Abstract
Individuals with HIV infection and two apolipoprotein L1 gene (APOL1) risk variants frequently develop nephropathy. Here we tested whether non-HIV viral infections influence nephropathy risk via interactions with APOL1 by assessing APOL1 genotypes and presence of urine JC and BK polyoma virus and plasma HHV6 and CMV by quantitative polymerase chain reaction. We analyzed 300 samples from unrelated and related first-degree relatives of African Americans with non-diabetic nephropathy using linear and non-linear mixed models to account for familial relationships. The four groups evaluated were APOL1 0/1 versus 2 risk alleles, with or without nephropathy. Urine JCV and BKV were detected in 90 and 29 patients while HHV6 and CMV were rare. Adjusting for family age at nephropathy, gender and ancestry, presence of JCV genomic DNA in urine and APOL1 risk alleles were significantly negatively associated with elevated serum cystatin C, albuminuria (albumin to creatinine ratio over 30 mg/g), and kidney disease defined as an eGFR under 60 ml/min per 1.73 m2 and/or albuminuria in an additive (APOL1 plus JCV) model. BK viruria was not associated with kidney disease. Thus, African Americans at increased risk for APOL1-associated nephropathy (two APOL1 risk variants) with JC viruria had a lower prevalence of kidney disease, suggesting that JCV interaction with APOL1 genotype may influence kidney disease risk.
Keywords: APOL1, BK polyomavirus, HIV, JC polyomavirus, kidney disease, proteinuria
Introduction
Two coding risk variants in the apolipoprotein L1 nephropathy susceptibility gene (APOL1) on chromosome 22q demonstrate among the most impressive genetic association in common complex disease.[1;2] The odds ratios (OR) for APOL1 association with HIV-associated nephropathy (HIVAN), idiopathic focal segmental glomerulosclerosis (FSGS), and non-diabetic end-stage renal disease (ESRD) are 29, 17, and 7.3, respectively.[1;3] Not all individuals inheriting two risk variants will develop nephropathy.[4] This led to the concept of second hits, where gene-gene or gene-environment interactions are required to initiate nephropathy.[5;6]
HIVAN develops in approximately 50% of African Americans with untreated HIV infection who possess two APOL1 risk variants.[3] The kidney serves as a reservoir for HIV replication.[7] Accelerated and aberrant replacement by partially differentiated podocytes develops in HIVAN, and renal histology reveals FSGS collapsing variant, the most aggressive form of FSGS.[8] Individuals without HIV infection also develop APOL1-associated nephropathy; however, the renal histologic patterns frequently differ. Some patients develop focal global glomerulosclerosis with interstitial fibrosis and vascular changes, [9] while others develop FSGS. It appears likely that specific second hits may ultimately determine renal histopathology.[4];[5]
Non-HIV viral infections, particularly those with the potential for renal or uroepithelial infection (JC and BK polyoma virus) or lymphotropic effects (Human Herpes Virus-6 [HHV6] and Cytomegalovirus [CMV]) like HIV, could serve as modifiable environmental factors that interact with APOL1 to initiate nephropathy.[4] To test this hypothesis, 300 samples of unrelated and related first-degree relatives of African Americans with non-diabetic forms of ESRD were evaluated for the presence of BK (BKV) and JC (JCV) polyoma virus genomic DNA in urine, and HHV6 and CMV genomic DNA in plasma; 300 was chosen based on available funds. Analyses were performed to test for association between active viral replication, and measures of albuminuria and glomerular filtration rate conditioning on APOL1 genotype.
Results
Table 1 contains demographic and clinical results in 300 informative first-degree relatives of African American patients with ESRD selected from among 835 Wake Forest School of Medicine Institutional Review Board-approved “Natural History of APOL1-associated Nephropathy Study” participants, based on the number of APOL1 risk variants. The four groups evaluated were APOL1 0/1 versus 2 risk alleles, with versus without nephropathy. The analyzed dataset included 199 unrelated singletons, 38 pairs (sibpairs and parent-offspring), and 11 trios (10 were siblings; 1 composed of 2 siblings and a parent). The sample was not selected to test for APOL1 association with nephropathy as APOL1 risk variants were previously shown to predict kidney disease in these families and in population-based samples.[10];[11] All subjects provided written informed consent. Informative individuals with two APOL1 risk variants were included if they had (a) urine albumin:creatinine ratio (ACR) <30 mg/g and MDRD estimated glomerular filtration rate (eGFR) >60 ml/min/1.73m2 and were no younger than 5 years below the age at ESRD in their family proband (N=87), or (b) eGFR <60 ml/min/1.73m2 or urine ACR >80 mg/g (N=43; due to variability in UACR, values >80 [not >30] were selected to minimize the likelihood of normalization of UACR on repeat testing). The presence of sibpairs and parent-offspring pairs in our data implies that the independence assumption underlying the Wilcoxon two-sample test is violated; however, this test is robust to this type of deviation.[12] We note that the results shown under the association with APOL1 column in Table 2 are adjusted for the familial correlation and led to the same statistical inference as the Wilcoxon two-sample test. This is true for the binary and the continuous outcomes even after appropriate transformations were applied.
Table 1.
Demographic and laboratory characteristics of participants, based on APOL1 risk status
| Variable | 0 or 1 APOL1 risk alleles (N=170) | 2 APOL1 risk alleles (N=130) | P-value4 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| N | Mean | Std | Median | N | Mean | Std | Median | ||
| African ancestry (%) | 161 | 0.81 | 0.10 | 0.83 | 127 | 0.82 | 0.09 | 0.83 | 0.4411 |
| Age (years) | 170 | 54.69 | 11.71 | 55.00 | 130 | 47.14 | 12.96 | 49.00 | <.0001 |
| Age difference (years)2 | 167 | 6.78 | 9.11 | 9.00 | 128 | 3.84 | 7.05 | 4.00 | 0.0017 |
| Female (%) | 98 | 57.7% | 80 | 61.5% | 0.4965 | ||||
| BMI (kg/m2)\ | 166 | 34.04 | 10.59 | 32.09 | 127 | 32.64 | 9.42 | 30.56 | 0.2213 |
| Cystatin-C (mg/L) | 170 | 1.03 | 0.52 | 0.88 | 130 | 0.86 | 0.35 | 0.79 | 0.0003 |
| Cystatin-C >0.96 mg/L | 72 | 42.4% | 33 | 25.4% | 0.0023 | ||||
| MDRD GFR (ml/min/1.73m2) | 170 | 82.76 | 28.66 | 83.60 | 130 | 91.50 | 26.49 | 91.20 | 0.01 |
| MDRD GFR <60 ml/min/1.73m2 | 35 | 20.6% | 14 | 10.8% | 0.0226 | ||||
| CKD-E GFR (ml/min/1.73m2) | 170 | 81.40 | 29.10 | 82.70 | 130 | 91.40 | 26.80 | 93.00 | 0.0059 |
| CKD-E GFR <60 ml/min/1.73m2 | 40 | 23.5% | 17 | 13.1% | 0.0222 | ||||
| Hypertension (%) | 86 | 51.8% | 58 | 45.7% | 0.2977 | ||||
| Diabetes (%) | 22 | 12.9% | 8 | 6.2% | 0.0522 | ||||
| UACR (mg/g) | 170 | 347.22 | 1090.00 | 12.75 | 130 | 84.58 | 198.14 | 7.80 | 0.0339 |
| UACR >30 mg/g (%) | 70 | 41.2% | 38 | 29.2% | 0.0327 | ||||
| UACR >300 mg/g (%) | 28 | 16.5% | 13 | 10.0% | 0.1059 | ||||
| Kidney disease1 (%) | 83 | 48.8% | 43 | 33.1% | 0.0062 | ||||
| Viral load JCV3 | 55 | 5.26 | 1.49 | 5.30 | 35 | 4.94 | 1.56 | 5.09 | 0.2978 |
| Viral load BKV3 | 15 | 4.00 | 0.94 | 3.99 | 14 | 4.04 | 0.90 | 4.07 | 0.532 |
Kidney disease is defined as MDRD GFR <60 ml/min/1.73m2 and/or urine albumin:creatinine ratio >30 mg/g.
Age difference between participants and the ESRD proband in their family.
Viral load (Log10 copies/mL) in subjects with active viral replication.
P-values obtained from chi-square tests and Wilcoxon two-sample tests, as appropriate (unadjusted for relatedness). Familial-adjusted pvalues are in Table 2, second column (unadjusted).
BMI body mass index; MDRD Modification of Diet in Renal Disease; GFR glomerular filtration rate; CKD-E CKD EPI; UACR urine albumin:creatinine ratio; JCV JC polyoma virus; BKV BK polyoma virus
Table 2.
Association between renal parameters and JC viruria, APOL1, and additive effect of (JC viruria + APOL1 [recessive])
| Outcome | Adjustment | Association with JCV | Association with APOL1 (rec) | Association with (JCV+APOL1)1 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Estimate | SE | P-value | Estimate | SE | P-value | Estimate | SE | P-value | ||
| Log(UACR+1) | None | −0.42 | 0.24 | 0.0887 | −0.59 | 0.23 | 0.0129 | −0.51 | 0.17 | 0.0047 |
| Log(UACR+1) | Full | −0.27 | 0.23 | 0.2438 | −0.86 | 0.21 | 0.0002 | −0.59 | 0.16 | 0.0006 |
| Log(Cystatin-C) | None | −0.06 | 0.04 | 0.2136 | −0.15 | 0.04 | 0.0007 | −0.11 | 0.03 | 0.0016 |
| Log(Cystatin-C) | Full | −0.05 | 0.05 | 0.2501 | −0.16 | 0.04 | 0.0004 | −0.11 | 0.03 | 0.0011 |
| MDRD GFR | None | 0.45 | 3.38 | 0.8935 | 7.38 | 3.26 | 0.0277 | 4.02 | 2.45 | 0.1058 |
| MDRD GFR | Full | 0.26 | 3.46 | 0.9401 | 7.57 | 3.31 | 0.0265 | 4.07 | 2.46 | 0.1044 |
| CKD-EPI GFR | None | −0.32 | 3.38 | 0.9252 | 8.64 | 3.29 | 0.0111 | - | - | - |
| CKD-EPI GFR | Full | 0.05 | 3.48 | 0.9887 | 8.51 | 3.35 | 0.0142 | 4.41 | 2.48 | 0.0818 |
| Kidney disease | None | −0.46 | 0.28 | 0.1039 | −0.73 | 0.26 | 0.0053 | −0.6 | 0.2 | 0.0025 |
| Kidney disease | Full | −0.51 | 0.36 | 0.1556 | −1.68 | 0.36 | 1.5E-06 | −1.12 | 0.26 | 8.2E-06 |
| UACR >30 mg/g | None | −0.51 | 0.29 | 0.077 | −0.59 | 0.26 | 0.0272 | −0.55 | 0.2 | 0.0067 |
| UACR >30 mg/g | Full | −0.42 | 0.35 | 0.2356 | −1.26 | 0.33 | 0.0002 | −0.87 | 0.25 | 0.0005 |
Test was performed only when equality of the independent APOL1 and JCV effects were demonstrated (- denotes inequality);
p-values reflect the additive (JCV + APOL1) variable for association with each outcome, based on the Estimate [beta coefficient]
JCV – JC polyoma virus; SE – Standard Error; rec – recessive genetic model; MDRD Modification of Diet in Renal Disease; GFR glomerular filtration rate (units are ml/min/1.73m2); UACR urine albumin:creatinine ratio
Kidney disease defined as MDRD GFR <60 ml/min/1.73m2 and/or urine albumin:creatinine >30 mg/g
Full model adjusted for ancestry, gender and age difference between subject and the ESRD proband in their family
Linear and generalized linear mixed models were fitted to adjust for familial relationships using the expected kinship matrix. The ‘burden’ test (JCV+APOL1) was conducted when we could not reject the null hypothesis that JCV and APOL1 had the same effect on the outcome.
After selecting all 130 individuals with two APOL1 risk variants that could be confidently classified as either having or lacking nephropathy, we added the remaining 170 subjects with zero/one risk variants to complete the sample of 300. Among the 170 subjects, 50% were selected to have no risk alleles and 50% to have one risk allele; approximately 50% of each genotype selected to have nephropathy and 50% to lack nephropathy. Those with nephropathy included the highest urine ACR and/or lowest eGFR for each genotype; those without nephropathy included those with urine ACR <30 mg/g and eGFR >60 ml/min/1.73m2 closest to (or older than) the age at ESRD in family probands. Higher mean UACR and plasma cystatin C and lower eGFR in affected cases within each genotype reflect the selection criteria. Albuminuria was also higher and eGFR lower among cases with kidney disease and either zero or one APOL1 risk variants, relative to cases with two risk variants. This finding was expected and reflects that 77% of relatives had zero or one APOL1 risk variant and fewer individuals with the low risk genotypes were included. Therefore, there was a larger pool for selecting subjects with nephropathy in the low risk genotype groups. As this was not a random sample, subjects with 0/1 APOL1 risk variants selected for viral testing were expected to differ from those not selected (Supplementary Table 1).
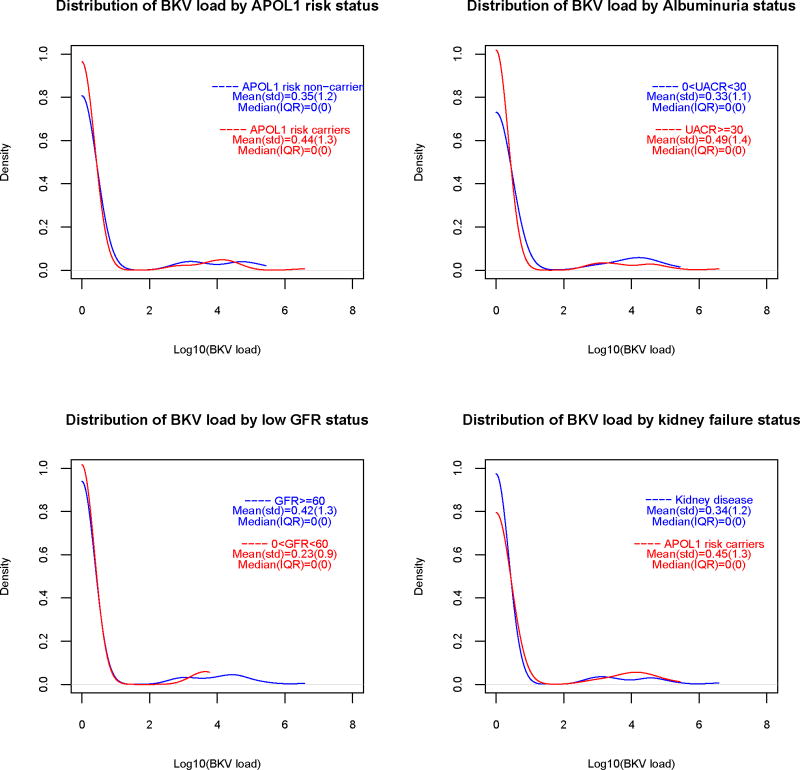
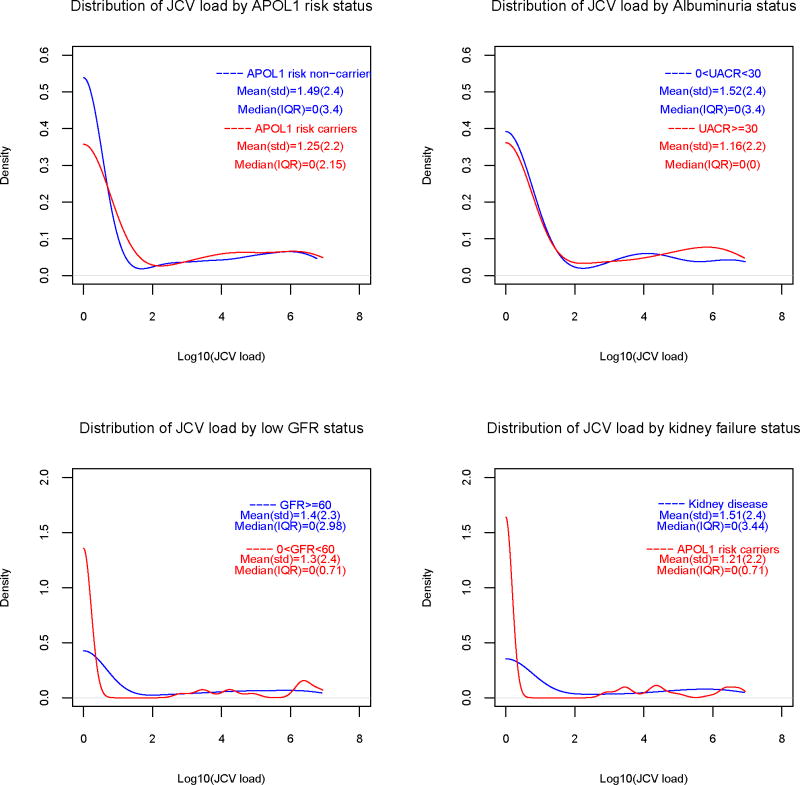
Urine JCV and BKV genomic DNA was detected in 90 (30%) and 29 (9.7%) of subjects, respectively. Table 1 presents the viral loads (Log10 copies/mL) in subjects positive for urine viral genomic DNA. Figure 1 contains smoothed distributions of viral loads, stratified by GFR and UACR, kidney disease and APOL1 risk status. Only two subjects had simultaneous JCV and BKV viruria. Plasma HHV6 and CMV genomic DNA were only detected in 2 and 1 individuals respectively. As such, analyses were not performed for HHV6 or CMV.
Figure 1.
1a. Smoothed distribution of urine JC virus loads, by APOL1 genotype and eGFR (≥60 vs. <60 ml/min/1.73m2), albuminuria (UACR <30 vs. ≥30 mg/g) and kidney disease status.
1b. Smoothed distribution of urine BK virus loads, by APOL1 genotype and eGFR (≥60 vs. <60 ml/min/1.73m2), albuminuria (UACR <30 vs. ≥30 mg/g) and kidney disease status.
Table 2 reveals association analyses between albuminuria and several measures of kidney function, based upon urine JCV replication alone, and the combination of APOL1 genotypes + JCV replication (as stated, APOL1 genotypes alone were not associated with kidney disease in this selected sample, but were in the full sample).[11] Association analyses between JCV alone and all renal parameters were non-significant in the unadjusted and fully adjusted models (adjusted for family age at ESRD, sex, and ancestry). Considering the additive interaction between presence of urine JCV + APOL1 genotypes in a recessive model based on the number of risk variants, elevated plasma cystatin C concentration, albuminuria, and the combined kidney disease variable were significantly less common in those with two APOL1 risk variants who had JCV replication in the urine. Table 3 displays similar association analyses with urine BKV. Here, detection of BKV alone, as for JCV alone, did not impact the risk of renal disease. When considering APOL1 + urine BKV in an additive fashion in fully adjusted models, no significant effect was observed with any of the renal parameters.
Table 3.
Association between renal parameters and BK viruria, APOL1, and additive effect of (BK viruria + APOL1 [recessive])
| Outcome | Adjustment | Association with BKV | Association with APOL1 (rec) | Association with (BKV+APOL1)1 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Estimate | SE | P-value | Estimate | SE | P-value | Estimate | SE | P-value | ||
| Log(UACR+1) | None | 0.04 | 0.07 | 0.5433 | −0.15 | 0.04 | 0.0009 | - | - | - |
| Log(UACR+1) | Full | 0.06 | 0.07 | 0.3856 | −0.16 | 0.04 | 0.0004 | - | - | - |
| Log(Cystatin-C) | None | 0.02 | 0.37 | 0.9504 | −0.55 | 0.23 | 0.019 | −0.39 | 0.19 | 0.048 |
| Log(Cystatin-C) | Full | 0.16 | 0.36 | 0.6528 | −0.86 | 0.22 | 0.0002 | - | - | - |
| MDRD GFR | None | −2.92 | 5.2 | 0.5768 | 7.44 | 3.25 | 0.0261 | 4.43 | 2.72 | 0.1094 |
| MDRD GFR | Full | −4.33 | 5.35 | 0.4217 | 7.76 | 3.31 | 0.0231 | 4.26 | 2.76 | 0.1289 |
| CKD-EPI GFR | None | −2.47 | 5.19 | 0.6365 | 8.75 | 3.28 | 0.0099 | 5.44 | 2.74 | 0.0516 |
| CKD-EPI GFR | Full | −3.73 | 5.37 | 0.4908 | 8.68 | 3.34 | 0.0124 | 5.04 | 2.78 | 0.0758 |
| Kidney disease | None | 0.35 | 0.43 | 0.4111 | −0.72 | 0.26 | 0.0068 | - | - | - |
| Kidney disease | Full | 1.05 | 0.54 | 0.0554 | −1.74 | 0.37 | 1.3E-06 | - | - | - |
| UACR > 30 mg/g | None | 0.49 | 0.43 | 0.2568 | −0.58 | 0.27 | 0.0337 | - | - | - |
| UACR > 30 mg/g | Full | 1.09 | 0.53 | 0.0414 | −1.33 | 0.34 | 0.0001 | - | - | - |
Test was performed only when equality of the independent APOL1 and JCV effects were demonstrated (- denotes inequality);
p-values reflect the additive (BKV + APOL1) variable for association with each outcome, based on the Estimate [beta coefficient]
BKV – BK polyoma virus; SE – Standard Error; rec – recessive genetic model; MDRD Modification of Diet in Renal Disease; GFR glomerular filtration rate (units are ml/min/1.73m2); UACR urine albumin:creatinine ratio
Kidney disease defined as MDRD GFR <60 ml/min/1.73m2 and/or urine albumin:creatinine >30 mg/g
Full model adjusted for ancestry, gender and age difference between subject and the ESRD proband in their family
Linear and generalized linear mixed models were fitted to adjust for familial relationships using the expected kinship matrix. The ‘burden’ test (BKV+APOL1) was conducted when we could not reject the null hypothesis that BKV and APOL1 had the same effect on the outcome.
To ensure that results were not biased based upon the sample selection process; formal multiplicative interaction and stratified analyses comparing individuals with 2 copies of the APOL1 risk variants to those with 0 or 1 copy were performed. Association was assessed between the dichotomous variable of kidney disease (yes, no) and continuous outcomes (eGFR, UACR and serum cystatin C) with presence of JCV (Supplementary Table 2) and presence of BKV (Supplementary Table 3), adjusted for age, gender and admixture. Analyses in APOL1 two risk allele carriers demonstrated trends toward negative association between JCV and Log(UACR+1) (parameter estimate [PE] −0.49; p=0.1488), kidney disease (PE −0.61; p=0.1902) and UACR >30 mg/g (PE −0.86; p=0.0705); results supported those in the full sample of 300 despite including only 130 participants.
Discussion
This is the first evaluation of potential roles for non-HIV infections as second hits for initiation of APOL1-associated nephropathy. In this sample of first-degree relatives of African Americans with non-diabetic ESRD, 30% and 9.7%, respectively, had detectable viral genomic DNA, and presumably active urinary tract replication of JCV and BKV, respectively. Accounting for gender, African ancestry proportion and the age difference between the selected individual and the proband, significant negative associations were observed between the combined variable (urine JCV + APOL1) with increasing albuminuria and reduced kidney function. Like HIV, polyoma viruses can maintain a renal/uroepithelial reservoir of infection.[13] These data support a role for urinary tract JC polyoma virus infection in susceptibility to APOL1-associated nephropathy and may provide important clues toward potential mechanisms of disease. APOL1 variants could also cause both kidney disease and suppress JCV replication. Although JCV in urine does not directly prove infection, median viral loads in these subjects were generally high suggesting true infection (Log10 copies/ml were 5.6 with no risk alleles; 4.95 with one risk allele; 5.08 with two risk alleles). Another possible explanation for detectable JCV in urine could be increased numbers of cells or active replication/shedding of latently infected cells (as with increased turnover of collecting duct and urothelium). Urine was not centrifuged post-collection and samples were collected using a single protocol. Samples were gently mixed to allow homogeneous aliquots to be frozen at −80°C and frozen samples were shipped to Viracor-IBT Laboratories where pre-nucleic acid extraction techniques were not applied.
These results were felt to be somewhat paradoxical and replication is necessary when appropriate samples become available. We postulated that polyoma virus in the urinary tract would likely interact with APOL1 to increase nephropathy risk in individuals’ possessing two risk variants. The absence of an effect with BKV (and low frequencies of CMV and HHV6 viral detection) provided reassurance that this was an effect specific for JCV. We may have lacked power to detect the BK effect given the small number of BK infected individuals. BK and JC viruria are typically present in ~0–20% and ~20–30% of immune-competent individuals, respectively.[14;15] Among immune-compromised subjects, BK viruria increases to 10–60%, without an increase for JCV.[14;15] However, JC viruria increases with age.[16] Pires et al. reported significantly lower frequencies of JC viruria in Brazilians with ESRD (3.9%), relative to non-nephropathy controls (20.1%; p<0.0001).[17] The authors considered the impact of high urinary urea concentrations and reductions in renal mass reflected in diminished renal JCV loads. However, we observed JC viruria less often in individuals with mild asymptomatic and late stage nephropathy. Pires et al.[17] did not assess interactive effects of APOL1 on relationships between ESRD and JCV in their patients.
Primary infection with human polyoma viruses occurs early in life; 58% and 82% of blood donors have antibodies against JCV and BKV, respectively.[18] Infection is thought to disseminate to the kidney via the bloodstream (viremia), where it remains latent. Supporting this hypothesis, 10–41% of normal individuals have BKV and JCV sequences in renal tissue.[19;20] The role of JCV as a nephropathic virus is less clear, including in immunosuppressed transplant patients.[21–23]
Evidence supports that JCV and BKV infection inhibit each other’s replication.[21] As such, although 129 of these 300 study subjects had BKV or JCV detected in the urine, only 2 had simultaneous detection of JCV and BKV. JCV appears in the urine as early as 5 days after kidney transplantation, where it can take 3–6 months for BKV replication.[24] Prior colonization with JCV may inhibit subsequent infection with other more nephropathic viruses, perhaps including other polyoma viruses (e.g., Merkel Cell polyoma virus). Unlike BV virus, JCV does not enter renal tubule cells by the caveolae pathway.[25] However, after entry via clathrin coated pits, JCV is found in caveolin-1-decorated vesicles suggesting that it sorts to the caveolin pathway after entry.[26] If this interaction perturbs caveolin-1 recycling to the cell surface, a molecular model can be envisioned in which JCV retains caveolin-1 in the cell and reduces the efficiency of a subsequent BKV infection. Thus, we speculate that the presence of JCV may be renal protective via inhibition of other viral infections. A second possibility that could explain this observation is JCV could impact gene transcription profiles in kidney cells, [27] which may affect pathways of apoptosis or autophagy with which APOL1 is reportedly involved.
This report has limitations and we await availability of samples for replication. Kidney disease in relatives with two APOL1 risk variants was generally mild and reflected albuminuria to a greater extent than reduced eGFR; few subjects had a markedly low eGFR. However, APOL1 is strongly associated with kidney disease in African American Study of Kidney Disease and Hypertension participants with baseline proteinuria (odds ratio >6), reducing this concern.[9] We were unable to assess potential effects of JCV on transcriptional profiles in renal cells from these individuals since we lack their kidney tissue. Evaluation of kidney tissue would also allow us to detect latent renal infection. JCV DNA quantification fluctuates over time, an effect present in all cross-sectional studies.[27] In addition to the hypotheses we propose to explain our results, it is possible that APOL1 (directly or via association with other genes) impacts the immune system’s ability to suppress JCV. We did not test for HIV infection, but feel it is unlikely that HIV played a major role. HIV seropositivity occurs in <1.8% of African Americans (http://www.cdc.gov/hiv/topics/surveillance/resources/slides/race-ethnicity/index.htm). All participants were queried regarding health status and medications. HIV infection was reported by 13 of 835 study participants (1.56%); 5 were in this report (1 with urine JCV and BKV, 2 with urine JCV, 1 with urine BKV, 1 negative for both viruses). Thirteen of the 300 subjects reported prior hepatitis B or hepatitis C infection. JCV replication in plasma was not assessed among those with urinary tract replication as JCV viremia is rare. Finally, the combined variable (APOL1+JCV) was created whenever we failed to reject the null hypothesis that the effect sizes associated with APOL1 and JCV were equal. This could result in a type I error if there was insufficient power to reject this hypothesis. In this case, the model that combines urine JCV with APOL1 to create a single variable might not provide the best fit to the data. However, stratified analyses continued to suggest a consistent negative interaction between APOL1 and JCV.
This study tested for four viral infections that could serve as environmental exposures or second hits for APOL1-associated nephropathy. Accounting for APOL1, results demonstrated reduced rates of albuminuria and kidney disease in African Americans with active JCV urinary tract replication. These data suggest that JCV may inhibit urinary tract infection with other more nephrotoxic viruses or impact renal gene expression thereby protecting from development of APOL1-associated nephropathy. It is critical that we detect environmental second hits that interact with genetic risk variants to produce non-diabetic kidney disease.[28] These factors may be modifiable and could lead to novel treatments for non-diabetic ESRD, a refractory family of genetic disorders that are strongly APOL1-associated.
Methods
Study Population
The “Natural History of APOL1-associated Nephropathy Study” has recruited 835 African American children/siblings from 487 families with an index case having ESRD attributed to hypertension, FSGS, HIV-infection, or unknown cause in non-diabetic subjects.[11;29] Parents were not recruited, nor were relatives with ESRD (N=8). Relatives were eligible if they were no more than 15 years below the age at ESRD in their families’ index case. Those older than the age at ESRD in index cases were included. Relatives were genotyped for APOL1 nephropathy risk variants and genome-wide ancestry informative markers, used for estimation of individual ancestry proportions. Fasting serum, ethylenediaminetetraacetic acid plasma, urine, buffy coat and DNA samples were collected and phenotyping performed for sub-clinical kidney disease and associated risk factors (e.g., blood pressure, fasting blood sugar, body mass index).[29]
The 300 subjects selected for this study were chosen to provide clear clinical phenotypes. This included assessment of the number of APOL1 risk variants (zero, one or two), followed by comparison of presence of kidney disease based on urine albumin:creatinine ratio (UACR) >80 mg/g and/or estimated glomerular filtration rate (eGFR) <60 ml/min per 1.73 m2 in those older than or closest to the age at ESRD in the index case from their family. UACR >80 mg/g was used to reflect kidney disease, since minimally elevated UACR values between 30–80 were felt more likely to revert toward normal in the future and be less likely to reflect CKD. To minimize misclassification, control subjects without nephropathy (UACR <30 mg/g or eGFR >60 ml/min per 1.73m2) with each genotype could not be younger than 5 years below the age at ESRD in their family index case. Of the 300 subjects, 199 were from families with one participant, 34 from families with two participants (N=68), and 11 from families with three participants (N=33). All subjects with two APOL1 risk genotypes who met these criteria were included; remaining samples were comprised of nearly equal numbers of relatives with zero or one risk variant meeting nephropathy or non-nephropathy criteria. Since there was a larger pool of subjects with zero or one risk variant (and fewer individuals were selected in low risk groups), subjects with nephropathy in the zero and one genotype groups had higher albuminuria and lower eGFR, relative to those with two APOL1 risk variants.
Laboratory Evaluation
Serum creatinine concentrations were measured using creatinase enzymatic spectrophotometry, blood urea nitrogen using the urease enzymatic assay, cystatin C using an immunoassay, and UACR by microalbumin immunoturbidimetric methods at Laboratory Corporation of America (LabCorp; Burlington, North Carolina; www.labcorp.com). eGFR was computed using the four-variable MDRD equation and CKD-EPI equations.[30;31] UACR values ≥30 mg/g or MDRD GFR values <60 ml/minute per 1.73m2 were considered evidence of kidney disease. Normal values for cystatin C were <0.95 mg/L.
Nucleic acid extraction and quantitative PCR (qPCR) for CMV, BKV, JCV and HHV-6
Viral quantification was performed by Viracor-IBT Laboratories. Nucleic acid extraction of plasma (0.1 mL) and urine (0.5 mL) were performed by the bioMérieux NucliSENS easyMAG (reagent catalogue nos. 280130, 280131, 280132, 280133, and 280134). An internal control (bacteriophage lambda) was added to each sample prior to extraction. Following extraction, samples were amplified in a qPCR reaction using TaqMan Fast Advanced Master Mix (Applied Biosystems, Inc., Foster City, CA, catalogue no. 4444557). Thermocycling was performed in an Applied Biosystems 7500 Fast with the (50°C, 2 min; 95°C, 20 sec; 40 cycles of 95°C, 3 sec and 60°C, 30 sec). The qPCR reaction volume was 30 RL. Reactions were analyzed by ABI Prism SDS version 1.4.0.25 software. For standard curve preparation, a plasmid clone containing viral target regions was prepared and quantified by a picogreen dye-binding assay (Quant-iT Picogreen dsDNA assay, catalogue no. P11496, Invitrogen Inc., Carlsbad, CA). Serial plasmid DNA dilutions (5 – 5 × 106 copies) were used to construct a standard curve. The number of viral DNA copies/mL in plasma and urine samples was calculated using the CT of the sample, standard curve parameters, extracted sample volume, elution volume and volume of elution amplified. Viral DNA copies/mL values were not reported unless the internal control value was within a pre-determined range.
Genotype Analysis
DNA extraction from whole blood was performed using the PureGene system (Gentra Systems, Minneapolis, MN). Two single nucleotide polymorphisms (SNPs) in the APOL1 G1 nephropathy risk variant (rs73885319; rs60910145) and an indel for the G2 risk variant (rs71785313) were genotyped using a custom assay designed in the Wake Forest School of Medicine Center of Genomics and Personalized Medicine on the Sequenom (San Diego, California). In addition, 106 di-allelic ancestry informative markers (AIMs) were genotyped to determine population substructure. African ancestry proportion estimates were obtained for 44 Yoruba (YRI), 39 European American controls (used as ancestral populations) and the 826 African American study participants. The maximum likelihood approach of Tang et al. [32] as coded in the package FRAPPE was used to obtain the proportion of African and European ancestry for each individual.
Statistical Methods
The Wilcoxon two-sample test for comparing the distribution of all continuous outcomes between individuals who carry two APOL1 risk variants and those with zero or one copy (recessive model) was employed. Association with categorical outcomes was tested using the chi-square test with appropriate degree of freedom. The Box-Cox method identified the appropriate transformation of each outcome variable best approximating the distributional assumptions of conditional normality and homogeneity of variance of the residuals.[33] The logarithm transformation was applied for urine ACR and cystatin C. We added 1 to all urine ACR values before taking the log to prevent SAS from generating missing values in case a participant had a urine ACR value of 0. The African ancestry component was included as a covariate in the fully adjusted models to control for the possible confounding effect of admixture.
Linear mixed models were fitted for the continuous outcomes and non-linear mixed models for the dichotomous outcomes to test for association between the presence of BKV/JCV genomic DNA and each outcome. The mixed model framework allowed us to account for the familial relationship using the expected kinship coefficient matrix. This framework has been shown to provide valid inference when the observed data is a combination of unrelated and related individuals.[34] We could not compute the observed matrix since we had genotyped data available only on the APOL1 markers and the 106 AIMs. These mixed models can be fitted using maximum likelihood estimation thus providing the log-likelihood for each model, which we used to test the null hypothesis that the parameter estimate associated with replication of each virus was equal to the parameter estimate associated with APOL1 risk variants coded as a recessive model (based upon the number of APOL1 variants). This test is computed as twice the difference between the log-likelihoods of the reduced and the full models. Failure to reject this null hypothesis meant that viral replication and the APOL1 risk variant could be combined to create a single variable (APOL1+JCV) or (APOL1+BKV) both ranging between 0 and 2, where APOL1 coded under a recessive mode of inheritance is added to the indicator variable used to denote when the virus was detected. Results for the APOL1+JCV (Table 2) and APOL1+BKV variable (Table 3) are provided only in cases where the null hypothesis could not be rejected. APOL1 associations with FSGS and non-diabetic ESRD best fit autosomal recessive inheritance.[1;2] Therefore, association analyses between APOL1 nephropathy risk variants, nephropathy status in categorical and continuous evaluation of renal phenotypes, and presence of urine JCV and BKV genomic DNA were performed using the recessive model. Individuals who carry two copies of the APOL1 risk variant and had evidence of viral replication were assigned a risk score of 2. Those who had one risk factor but not the other received a risk score of 1. That is, individuals who either had two copies of APOL1 risk variants but lacked viral replication, or had viral replication and 0 or 1 APOL1 risk variants received a risk score of 1. Finally, those who lacked viral replication and had 0 or 1 risk variants were coded as 0. We considered three models: (1) an unadjusted model where we tested for the association between each virus and each outcome alone, (2) an APOL1 adjusted model where we included APOL1 as a covariate in the base model, and (3) a fully adjusted model where African ancestry, gender, and the age difference between the study participant and age at dialysis initiation in the family proband. Results from the unadjusted model are not shown since (1) they did not vary greatly from the other 2 models, and (2) the tests of the equality of effect between viral replication and APOL1 risk could not be performed in this model.
Acknowledgments
This work was supported by NIH grants R01 HL56266, RO1 DK070941, and RO1 DK084149 (BIF).
Footnotes
Disclosures:
None.
The authors report no conflicts of interest.
Reference List
- 1.Genovese G, Friedman DJ, Ross MD, et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. 2010;329:841–845. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tzur S, Rosset S, Shemer R, et al. Missense mutations in the APOL1 gene are highly associated with end stage kidney disease risk previously attributed to the MYH9 gene. Hum Genet. 2010;128:345–350. doi: 10.1007/s00439-010-0861-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kopp JB, Nelson GW, Sampath K, et al. APOL1 Genetic Variants in Focal Segmental Glomerulosclerosis and HIV-Associated Nephropathy. Journal of the American Society of Nephrology. 2011;22:2129–2137. doi: 10.1681/ASN.2011040388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Freedman BI, Kopp JB, Langefeld CD, et al. The Apolipoprotein L1 (APOL1) Gene and Nondiabetic Nephropathy in African Americans. J Am Soc Nephrol. 2010;21:1422–1426. doi: 10.1681/ASN.2010070730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bostrom MA, Freedman BI. The spectrum of MYH9-associated nephropathy. Clin J Am Soc Nephrol. 2010;5:1107–1113. doi: 10.2215/CJN.08721209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bostrom MA, Kao WH, Li M, et al. Genetic Association and Gene-Gene Interaction Analyses in African American Dialysis Patients With Nondiabetic Nephropathy. Am J Kidney Dis. 2011 doi: 10.1053/j.ajkd.2011.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Winston JA, Bruggeman LA, Ross MD, et al. Nephropathy and establishment of a renal reservoir of HIV type 1 during primary infection. N Engl J Med. 2001;344:1979–1984. doi: 10.1056/NEJM200106283442604. [DOI] [PubMed] [Google Scholar]
- 8.D’Agati V, Suh JI, Carbone L, et al. Pathology of HIV-associated nephropathy: a detailed morphologic and comparative study. Kidney Int. 1989;35:1358–1370. doi: 10.1038/ki.1989.135. [DOI] [PubMed] [Google Scholar]
- 9.Lipkowitz M, Freedman BI, Langefeld CD, et al. Apolipoprotein L1 gene variants associate with hypertension-attributed nephropathy and the rate of the kidney function decline in African Americans. Kidney Int. 2012 doi: 10.1038/ki.2012.263. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedman DJ, Kozlitina J, Genovese G, et al. Population-based risk assessment of APOL1 on renal disease. J Am Soc Nephrol. 2011;22:2098–2105. doi: 10.1681/ASN.2011050519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freedman BI, Langefeld CD, Turner J, et al. Association of APOL1 variants with mild kidney disease in the first-degree relatives of African American patients with non-diabetic end-stage renal disease. Kidney Int. 2012;82:805–811. doi: 10.1038/ki.2012.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hollander M, Gordon P, Lin PE. Robustness of the Wilcoxon Test to a certain dependency between samples. The Annals of Statistics. 1974;2:177–181. [Google Scholar]
- 13.McCance DJ. Persistence of animal and human papovaviruses in renal and nervous tissues. Prog Clin Biol Res. 1983;105:343–357. [PubMed] [Google Scholar]
- 14.Knowles WA. Discovery and epidemiology of the human polyomaviruses BK virus (BKV) and JC virus (JCV) Adv Exp Med Biol. 2006;577:19–45. doi: 10.1007/0-387-32957-9_2. [DOI] [PubMed] [Google Scholar]
- 15.Doerries K. Human polyomavirus JC and BK persistent infection. Adv Exp Med Biol. 2006;577:102–116. doi: 10.1007/0-387-32957-9_8. [DOI] [PubMed] [Google Scholar]
- 16.Kitamura T, Aso Y, Kuniyoshi N, et al. High incidence of urinary JC virus excretion in nonimmunosuppressed older patients. J Infect Dis. 1990;161:1128–1133. doi: 10.1093/infdis/161.6.1128. [DOI] [PubMed] [Google Scholar]
- 17.Pires EP, Bernardino-Vallinoto CV, Alves DM, et al. Prevalence of infection by JC and BK polyomaviruses in kidney transplant recipients and patients with chronic renal disease. Transpl Infect Dis. 2011;13:633–637. doi: 10.1111/j.1399-3062.2011.00614.x. [DOI] [PubMed] [Google Scholar]
- 18.Egli A, Infanti L, Dumoulin A, et al. Prevalence of polyomavirus BK and JC infection and replication in 400 healthy blood donors. J Infect Dis. 2009;199:837–846. doi: 10.1086/597126. [DOI] [PubMed] [Google Scholar]
- 19.Chesters PM, Heritage J, McCance DJ. Persistence of DNA sequences of BK virus and JC virus in normal human tissues and in diseased tissues. J Infect Dis. 1983;147:676–684. doi: 10.1093/infdis/147.4.676. [DOI] [PubMed] [Google Scholar]
- 20.Tominaga T, Yogo Y, Kitamura T, Aso Y. Persistence of archetypal JC virus DNA in normal renal tissue derived from tumor-bearing patients. Virology. 1992;186:736–741. doi: 10.1016/0042-6822(92)90040-v. [DOI] [PubMed] [Google Scholar]
- 21.Cheng XS, Bohl DL, Storch GA, et al. Inhibitory interactions between BK and JC virus among kidney transplant recipients. J Am Soc Nephrol. 2011;22:825–831. doi: 10.1681/ASN.2010080877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kantarci G, Eren Z, Demirag A, et al. JC virus-associated nephropathy in a renal transplant recipient and comparative analysis of previous cases. Transpl Infect Dis. 2011;13:89–92. doi: 10.1111/j.1399-3062.2010.00567.x. [DOI] [PubMed] [Google Scholar]
- 23.Lopez V, Gutierrez C, Sola E, et al. Does JC polyomavirus cause nephropathy in renal transplant patients? Transplant Proc. 2010;42:2889–2891. doi: 10.1016/j.transproceed.2010.07.061. [DOI] [PubMed] [Google Scholar]
- 24.Saundh BK, Tibble S, Baker R, et al. Different patterns of BK and JC polyomavirus reactivation following renal transplantation. J Clin Pathol. 2010;63:714–718. doi: 10.1136/jcp.2009.074864. [DOI] [PubMed] [Google Scholar]
- 25.Querbes W, O’Hara BA, Williams G, Atwood WJ. Invasion of host cells by JC virus identifies a novel role for caveolae in endosomal sorting of noncaveolar ligands. J Virol. 2006;80:9402–9413. doi: 10.1128/JVI.01086-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moriyama T, Marquez JP, Wakatsuki T, Sorokin A. Caveolar endocytosis is critical for BK virus infection of human renal proximal tubular epithelial cells. J Virol. 2007;81:8552–8562. doi: 10.1128/JVI.00924-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eash S, Manley K, Gasparovic M, et al. The human polyomaviruses. Cell Mol Life Sci. 2006;63:865–876. doi: 10.1007/s00018-005-5454-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skorecki KL, Wasser WG. Hypertension-misattributed kidney disease in African Americans. Kidney Int. 2013;83:6–9. doi: 10.1038/ki.2012.369. [DOI] [PubMed] [Google Scholar]
- 29.Freedman BI, Langefeld CD, Murea M, et al. Apolipoprotein L1 nephropathy risk variants associate with HDL subfraction concentration in African Americans. Nephrol Dial Transplant. 2011;26:3805–3810. doi: 10.1093/ndt/gfr542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levey AS, Bosch JP, Lewis JB, et al. A more accurate method to estimate glomerular filtration rate from serum creatinine: a new prediction equation. Modification of Diet in Renal Disease Study Group. Ann Intern Med. 1999;130(6):461–470. doi: 10.7326/0003-4819-130-6-199903160-00002. [DOI] [PubMed] [Google Scholar]
- 31.Levey AS, Stevens LA, Schmid CH, et al. A New Equation to Estimate Glomerular Filtration Rate. Annals of Internal Medicine. 2009;150:604–612. doi: 10.7326/0003-4819-150-9-200905050-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang H, Peng J, Wang P, Risch NJ. Estimation of individual admixture: analytical and study design considerations. Genet Epidemiol. 2005;28:289–301. doi: 10.1002/gepi.20064. [DOI] [PubMed] [Google Scholar]
- 33.Box GEP, Cox DR. An analysis of tranformations. Journal of the Royal Statistical Society, Series B. 1964;26:211–246. [Google Scholar]
- 34.Manichaikul A, Chen WM, Williams K, et al. Analysis of family- and population-based samples in cohort genome-wide association studies. Hum Genet. 2012;131:275–287. doi: 10.1007/s00439-011-1071-0. [DOI] [PMC free article] [PubMed] [Google Scholar]