pathways. In contrast to solid tumors, loss of EZH2 from hematopoietic cells has been linked to malignancies (Fig. 1). The challenge will be to understand not only cell-specific functions of EZH2, but also the extent to which it relies on its enzymatic activity versus its ability to serve as a transcriptional co-factor.
Keywords: EZH2, Histone methyltransferase, Breast
1. EZH1 and EZH2, histone methyltransferases and beyond
Enhancer of zeste homolog 2 (EZH2) is a bona fide H3K27 methyltransferase and an integral component of the polycomb repressive complex (PRC) 2. Its enzymatic function catalyzes the establishment of trimethyl groups on lysine 27 of histone H3 (H3K27me3), which is generally associated with the repression of large gene sets (Sauvageau and Sauvageau, 2010). Studies of Ezh2-null embryonic stem cells (ESCs) have demonstrated the existence of residual H3K27me3, which was attributed to the presence of the methyltransferase EZH1 (Shen et al., 2008), suggesting that these two enzymes can, at least partially, compensate for each other. Genetic loss-of-function studies have demonstrated a role for EZH2 in the establishment and physiology of several cell types and tissues, such as skin (Ezhkova et al., 2011), heart (Shen et al., 2008) and the mammary gland (Pal et al., 2013; Laible et al., 1997). In general, genetic loss of Ezh2 has less severe consequences than the loss of other PRC components, which has been attributed to the compensatory presence of EZH1. For example, in skin, loss of Ezh2 only has only marginal consequences on epidermal development and significant defects are only observed in the combined absence of EZH1 and EZH2 (Ezhkova et al., 2011). While in most cell types, the absence of only EZH1 does not provoke a visible pathology, it is required for the maintenance of hematopoietic stem cells (Hidalgo et al., 2012).
Since EZH1 and EZH2 display histone methyltransferase activity, epigenetic mechanisms are frequently invoked in explaining developmental and physiological consequences obtained upon their loss or overexpression. However, recent evidence suggests that EZH2 can also control gene expression by serving as a transcription co-activator of steroid hormone receptors (Xu et al., 2012) and by methylating cellular proteins and thereby controlling their half-life (Lee et al., 2012). In general studies on Ezh2-null cells have not provided a comprehensive evaluation of the global and cell-specific H3K27me3 status and thus did not necessarily link EZH2 to epigenetic control mechanisms. It is well possible that non-canonical functions of EZH2 are a major venue to control specific target genes. The histone methyltransferase EZH1 also appears to strive beyond its enzymatic activity and binds to transcriptional start sites to promote RNA polymerase II elongation (Mousavi et al., 2012). Knock-in mutations that specifically ablate only the methyltransferase activity of these proteins should provide a better understanding of their function (see Fig. 1).
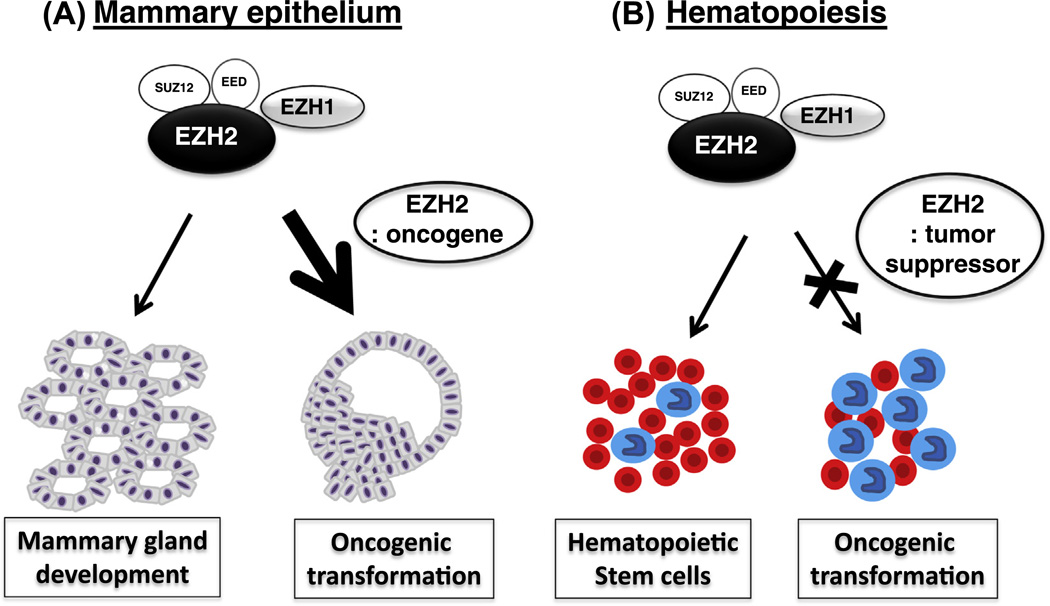
Fig. 1.
Epigenetic regulation of histone methyltransferase EZH2 in development and cancer. (A) EZH1/2 controls the homeostasis of mammary stem cells and development. Overexpression of EZH2 is correlated with breast cancer progression and poor prognosis. (B) EZH2 controls the homeostasis of hematopoietic stem cells. Loss of EZH2 from hematopoietic cells has been linked to malignancy.
2. EZH2 in mammary gland development
Development of the mammary gland during pregnancy is a coordinated process including the proliferation of mammary alveolar cells followed by their differentiation to produce milk (Hennighausen and Robinson, 2005). These processes are driven largely by progesterone and prolactin through their respective receptors that, in turn, elicit a strict temporal activation of genetic programs. Extensive insight into the mechanisms by which prolactin controls alveolar proliferation and differentiation have been obtained from mice carrying hypomorphic Stat5 germline mutations as well as mammary epithelium-specific Stat5-null mice (Miyoshi et al., 2001; Cui et al., 2004; Yamaji et al., 2009, 2013). The transcription factors STAT5a and STAT5b (referred to as STAT5) are key to convey prolactin signaling in mammary epithelium during pregnancy and the establishment of functional alveoli. Recent studies determined that distinct thresholds of STAT5 are required to induce unique biological programs, which help to explain the differential activation of genetic programs during pregnancy. While low levels of STAT5 are sufficient for the establishment and proliferation of alveolar cells, differentiation and milk production are only obtained in the presence of high STAT5 concentrations (Yamaji et al., 2013). These programs are initiated by the binding of STAT5 to respective target genes and the establishment of trimethyl groups on lysine 4 of histone 3 (H3K4me3) epigenetic marks.
While transcription factors, such as STAT5, and the accompanying formation of H3K4me3 marks, are keys to the execution of hormone-induced biological programs, science surrounding the biological significance of epigenetic marks is still in its infancy. A major step forward came from a recent study by Pal and colleagues, who used mammary epithelium-specific gene knockout mice to explore the role of EZH2, and thus the accompanied H3K27me3, in the developing mammary gland during pregnancy (Pal et al., 2013). Their interest in EZH2 stemmed from the observation that mammary stem cells (MaSCs), committed luminal progenitors and mature luminal cells isolated from virgin mice displayed unique and distinct H3K27me3 patters. Most notably, the degree of H3K27me3 was lower in the MaSC/basal cell population as compared to luminal progenitors and mature luminal cells. Moreover, during pregnancy preferential loss of H3K27me3 marks were observed over genes that are normally activated by STAT5. In contrast, gene classes that are potentially associated with mammary morphogenesis acquired H3K27me3 marks during pregnancy. Their data argued that an evolving histone methylation landscape is likely key to expression changes that accompany cell lineage progression and the establishment of mature luminal cells. They tested the causal relationship between H3K27me3 and the progression of mammary lineages during pregnancy through inactivation of the Ezh2 gene in mammary stem and progenitor cells using Cre-mediated gene recombination. Loss of EZH2 function resulted in a paucity of mammary stem and progenitor cells with the consequence of delayed ductal morphogenesis, as indicated by the inability of ducts to fill the mammary fat the persistence of terminal end buds during puberty. Moreover, in vitro culture experiments revealed an at least 10-fold reduction in the clonogenic capacity of Ezh2-mutant MaSCs. Most dramatically, alveolar development during pregnancy was severely impaired and mutant mice failed to lactate, both on a pure FVB/N and a mixed FVB/N-B6 background. Impaired development was associated with deregulated expression of bona fide EZH2 target genes, such as Ink4a and Cdkn1a.
3. EZH2 in breast cancer
The Ezh2 gene is amplified in a variety human cancers, including those of the bladder, breast, colon and prostate and in lymphoma (Sauvageau and Sauvageau, 2010) and elevated EZH2 levels are deemed a good indicator for aggressiveness and poor outcome in breast cancer (Kleer et al., 2003). Increased EZH2 concentrations were also detected in a precancerous state in morphologically normal breast epithelium, suggesting that an increase of EZH2 is possibly an early event in the development of breast cancers (Ding et al., 2006). These correlative in vivo studies were supported by tissue culture experiments where EZH2 overexpression in immortalized human mammary epithelial cell lines promotes anchorage-independent growth and cell invasion (Kleer et al., 2003). Additional experiments were performed in transgenic mice overexpressing Ezh2 under control of the MMTV-LTR (Li et al., 2009). While overexpression of the Ezh2 transgene resulted in distinct intraductal epithelial hyperplasias, no frank tumors were observed (Li et al., 2009), suggesting that EZH2 is not the primary driver of cellular transformation but rather a modulator. Since the status of H3K27me3 in mammary tissue of these mice has not been evaluated it is not clear to what extent EZH2 overexpression affects epigenetic programming or non-canonical pathways.
Although breast cancer is in general associated with elevated EZH2 levels in transformed epithelium, loss of Ezh2 might be linked to some subtypes of breast cancer. Notably, gene expression profiles of the Ezh2-null MaSCs/basal cell population displayed similarities with signatures of claudin-low breast cancers (Pal et al., 2013). The claudin-low subtype is associated with metaplastic features and displays lower EZH2 levels (Keller, Arendt .PNAS. 2012), which reflects aberrant differentiation. Challenging mammary-specific Ezh2-null mice with oncogenic hits and an analysis of genome-wide H3K27me3 patters is needed to deliver a growth advantage to cells.
4. EZH2 and DNA methylation
EZH2 catalyzes H3K27 trimethylation, which leads to subsequent recruitment of other polycomb complexes and also to the conscription of DNA methyltransferases and histone deacetylases, which in turn results in transcriptional repression and chromatin compaction (Sauvageau and Sauvageau, 2010).In general, cancer cells display global DNA hypomethylation of specific gene loci, thereby enabling stable repression of tumor suppressor genes (Christman, 2002). Since EZH2 can physically contact DNA methyltransferases and recruit them to target genes to repress gene expression (Vire et al., 2006), it can be argued that EZH2 overexpression could repress tumor suppressor genes. However, it is unclear whether EZH2 is associated with DNA methylation at target sites or not and several studies have reported conflicting results. One report showed that human mammary epithelial cells with up-regulated EZH2 present PcG-dependent hypermethylation of the HOXA9 locus (Reynolds et al., 2006). In prostate cancer cells, downregulation of EZH2 restores expression of H3K27me3 target genes without affecting promoter DNA methylation and with no effect on silencing by DNA methylation (Kondo et al., 2008). However, it remains unclear whether EZH2 deregulation in cancer results in aberrant H3K27me3, and this conflict might be explained by tissue- and cancer-specific differences related to activation of specific silencing pathways.
5. EZH2-regulated genetic programs in breast cancer cells
EZH2 has been linked to different signaling pathways, each possibly contributing uniquely to the establishment and progression of breast cancer by regulating large sets of target genes (Yoo and Hennighausen, 2012). Most of the available data sets were derived from in vitro studies using tissue culture cells that had been manipulated with siRNA technology to modulate gene expression. Thus, the relevance of these studies to cancers in situ still needs to be established.
A number of genes, including RAD51 (Chang et al., 2011), CDH1 (Cao et al., 2008) and CDKN1C (Yang et al., 2009), HOTAIR (Gupta et al., 2010) are EZH2 targets in primary breast cancer tissues. EZH2 mediated downregulation of RAD51 leads to accumulation of recurrent RAF1 gene amplification in breast tumor initiating cells (BTICs), which activates p-ERK-β-catenin signaling to promote BTIC expansion (Chang et al., 2011). EZH2 also mediates transcriptional silencing of CDH1 (E-cadherin) through H3K27me3 (Cao et al., 2008). E-cadherin maintains epithelial cellular adhesion and integrity, and loss of E-cadherin expression is associated with invasiveness. CDKN1C (encoding tumor suppressor p57KIP2) is a cyclin-dependent kinase (CDK) inhibitor. Expression of the CDKN1C gene is regulated through H3K27me3 of its promoter (Yang et al., 2009). Reduction of p57KIP2 leads to an accelerated G1-S transition, which has been suggested to support breast cancer progression.
One study revealed elevated levels of HOTAIR, a large intervening non-coding RNA (lincRNA), in primary breast tumors and metastases (Gupta et al., 2010). Enforced expression of HOTAIR is induced to altered H3K27me3, gene expression, and increased breast cancer invasiveness and metastasis in a manner dependent on EZH2, whereas loss of HOTAIR inhibits cancer invasiveness. These data point to a modulating role of lincRNAs in modulating EZH2 in breast cancer.
In addition, several genes, including RUNX3 (Fujii et al., 2008), FOXC1 (Du et al., 2012) and ADRB2 (Yu et al., 2007), have been found to be EZH2 targets in breast cancer cell lines. An inverse correlation between EZH2 and RUNX3 gene expression has been reported in a breast cancer cell line (Fujii et al., 2008). Lower RUNX3 concentrations result in lower CDKN1A (p21WAF1/Cip1) levels, which do not block the cell cycle (Chi et al., 2005). EZH2 negatively regulates FOXC1 gene expression through H3K27me3 of the FOXC1 promoter. Increased FOXC1 expression leads to reduced breast cancer cell migration and invasion (Du et al., 2012). EZH2 may mediate increased invasiveness and metastasis by silencing a number of downstream targets including the β-2-adrenergic receptor (ADRB2) (Yu et al., 2007). ADRB2 has been implicated to play a role in cell growth, adhesion, and transformation (Bos, 2005). Although these studies suggest that EZH2 could have multi-faceted roles in breast cancer progression, additional in vivo studies on mice and primary human tissue are needed.
6. Linking EZH2 and BRCA1
Polycomb group (PcG) proteins control stem cell homeostasis through chromatin silencing and other non-histone functions (Sauvageau and Sauvageau, 2010). Poorly differentiated breast tumors present an embryonic stem cell-like signature characterized by the expression of pluripotency factors such as Nanog, Oct4, Sox2, and c-Myc as well as repression of PcG targets. This signature is strongly associated with high-grade estrogen-receptor (ER) negative tumors, often of the basal-like subtype and poor clinical outcome (Ben-Porath et al., 2008). Higher EZH2 expression is more associated with the aggressive basal-like subtype and poor metastasis-free survival (Kleer et al., 2003). BRCA1 mutations are a cause of basal-like breast carcinomas, and, consistent with their basal-like characteristics, BRCA1-deficient breast tumors exhibit aggressive behavior and are associated with poor survival.
Although the modulatory role of EZH2 in BRCA1-mutant breast tumors is not well defined, it has been proposed that EZH2 overexpression inhibits BRCA1 phosphorylation and promotes cell proliferation through increases in Cdc25C, which is crucial for entry into mitosis (Gonzalez et al., 2009). It appears that BRCA1-deficient mouse mammary tumor cells are selectively sensitive to an EZH2 inhibitor (Puppe et al., 2009). Cells derived from BRCA1-deficient mice mammary tumors are 20-fold more sensitive to the EZH2 inhibitor (DZNep) than that of cells derived from BRCA1-proficient mammary tumor cells. Furthermore, EZH2 can function in a context-dependent manner (Lee et al., 2011). Intriguingly, EZH2 confers a transactivation function on a set of nuclear factor (NF)-κB targets such as interleukin-6 and tumor necrosis factor, which are important in basal-like breast cancer, but act oppositely to repress NF-κB targets in ER-positive breast cancers (Lee et al., 2011). These data suggest that inhibiting EZH2 could potentially be as a tailored strategy for treating sporadic basal-like breast tumors with decreased BRCA1 activity.
7. EZH2 mutations in hematopoietic malignancies
In contrast to the concept that increased EZH2 activity has oncogenic functions, inactivating somatic EZH2 mutations are found in B-cell lymphoma (Morin et al., 2010). Replacement of a single tyrosine in the SET domain of the EZH2 protein (Tyr641) occurs in 7–21% of specific B-cell lymphomas. Although initially thought to result in a loss of catalytic activity, several studies have shown that Y641 mutants have very limited ability to perform the H3K27 mono methylation reaction but have enhanced catalytic efficiency for subsequent reactions (Sneeringer et al., 2010). However, the vast majority of Ezh2 alterations in myeloid disorders are comprised of deletions, missense and frame-shift mutations (Ernst et al., 2010). These mutations are predicted to result in the complete loss of histone methyltransferase activity. The finding of inactivating Ezh2 mutations opens a new perspective on the tumor suppressive role of EZH2 in other cancers (Fig. 2).
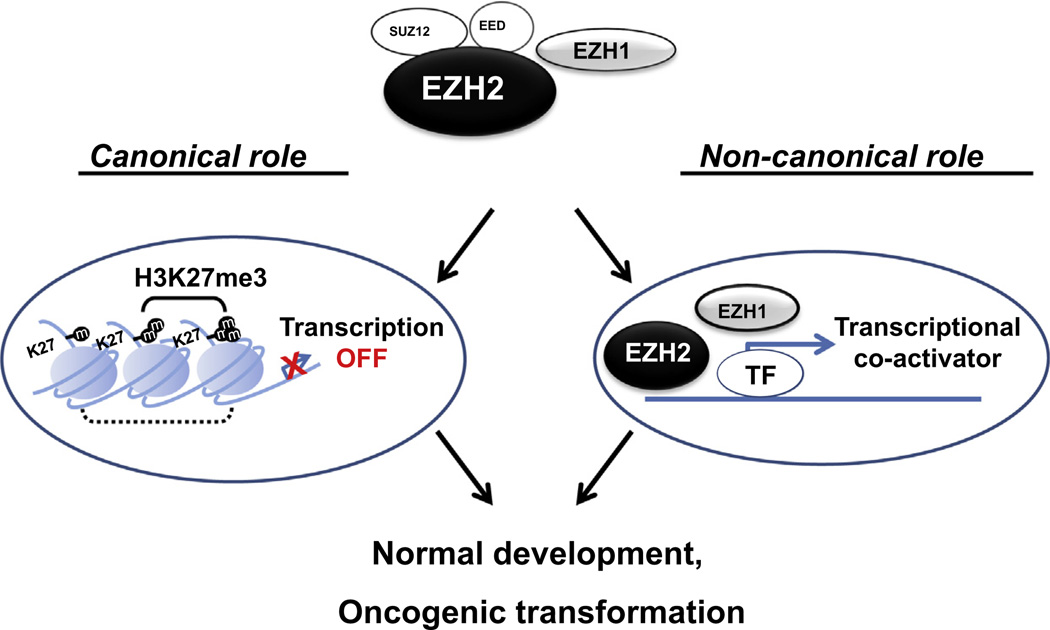
Fig. 2.
Functions of EZH2 in modulating polycomb target genes. PRC2 complexes, which include EZH1 and EZH2, catalyze H3K27 di- and tri-methylation, and leads to transcriptional repression. Furthermore, EZH1 and EZH2 also can act as transcriptional co-activator to promote the expression of target genes.
8. Perspective
Mutations in genes encoding signaling proteins, such as a transcription factors, affect a limited number of pathways, but mutations in epigenetic regulators can affect developmental programs on a large scale and alter the expression of many diverse gene sets. Although the deregulation of H3K27 methyltransferases and demethylases has been associated with cancer aggressiveness, there is no clear genetic evidence that they can serve as molecular tumor markers. Decisive genetic experiments must be conducted to link these histone modifying enzymes to the H3K27 methylation status in relevant cancer models. Equally important, it is necessary to pursue non-canonical functions of these proteins, such as their ability to function as transcriptional co-activators. The identification of epigenetic changes in cancer cells in vivo would be another step in the development of novel strategies for cancer treatment.
Acknowledgements
This work was supported in part by the intramural research program (IRP) of the NIDDK at the National Institutes of Health (NIH), USA, the World Class University Program, Ministry of Education, Science and Technology, through the National Research Foundation of Korea, South Korea (R31-100069); WCU Research Center, Dankook University, and Research Institute of Medical Sciences, Chonnam National University.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
References
- Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos JL. Linking Rap to cell adhesion. Curr. Opin. Cell Biol. 2005;17:123–128. doi: 10.1016/j.ceb.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Cao Q, Yu J, Dhanasekaran SM, Kim JH, Mani RS, Tomlins SA, Mehra R, Laxman B, Cao X, Kleer CG, Varambally S, Chinnaiyan AM. Repression of E-cadherin by the polycomb group protein EZH2 in cancer. Oncogene. 2008;27:7274–7284. doi: 10.1038/onc.2008.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CJ, Yang JY, Xia W, Chen CT, Xie X, Chao CH, Woodward WA, Hsu JM, Hortobagyi GN, Hung MC. EZH2 promotes expansion of breast tumor initiating cells through activation of RAF1-beta-catenin signaling. Cancer Cell. 2011;19:86–100. doi: 10.1016/j.ccr.2010.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi XZ, Yang JO, Lee KY, Ito K, Sakakura C, Li QL, Kim HR, Cha EJ, Lee YH, Kaneda A, Ushijima T, Kim WJ, Ito Y, Bae SC. RUNX3 suppresses gastric epithelial cell growth by inducing p21(WAF1/Cip1) expression in cooperation with transforming growth factor {beta}-activated SMAD. Mol. Cell. Biol. 2005;25:8097–8107. doi: 10.1128/MCB.25.18.8097-8107.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christman JK. 5-Azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene. 2002;21:5483–5495. doi: 10.1038/sj.onc.1205699. [DOI] [PubMed] [Google Scholar]
- Cui Y, Riedlinger G, Miyoshi K, Tang W, Li C, Deng CX, Robinson GW, Hennighausen L. Inactivation of Stat5 in mouse mammary epithelium during pregnancy reveals distinct functions in cell proliferation, survival, and differentiation. Mol. Cell. Biol. 2004;24:8037–8047. doi: 10.1128/MCB.24.18.8037-8047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Erdmann C, Chinnaiyan AM, Merajver SD, Kleer CG. Identification of EZH2 as a molecular marker for a precancerous state in morphologically normal breast tissues. Cancer Res. 2006;66:4095–4099. doi: 10.1158/0008-5472.CAN-05-4300. [DOI] [PubMed] [Google Scholar]
- Du J, Li L, Ou Z, Kong C, Zhang Y, Dong Z, Zhu S, Jiang H, Shao Z, Huang B, Lu J. FOXC1, a target of polycomb, inhibits metastasis of breast cancer cells. Breast Cancer Res. Treat. 2012;131:65–73. doi: 10.1007/s10549-011-1396-3. [DOI] [PubMed] [Google Scholar]
- Ernst T, Chase AJ, Score J, Hidalgo-Curtis CE, Bryant C, Jones AV, Waghorn K, Zoi K, Ross FM, Reiter A, Hochhaus A, Drexler HG, Duncombe A, Cervantes F, Oscier D, Boultwood J, Grand FH, Cross NC. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat. Genet. 2010;42:722–726. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- Ezhkova E, Lien WH, Stokes N, Pasolli HA, Silva JM, Fuchs E. EZH1 and EZH2 cogovern histone H3K27 trimethylation and are essential for hair follicle homeostasis and wound repair. Genes Dev. 2011;25:485–498. doi: 10.1101/gad.2019811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S, Ito K, Ito Y, Ochiai A. Enhancer of zeste homologue 2 (EZH2) down-regulates RUNX3 by increasing histone H3 methylation. J. Biol. Chem. 2008;283:17324–17332. doi: 10.1074/jbc.M800224200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez ME, Li X, Toy K, DuPrie M, Ventura AC, Banerjee M, Ljungman M, Merajver SD, Kleer CG. Downregulation of EZH2 decreases growth of estrogen receptor-negative invasive breast carcinoma and requires BRCA1. Oncogene. 2009;28:843–853. doi: 10.1038/onc.2008.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennighausen L, Robinson GW. Information networks in the mammary gland. Nat. Rev. Mol. Cell Biol. 2005;6:715–725. doi: 10.1038/nrm1714. [DOI] [PubMed] [Google Scholar]
- Hidalgo I, Herrera-Merchan A, Ligos JM, Carramolino L, Nunez J, Martinez F, Dominguez O, Torres M, Gonzalez S. Ezh1 is required for hematopoietic stem cell maintenance and prevents senescence-like cell cycle arrest. Cell Stem Cell. 2012;11:649–662. doi: 10.1016/j.stem.2012.08.001. [DOI] [PubMed] [Google Scholar]
- Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, Ghosh D, Sewalt RG, Otte AP, Hayes DF, Sabel MS, Livant D, Weiss SJ, Rubin MA, Chinnaiyan AM. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc. Natl. Acad. Sci. USA. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo Y, Shen L, Cheng AS, Ahmed S, Boumber Y, Charo C, Yamochi T, Urano T, Furukawa K, Kwabi-Addo B, Gold DL, Sekido Y, Huang TH, Issa JP. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat. Genet. 2008;40:741–750. doi: 10.1038/ng.159. [DOI] [PubMed] [Google Scholar]
- Laible G, Wolf A, Dorn R, Reuter G, Nislow C, Lebersorger A, Popkin D, Pillus L, Jenuwein T. Mammalian homologues of the Polycomb-group gene Enhancer of zeste mediate gene silencing in Drosophila heterochromatin and at S. cerevisiae telomeres. EMBO J. 1997;16:3219–3232. doi: 10.1093/emboj/16.11.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ST, Li Z, Wu Z, Aau M, Guan P, Karuturi RK, Liou YC, Yu Q. Context-specific regulation of NF-kappaB target gene expression by EZH2 in breast cancers. Mol. Cell. 2011;43:798–810. doi: 10.1016/j.molcel.2011.08.011. [DOI] [PubMed] [Google Scholar]
- Lee JM, Lee JS, Kim H, Kim K, Park H, Kim JY, Lee SH, Kim IS, Kim J, Lee M, Chung CH, Seo SB, Yoon JB, Ko E, Noh DY, Kim KI, Kim KK, Baek SH. EZH2 generates a methyl degron that is recognized by the DCAF1/DDB1/CUL4 E3 ubiquitin ligase complex. Mol. Cell. 2012;48:572–586. doi: 10.1016/j.molcel.2012.09.004. [DOI] [PubMed] [Google Scholar]
- Li X, Gonzalez ME, Toy K, Filzen T, Merajver SD, Kleer CG. Targeted overexpression of EZH2 in the mammary gland disrupts ductal morphogenesis and causes epithelial hyperplasia. Am. J. Pathol. 2009;175:1246–1254. doi: 10.2353/ajpath.2009.090042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi K, Shillingford JM, Smith GH, Grimm SL, Wagner KU, Oka T, Rosen JM, Robinson GW, Hennighausen L. Signal transducer and activator of transcription (Stat) 5 controls the proliferation and differentiation of mammary alveolar epithelium. J. Cell Biol. 2001;155:531–542. doi: 10.1083/jcb.200107065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, Paul JE, Boyle M, Woolcock BW, Kuchenbauer F, Yap D, Humphries RK, Griffith OL, Shah S, Zhu H, Kimbara M, Shashkin P, Charlot JF, Tcherpakov M, Corbett R, Tam A, Varhol R, Smailus D, Moksa M, Zhao Y, Delaney A, Qian H, Birol I, Schein J, Moore R, Holt R, Horsman DE, Connors JM, Jones S, Aparicio S, Hirst M, Gascoyne RD, Marra MA. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat. Genet. 2010;42:181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousavi K, Zare H, Wang AH, Sartorelli V. Polycomb protein Ezh1 promotes RNA polymerase II elongation. Mol. Cell. 2012;45:255–262. doi: 10.1016/j.molcel.2011.11.019. [DOI] [PubMed] [Google Scholar]
- Pal B, Bouras T, Shi W, Vaillant F, Sheridan JM, Fu N, Breslin K, Jiang K, Ritchie ME, Young M, Lindeman GJ, Smyth GK, Visvader JE. Global changes in the mammary epigenome are induced by hormonal cues and coordinated by Ezh2. Cell Rep. 2013;3:411–426. doi: 10.1016/j.celrep.2012.12.020. [DOI] [PubMed] [Google Scholar]
- Puppe J, Drost R, Liu X, Joosse SA, Evers B, Cornelissen-Steijger P, Nederlof P, Yu Q, Jonkers J, van Lohuizen M, Pietersen AM. BRCA1-deficient mammary tumor cells are dependent on EZH2 expression and sensitive to Polycomb Repressive Complex 2-inhibitor 3-deazaneplanocin A. Breast Cancer Res. 2009;(11):R63. doi: 10.1186/bcr2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds PA, Sigaroudinia M, Zardo G, Wilson MB, Benton GM, Miller CJ, Hong C, Fridlyand J, Costello JF, Tlsty TD. Tumor suppressor p16INK4A regulates polycomb-mediated DNA hypermethylation in human mammary epithelial cells. J. Biol. Chem. 2006;281:24790–24802. doi: 10.1074/jbc.M604175200. [DOI] [PubMed] [Google Scholar]
- Sauvageau M, Sauvageau G. Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell. 2010;7:299–313. doi: 10.1016/j.stem.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, Yuan GC, Orkin SH. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol. Cell. 2008;32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneeringer CJ, Scott MP, Kuntz KW, Knutson SK, Pollock RM, Richon VM, Copeland RA. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc. Natl. Acad. Sci. USA. 2010;107:20980–20985. doi: 10.1073/pnas.1012525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, Bollen M, Esteller M, Di Croce L, de Launoit Y, Fuks F. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- Xu K, Wu ZJ, Groner AC, He HH, Cai C, Lis RT, Wu X, Stack EC, Loda M, Liu T, Xu H, Cato L, Thornton JE, Gregory RI, Morrissey C, Vessella RL, Montironi R, Magi-Galluzzi C, Kantoff PW, Balk SP, Liu XS, Brown M. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science. 2012;338:1465–1469. doi: 10.1126/science.1227604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji D, Na R, Feuermann Y, Pechhold S, Chen W, Robinson GW, Hennighausen L. Development of mammary luminal progenitor cells is controlled by the transcription factor STAT5A. Genes Dev. 2009;23:2382–2387. doi: 10.1101/gad.1840109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji D, Kang K, Robinson GW, Hennighausen L. Sequential activation of genetic programs in mouse mammary epithelium during pregnancy depends on STAT5A/B concentration. Nucl. Acids Res. 2013;41:1622–1636. doi: 10.1093/nar/gks1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Karuturi RK, Sun F, Aau M, Yu K, Shao R, Miller LD, Tan PB, Yu Q. CDKN1C (p57) is a direct target of EZH2 and suppressed by multiple epigenetic mechanisms in breast cancer cells. PLoS ONE. 2009;4:e5011. doi: 10.1371/journal.pone.0005011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo KH, Hennighausen L. EZH2 methyltransferase and H3K27 methylation in breast cancer. Int. J. Biol. Sci. 2012;8:59–65. doi: 10.7150/ijbs.8.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Cao Q, Mehra R, Laxman B, Tomlins SA, Creighton CJ, Dhanasekaran SM, Shen R, Chen G, Morris DS, Marquez VE, Shah RB, Ghosh D, Varambally S, Chinnaiyan AM. Integrative genomics analysis reveals silencing of beta-adrenergic signaling by polycomb in prostate cancer. Cancer Cell. 2007;12:419–431. doi: 10.1016/j.ccr.2007.10.016. [DOI] [PubMed] [Google Scholar]