Abstract
Tissue specific stem cell (TSC) number is tightly regulated in normal individuals but can change following severe injury. We previously showed that tracheobronchial epithelial TSC number increased after severe naphthalene (NA)-injury and then returned to normal. The present study focused on the fate of the supernumerary TSC and the signals that regulate TSC pool size. We used the Keratin 5-rTA/Histone 2B:GFP model to purify basal cells that proliferated infrequently (GFPbright) or frequently (GFPdim) after NA-injury. Both populations contained TSC but TSC were 8.5-fold more abundant in the GFPbright population. Interestingly, both populations also contained a unipotential basal progenitor (UPB), a mitotic basal cell subtype whose daughters were terminally-differentiated basal cells. The ratio of TSC to UBP was 5:1 in the GFPbright population and 1:5 in the GFPdim population. These data suggested that TSC proliferation in vivo promoted TSC-to-UPB differentiation. To evaluate this question, we cloned TSC from the GFPbright and GFPdim populations and passaged the clones 7 times. We found that TSC number decreased and UPB number increased at each passage. Reciprocal changes in TSC and UPB frequency were more dramatic in the GFPdim lineage. Gene expression analysis showed that β-catenin and Notch pathway genes were differentially expressed in freshly-isolated TSC derived from GFPbright and GFPdim populations. We conclude that: 1) TSC and UPB are members of a single lineage; 2) TSC proliferation in vivo or in vitro promotes TSC-to-UPB differentiation; and 3) an interaction between the β-catenin and Notch pathways regulates the TSC-to-UPB differentiation process.
Keywords: multipotential stem cells, unipotential basal progenitor, terminal differentiation, Wnt/β-catenin, Notch
Introduction
A tissue-specific stem cell (TSC) is defined, most simply, as a cell that can self-renew (e.g. replicate itself through cell division) and is multipotential (e.g. generates each of the differentiated cell types found in its native tissue 1. Lineage-tracing of tracheobronchial epithelial (TBE) basal cells in vivo identified a multipotential basal cell subtype that was a progenitor for tracheal Clara-like and ciliated cells 2–4. Subsequent cell purification and functional analysis demonstrated that the TSC was a basal cell subtype 4, 5 and we showed that the TSC was a CD49fbright/Sca1+/ALDH+ basal cell subtype 5. Functional analysis demonstrated that the TSC generated a unique clone, the rim clone, and that daughter-TSC were sequestered in the rim-domain. Serial passage studies demonstrated that TSC maintained their self-renewal and differentiation potential over at least 5 generations 5.
The in vivo lineage tracing studies also identified a unipotential basal progenitor (UPB), which generated only basal cell progeny. Our cell purification studies demonstrated that the UPB was CD49fbright/Sca1+ and that it generated a distinct clone type, the non-rim clone. UPB-derived non-rim clones could not be passaged. Thus, the UPB generated basal cell daughters that were terminally-differentiated.
We previously showed that only 10% of TSC proliferated in the steady state. This low mitotic index reflects the long half-life of TBE cells 6, 7. Thus, the TSCs are typically evaluated after injury. Our preferred injury-model utilizes naphthalene (NA), which is metabolized to a cytotoxic epoxide in cells that express cytochrome P450-2F2 or −2B28, 9. TBE Clara-like cells express these enzymes and are ablated after high-dose NA treatment. We showed that NA-injury caused 56% of TSC to proliferate and increased TSC number 3-fold 5. By recovery day 40, the TSC mitotic index and frequency returned to normal. This study demonstrated that injury resulted in generation of supernumerary TSC and suggested that these cells were lost during TBE regeneration.
TBE progenitor cells proliferate at different frequencies in the steady state and after injury 10. Mitotic frequency can be evaluated in vivo using the label-retention assay 11, 12. Herein, mitotic cell DNA is labeled with a nucleotide analogue or chromatin is labeled using the TRE-Histone 2B:GFP transgene 13. In the latter assay, a cell that divides infrequently retains the GFP-label and is identified as a GFP+ cell using histological methods or a GFPbright cell using FLOW cytometry (FLOW). Similarly, a cell that proliferates frequently dilutes the GFP-label and is identified as a GFP− cell on histological sections or a GFPdim cell by FLOW. The DNA and chromatin labeling approaches yielded similar results when compared using hair follicle histological sections 14. However, the chromatin labeling method allowed isolation of viable-cells and subsequent analysis of their proliferation and differentiation potential using in vitro functional assays.
TSC proliferation and differentiation are regulated by multiple, interacting signaling pathways 15–17. Previous studies demonstrated that the β-catenin pathway regulates bronchiolar TSC pool size 18–20 and that Notch signaling regulates bronchiolar TSC differentiation 21, 22. We reported β-catenin pathway activation in the NA-injured TBE 23, 24 and others demonstrated that β-catenin was necessary for TBE repair 25. β-catenin target genes include Notch pathway-components, which in turn regulate cell-cell interactions 26, 27. Our analysis of mosaic TBE cell cultures, including adjacent wild type and β-catenin stabilized clones 23 or wild type and β-catenin knockout clones 28 suggested that β-catenin regulated cell-cell interactions which in turn led to Clara-like and ciliated cell differentiation. Similarly, Notch pathway gene expression was identified in spheroid cultures of TBE basal cells 29. Genetic studies indicated that interactions between adjacent cells expressing a Notch-receptor and those expressing a Jagged-ligand led to TBE stratification, a form of epithelial differentiation. Thus, interactions between the β-catenin and Notch pathway were implicated in TSC differentiation.
The goal of this study was to determine the cellular and molecular mechanisms regulating normalization of TSC pool-size after NA-injury. First, we validated the Keratin 5-rTA/TRE-Histone2B:GFP system for analysis of TBE basal cells. Second, we used FLOW to purify GFPbright and GFPdim cells and evaluated TSC and UPB frequency within each population. Third, we established TSC-lineages from the GFPbright and GFPdim populations and determined the phenotype of the TSC clone-initiating cell. Fourth, we used serial passage to evaluate the impact of repeated TSC proliferation on TSC and UPB number, and on TSC mitotic and differentiation potential. Finally, we used gene expression analysis to identify signals that regulate TSC pool-size.
Materials and Methods
Materials
Detailed in the Online Supplement.
Animals
Mice were maintained in an AAALAC-approved facility and screened for pathogens on a quarterly basis. All procedures involving animal use were reviewed and approved by the National Jewish Health Institutional Animal Care and Use Committee. K5-rtTA 30 and TRE-histone 2B 13 monotransgenic (MTg) mice and K5rTA/TRE-H2B:GFP (BiTg) were used for the label-retention studies. C57/Bl6 mice were used for isolation of tracheal ‘filler’ cells and Fvb/n mice were used to analyze mitotic index after NA injury. Mice were treated with 250 mg/kg NA and/or fed dox chow. Sample size is indicated in Results.
Immunofluorescence studies
Tracheal tissue sections (5 µm) were generated from paraffin-embedded or frozen tissues and processed as described previously 24. All antibodies (K5, CCSP and ACT), and staining methods were previously described 10. See Supplement for GFP source and methods. Images were acquired using an upright Zeiss Imager Z1 fluorescent microscope and AxioVision software (Carl Zeiss) or an inverted Zeiss 200M confocal microscope using Intelligent Imaging Innovations Inc. software. Cell type frequency was quantified as previously indicated 24 and values were expressed as a percent of total epithelial cells. N=6 tracheas were quantified/group.
FLOW cytometry
Tracheal epithelial cells were recovered by digestion with dispase/ collagenase/trypsin (DCT) as previously described 5. A Moflo high-speed cell sorter (Dako Cytomation) was used to identify: 1) cells that were CD45-/CD31-/TER119-/DAPI- cells, 2) quantify GFP-fluorescence intensity in each cell; and 3) to separate GFPbright and GFPdim cells according to CD49f and Sca1 expression.
Clone forming cell frequency (CFCF) analysis
Freshly-isolated cells were cultured on irradiated NIH3T3 feeder layer in MTEC-plus medium as previously described 5 and analyzed using the limiting dilution method 31. Specifically, CD45-/CD31-/TER119-/DAPI-/GFPbright and CD45-/CD31-/TER119-/DAPI-/GFPdim cells were directly deposited to the wells of 96-well plates using the CyCLONE automated cloner. Two-fold decrements from 200 cells/well to 1 cell/well were used. Each well was scored as positive or negative for rim or non-rim clones. Linear regression analysis was used to determine the rim-CFCF and non-rim-CFCF for each cell population.
Serial passage analysis
P0 rim clones were derived from GFPbright or GFPdim cells were harvested by trypsinization. One thousand cells were plated in each well of a 6-well plate. Each well contained an irradiated NIH3T3 feeder layer. The cells were cultured in dox-free MTEC-plus medium for 14-days. On day 14 the number of rim and non-rim clones was quantified. This process was repeated with P1-P6 rim-clones.
Real time PCR analysis
Total RNA was prepared from flow-sorted cells using a RNAeasy kit (Qiagen). cDNA was prepared and assayed as previously described using Assays-on-Demand™ 32. The expression level of mRNA was quantified using an ABI Prism 7000 Sequence Detection System™ and values were calculated by the ΔΔCT method using tracheal calibrator 19.
Statistical analysis
Results are presented as average ± standard error mean (SEM). Differences were calculated by Students t-test, one or two-way ANOVA using Graph-pad prism software.
Results
Validation of the Keratin 5-rTA/TRE-Histone2B:GFP system for analysis of TBE basal cells
We used the Keratin 5-rTA/TRE-H2B:GFP bitransgenic mouse model (BiTg, Fig 1A) to distinguish TBE basal cells that proliferated infrequently and or frequently. Dox-dependence and cell type specificity were evaluated in BiTg animals and monotransgenic (MTg) controls that were fed standard (Fig 1B) or dox (Fig 1C) chow for 6 days. Transgene activity was evaluated by dual immunofluorescence (DIF) detection of GFP and Keratin 5 (K5). H2B:GFP positive (GFP+) nuclei were not detected in MTg animals that were fed either standard or dox chow (SFig 1A and B) or in BiTg mice that were fed standard chow (Fig 1D). In contrast, dox exposure of BiTg mice resulted in expression of nuclear GFP (Fig 1E) and 70% of K5+ cells were GFP+. These data indicated that GFP expression was dependent on the presence of both transgenes and on dox exposure, and that most basal cells were labeled with GFP.
Figure 1. The chromatin label-retention system is cell type-specific and doxycycline (dox) dependent.
(A) Schematics depicting transgene structure. Keratin 5 (K5), tetracycline (TET), green fluorescent protein (GFP). (B–C) Images depicting expression of Histone 2B:GFP (H2B:GFP) when mice are fed standard chow (B) or dox chow (C). (D–E) Dual immunofluorescence analysis (DIF) of the tracheobronchial epithelium (TBE) from animals that were fed standard chow (D) or dox chow (E). K5-red, H2B:GFP-green, and DAPI (blue). (F) Experimental design. NA-naphthalene. DIF analysis of TBE from mice that were recovered to day 6 (G) or day 40 (H). Arrows-H2B:GFP+ nuclei (n=6 tracheas).
NA-injury is associated with decreased in food and water intake for 48 hrs 10, 23. Therefore, BiTg mice were fed dox chow 3-days prior to and 3 days after NA-injury (Fig 1F). A bromo-deoxyuridine (BrdU) pulse-labeling study indicated that the mitotic index increased 2–6 days after NA injury and returned to normal levels by recovery day 40 (SFig 2). Consequently, the GFP-label was chased for 3 days (tissue recovery on day 6) or 34 days (tissue recovery on day 40). Numerous GFP+ cells were detected in BiTg mice that were fed dox chow, treated with NA, and recovered after 3 days (Fig 1G). On post-naphthalene injury day 6, 90% of K5+ cells were GFP+. In contrast, a rare subset of TBE cells were GFP+ on day 40 (Fig1H). These data indicate that the BiTg model allows identification of TBE cells that proliferate frequently or infrequently after NA-injury.
Chromatin-labeling recapitulates DNA-labeling
To determine if the chromatin (GFP) and DNA (BrdU) label-retaining systems detected the same TBE cells, we treated BiTg mice with dox and NA and pulse-labeled DNA with BrdU on days 1–3 (SFig 3A). On day 40, GFP and BrdU were detected by DIF (n=6, SFig 3B). The αGFP antibody was titrated so that the frequency of GFP-immunopositive cells was equal to the number of cells detected using endogeneous GFP fluorescence (SFig 4). Quantification demonstrated that 7.87 ± 0.6% TBE cells were GFP+ and that 2.21 ± 0.45% of TBE cells were BrdU+ on day 40. Importantly, 90.07 ± 4.87% BrdU+ cells were also GFP+ (SFig 3C). These data demonstrated that the chromatin labeling model recapitulated DNA labeling system.
GFP+ cells are distributed throughout the cartilaginous region of the TBE
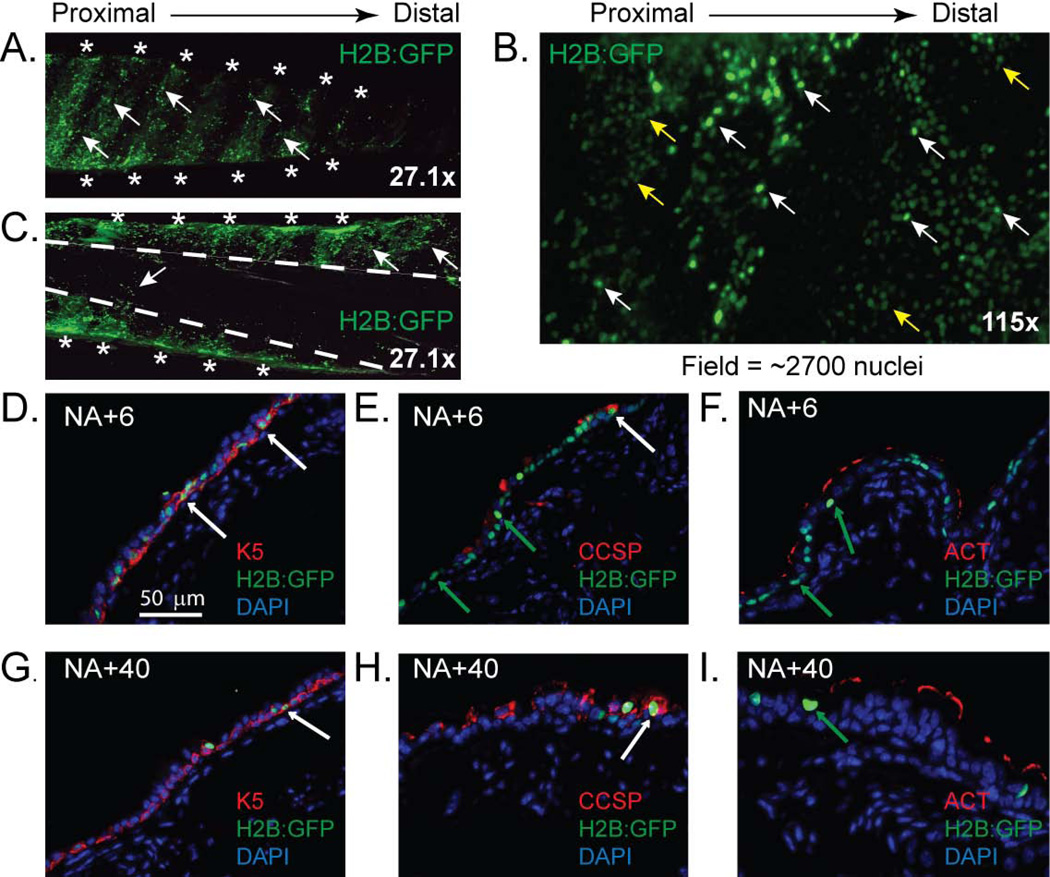
A previous study reported that acid injury resulted in long-lived BrdU+ cells that were restricted to the submucosal gland duct junction (SGDJ) and to the intercartilaginous (ICR) zones of the TBE 33. Thus, the SGDJ and ICR were identified as TSC niche. To determine if GFP+ cells were spatially-restricted after NA injury, we evaluated whole-mount tracheal preparations from BiTg mice that were treated with dox and NA and recovered 40 days. GFP+ cells were not detected in the SGDJ (SFig 5). However, GFP+ cells were detected throughout the ventral (cartilaginous) region of the trachea (Fig 2A) including the ICR and midcartilaginous (MCR) zones (Fig 2B). Very few GFP+ cells were detected in the dorsal (membranous) region of the trachea (Fig 2C). Thus, NA injury generated GFP+ cells that were more uniformly distributed than demonstrated in the acid injury model.
Figure 2. Localization and phenotype of Histone 2B:GFP positive (H2B:GFP+) cells.
(A–C) Whole mount images of trachea recovered from doxycycline (dox) and naphthalene (NA)-treated mice on recovery day 40. (A, C) external and (B) luminal surface. Asterisks: cartilage rings. Dashed lines: border between cartilaginous and membranous regions. Arrows: white-GFP+; yellow-GFPlow nuclei. (D–I) Dual immunofluorescence analysis of H2B:GFP+ cell phenotype on recovery day 6 (D–F) or 40 (G–I). Arrows: white H2B:GFP+ cells that co-express Keratin (K) 5 (D, G) or CCSP (E, H); green: H2B:GFP+ cells that do not express CCSP (E, H) or ACT (F, I) (n=6 tracheas).
We then determined the frequency of GFP+ in dox and NA-treated BiTg mice. GFP+ cell frequency was determined after a 3 days (post-injury day 6, Fig 2D–F) or 34 days (post-injury day 40, Fig 2G–I) chase. On day 6, 68.39 ± 4.66% of TBE cells were GFP+. On day 40, 7.4 ± 2.94% of TBE cells were GFP+. These cells were detected in both the ICR and MCR zones.
We used DIF analysis of GFP and K5, Clara cell secretory protein (CCSP) or acetylated tubulin (ACT) to determine the molecular phenotype of the GFP+ cells. On day 6, 91.34 ± 5.76% of GFP+ cells were K5+ (Fig 2D) and 3.12 ± 1.87% of GFP+ cells were CCSP+ (Fig 2E). No GFP+/ACT+ cells were detected on day 6 (Fig 2F). On recovery day 40, 95.47 ± 3.83% GFP+ cells were K5+ (Fig 2G) but only 10.13 ± 1.39% of K5+ cells were GFP+. Thus, the basal cell population contained cells that proliferated frequently (GFP−) or infrequently (GFP+). We also showed that 2.69 ± 0.85% of GFP+ cells were CCSP+ (Fig 2H) on day 40. No GFP+/ACT+ cells were detected on day 40 (Fig 2I). Thus, the GFP+ population contained 2 cell types, basal and Clara-like cells, on day 40.
GFPbright and GFPdim populations contain functional TSC
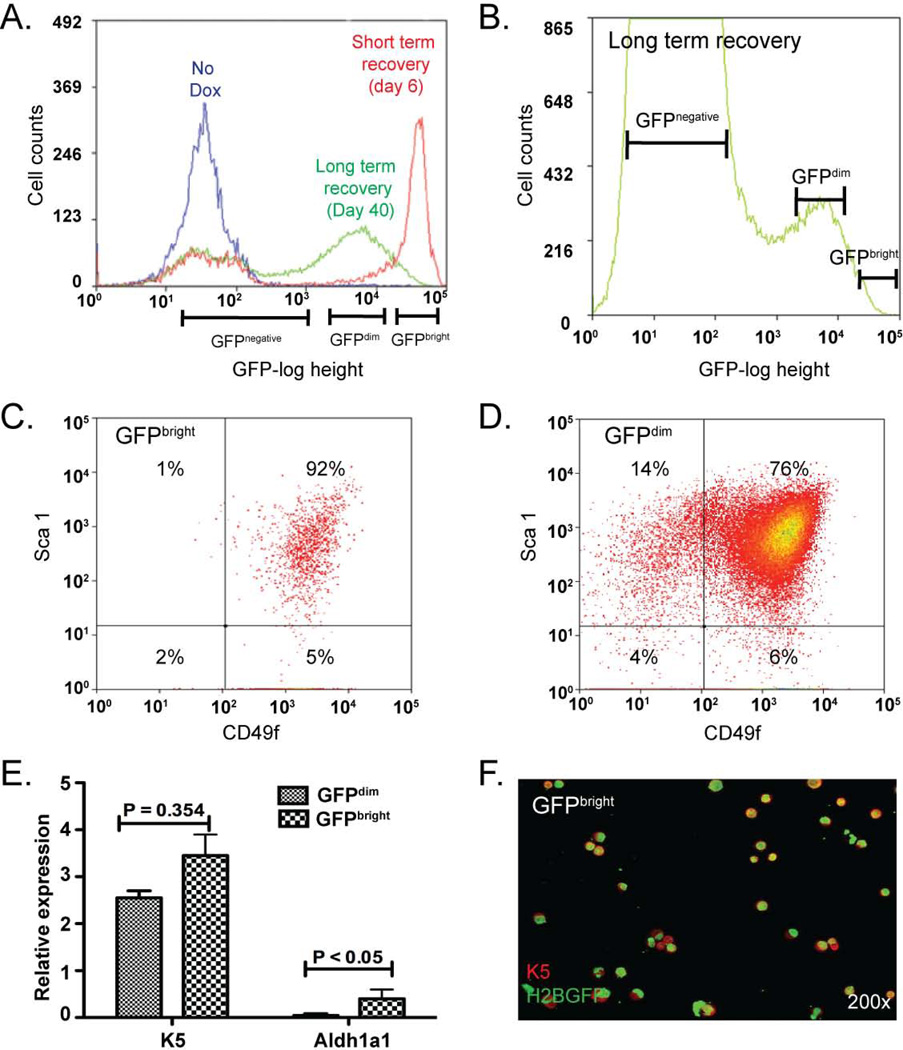
GFP intensity was evaluated by FLOW in MTg and BiTg mice that were treated as previously indicated (Fig 1F). The 6-day chase was termed ‘short term recovery’ whereas the 40-day chase period was termed ‘long-term recovery’. Tracheal tissue was digested to a single cell suspension and live epithelial cells 5 (CD45-, CD31-, TER119- and DAPI-) were identified. Epithelial cells were then separated according to GFP fluorescence intensity (GFI, Fig 3A). Cells from MTg mice were GFP− and had a GFI of <103 units. All GFP+ cells recovered from short term recovery mice were GFPbright (Fig 3A) and had a GFI of >104 units. Long term recovery resulted in two GFP+ subpopulations (Fig 3A, B). GFPbright cells were 0.5–0.8% of epithelial cells (0.08–0.1% of total tracheal cells). GFPdim cells had a GFI of 103–104 units and were 3–6% of epithelial cells (0.5–2% of total).
Figure 3. Isolation of viable Histone 2B:GFPbright (GFPbright) cells.
(A–B) FLOW cytometry histograms depicting cell count vs. GFP fluorescence intensity for CD45-/CD31-/TER119-/DAPI- cells derived from bitransgenic (BiTg) mice fed standard chow (no Dox, blue line) or BiTg mice fed doxycycline (dox) chow and treated with naphthalene (NA). Cells from dox/NA treated mice were evaluated on recovery day 6 (short term recovery, red line) or day 40 (long term recovery, green line). (C–D) Flow cytometry bivariate plots depicting the Sca1 and CD49f surface phenotype of GFPbright (C) and GFPdim (D) cells. (n = 7 analyses). (E) Real time RT-PCR analysis of Keratin (K) 5 and aldehyde dehydrogenase 1a1 (Aldh1a1) mRNAs in GFPbright (small check) and GFPdim (large check) populations. Mean ± SEM (n=4). (F) Dual immunofluorescence analysis of Keratin (K) 5 and GFP expression in cytospin preparations of GFPbright cells. (n= 4 analyses).
We previously demonstrated that the TSC was CD49fbright/Sca1+/Aldh+. To determine if the GFPbright and GFPdim populations contained TSC-like cells, we first evaluated expression of the surface markers CD49f and Sca1 by FLOW. GFPbright cells were a homogenous population of CD49fbright/Sca1+ cells (Fig 3C) that expressed K5 mRNA (Fig 3E) and protein (Fig 3F). GFPbright cells did not express detectable levels of CCSP or the ciliated cell marker FoxJ1 (SFig 6A, B). In contrast, the GFPdim population was a mixture of CD49fbright/Sca1+ and CD49fnegative/Sca1+ cells (Fig 3D). GFPdim cells expressed K5 protein (Fig 3E), as well as CCSP, and FoxJ1 mRNAs (SFig 6A, B).
We were unable to evaluate ALDH activity because the green channel was occupied by GFP and the only ALDH substrate (Aldefluor™) is green. Consequently, we evaluated expression of Aldh1a1 mRNA, which encodes the enzyme responsible for Aldefluor™ metabolism 34. Aldh1a1 mRNA was significantly enriched in the GFPbright cells compared to GFPdim cells (Fig 3E). In aggregate, the FLOW and gene expression analysis suggested that both the GFPbright and GFPdim populations contained TSC and that some TSC proliferated more frequently than others after NA injury.
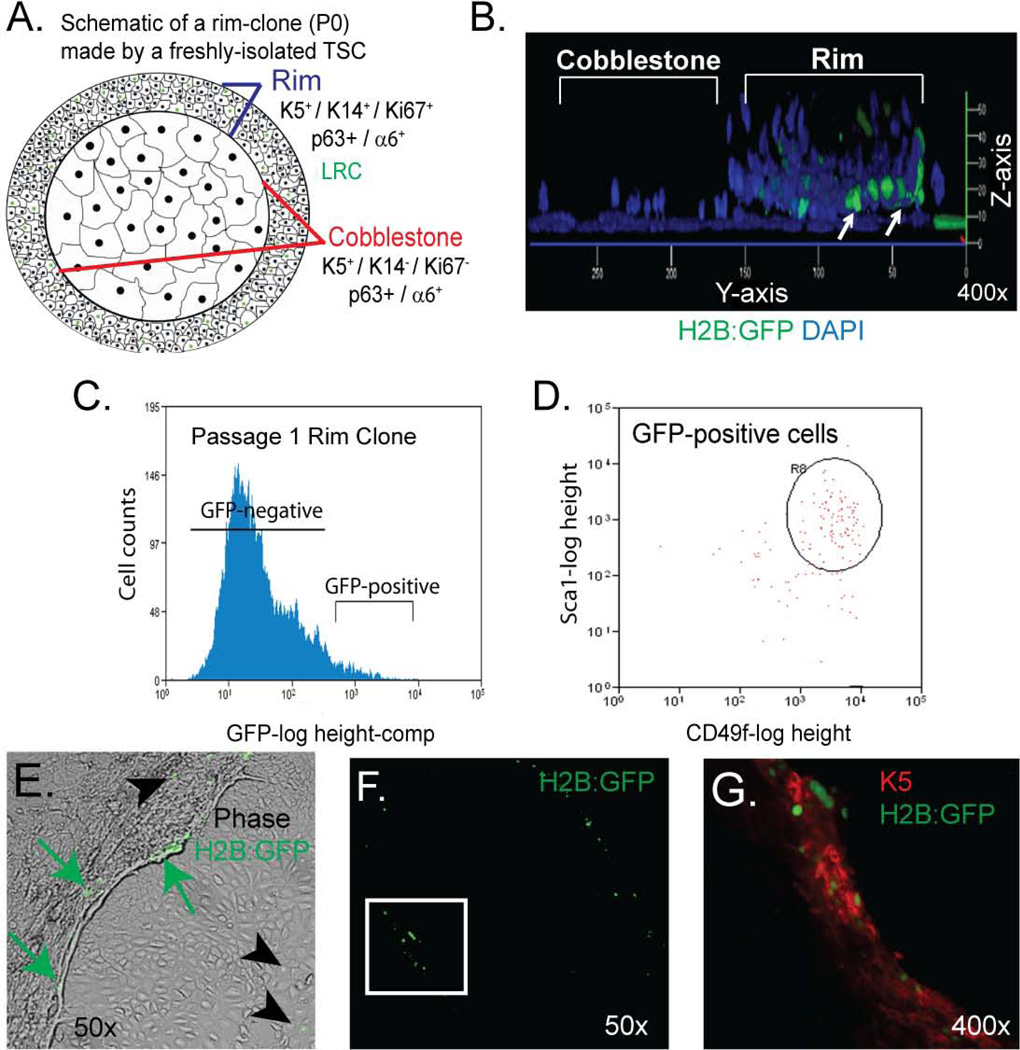
We previously demonstrated that the TSC generates a unique clone type, the rim clone, in vitro (see schematic Fig 4A). These clones are composed of two domains. The rim domain is a three dimensional structure 5 (Fig 4B) that is composed of abundant, highly-mitotic, K5+/K14+/p63+/integrin α6+ basal cells and rare, mitotically-quiescent cells (Fig 4A, B). The cobblestone domain is composed of non-mitotic K5+/K14-/p63+/integrin α6+ basal cells (Fig 4A).
Figure 4. The GFPbright and GFPdim populations contain functional tissue specific stem cells (TSC).
(A) Schematic of TSC-derived rim clone. (B) Confocal 3-dimencional projection of a rim clone generated by a GFPbright cell that was cultured in doxycycline (dox)-free medium. Representative of 10 rim clones. (C) FLOW cytometry histogram depicting cell count vs. GFP fluorescence intensity for Passage (P) 1 rim clones that were derived from GFPbright cells and cultured in dox-free medium. Representative of 3 analyses. (D) Flow cytometry bivariate plots depicting the Sca1 and CD49f surface phenotype of GFPpositive cells indicated in panel C. Representative of 3 analyses. (E) Merged phase contrast and fluorescence images of a P2 rim clone that was cultured in dox-free medium. Arrows: GFP+ cells. Arrowheads GFP− cells. Representative of 3 analyses. (F–G) Dual immunofluorescence analysis of GFP+ and Keratin 5 (K) 5. Green (GFP) channel only at 50X (F). Green (GFP) and red channels (K5) at 400x (G). (n = 3 analyses).
In the present study, we used the rim clone formation assay to determine if the GFPbright and GFPdim populations contained cells that were functionally similar to the TSC. Surprisingly, both the GFPbright and the GFPdim population generated rim clones that harbored GFP+ cells in the rim domain (Fig 4B). These data recapitulated our previous BrdU label retaining study and demonstrated that the GFPbright and the GFPdim populations contained cells that were functionally similar to the TSC.
GFP+ rim clone cells are TSC
We next determined if the GFP+ cells detected in rim clones were TSC as defined initially by the ability to generate a rim clone. BiTg mice were treated with dox and NA (Fig 1F). GFPbright and GFPdim cells were isolated by FLOW (Fig 3B) and cultured under rim clone forming conditions in dox-free medium for 2 weeks. Cells from these P0 clones were then separated into GFPbright and GFPdim populations. FLOW revealed significant dilution of GFP fluorescence (Fig 4C) indicating that TSC in both the GFPbright and GFPdim populations could proliferate in vitro. These data confirmed previous studies indicating that BrdU label retention was not a consequence of BrdU-induced cellular senescence 5.
Microscopic analysis of P0 rim clones showed that 0.3–0.5% of P0 rim clone cells were GFP+. FLOW analysis demonstrated that the P0-GFPbright cells were CD49fbright/Sca1+ TSC-like cells (Fig 4D). We then isolated the GFPbright/CD49fbright/Sca1+ cells and cultured them under rim clone forming conditions in dox-free medium. These cells generated P1 rim clones that contained GFP+ cells that were located in the rim domain (Fig 4E) and were K5+ (Fig 4F, G). Cells that diluted the GFP-label were detected in the rim and cobblestone domains of P1 clones (Fig 4E). GFP− cells did not generate clones. Since P0 and P1 GFPbright cells are the only rim clone forming cell type and the TSC was the only rim clone forming basal cell subtype 5, these data indicated that: 1) the GFPbright cell and the TSC were synonymous cell types; and 2) the TSC was responsible for rim clone formation after passage in vitro.
TSC that proliferate frequently in vivo have decreased mitotic potential in vitro
The demonstration that both GFPbright and GFPdim cells generated rim clones (Fig 4A, B) indicated that TSC proliferate at different frequencies in vivo. To determine if in vivo proliferation affected mitotic potential, we quantified TSC number in freshly-isolated GFPbright and GFPdim cells using the limiting dilution method 31. The rim-clone forming cell frequency (rim-CFCF) for GFPbright cells was 4.64 ± 1.03. This value was 8.5-fold greater (p<0.001) than that of the GFPdim population (Fig 5A). These data indicated that TSC proliferation in vivo led to decreased mitotic potential in vitro.
Figure 5. Tissue specific stem cells (TSC) that proliferate frequently in vivo have decreased mitotic potential in vitro.
(A) Rim (white bars) and non-rim (hatched bars) clone forming cell frequency (CFCF) of freshly-isolated GFPbright and GFPdim cells (n=4) (B) Schematic of a non-rim clone. (C) Merged phase contrast and fluorescence images of a passage (P) 0 rim clone that was cultured in dox-free medium. Representative of 3 analyses. (D) Frequency of rim and non-rim clone formation as a function of passage. Green circles: GFPbright rim clones. Green squares: GFPbright non-rim clones; Red triangles: GFPdim rim clones; Red inverted triangles: GFPdim non-rim clones. Mean ± SEM (n=6). (E) Phase-contrast image of a P6 clone. Arrow: edge of the clone indicating the absence of a rim. (F) Western blot analysis of p53 protein level in rim clones from P3-P6. Lanes 1, 3, 5 and 7: clones derived from GFPdim cells. Lanes 2, 4, 6 and 8: clones generated by GFPbright cells. Lane 9, normal NIH3T3 cells. Lane 10, irradiated NIH3T3 cells, a positive control. Actin was used as the loading control.
Our previous work also showed that non-TSC basal cells generated flat clones that were composed of K5+/K14-/p63+/integrin α6+ cells (see schematic, Fig 5B). These “non-rim” clones lacked GFP+ cells (Fig 5C) and could not be passaged. Thus, the non-rim clone forming cell was a unipotential basal cell progenitor (UPB) that generated terminally differentiated basal cells. To determine if proliferation of TSC in vivo was associated with generation of UPB, we evaluated non-rim clone formation in vitro. The non-rim CFCF for freshly-isolated GFPbright cells was 0.83 ± 0.067 (Fig 5A). This value was 5.6-fold less (p<0.0001) than that for GFPdim cells (Fig 5A). Non-rim clones from both GFPbright and GFPdim cells lacked GFP+ cells. These data suggested that TSC proliferation in vivo was associated with generation of the non-rim clone-forming UPB.
TSC are lost through terminal differentiation
To further evaluate the relationship between TSC proliferation and its mitotic potential, two lineages of rim clones were evaluated, one derived from freshly-isolated GFPbright cells and one generated from GFPdim cells. These P0 clones were then sub-cultured under rim clone-forming conditions and the number and type of clones was quantified at P1. We repeated this process for 6 consecutive passages (P2-P7).
At P1, rim clone cells from the GFPbright population generated 40 ± 7.82 rim clones/1000 cells. This value decreased as a function of passage and was 0 at P7 (Fig 5D). At P1, rim clone cells from the GFPdim population generated 10 ± 3.2 rim clones/1000 cells. This value also decreased as a function of passage and was 0 at P7. Rim clone generation for GFPbright cells was significantly greater than for GFPdim cells at passages 1–3 (p=0.01). In addition, the decline in rim clone formation by GFPbright cells was significantly less than that of the GFPdim cells (p=0.01). These data indicated that repeated TSC proliferation in vivo depleted TSC mitotic potential.
To investigate the relationship between the TSC and the UPB, we determined the frequency of non-rim clone forming cells in the GFPbright and GFPdim populations as a function of passage. At P1, rim clone cells from the GFPbright population generated 2 ± 0.32 non-rim clones/1000 cells (Fig 5D). This value increased significantly (p<0.01) to 38 ± 5.49 non-rim clones/1000 cells at P7 (Fig 5D). At P1, rim clone cells from the GFPdim population generated 16 ± 2.33 non-rim clones/1000 cells. This value increased significantly (p=0.001) to 45 ± 6.42 non-rim clone cells/1000 cells at P7. GFPbright cells generated significantly fewer non-rim clones than GFPdim cells at passages 1–7 (p=0.02). However, the frequency of non-rim clone forming cells in the GFPbright and GFPdim cells increased in parallel.
Generation of non-rim clones by rim-clone cells and the reciprocal relationship between rim and non-rim frequency (Fig 5D) suggested that TSC terminated as UPB. However, it has been shown that ex vivo expansion of mesenchymal stem cells decreased their clonogenicity and that this was the result of cellular senescence 35. To test the possibility that rim clone cells become senescent during passage, we stained passage 3–6 non-rim clones (Fig 5E) for a late senescence-marker, senescence associated β-galactosidase activity (SA-βgal). No SA-βgal was detected. In addition, increased p53 expression has been associated with early cellular senescence 36, 37. Accordingly, we measured p53 protein in passage 3–6 rim clones, by western blot (Fig 5F). This study demonstrated that p53 protein level did not change with increasing passage number. Collectively, these data suggest that non-rim clone cells were composed of terminally differentiated basal cells.
TSC proliferation in vivo does not change differentiation potential
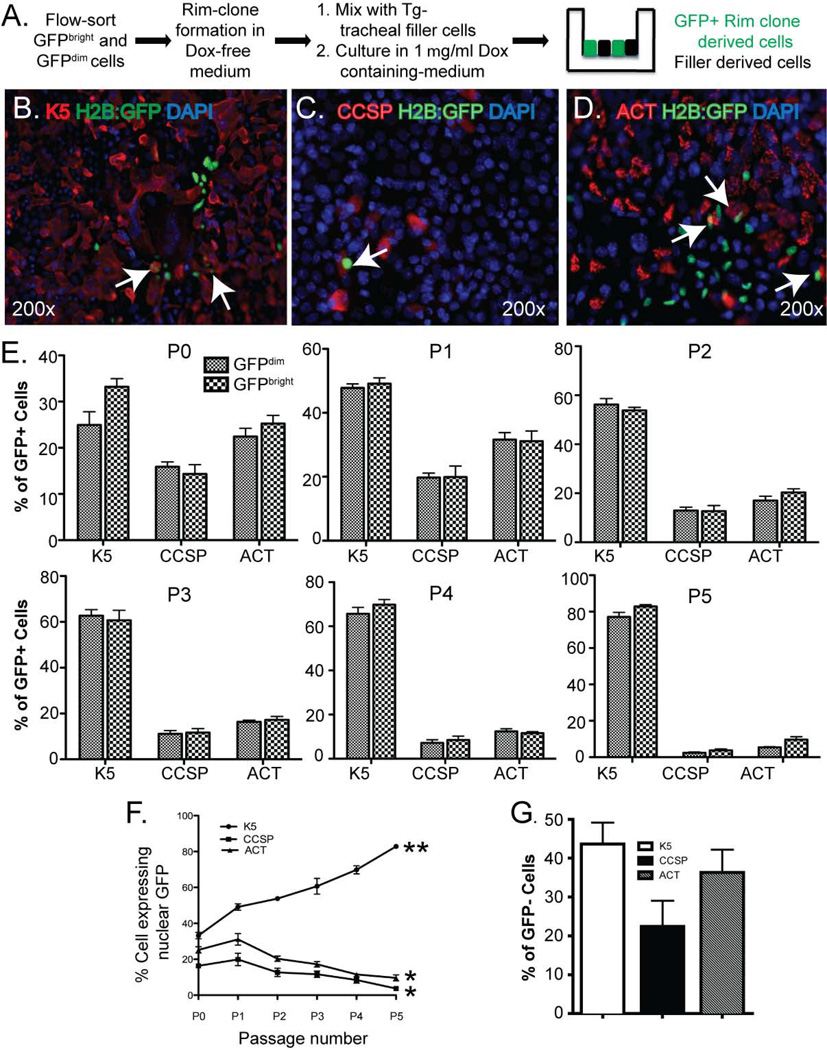
We used tracheal air-liquid interface (ALI) cultures to determine the differentiation potential of TSC derived from the GFPbright and GFPdim populations. BiTg mice were treated with dox and NA (see Fig 1F), GFPbright and GFPdim cells were isolated (see Fig 3B), and TSC were recovered by culture under rim clone forming conditions (Fig 6A). Since we previously showed that multipotential differentiation of TSC was dependent on factors provided by unfractionated tracheal “filler” cells, we mixed the TSC with transgene-negative filler cells at 1:10 ratio (Fig 6A). GFP+ cells were not detected in co-cultures that were maintained in dox-free medium (SFig 7A). In contrast, cultures that were maintained in medium supplemented with 2µg/ml dox contained numerous GFP+, TSC-derived cells (SFig 7B). Consequently, we were able to compare the differentiation potential of TSC with that of filler cells.
Figure 6. Tissue specific stem cell (TSC) proliferation in vivo does not alter differentiation potential.
(A) Schematic of the experimental design. (B–D) Dual immunofluorescence analysis of GFP (green) and Keratin (K) 5 (red) (B), GFP (green) and CCSP (red) (C), or GFP (green) and ACT (red) (D). Nuclei are counterstained with DAPI (blue). All images are enface views of the apical surface. Arrows: differentiated GFP+ cells. Representative of 3 analyses. (E) Differentiation potential of rim clone cells as a function of passage (P). Mean ± SEM (n=3). (F) Rate of differentiation to K5+, CCSP+, and ACT+ cells with increasing passage number. Data summarize those in Panel E. Mean ± SEM (n=3). (G) Frequency of filler cell differentiation to K5+, CCSP+, and ACT+ cells.
The ALI cultures were allowed to differentiate for 9 days, the time point when Clara-like and ciliated cells are routinely detected 2, 5, 23. On day 9, the cultures were fixed and stained with αK5, αCCSP and αACT and imaged using wide-field microscopy (Fig 6B–D) and confocal microscopy (SFig 8). The number of basal, Clara-like, and ciliated cells within the TSC-derived GFP+ population was quantified. Both the GFPbright and the GFPdim TSC produced basal (K5+), Clara-like (CCSP+) and ciliated (ACT+) cells (Fig 6E) at P0. The frequency with which GFPbright and GFPdim TSC generated differentiated basal, Clara-like, and ciliated cells did not differ (Fig 6E) and was equivalent to that of the filler cells (Fig 6G). These results demonstrated that TSC in the GFPbright and GFPdim populations were multipotential.
To determine if the differentiation potential of GFPbright and GFPdim TSC varied with passage, we carried out the co-culture studies using P1 to P5 TSC (Fig 6E). Both the GFPbright and GFPdim TSC differentiated to K5+, CCSP+ and ACT+ cells at each passage and the frequency with which differentiated cell types were generated did not differ between for the GFPbright and GFPdim rim clone cells at any passage.
However, the frequency of K5+ cells increased significantly between P0 and P5 (Fig 6F) and differentiation to CCSP+ or ACT+ cells decreased significantly between P0 and P5 (p<0.01). At P0, 32.21±4.07% of GFP+ cells were K5+ compared to 82.68 ± 7.49% at P5 (p<0.0001). Given that the UBP number increased between P0 and P6 (Fig 5B) and the frequency of differentiated K5+ basal cells increased as a function of passage, these data indicated that most P5 rim clone cells were UPB and that these cells generated terminally-differentiated basal cells. Overall the clone formation and differentiation analyses indicated that UPB clone cells were the terminally differentiated product of the TSC.
The Wnt/β-catenin pathway regulates TSC function in vivo
The Wnt/β-catenin signaling plays important roles in self-renewal and differentiation of the hematopoietic, intestinal, hair follicle and neural stem cells 38–41. To determine if this pathway is active in TSC, we isolated TSC from β-catenin reporter mice TOPGal (C57Bl/6 congenic) mice 42 and generated rim clones. X-gal staining demonstrated extensive transgene activation in the rim domain, which sequesters the TSC (SFig 9A).
Our reporter transgene analysis suggested that the Wnt/βcatenin dependent gene expression might be involved in TSC maintenance. To explore this possibility we treated BiTg mice with dox and NA (Fig 1F), purified GFPbright and GFPdim cells by FLOW (Fig 3B), and measured Wnt/βcatenin pathway gene expression by RT-PCR (n=3 fresh cell isolates, Fig 7A). Wnt/βcatenin pathway genes were expressed both in GFPbright and GFPdim cells. Expression of several β-catenin pathway genes, DKK2, Cyclin D1, GSK3β, APC, Axin-2 and β-catenin, did not differ significantly in GFPbright and GFPdim cells (Fig 7I and SFig 9B).
Figure 7. Wnt/β-catenin pathway genes are differentially expressed in GFPbright and GFPdim cells.
(A–H) Quantitative RT-PCR analysis of Wnt/β-catenin pathway gene expression in freshly isolated GFPbright (closed bar) and GFPdim (open bar) cells. Mean ± SD (n = 3). (I) A summary of the gene expression data is shown in the Venn diagram. Genes that are significantly (p < 0.05) over expressed in the GFPbright or in the GFPdim cells are shown in the left and right respectively. Genes that are not significantly different between the two populations are shown in the overlapping area.
However, three Wnt/βcatenin pathway inhibitors, SFRP1, SFRP2 and Psen1, and the Wnt/β-catenin target gene cMyc were over-expressed in the GFPbright population (Fig 7A–D). On the contrary, two co-activators of Wnt/β-catenin dependent gene transcription, LEF1 and TCF7, were over expressed in the GFPdim cells (Fig 7E, F). The Wnt pathway inhibitor DKK3 and the target gene MSX1 were also over-expressed in GFPdim cells (Fig 7G, H). DKK1 was not detected in either population. These data are summarized in Fig 7I. Taken together, this study demonstrates that the differential over-expression of Wnt/β-catenin pathway antagonists in GFPbright cells and Wnt/β-catenin co-activators in GFPdim cells.
Discussion
Summary
We previously demonstrated that TSC number increased after NA-injury in vivo and that TSC pool-size returned to normal on recovery day 40. The goal of this study was to determine the cellular and molecular mechanisms regulating normalization of TSC number after injury. Our in vivo and in vitro studies support the conclusion that NA-injury stimulates expansion of the TSC pool and that these supernumerary TSC are eliminated through terminal differentiation to a UPB. Gene expression analysis indicates that TSC-to-UPB differentiation involves interactions between the Wnt/β-catenin and Notch pathways.
Refinement of the TBE basal cell lineage
Lineage tracing of TBE basal cells in vivo supported the concept that a basal cell functioned as a TSC. However, some groups suggested that all TBE basal cells were TSC 4, 29, 43 while our study indicated that a TSC were a subtype of basal cells 3. We subsequently purified this basal cell subtype and demonstrated that these cells function as self-renewing, multipotential TSC 5. Our lineage-tracing analyses 2,3, 44 also detected unipotential and bipotential basal cells and suggested that there might be additional basal cell subtypes. The present study strongly suggests that the TSC is a progenitor for the UPB and that the UPB generates terminally-differentiated basal cells. Thus, the TSC, UPB, and the terminally-differentiated basal cell are likely to be members of a single lineage rather than independent progenitor cell pools. Additional studies are needed to determine the relationship between the TSC and the bipotential basal cell progenitors.
Wnt/βcatenin and Notch signaling in TSC proliferation and differentiation
We previously showed that the rim domain of the TSC clone sequesters rare mitotically-quiescent cells and abundant mitotically-active cells 5. In the present study we demonstrated that the quiescent cells are indeed TSC (Fig 4). Thus, the TSC proliferates infrequently but is surrounded by cells that proliferate repeatedly. Collectively, our data indicate that the TSC is insulated from the promitotic signals.
Demonstration of TOPGal transgene activity in TSC clones (SFig 9A) suggested that Wnt/β-catenin signaling regulated proliferation of non-TSC cells. This notion was supported by the finding that freshly-isolated GFPdim cells over-expressed Lef1 and TCF7, transcriptional co-factors that are necessary for β-catenin dependent gene expression. Since the GFPdim population contains few TSC, these data suggest that Wnt/β-catenin acts through Lef1 and/or TCF7 to promote proliferation of non-TSC. We also show that freshly-isolated GFPbright cells, which are enriched with TSC, over-express the Wnt antagonists SFRP1, SFRP2, and Psen1. These data suggest that Wnt/β-catenin signaling is inhibited in the TSC. Although Wnt/β-catenin signaling has been implicated in TSC proliferation in the gut and other tissues 45–47, our data suggest a different role for Wnt/β-catenin signaling in the tracheobronchial TSC.
Analysis of TSC that were propagated as spheroids indicated that Notch signaling promoted differentiation 29. Interestingly, SFRPs and Psen1 activate Notch signaling, in some systems 48–50. Since we now show that the TSC and the UPB are members of a single lineage, it is interesting to speculate that TSC-derived SFRP1, SFRP2, and Psen1 promote TSC-to-UPB differentiation. This cell-cell interaction may be reinforced by MSX1, a homeobox transcription factor that regulates expression of SFRPs and other Wnt antagonists including the DKKs 51. In summary, we propose that TSC-to-UPB differentiation is regulated by cell-cell interactions that are mediated by cross-talk between Wnt/β-catenin and Notch signaling pathways.
Limited TSC proliferation and differentiation potential
We demonstrated that TSC proliferation in vivo decreases mitotic-potential but not differentiation-potential and that repeated proliferation in vitro decreases both mitotic and differentiation potential. These data are at odds with the commonly held concept that the TSC maintains its functional properties throughout an individual’s life span.
The paradigms that guide TSC concepts are based on analysis of TSC in tissues in which most terminally-differentiated cells are short-lived. TSC in these tissues proliferate on a daily to weekly basis and their functional properties led to establishment of the “conservation of mitotic and differentiation potential paradigm”. In contrast with the gut and other high turn-over tissues, differentiated cells within the TBE and bronchiolar epithelium are long lived 7, 52–54 and only ~10% of TSC proliferate in the normal tissue 5. Consequently, it is likely that TSC proliferation and differentiation play a minor role in TBE homeostasis in mice, as previously suggested for bronchiolar TSC 55. Thus, we propose that TBE-TSC activation is reserved for devastating injuries that deplete non-TSC progenitor pools. Our studies and those of others identified numerous TBE progenitor pools including the Clara-like cell 56, a facultative basal progenitor 10, and the UPB. Although much remains to be learned about TBE progenitor cells and their interactions, we suggest that the finite proliferation and differentiation potential of the TSC is buffered by these progenitor cell types.
Implications for finite TSC proliferation- and differentiation potential in chronic lung disease
In contrast with mice, the lungs of humans are constantly exposed to injurious environmental agents. This situation begs the question, “Does TSC-depletion contribute to age-related decrements in lung function?” Our published study indicated that a single injury-cycle in mice stimulated TSC proliferation and that TSC numbers returned to normal on recovery day 40 5. However, preliminary studies indicate that a second NA-injury cycle results in depletion of the TSC pool. Thus, the injuries that occur during a typical human life-span may deplete the TBE-TSC pool and contribute chronic lung disease initiation and/or progression.
Conclusion and Summary
We conclude that the supernumerary TSC are lost through terminal differentiation and that this process is regulated by an interaction between the Wnt/β-catenin and Notch signaling pathways.
Supplementary Material
Acknowledgements
This work was supported by NIH awards: RC1 (HL099461, SDR), NIH RO1 (HL075585, SDR), Supplement (HL075585-S1, SDR) and K18 (HL107686, MG).
Footnotes
Author Contributions: Moumita Ghosh: conception and design; financial support; collection and/or assembly of data; data analysis and interpretation; manuscript writing; final approval of manuscript. Russell W. Smith: collection of data, final approval of manuscript. Christine M. Runkle: Collection of data, final approval of manuscript. Douglas A. Hicks: Collection of data; final approval of manuscript. Karen M. Helm: conception and design; data analysis and interpretation; collection of data; final approval of manuscript. Susan D. Reynolds: conception and design; financial support; collection and/or assembly of data; data analysis and interpretation; manuscript writing; final approval of manuscript.
Disclaimers:
None.
References
- 1.Morrison SJ. Stem cell potential: can anything make anything? Curr Biol. 2001;11:R7–R9. doi: 10.1016/s0960-9822(00)00033-6. [DOI] [PubMed] [Google Scholar]
- 2.Ghosh M, Brechbuhl HM, Smith RW, et al. Context-dependent differentiation of multipotential keratin 14-expressing tracheal Basal cells. Am J Respir Cell Mol Biol. 45:403–410. doi: 10.1165/rcmb.2010-0283OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hong KU, Reynolds SD, Watkins S, et al. In vivo differentiation potential of tracheal basal cells: Evidence for multipotent and unipotent subpopulations. Am J Physiol Lung Cell Mol Physiol. 2003 doi: 10.1152/ajplung.00155.2003. [DOI] [PubMed] [Google Scholar]
- 4.Rock JR, Onaitis MW, Rawlins EL, et al. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc Natl Acad Sci U S A. 2009;106:12771–12775. doi: 10.1073/pnas.0906850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ghosh M, Helm KM, Smith RW, et al. A Single Cell Functions as a Tissue-Specific Stem Cell and the In Vitro Niche-Forming Cell. Am J Respir Cell Mol Biol. doi: 10.1165/rcmb.2010-0314OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rawlins EL, Hogan BL. Epithelial stem cells of the lung: privileged few or opportunities for many? Development. 2006 doi: 10.1242/dev.02407. [DOI] [PubMed] [Google Scholar]
- 7.Rawlins EL, Hogan BL. Ciliated epithelial cell lifespan in the mouse trachea and lung. Am J Physiol Lung Cell Mol Physiol. 2008;295:L231–L234. doi: 10.1152/ajplung.90209.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Plopper CG, Chang AM, Pang A, et al. Use of microdissected airways to define metabolism and cytotoxicity in murine bronchiolar epithelium. Exp Lung Res. 1991;17:197–212. doi: 10.3109/01902149109064411. [DOI] [PubMed] [Google Scholar]
- 9.Plopper CG, Cranz DL, Kemp L, et al. Immunohistochemical demonstration of cytochrome P-450 monooxygenase in Clara cells throughout the tracheobronchial airways of the rabbit. Exp Lung Res. 1987;13:59–68. doi: 10.3109/01902148709064309. [DOI] [PubMed] [Google Scholar]
- 10.Cole BB, Smith RW, Jenkins KM, et al. Tracheal Basal Cells. A Facultative Progenitor Cell Pool. Am J Pathol. doi: 10.2353/ajpath.2010.090870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cotsarelis G, Cheng SZ, Dong G, et al. Existence of slow-cycling limbal epithelial basal cells that can be preferentially stimulated to proliferate: implications on epithelial stem cells. Cell. 1989;57:201–209. doi: 10.1016/0092-8674(89)90958-6. [DOI] [PubMed] [Google Scholar]
- 12.Cotsarelis G, Sun TT, Lavker RM. Label-retaining cells reside in the bulge area of pilosebaceous unit: implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell. 1990;61:1329–1337. doi: 10.1016/0092-8674(90)90696-c. [DOI] [PubMed] [Google Scholar]
- 13.Tumbar T, Guasch G, Greco V, et al. Defining the epithelial stem cell niche in skin. Science. 2004;303:359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greco V, Chen T, Rendl M, et al. A two-step mechanism for stem cell activation during hair regeneration. Cell Stem Cell. 2009;4:155–169. doi: 10.1016/j.stem.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuchs E, Nowak JA. Building epithelial tissues from skin stem cells. Cold Spring Harb Symp Quant Biol. 2008;73:333–350. doi: 10.1101/sqb.2008.73.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuchs E, Horsley V. More than one way to skin. Genes Dev. 2008;22:976–985. doi: 10.1101/gad.1645908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watt FM. Epidermal stem cells: markers, patterning and the control of stem cell fate. Philos Trans R Soc Lond B Biol Sci. 1998;353:831–837. doi: 10.1098/rstb.1998.0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mucenski ML, Nation JM, Thitoff AR, et al. Beta-catenin regulates differentiation of respiratory epithelial cells in vivo. Am J Physiol Lung Cell Mol Physiol. 2005;289:L971–L979. doi: 10.1152/ajplung.00172.2005. [DOI] [PubMed] [Google Scholar]
- 19.Reynolds SD, Zemke AC, Giangreco A, et al. Conditional Stabilization of {beta}-Catenin Expands the Pool of Lung Stem Cells. Stem Cells. 2008 doi: 10.1634/stemcells.2008-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Teisanu RM, Lagasse E, Whitesides JF, et al. Prospective isolation of bronchiolar stem cells based upon immunophenotypic and autofluorescence characteristics. Stem Cells. 2009;27:612–622. doi: 10.1634/stemcells.2008-0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guha A, Vasconcelos M, Cai Y, et al. Neuroepithelial body microenvironment is a niche for a distinct subset of Clara-like precursors in the developing airways. Proc Natl Acad Sci U S A. 109:12592–12597. doi: 10.1073/pnas.1204710109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morimoto M, Nishinakamura R, Saga Y, et al. Different assemblies of Notch receptors coordinate the distribution of the major bronchial Clara, ciliated and neuroendocrine cells. Development. 139:4365–4373. doi: 10.1242/dev.083840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brechbuhl HM, Ghosh M, Smith MK, et al. beta-catenin dosage is a critical determinant of tracheal basal cell fate determination. Am J Pathol. 179:367–379. doi: 10.1016/j.ajpath.2011.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith RW, Hicks DA, Reynolds SD. Roles for beta-catenin and doxycycline in the regulation of respiratory epithelial cell frequency and function. Am J Respir Cell Mol Biol. 46:115–124. doi: 10.1165/rcmb.2011-0099OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giangreco A, Lu L, Vickers C, et al. beta-Catenin determines upper airway progenitor cell fate and preinvasive squamous lung cancer progression by modulating epithelial-mesenchymal transition. J Pathol. 226:575–587. doi: 10.1002/path.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ambler CA, Watt FM. Expression of Notch pathway genes in mammalian epidermis and modulation by beta-catenin. Dev Dyn. 2007;236:1595–1601. doi: 10.1002/dvdy.21151. [DOI] [PubMed] [Google Scholar]
- 27.Katoh M. Notch ligand, JAG1, is evolutionarily conserved target of canonical WNT signaling pathway in progenitor cells. Int J Mol Med. 2006;17:681–685. [PubMed] [Google Scholar]
- 28.Smith MK, Koch PJ, Reynolds SD. Direct and indirect roles for beta-catenin in facultative basal progenitor cell differentiation. Am J Physiol Lung Cell Mol Physiol. 302:L580–L594. doi: 10.1152/ajplung.00095.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rock JR, Gao X, Xue Y, et al. Notch-dependent differentiation of adult airway basal stem cells. Cell Stem Cell. 8:639–648. doi: 10.1016/j.stem.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diamond I, Owolabi T, Marco M, et al. Conditional gene expression in the epidermis of transgenic mice using the tetracycline-regulated transactivators tTA and rTA linked to the keratin 5 promoter. J Invest Dermatol. 2000;115:788–794. doi: 10.1046/j.1523-1747.2000.00144.x. [DOI] [PubMed] [Google Scholar]
- 31.Taswell C. Limiting dilution assays for the determination of immunocompetent cell frequencies. I. Data analysis. J Immunol. 1981;126:1614–1619. [PubMed] [Google Scholar]
- 32.Heid CA, Stevens J, Livak KJ, et al. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 33.Borthwick DW, Shahbazian M, Krantz QT, et al. Evidence for stem-cell niches in the tracheal epithelium. Am J Respir Cell Mol Biol. 2001;24:662–670. doi: 10.1165/ajrcmb.24.6.4217. [DOI] [PubMed] [Google Scholar]
- 34.Moreb JS. Aldehyde dehydrogenase as a marker for stem cells. Curr Stem Cell Res Ther. 2008;3:237–246. doi: 10.2174/157488808786734006. [DOI] [PubMed] [Google Scholar]
- 35.Russell KC, Lacey MR, Gilliam JK, et al. Clonal analysis of the proliferation potential of human bone marrow mesenchymal stem cells as a function of potency. Biotechnol Bioeng. 108:2716–2726. doi: 10.1002/bit.23193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmid G, Kramer MP, Maurer M, et al. Cellular and organismal ageing: Role of the p53 tumor suppressor protein in the induction of transient and terminal senescence. J Cell Biochem. 2007;101:1355–1369. doi: 10.1002/jcb.21383. [DOI] [PubMed] [Google Scholar]
- 37.Oren M. Decision making by p53: life, death and cancer. Cell Death Differ. 2003;10:431–442. doi: 10.1038/sj.cdd.4401183. [DOI] [PubMed] [Google Scholar]
- 38.Katoh M. WNT signaling in stem cell biology and regenerative medicine. Curr Drug Targets. 2008;9:565–570. doi: 10.2174/138945008784911750. [DOI] [PubMed] [Google Scholar]
- 39.Alonso L, Fuchs E. Stem cells in the skin: waste not, Wnt not. Genes Dev. 2003;17:1189–1200. doi: 10.1101/gad.1086903. [DOI] [PubMed] [Google Scholar]
- 40.Haegebarth A, Clevers H. Wnt signaling, lgr5, and stem cells in the intestine and skin. Am J Pathol. 2009;174:715–721. doi: 10.2353/ajpath.2009.080758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Staal FJ, Luis TC. Wnt signaling in hematopoiesis: crucial factors for self-renewal, proliferation, and cell fate decisions. J Cell Biochem. 109:844–849. doi: 10.1002/jcb.22467. [DOI] [PubMed] [Google Scholar]
- 42.DasGupta R, Fuchs E. Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation. Development. 1999;126:4557–4568. doi: 10.1242/dev.126.20.4557. [DOI] [PubMed] [Google Scholar]
- 43.Rock JR, Randell SH, Hogan BL. Airway basal stem cells: a perspective on their roles in epithelial homeostasis and remodeling. Dis Model Mech. 3:545–556. doi: 10.1242/dmm.006031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hong KU, Reynolds SD, Watkins S, et al. Basal cells are a multipotent progenitor capable of renewing the bronchial epithelium. Am J Pathol. 2004;164:577–588. doi: 10.1016/S0002-9440(10)63147-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pinto D, Clevers H. Wnt control of stem cells and differentiation in the intestinal epithelium. Exp Cell Res. 2005;306:357–363. doi: 10.1016/j.yexcr.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 46.Wilusz M, Majka M. Role of the Wnt/beta-catenin network in regulating hematopoiesis. Arch Immunol Ther Exp (Warsz) 2008;56:257–266. doi: 10.1007/s00005-008-0029-y. [DOI] [PubMed] [Google Scholar]
- 47.Lewis HC, Molbak K, Reese C, et al. Pigs as source of methicillin-resistant Staphylococcus aureus CC398 infections in humans, Denmark. Emerg Infect Dis. 2008;14:1383–1389. doi: 10.3201/eid1409.071576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Esteve P, Sandonis A, Cardozo M, et al. SFRPs act as negative modulators of ADAM10 to regulate retinal neurogenesis. Nat Neurosci. 14:562–569. doi: 10.1038/nn.2794. [DOI] [PubMed] [Google Scholar]
- 49.Veeraraghavalu K, Choi SH, Zhang X, et al. Presenilin 1 mutants impair the self-renewal and differentiation of adult murine subventricular zone-neuronal progenitors via cell-autonomous mechanisms involving notch signaling. J Neurosci. 30:6903–6915. doi: 10.1523/JNEUROSCI.0527-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.De Strooper B, Annaert W, Cupers P, et al. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature. 1999;398:518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- 51.Revet I, Huizenga G, Koster J, et al. MSX1 induces the Wnt pathway antagonist genes DKK1, DKK2, DKK3, and SFRP1 in neuroblastoma cells, but does not block Wnt3 and Wnt5A signalling to DVL3. Cancer Lett. 289:195–207. doi: 10.1016/j.canlet.2009.08.019. [DOI] [PubMed] [Google Scholar]
- 52.Blenkinsopp WK. Proliferation of respiratory tract epithelium in the rat. Exp Cell Res. 1967;46:144–154. doi: 10.1016/0014-4827(67)90416-8. [DOI] [PubMed] [Google Scholar]
- 53.Bowden DH. Cell turnover in the lung. Am Rev Respir Dis. 1983;128:S46–S48. doi: 10.1164/arrd.1983.128.2P2.S46. [DOI] [PubMed] [Google Scholar]
- 54.Kauffman SL. Cell proliferation in the mammalian lung. Int Rev Exp Pathol. 1980;22:131–191. [PubMed] [Google Scholar]
- 55.Giangreco A, Arwert EN, Rosewell IR, et al. Stem cells are dispensable for lung homeostasis but restore airways after injury. Proc Natl Acad Sci U S A. 2009;106:9286–9291. doi: 10.1073/pnas.0900668106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Evans MJ, Shami SG, Cabral-Anderson LJ, et al. Role of nonciliated cells in renewal of the bronchial epithelium of rats exposed to NO2. Am J Pathol. 1986;123:126–133. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.