Abstract
Objective
Information overload is a significant problem facing online clinical trial searchers. We present eTACTS, a novel interactive retrieval framework using common eligibility tags to dynamically filter clinical trial search results.
Materials and Methods
eTACTS mines frequent eligibility tags from free-text clinical trial eligibility criteria and uses these tags for trial indexing. After an initial search, eTACTS presents to the user a tag cloud representing the current results. When the user selects a tag, eTACTS retains only those trials containing that tag in their eligibility criteria and generates a new cloud based on tag frequency and co-occurrences in the remaining trials. The user can then select a new tag or unselect a previous tag. The process iterates until a manageable number of trials is returned. We evaluated eTACTS in terms of filtering efficiency, diversity of the search results, and user eligibility to the filtered trials using both qualitative and quantitative methods.
Results
eTACTS (1) rapidly reduced search results from over a thousand trials to ten; (2) highlighted trials that are generally not top-ranked by conventional search engines; and (3) retrieved a greater number of suitable trials than existing search engines.
Discussion
eTACTS enables intuitive clinical trial searches by indexing eligibility criteria with effective tags. User evaluation was limited to one case study and a small group of evaluators due to the long duration of the experiment. Although a larger-scale evaluation could be conducted, this feasibility study demonstrated significant advantages of eTACTS over existing clinical trial search engines.
Conclusion
A dynamic eligibility tag cloud can potentially enhance state-of-the-art clinical trial search engines by allowing intuitive and efficient filtering of the search result space.
Keywords: Information Storage and Retrieval, Clinical Trials, Dynamic Information Filtering, Interactive Information Retrieval, Tag Cloud, Association Rules, Eligibility Criteria
1. Introduction
Randomized controlled trials generate high-quality medical evidence for disease treatment and therapeutic development but still face longstanding recruitment problems. In fact, more than 90% of trials are delayed because of difficulties recruiting eligible patients [1–3]. Using Web applications, health consumers are becoming increasingly comfortable searching online for clinical research opportunities [4]. However, information overload is a common and significant problem with most existing clinical trial search engines (e.g., ClinicalTrials.gov [5], UK Clinical Trials Gateway [6]). For example, searching “diabetes mellitus, type II” on ClinicalTrials.gov returns a list of more than 5,000 trials (as of April 2013), which are sorted just by their probabilistic relevance to the search terms, with those containing the query in the title ranked highest [7]. Supplying additional parameters, such as location or study type, can only modestly improve search specificity, especially for searches of eligibility criteria. Moreover, identifying terms that are effective at retrieving relevant trials can be difficult for the average user [8].
One major limitation of existing clinical trial search engines is the underutilization of free-text eligibility criteria. This is mostly due to varied and complicated semantic structures (e.g., inclusion vs. exclusion and negation) that make it difficult to define standardized parsers as well as user-friendly representations to exploit in search applications [9–11]. Yet, we hypothesize that filtering clinical trials by eligibility criteria can greatly increase the specificity of the search engines.
1.1. Objective
This article presents eTACTS (eligibility TAg cloud-based Clinical Trial Search), a faceted search method to filter the list of clinical trials returned by any type of initial search (e.g., simple free-text query terms, advanced form-based). In particular, the resulting trials are indexed through a small number of facets, each defining a distinct property of the text, and users can select facets to filter the search results [12–15]. eTACTS defines eligibility tags as facets for the clinical trial search results. An eligibility tag is a meaningful multi-word pattern, e.g., “breast carcinoma”, “active malignancy”, that frequently appears within the free-text eligibility criteria of clinical trials [16]. Eligibility tags are presented to users as a dynamic tag cloud to assist with iterative filtering of the resulting trials. A tag cloud is a visual representation of key concepts associated with textual documents. In this domain, individual tags are displayed as hyperlinks to a set of clinical trials that contain the tags in their eligibility criteria, with each tag's “importance” or relative frequency indicated by a mix of font size and color. When the user selects a tag, the cloud is updated according to the tag distribution in the remaining trials, which contain all of the selected tags in their eligibility criteria. By using common tags, we allow the users to quickly identify common and intuitive facets that lead to efficient and effective result filtering [16].
In this paper, we (1) describe the design of a novel interactive clinical trial search framework named eTACTS; (2) demonstrate that a dynamic tag cloud can efficiently reduce the trial search results based on interactive search parameters expressed by eligibility tags; (3) demonstrate that eTACTS helps users discover trials not highlighted by conventional search engines; and (4) demonstrate that searching by eTACTS effectively produce more relevant results than other available search engines.
1.2. Related Work
Prior studies proposed automatic techniques to transform clinical trial specifications into a computable form that can be efficiently reused for classification, clustering, and retrieval [17–22]. A number of efforts also focused on formally representing free-text clinical trial eligibility criteria for computational processing [10, 16, 23–27]. Consequently, several projects are underway to improve clinical trial recruitment with Web-based information technologies [28–30]. These methods either help clinicians find relevant trials for their patients [31] or help patients identify trials themselves [5, 6, 32–38]. Some tools provide general search facilities that query public trial repositories (e.g., ClinicalTrials.gov [5], UK Clinical Trials Gateway [6], Search Clinical Trials [34], TrialReach [35], ASCOT [38]). Others employ user provided medical history to recommend suitable trials (e.g., PatientsLikeMe [33], Corengi for “type II diabetes” trials [37]) or match users with research coordinators (e.g., ResearchMatch [36]). Alternatively, TrialX employs a question/answer mechanism (i.e., AskDory!) to provide users with a list of actively-recruiting trials, whose recruiters users can then call to verify eligibility [32].
Most of these systems use only pre-structured information (e.g., condition, location, title) or limited manual annotations of the eligibility criteria for clinical trial searches. Only ASCOT [38] provides searches with discriminative power based on automatic processing of eligibility criteria. In particular, ASCOT annotates each clinical trial with the Unified Medical Language System (UMLS) [39] terms extracted from its eligibility criteria. The annotations related to the trials retrieved by an initial search are then displayed as a list beside the search results, allowing the user select those he/she considers effective at reducing the number of results. The most frequent annotations in the clinical trial repository are also provided as a static tag cloud (i.e., related neither to the initial search nor to the user interaction) to initially filter the results.
Interactive information retrieval has gained popularity lately [40] and presenting tag clouds has become a well-established data visualization technique [41–43]. While some criticisms have been raised by the Internet community about the use of tag clouds in general domain and social applications [44–46], they were effectively used as a data-driven aid for users searching and browsing pertinent information in more specific scenarios, e.g., to discern credible content in online health message forums [47], music [48] and image retrieval [49]. Our method differs from ASCOT in that the cloud of eligibility tags, which gets updated after each user tag selection, is the main filtering tool. Additionally, while ASCOT mines annotations from each trial independently, we use a controlled vocabulary composed only of frequent and common tags that are mined across multiple trials. This leads to a higher level and more intuitive representation designed to simplify searches and to help users interact with the search system.
2. Material and Methods
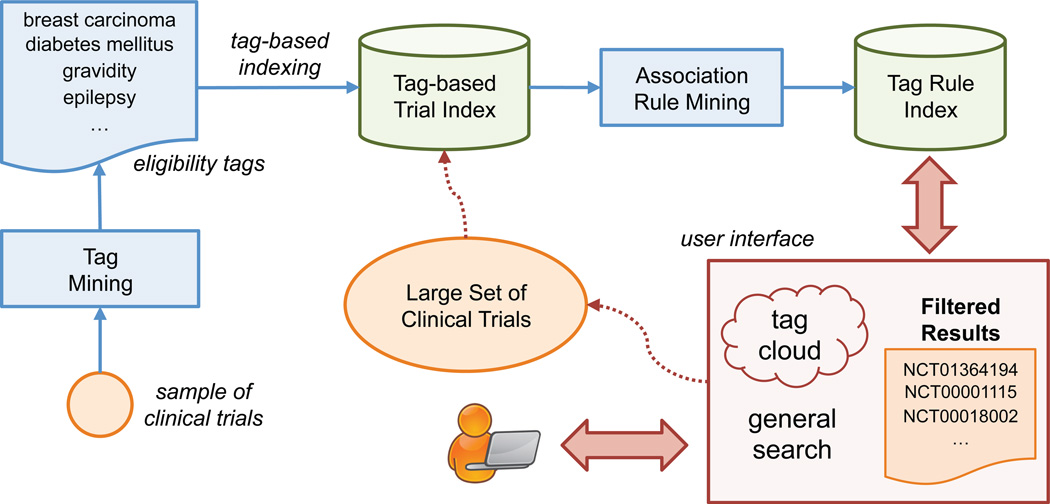
The eTACTS framework consists of two components (see Figure 1): (1) eligibility tag mining and clinical trial indexing, and (2) online tag cloud-based dynamic trial search. In the following sections, we present the main components of the proposed framework and the evaluation design. The detailed design and evaluation for unsupervised tag mining and eligibility criteria indexing were reported previously [16] and hence will only be briefly reviewed here.
Figure 1.
Overview of the eTACTS framework.
2.1. Tag Mining and Eligibility Criteria Indexing
eTACTS automatically mines tags from the free-text eligibility criteria of a representative set of clinical trials. Text processing techniques are used to extract relevant n-grams from each criterion, where the n-gram relevance is defined by the grammatical role of the words, limited presence of stop words, and matching of at least one word with the UMLS lexicon. Terms that match the UMLS are also normalized into preferred UMLS terms. Only the most frequent n-grams of the collection are retained as potential tags. This set is then automatically polished — not-discriminative n-grams and irrelevant substrings are removed — to obtain the final controlled vocabulary of eligibility tags. At indexing time, each clinical trial available in the repository is annotated with only those tags extracted from their eligibility criteria.
Tags are mined and assigned to trials regardless of their role being inclusion or exclusion. In fact, with eTACTS, tags are meant to identify high-level concepts mentioned in the text rather than structured semantic patterns (e.g., “concept-X greater than N”, “not concept-X”) in eligibility criteria. While distinguishing between inclusion and exclusion roles can be useful with semantic patterns, it is not always useful for tags. For example, a tag appearing frequently in clinical trial eligibility criteria is “body mass index” (BMI), which is usually followed by a value (e.g., “inclusion: BMI > 40”). Without indexing the entire pattern, distinguishing BMI between inclusion/exclusion would be misleading in the filtering process. In fact, for example, a user could select “inclusion: BMI” aiming to find trials enrolling participants with a high BMI value; however, in this way, the user might miss those trials where the same concept is expressed as an exclusion criterion (e.g., “exclusion: BMI ≤ 40”). In contrast, tags related to medical conditions, e.g., “breast cancer”, might benefit from identifying their role. Nevertheless, in this study we treated all tags identically because our objective was to assess the general feasibility of our approach, which is based on the hypothesis that natural language processing-based semantic pattern recognition and processing (which can be error-prone and lead to noisy representations) are not necessary for information filtering, which is our focus.
2.2. Tag Cloud-based Retrieval
The objective of tag-based retrieval is to refine the results of a simple search. The key feature of eTACTS is to present to users a tag cloud highlighting the most relevant eligibility tags. In this domain, the relevance of a tag is defined as the combination between its frequency in the resulting set and its relatedness to the tags currently picked by the user (i.e., how many times a tag co-occurs with the tags already chosen in the filtering process). In the cloud, greater tag relevance is represented by increased point size and color contrast. Figure 2 shows an example of general eligibility tag cloud associated with the query “diabetes mellitus”.
Figure 2.
Eligibility tag cloud derived from results associated to the search “diabetes mellitus”.
A user initiates a search using simple query free-text terms (e.g., “breast cancer”, “diabetes new york”) or more advanced forms (e.g., the advanced search of ClinicalTrials.gov), which can potentially return a large number of clinical trials. The initial tag cloud reflects the most frequent tags in the eligibility criteria of the resulting trials. In order to ensure the readability of the cloud and to provide a manageable number of selection options to the users, we configured eTACTS to display 30 tags per cloud. The user then selects a tag from the cloud to filter the resulting trials. At this point, eTACTS retains only those trials having all the chosen tags, which are shown beside the cloud. A user can remove a tag from this list of selected tags so that eTACTS reverts to the previous cloud, which does not contain the removed tag, and trial results. The user repeats this process until a manually reviewable number of trials remains.
2.2.1. Tag-cloud updating via Association Rules
At each iteration, eTACTS automatically updates the tag cloud to represent the current resulting trials. Different techniques can be used to drive the choice of which new tags to display (e.g., sampling the most frequent tags in the resulting trials or sampling based on tag co-occurrences). We applied statistics based on association rules among the tags. In particular, let D be a set of trials where each trial is represented as a sequence of eligibility tags; an association rule is an implication of the form X Y, where X and Y are mutually exclusive sets of tags, i.e., X ∩ Y = ∅. The rule X Y holds in the collection with confidence c if c% of trials in D that contain X also contain Y. The rule X Y has support s in the collection if s% of trials in D contain X ∪ Y [50]. In this domain, an example of association rule is (“gender = female”, “breast carcinoma”) (“negative pregnancy test”).
Mining association rules in a collection of documents reduces to retain all rules having support and confidence greater than specified minimum support and minimum confidence, respectively. The algorithm consists of two steps: (1) automatically generate all the sets of tags with minimum support; and (2) from each set, choose the rules that have minimum confidence. Tag sets are generated using the FP-growth algorithm, which maps the collection of trials to an extended frequent-pattern tree and processes this tree recursively to grow all the sets [51]. This is more efficient than combinatorial approaches, such as the Apriori algorithm [50], which iterate multiple times across the collection.
The association rules were mined offline from the eligibility criteria index and then used to inform the choice of tags to be displayed in the cloud. Because we wanted to maximize the number of tag combinations covered by the mined rules, we set minimum support and confidence to low values, 1 and 30, respectively. Given a set of tags selected by the user at a certain status, the new tag cloud is composed of the tags having the greatest confidence with that selection in the association rules. If too few rules are available for a certain tag combination (i.e., there are fewer than 30 tags related to that combination satisfying the minimum confidence requirement), the algorithm integrates the cloud with the most frequent tags in the resulting set. However, these tags are assigned a lower relevance — and thus displayed with a smaller font — than those derived from the association rules. Consequently, if no rules are available for a particular tag combination, the tag cloud is simply composed of the most frequent tags of the resulting set, as it is at the beginning of the process.
2.3. Evaluation Design
We intend eTACTS to work with any clinical trial repository and any type of initial search. As an applicative example, in this evaluation the data collection consisted of the 141,291 trials available at ClinicalTrials.gov as of February 2013. We first mined a vocabulary of 260 frequent (i.e., appearing in at least 2% of all the trials) eligibility tags (see Appendix A) from 65,000 randomly sampled clinical trials [16], and we used these tags to index all the trials of the repository. We then developed a Web interface linked to the ClinicalTrials.gov API to enhance its search facilities with an interactive tag cloud; in that way, we evaluated eTACTS’ ability to refine the results returned by a general search performed on ClinicalTrials.gov. The interface allows users to perform simple free-text searches as well as searches based on the ClinicalTrials.gov advanced form (which is embedded in the architecture), and to use the tag cloud to filter the resulting trials. The Web-based eTACTS implementation used in this study is available at: http://is.gd/eTACTS.
Evaluation of algorithms for clinical trial retrieval is difficult due to the lack of a well-established gold standard reference. To overcome this difficulty, we used mixed-methods to design our evaluation. We first performed automatic quantitative evaluations of the scalability of eTACTS in filtering the initial search results and of the diversity of the filtered trials with respect to the initial results. Additionally, we performed qualitative evaluations using surveys to measure the user-perceived usefulness and usability of the current version of eTACTS.
2.3.1. Reduction of the Result List
This experiment aimed to test the feasibility of reducing the search resulting documents to a manually reviewable list by selecting a limited number of tags. We chose 50 medical conditions (see Appendix B) that each had more than 1,000 associated trials in ClinicalTrials.gov. For each condition, we performed 500 distinct simulations — that is, 25,000 total simulations — based on random tag clicks to filter the result sets until only one trial was left. This simulates a binary search through the initial resulting set [52], with every tag click reducing the number of remaining trials. We measured the number of tags required to reach a preset threshold for the number of resulting trials (i.e., at most 3, 5, 10, 20, 50 trials) and the number of trials returned after a certain number of selected tags.
We compared eTACTS based on association rules (i.e., “a-rule”) with other options to manage the tags displayed in the clouds. In particular, we included tag sampling based on the most frequent tags in the resulting sets (i.e., “tag-most”) as well as tag sampling based on the context of the last tag selected (i.e., “jc-context”). In this case, the cloud was composed of the tags that mostly co-occurred in the repository with the last tag clicked; co-occurrence scores were based on Jaccard coefficients (mined offline), which measure the number of times two tags co-occur over all trials, normalized by the number of times each tag appears [52].
2.3.2. Diversity of the Filtered Results
Because conventional search engines display ranked results page by page (e.g., ClinicalTrials.gov displays 20 results per page), users generally review just the first page. Therefore, we hypothesize that returning different trials in the first page from those displayed by the initial search is a necessary though insufficient requirement to show improvement over existing trial search engines [53, 54].
This experiment measured the diversity of the results with respect to the original search in terms of the first 20 trials. To determine variation, we sampled 10 common medical conditions and 10 common locations (see Appendix C) and issued all 100 possible term-combination queries (i.e., “condition” AND “location”). For each query, we simulated each available tag click from the first tag cloud (i.e., 30 tags per query for a total of 3,000 simulations). We then measured the number of common trials that appeared in both the first 20 results of the original search and the first 20 results of the filtered search obtained after each tag click. Since eTACTS does not provide any specific rank measures for the filtered clinical trials, we sorted them according to their rank in the initial search. We compared the results with filters based on age, gender, and study type (and their combinations) from the advanced search of ClinicalTrials.gov. To do this, we filtered each initial query result according to all available filter options one by one.
2.3.3. A Scenario-based User Study Comparing Multiple Clinical Trial Search Engines
To measure the quality of the filtered trials, we recruited 12 users to search for clinical trials using the combined query terms (as free-text search): “diabetes mellitus type II” and “New York”. We applied a scenario-based evaluation using simulated user profiles [55, 56]. In particular, we created four mock patient profiles that could simulate the symptoms and behaviors typical in patients with “diabetes mellitus type II”. These mock characteristics were generated by manually analyzing the eligibility criteria of 50 random trials on ClinicalTrials.gov with “diabetes mellitus type II” as the query condition. Common characteristics were defined by the most frequent UMLS terms that were found in the text. Figure 3 shows an example of a mock patient. In this study, we assigned each mock patient three times among the 12 independent test users. The pool of participants was composed of four biomedical informatics students, three physicians, three clinical research coordinators, and two database administrators. During the evaluation, each tester played the role of the assigned mock patient and searched for trials that matched that patient’s characteristics1.
Figure 3.
Example of a mock patient profile adopted in the user evaluation.
Each tester compared eTACTS with other five search engines: ClinicalTrials.gov simple search, ClinicalTrials.gov advanced search with age and gender (i.e., “advanced ClinicalTrials.gov”), ASCOT, Corengi, and PatientsLikeMe. Each of these systems displays the eligibility criteria of trials resulting from a search. The evaluation focused on entering information into each system, navigating the site, and determining trial eligibility. As the most relevant measure, we asked users to manually review the top five trials in each ranking list and to determine if their mock patients were eligible for those trials. The entire process required about 45 minutes. To measure the user-perceived usefulness and usability of each of the five systems, we also modeled a final survey on existing studies that evaluate clinical information systems [57]. This survey was given to the test users at the end of the process (see Appendix D). In the survey, we chose to not evaluate the usability of “advanced ClinicalTrials.gov” but only of “ClinicalTrials.gov” to avoid creating confusion in the participants and redundancy in the results.
3. Results
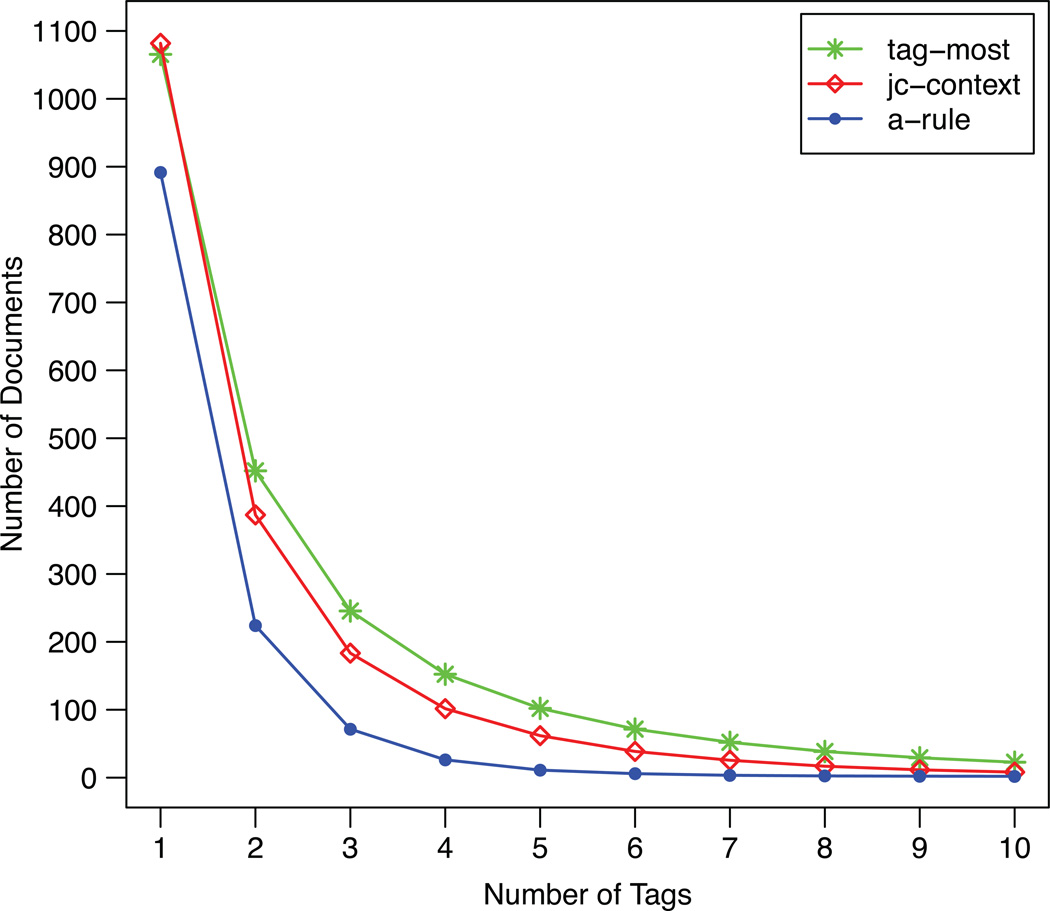
Table 1 reports the number of tags required to reduce the resulting trials to a preset number. For each query condition, we measured minimum, maximum, and mean number of tags over the 500 random simulations and reported the results averaged over all queries. In addition, Figure 4 reports the mean number of documents returned after a certain number of selected tags. The tag cloud based on association rules achieved the best results by allowing users to obtain a manually reviewable number of trials with fewer tag selections. Additionally, Table 2 presents examples of the eTACTS filtering process performed with random tag selection on four query conditions. The resulting set can easily be reduced by specifying eligibility characteristics; the more specific the tags, the faster the number of remaining trials decreased. Table 3 reports the mean number of trials shared with the initial search in the first 20 results as well as the average number of trials returned by each filter. eTACTS generally retains only 25% of the trials displayed in the first page by the initial search, leading to more diverse results. This measure also decreases when other tags are applied, converging to zero at about the third tag selection.
Table 1.
Minimum, maximum, and mean number of tags a user must select to not exceed a preset number of filtered trials averaged over 50 query conditions. Each query was tested with 500 simulations based on random tag clicks. We compare different strategies for tag cloud updating: “tag-most” samples the most frequent tags in the resulting trials; “jc-context” samples the tags mostly co-occurring with the last tag selected; “a-rule” samples the tags based on association rules. For each metric and experiment, best results are reported in bold.
| Number of Tag Selections | ||||
|---|---|---|---|---|
| Filtered Trial Limit | Algorithm | MIN | MAX | MEAN |
| tag-most | 7.38 | 31.44 | 17.17 | |
| 3 | jc-context | 3.04 | 18.84 | 8.82 |
| a-rule | 2.44 | 14.92 | 5.52 | |
| tag-most | 6.86 | 26.00 | 14.55 | |
| 5 | jc-context | 2.92 | 15.80 | 8.04 |
| a-rule | 2.38 | 11.90 | 5.03 | |
| tag-most | 5.84 | 19.24 | 11.30 | |
| 10 | jc-context | 2.72 | 13.24 | 7.06 |
| a-rule | 2.22 | 9.52 | 4.47 | |
| tag-most | 4.94 | 14.54 | 8.89 | |
| 20 | jc-context | 2.48 | 11.74 | 6.09 |
| a-rule | 2.08 | 7.92 | 3.95 | |
| tag-most | 3.54 | 10.46 | 6.48 | |
| 50 | jc-context | 2.28 | 9.22 | 4.85 |
| a-rule | 1.98 | 6.46 | 3.30 | |
Figure 4.
The mean number of clinical trials returned after a certain number of selected tags averaged over 50 distinct query conditions (each tested with 500 simulations based on random tag clicks). We compare tag cloud updating strategies based on most frequent tag sampling (“tagmost”), last selected tag context (“jc-context”), and association rules (“a-rule”).
Table 2.
eTACTS filtering using the random tag selection. For example, a search of “breast cancer” returns 4,822 trials: of these, 836 contain tag 1 (“metastatic malignant neoplasm to brain”), 236 contain tags 1 and 2 (“pregnancy test negative”), 82 contain tags 1, 2, and 3 (“mental disorders”), and 10 contain tags 1, 2, 3, and 4 (“major surgery”). By selecting these four tags in this order, a user can reduce search results from 4,822 trials to 10.
| breast cancer | 4,822 trials | diabetes mellitus, type 2 | 4,015 trials |
|---|---|---|---|
| tag 1: metastatic malignant neoplasm to brain | 836 trials | tag 1: diabetes mellitus insulin-dependent | 1,448 trials |
| tag 2: pregnancy test negative | 236 trials | tag 2: body mass index | 584 trials |
| tag 3: mental disorders | 82 trials | tag 3: hypertensive disease | 392 trials |
| tag 4: major surgery | 10 trials | tag 4: body weight decreased | 45 trials |
| tag 5: hemoglobin | 8 trials | ||
| hypertensive disease | 4,106 trials | hiv infections | 4,827 trials |
| tag 1: uncontrolled hypertensive disease | 192 trials | tag 1: hiv seropositivity | 913 trials |
| tag 2: alcohol abuse | 87 trials | ||
| tag 2: diastolic blood pressure | 77 trials | tag 3: pregnant | 59 trials |
| tag 3: cardiac arrhythmia | 7 trials | tag 4: antiretroviral therapy | 8 trials |
Table 3.
Diversity of the filtered results in terms of mean number of trials in common with the first 20 displayed by ClinicalTrials.gov after the initial search. A small number of trials in common mean high diversity; the best system measures are reported in bold. Results are average over 100 distinct queries (“condition” AND “location”); for each query we applied different filters (all the combinations of available filtering values).
| Filter Type | Number of Resulting Trials |
Number of Trials Common to the Top 20 of the Initial Search |
|---|---|---|
| Original Search Results | 544.13 | n/a |
| Age only | 358.86 | 12.62 |
| Gender only | 511.85 | 18.71 |
| Study Type only | 271.44 | 9.95 |
| Age and Gender | 337.39 | 11.79 |
| Age and Study Type | 179.28 | 6.29 |
| Gender and Study Type | 255.31 | 9.30 |
| Age, Gender, and Study Type | 168.80 | 5.88 |
| eTACTS - 1 tag | 142.62 | 4.56 |
Table 4 presents the results of the user evaluation. “Q1” reports the retrieval performances in terms of the average percentage of top five ranked trials for which users considered their mock patient to be eligible. As it can be seen, eTACTS and ASCOT achieved the best results, reinforcing the idea that processing eligibility criteria helps volunteers to find trials for which they might be eligible. eTACTS significantly improved the search results of ClinicalTrials.gov as well as those of user profile-based engines. ASCOT and eTACTS obtained similar results in terms of percentage of trials where users considered their profile being eligible. However, when explicitly asked, 11 out of 12 users found eTACTS outperformed ASCOT in terms of usability of the cloud and relatedness of the displayed tags to the initial search. One user expressed no preference for either system. Additionally, eTACTS was the easiest tool to use among the system compared, with ClinicalTrials.gov ranking as second (i.e., “Q2” through “Q5”). Overall, users strongly preferred systems based on simple searches or result refining than systems based on matching medical profiles with relevant trials (as in Corengi and PatientsLikeMe).
Table 4.
User evaluation of six clinical trial search engines. We measured: (Q1) percentage of trials within the top five ranked by each clinical trial search engine for which users considered their mock patient eligible; (Q2) ease of entering information; (Q3) ease of site navigation; (Q4) ease of use without preliminary training; and (Q5) overall ease of use. For questions Q2 through Q5, "1" indicates best, "6" indicates worst, and ties were allowed. The usability of “advanced ClinicalTrials.gov” refers to the results of “ClinicalTrials.gov”. Results are the average of 12 participants, 4 mock patient profiles (each assigned 3 times), and initial search terms “diabetes mellitus type II” and “New York”. For each column, the best results are reported in bold.
| Search System | Q1 | Q2 | Q3 | Q4 | Q5 |
|---|---|---|---|---|---|
| eTACTS | 50.0% | 2.41 | 1.75 | 2.16 | 1.91 |
| ASCOT | 48.3% | 4.08 | 3.67 | 3.75 | 3.83 |
| advanced ClinicalTrials.gov | 34.9% | n/a | n/a | n/a | n/a |
| ClinicalTrials.gov | 23.3% | 2.91 | 2.17 | 2.67 | 2.91 |
| PatientsLikeMe | 13.3% | 3.00 | 3.17 | 2.75 | 3.33 |
| Corengi | 6.7% | 4.17 | 3.58 | 3.42 | 3.42 |
4. Discussion
eTACTS augments state-of-the-art clinical trial search engines with eligibility criteria-based dynamic filtering to provide users with a manually reviewable number of trials. eTACTS can benefit various types of user: e.g., volunteers and family members can select tags representing medical conditions, symptoms, or laboratory tests, whereas research coordinators can choose tags related to the specific task for which they need information (e.g., creation of a new trial). Given its generality, eTACTS can be applied to filter any type of search (e.g., condition, intervention, outcome, advanced form-based search) and can complement existing systems toward more specific search results.
Experimental evaluation using ClinicalTrials.gov led to three conclusions. First, eTACTS allows users to reduce the number of trials in search results from thousands of trials to 10, with an average of five tags. Since users are not required to select many tags to reach a manually reviewable number of trials, they might be more likely to repeat the refining process if satisfactory results were not obtained. In contrast, more time-consuming alternative methods (e.g., completing a user profile form) discourage users from repeating the process if initial results were not satisfactory.
Second, filtered results provided by eTACTS tend to differ from those highly ranked in the initial search. This allows users to discover trials not easily found using standard retrieval techniques. During our simulations, we found many trials currently recruiting participants ranked after the first hundred results. These trials are unlikely to be seen by users with a simple search, and result filtering is necessary. Refining search results by eligibility tags can speed up the search process while discovering new trials. Moreover, experiments on tag diversity also showed that eTACTS — and result filtering in general — can be necessary even when a location is added to the initial search. In fact, adding geographical details, such as New York and the United Kingdom, to the search produces results comprising, on average, over 500 trials, still too many for manual review. eTACTS can benefit this scenario as well by enabling users to also refine more specific hybrid searches (i.e., which are composed by different aspects). In fact, our recommended use of eTACTS is for users to start with an initial form-based search involving contextual details (e.g., status of the trial, title, geographic location) and then interact with the cloud to filter the current search results according to eligibility criteria.
Third, user evaluations showed that eTACTS provided more relevant top-ranked results than other available clinical trial search engines. The systems included in the comparison represent the state-of-the-art for clinical trial retrieval. The only relevant system excluded from the evaluation was TrialX, because it does not display eligibility criteria sections of the trials retrieved. Instead, TrialX only provides a brief summary for the resulting trials and requires the user to contact research coordinators for more information.
4.1. Limitations and Future Works
This study has a couple of limitations. The user evaluation provided in this article only aimed to prove the feasibility of the framework for a simple case study. In addition, the user evaluation only referred to a single medical condition, both in terms of search and user (i.e., potential non-healthy volunteer). While a pool of 10 – 12 participants in a user evaluation is generally enough to assess the usability of a system [58], a larger scale evaluation should be conducted. For this reason, we publicly released the system (as already mentioned, available at http://is.gd/eTACTS) in order to exploit log analysis and relevancy feedback techniques to model and analyze user actions and satisfaction on a larger scale. This will also permit analysis of the clinical quality and retrieval usefulness of the eligibility tags, which are automatically mined from the text and could include concepts irrelevant for retrieval purposes, and inform future design to improve its accessibility to health consumers with low health literacy levels.
Second, an aspect of the eTACTS framework that should be further evaluated is the addition of inclusion/exclusion status to those tags where appropriate (e.g., conditions and signs), which may result in a faster filtering process or more specific results.
5. Conclusions
This paper presented eTACTS, a novel clinical trial search framework that can potentially reduce the information overload for people searching for clinical trials online. eTACTS combines tag mining of free-text eligibility criteria and interactive retrieval based on dynamic tag clouds to reduce the number of resulting trials returned by a simple search. Evaluation on ClinicalTrials.gov showed the feasibility of this approach in terms of its scalability, diversity of the resulting information, and effectiveness in retrieving trials for which users are eligible.
Supplementary Material
Highlights.
A dynamic tag cloud may reduce information overload for clinical trial search
We present eTACTS, a novel method to dynamically filter clinical trial search results
eTACTs relies on tags automatically mined from the eligibility criteria
eTACTS is available at http://is.gd/eTACTS
eTACTS may be a valuable enhancement for available clinical trial search engines
Acknowledgments
The authors would like to thank the reviewers for their constructive and insightful comments as well as all the study participants in the user evaluations.
Funding: The research described was supported by grants R01LM009886 and R01LM010815 (PI: Weng) and T15LM007079 (PI: Hripcsak) from the National Library of Medicine, and grant UL1 TR000040 (PI: Ginsberg), funded through the National Center for Advancing Translational Sciences (NCATS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The user evaluation presented in this human subject study was approved by the Institutional Review Board of Columbia University Medical Center (IRB-AAAJ8850).
Competing Interests: None.
References
- 1.Califf RM, Filerman G, Murray R, Rosenblatt M. The clinical trials enterprise in the United States: A call for disruptive innovation; Envisioning a Transformed Clinical Trials Enterprise in the United States: Establishing an Agenda for 2020 (Workshop Summary); 2012. [PubMed] [Google Scholar]
- 2.Tassignon JP, Sinackevich N. Speeding the critical path. Appl Clin Trials. 2004 [Google Scholar]
- 3.Sullivan J. Subject recruitment and retention: barriers to success. Appl Clin Trials. 2004 [Google Scholar]
- 4.Atkinson NL, Massett HA, Mylks C, Mc Cormack LA, Kish-Doto J, Hesse BW, Wang MQ. Assessing the impact of user-centered research on a clinical trial eHealth tool via counterbalanced research design. J Am Med Inform Assoc. 2011;18(1):24–31. doi: 10.1136/jamia.2010.006122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. ClinicalTrials.gov. 2013 Apr; Available from: http://www.clinicaltrials.gov/.
- 6.UK Clinical Trials Gateway. 2013 Apr; Available from: http://www.ukctg.nihr.ac.uk/. [Google Scholar]
- 7.Ide NC, Loane RF, Demner-Fushman D. Essie: a concept-based search engine for structured biomedical text. J Am Med Inform Assoc. 2007;14(3):253–263. doi: 10.1197/jamia.M2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patel CO, Garg V, Khan SA. What do patients search for when seeking clinical trial information online? AMIA Ann Symp Proc. 2010;2010:597–601. [PMC free article] [PubMed] [Google Scholar]
- 9.Weng C, Tu SW, Sim I, Richesson R. Formal representation of eligibility criteria: a literature review. J Biomed Inform. 2010;43(3):451–467. doi: 10.1016/j.jbi.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weng C, Wu X, Luo Z, Boland MR, Theodoratos D, Johnson SB. EliXR: an approach to eligibility criteria extraction and representation. J Am Med Inform Assoc. 2011;18(Suppl 1):i116–i124. doi: 10.1136/amiajnl-2011-000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ross J, Tu S, Carini S, Sim I. Analysis of eligibility criteria complexity in clinical trials. AMIA Summits Transl Sci Proc. 2010;2010:46–50. [PMC free article] [PubMed] [Google Scholar]
- 12.Wei BF, Liu J, Zheng QH, Zhang W, Fu XY, Feng BQ. A Survey of Faceted Search. Journal of Web Engineering. 2013;12(1–2):41–64. [Google Scholar]
- 13.Devadason F, Intaraksa N, Patamawongjariya P, Desai K. Search interface design using faceted indexing for Web resources. ASIST. 2001;38:224–238. [Google Scholar]
- 14.Yee KP, Swearingen K, Li K, Hearst M. Faceted metadata for image search and browsing. ACM SIGCHI. 2003:401–408. [Google Scholar]
- 15.Yeh ST, Liu Y. Integrated Faceted Browser and Direct Search to Enhance Information Retrieval in Text-Based Digital Libraries. International Journal of Human-Computer Interaction. 2011;27(4):364–382. [Google Scholar]
- 16.Miotto R, Weng C. Unsupervised Mining of Frequent Tags for Clinical Eligibility Text Indexing. J Biomed Inform. 2013 doi: 10.1016/j.jbi.2013.08.012. (under second review) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Bruijn B, Carini S, Kiritchenko S, Martin J, Sim I. Automated information extraction of key trial design elements from clinical trial publications; AMIA Annu Symp Proc; 2008. pp. 141–145. [PMC free article] [PubMed] [Google Scholar]
- 18.Hernandez ME, Carini S, Storey MA, Sim I. An interactive tool for visualizing design heterogeneity in clinical trials; AMIA Annu Symp Proc; 2008. pp. 298–302. [PMC free article] [PubMed] [Google Scholar]
- 19.Chung GY. Sentence retrieval for abstracts of randomized controlled trials. BMC Med Inform Decis Mak. 2009;9:10. doi: 10.1186/1472-6947-9-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paek H, Kogan Y, Thomas P, Codish S, Krauthammer M. Shallow semantic parsing of randomized controlled trial reports; AMIA Annu Symp Proc; 2006. pp. 604–608. [PMC free article] [PubMed] [Google Scholar]
- 21.Xu R, Supekar K, Huang Y, Das A, Garber A. Combining text classification and Hidden Markov Modeling techniques for categorizing sentences in randomized clinical trial abstracts; AMIA Annu Symp Proc; 2006. pp. 824–828. [PMC free article] [PubMed] [Google Scholar]
- 22.Boland MR, Miotto R, Gao J, Weng C. Feasibility of Feature-based Indexing, Clustering, and Search of Clinical Trials on ClinicalTrials.gov: A Case Study of Breast Cancer Trials. Methods Inf Med. 2013;52(4) doi: 10.3414/ME12-01-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo Z, Miotto R, Weng C. A human-computer collaborative approach to identifying common data elements in clinical trial eligibility criteria. J Biomed Inform. 2013;46(1):33–39. doi: 10.1016/j.jbi.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tu SW, Peleg M, Carini S, Bobak M, Ross J, Rubin D, Sim I. A practical method for transforming free-text eligibility criteria into computable criteria. J Biomed Inform. 2011;44(2):239–250. doi: 10.1016/j.jbi.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parker CG, Embley DW. Generating medical logic modules for clinical trial eligibility criteria; AMIA Annu Symp Proc; 2003. p. 964. [PMC free article] [PubMed] [Google Scholar]
- 26.Aronson AR, Lang FM. An overview of MetaMap: historical perspective and recent advances. J Am Med Inform Assoc. 2010;17(3):229–236. doi: 10.1136/jamia.2009.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo Z, Duffy R, Johnson S, Weng C. Corpus-based approach to creating a semantic lexicon for clinical research eligibility criteria from UMLS. AMIA Summits Transl Sci Proc. 2010;2010:26–30. [PMC free article] [PubMed] [Google Scholar]
- 28.Atkinson NL, Saperstein SL, Massett HA, Leonard CR, Grama L, Manrow R. Using the Internet to search for cancer clinical trials: A comparative audit of clinical trial search tools. Contemporary Clinical Trials. 2008;29(4):555–564. doi: 10.1016/j.cct.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Metz JM, Coyle C, Hudson C, Hampshire M. An Internet-based cancer clinical trials matching resource. J Med Internet Res. 2005;7(3):e24. doi: 10.2196/jmir.7.3.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei SJ, Metz JM, Coyle C, Hampshire M, Jones HA, Markowitz S, Rustgi AK. Recruitment of patients into an Internet-based clinical trials database: the experience of OncoLink and the National Colorectal Cancer Research Alliance. J Clin Onc. 2004;22(23):4730–4736. doi: 10.1200/JCO.2004.07.103. [DOI] [PubMed] [Google Scholar]
- 31.Embi PJ, Jain A, Clark J, Bizjack S, Hornung R, Harris CM. Effect of a clinical trial alert system on physician participation in trial recruitment. Arch Intern Med. 2005;165(19):2272–2277. doi: 10.1001/archinte.165.19.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ask Dory! 2013 Apr; Available from: http://dory.trialx.com/ask/. [Google Scholar]
- 33.PatientsLikeMe. 2013 Apr; Available from: http://www.patientslikeme.com/. [Google Scholar]
- 34.SearchClinicalTrials. 2013 Apr; Available from: http://www.searchclinicaltrials.org/. [Google Scholar]
- 35.TrialReach. 2013 Apr; Available from: http://www.trialreach.com/. [Google Scholar]
- 36.ResearchMatch. 2013 Apr; Available from: https://http://www.researchmatch.org/. [Google Scholar]
- 37.Corengi. 2013 Apr; Available from: https://http://www.corengi.com/. [Google Scholar]
- 38.Korkontzelos I, Mu T, Ananiadou S. ASCOT: a text mining-based Web-service for efficient search and assisted creation of clinical trials. BMC Med Inform Decis Mak. 2012;12(Suppl. 1):S3. doi: 10.1186/1472-6947-12-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lindberg DA, Humphreys BL, Mccray AT. The Unified Medical Language System. Methods Inf Med. 1993;32(4):281–291. doi: 10.1055/s-0038-1634945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruthven I. Interactive information retrieval. Annu Rev of Information Science and Technology. 2008;42:43–91. [Google Scholar]
- 41.Hassan-Montero Y, Herrero-Solana V. Improving tag-clouds as visual information retrieval interfaces; Int Conf on Multidisciplinary Information Sciences and Technologies; 2006. [Google Scholar]
- 42.Kaser O, Lemire D. Tag-cloud drawing: algorithms for cloud visualization; Workshop on Tagging and Metadata for Social Information Organization (WWW 2007); 2007. [Google Scholar]
- 43.Murtagh F, Ganz A, Mckie S, Mothe J, Englmeier K. Tag clouds for displaying semantics: The case of filmscripts. Information Visualization. 2010;9(4):253–262. [Google Scholar]
- 44.Harris J. Word clouds considered harmful. 2011 Available at: http://www.niemanlab.org/2011/10/word-clouds-considered-harmful/ [Google Scholar]
- 45.Hearst M, Rosner D. Tag Clouds: Data Analysis Tool or Social Signaller?; Annu Hawaii Int Conf on System Sciences; 2008. p. 160. [Google Scholar]
- 46.Zeldman J. Remove forebrain and serve: Tag Clouds II. 2008 Available from: http://www.zeldman.com/daily/0505a.shtml. [Google Scholar]
- 47.O'grady L, Wathen CN, Charnaw-Burger J, Betel L, Shachak A, Luke R, Hockema S, Jadad AR. The use of tags and tag clouds to discern credible content in online health message forums. Int J Med Inform. 2012;81(1):36–44. doi: 10.1016/j.ijmedinf.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 48.Levy M, Sandler M. Music information retrieval using social tags and audio. IEEE Transactions on Multimedia. 2009;11(3):383–395. [Google Scholar]
- 49.Callegari J, Morreale P. Assessment of the utility of tag clouds for faster image retrieval; ACM Int Conf on MIR; 2010. pp. 437–440. [Google Scholar]
- 50.Rakesh A, Ramakrishnan S. Fast algorithms for mining association rules in large databases; Int Conf on Very Large Databases; 1994. pp. 487–499. [Google Scholar]
- 51.Han JW, Pei J, Yin YW, Mao RY. Mining frequent patterns without candidate generation: A frequent-pattern tree approach. Data Mining and Knowledge Discovery. 2004;8(1):53–87. [Google Scholar]
- 52.Manning CD, Raghavan P, Schutze H. New York: Cambridge University Press; 2008. Introduction to information retrieval; p. 482. [Google Scholar]
- 53.Santos RLT, Macdonald C, Ounis I. On the role of novelty for search result diversification. Information Retrieval. 2012;15(5):478–502. [Google Scholar]
- 54.Santos RLT, Macdonald C, Ounis I. Selectively diversifying Web search results. ACM CIKM. 2010:1179–1188. [Google Scholar]
- 55.Rogers ML, Patterson E, Chapman R, Render M. Usability testing and the relation of clinical information systems to patient safety. Advances in Patient Safety: From Research to Implementation (Volume 2: Concepts and Methodology) 2005 [PubMed] [Google Scholar]
- 56.Dong J, Kelkar K, Braun K. Getting the most out of personas for product usability enhancements; Int Conf on Usability and Internationalization; 2007. pp. 291–296. [Google Scholar]
- 57.Zheng K, Padma R, Johnson MP, Diamond HS. An Interface-driven analysis of user interactions with an Electronic Health Records system. J Am Med Inform Assoc. 2009;16(2):228–237. doi: 10.1197/jamia.M2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hwang W, Salvendy G. Number of People Required for Usability Evaluation: The 10 +/− 2 Rule. Communications of the ACM. 2010;53(5):130–133. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.