TO THE EDITOR
While only about 15% of non-Hodgkin lymphomas are T-cell lymphomas, the treatment of this subset of neoplasms remains a significant challenge in the field of hematological cancers owing to unclear mechanisms regulating malignant T-cell growth. Cutaneous T cell lymphoma (CTCL) identifies a group of extranodal T-cell lymphomas characterized by the infiltration of malignant CD4+ T-cells in the skin (Wong et al., 2011). Sézary syndrome (SS) is an aggressive subtype of CTCL defined by diffuse pruritic rash, lymphadenopathy and malignant T-cells in the peripheral blood. The mechanisms underlying the proliferation of neoplastic CD4+ T-cells in SS are not fully understood, but abnormal epigenetic regulation of gene expression including silencing of tumor suppressor genes (TSG) likely plays an important role (van Doorn et al., 2005). Epigenetic mechanisms are critical contributors in cancer initiation and progression through modulation of gene expression, and transcriptional repression of TSG via epigenetic mechanisms occurs in many cancers (Baylin and Jones, 2011).
Sterile alpha motif (SAM) and HD domain containing protein 1 (SAMHD1) is the first identified mammalian triphosphohydrolase that hydrolyzes deoxynucleoside triphosphates (dNTPs), implicating a role in nucleic acid metabolism (Goldstone et al., 2011). SAMHD1 acts as an HIV-1 restriction factor in myeloid-cells and quiescent CD4+ T-cells by diminishing the intracellular dNTP pool to a level that is insufficient for viral replication (Baldauf et al., 2012; Lahouassa et al., 2012). Non-dividing CD4+ T-cells, monocytes, macrophages and dendritic cells from healthy individuals express high levels of SAMHD1 protein and have significantly lower levels of intracellular dNTPs compared to activated CD4+ T-cells, while several leukemia/lymphoma CD4+ T-cell lines lack SAMHD1 protein expression and have increased dNTP levels necessary for cell division (Baldauf et al., 2012; Hrecka et al., 2011; Laguette et al., 2011). We found that promoter methylation represses SAMHD1 expression in human leukemia/lymphoma CD4+ T cell lines, while the SAMHD1 promoter is unmethylated in primary CD4+ T-lymphocytes from healthy donors that express high levels of SAMHD1 protein (de Silvaet al., 2013). The role of SAMHD1 in cancer remains unknown.
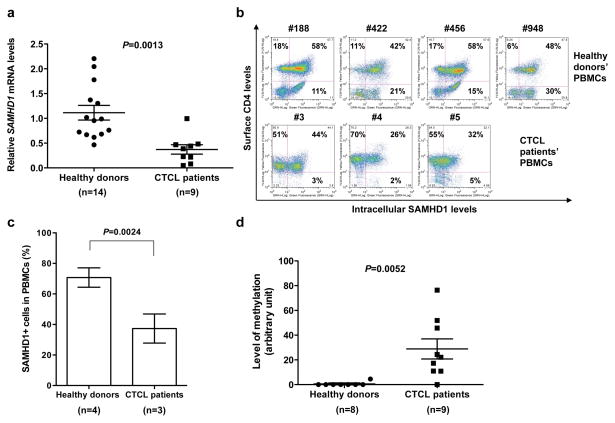
To explore the potential role of SAMHD1 in CTCL, we measured SAMHD1 mRNA levels in peripheral blood mononuclear cells (PBMCs) from 14 healthy donors and nine CTCL patients using a real-time quantitative-PCR (qPCR) assay (de Silva et al., 2013). These patients included 8 with SS and one with advanced stage mycosis fungoides (MF) (Table 1), which are two related subtypes of CTCL originating from CD4+ skin-homing T-cells (Wong et al., 2011). Interestingly, qPCR results revealed that PBMCs from the 8 SS patients and an advanced stage MF patient expressed on average 3-fold lower SAMHD1 mRNA levels (P=0.0013, two-sample t-test) compared to those from 14 healthy donors (Figure 1a and Table 1). These results indicate that SAMHD1 expression is significantly downregulated in PBMCs from SS patients relative to healthy individuals.
Table 1.
Clinical information of CTCL patients and relative levels of SAMHD1 mRNA and promoter methylation in PBMCs from the patients.
| Patient | CTCL subtype a | Disease stage | CD4+ cells (%) b | Absolute CD4 count | CD4: CD8 ratio | Sézary cell count (%) | Relative SAMHD1 mRNA levels c | Relative SAMHD1 promoter methylation levels d |
|---|---|---|---|---|---|---|---|---|
| #1 | SS | IVA | 98 | N/Ae | N/A | 29 | 0.079 | 30.74 |
| #2 | SS | IVA | 93 | N/A | N/A | 45 | 0.995 | 39.31 |
| #3 | SS | IVA2 | 80 | N/A | N/A | 31 | 0.217 | 0.000 |
| #4 | SS | IVA | 65 | N/A | N/A | N/A | 0.387 | 19.43 |
| #5 | SS | IVA2 | 78 | N/A | N/A | 61 | 0.469 | 135.79 |
| #6 | MF | IIB | 30 | 163 | 1.8 | N/A | 0.048 | 92.27 |
| #7 | SS | IVA | 85 | 1785 | 28 | N/A | 0.483 | 19.34 |
| #8 | SS | IVA2 | 88 | 3346 | 82 | N/A | 0.229 | 43.29 |
| #9 | SS | IVA | 82 | 1466 | 13 | N/A | 0.441 | 81.32 |
SS: Sézary syndrome, MF: mycosis fungoides. No patient in this group was treated with methotrexate. These patients have been on bexarotene and interferon at some time during their treatments.
CD4+ cells (%) indicate percentage of CD4-positive cells in patient PBMCs by flow cytometry analysis.
Relative levels of SAMHD1 mRNA in PBMCs from 9 CTCL patients (mean±SD is 0.372±0.285) compared to the average level of 14 healthy donors (1.113±0.553).
Relative levels of SAMHD1 promoter methylation in PBMCs from 9 CTCL patients (mean±SD is 51.27±43.32) compared to the average level of 8 healthy donors (1.000±1.703).
N/A, not available.
Figure 1. Comparison of SAMHD1 expression levels and the SAMHD1 promoter methylation in peripheral blood mononuclear cells (PBMCs) from CTCL patients and healthy donors.
(a) SAMHD1 mRNA levels in PBMCs from 14 healthy donors and 9 CTCL patients (refer to Table 1) were measured by qPCR and normalized to GAPDH levels. The average levels of SAMHD1 mRNA levels are shown. The error bars represent standard error of the mean. (b) Intracellular SAMHD1 and surface CD4 proteins in PBMCs from CTCL patients and healthy donors were detected by immunostaining and flow cytometry. Donor and patient numbers are indicated above the plots. The percentages of cells positive for CD4 only (upper left quadrant), SAMHD1 only (lower right quadrant), or double positive for CD4 and SAMHD1 (upper right quadrant) are indicated. (c) Flow cytometry-based quantification of SAMHD1-expressing (+) cells in PBMCs from healthy donors and CTCL patients. (d) Quantification of relative levels of SAMHD1 promoter methylation in PBMCs from 9 CTCL patients and 8 healthy donors. ImageJ program was used to quantify the intensity of the SAMHD1 promoter-specific PCR bands in Figure S2 and the relative level of methylation was calculated by setting the intensity of the HpaII-untreated sample as 100 and calculating the percentage intensity of the PCR band in the HpaII-treated samples relative to its matching untreated control. The error bars represent standard error of the mean.
Next we examined whether the downregulation of SAMHD1 mRNA levels would translate to reduced SAMHD1 protein levels in PBMCs of CTCL patients. To this end we measured expression levels of surface CD4 and intracellular SAMHD1 proteins in PBMCs from three CTCL patients with high circulating neoplastic T-cells (patients #3, #4, and #5 in Figure 1b and Table 1) and four healthy donors using immunostaining and flow cytometry (Figure 1b) (Descours et al., 2012). The limited sample size was due to available PBMCs only from three CTCL patients. Total SAMHD1-positive cells in PBMCs from CTCL patients (37±9%) were significantly lower (P=0.0024) than those from healthy donors (71±6%) (Figure 1c). Furthermore, the percentage of SAMHD1-expressing cells in CD4-negative PBMCs from CTCL patients (3±2%) was significantly lower than that from healthy donors (19±8%) (P=0.023). Analysis of SAMHD1 and CD4 double-positive cells in PBMCs from CTCL patients (34±9%) and healthy donors (52±8%) showed a statistical difference (P=0.042), suggesting significant downregulation of SAMHD1 protein expression in the neoplastic CD4+ cell subset from CTCL patients. Given that PBMCs from CTCL patients comprise mainly of CD4+ cells, we also compared the percentage of SAMHD1-expressing cells in the CD4+ gated population between healthy donor samples (81±3%) and CTCL patient samples (46±6%) and found a significant reduction (P=0.0024) in SAMHD1-expressing cells (supplemental Figure S1).
We hypothesized that the SAMHD1 promoter in PBMCs from the CTCL patients is methylated and thereby inhibits SAMHD1 expression. To compare the methylation status of the SAMHD1 promoter in PBMCs from the CTCL patients selected and healthy donors, genomic DNA of PBMCs was treated with the methylation-sensitive HpaII endonuclease, or left untreated, and then subjected to PCR amplification using SAMHD1 promoter-specific primers as described (de Silva et al., 2013). The SAMHD1 promoter contains five HpaII sites, and methylation of these sites prevents digestion by HpaII. The intact undigested sequence can serve as a template for PCR amplification to yield a 1.2-kb product. As an input control of genomic DNA, a 0.25-kb region within the GAPDH gene lacking HpaII sites was PCR amplified (Figure S2). The SAMHD1 promoter in PBMCs from eight of nine CTCL patients tested was methylated (Figure S2a, 1.2-kb bands). Strikingly, PBMCs from eight healthy donors showed that the SAMHD1 promoter was unmethylated (Figure S2b). The purity of the genomic DNA for complete enzyme digestion as well as the intact nature of the HpaII sites in the SAMHD1 promoter sequence in healthy donor and CTCL patient genomic DNA was confirmed by restriction digestion with MspI (an isoschizomer of HpaII), which cleaves the HpaII site irrespective of its methylation status (Figure S3).
Densitometry analysis of the PCR products in Figure S2 was used to quantify the relative level of methylation of the SAMHD1 promoter in PBMCs from CTCL patients and healthy donors. Our analysis revealed 51-fold higher average levels of methylation of the SAMHD1 promoter in PBMCs from 9 CTCL patients (P=0.0052) relative to 8 healthy donors (Figure 1d and Table 1). These results suggest a positive correlation between downregulation of SAMHD1 expression and methylation of the SAMHD1 promoter in PBMCs from CTCL patients. However, we observed a lack of correlation between reduced SAMHD1 expression and promoter methylation in patient #3, which suggests that besides promoter methylation, other transcriptional and epigenetic regulatory mechanisms such as microRNA and histone modifications, may also contribute to the regulation of SAMHD1 expression in CTCL patients.
SAMHD1 somatic mutations have been identified in patients with lung adenocarcinoma, medulloblastoma, glioblastoma, breast, pancreatic and colorectal cancers, albeit at a very low frequency (Imielinski et al., 2012; Jones et al., 2008; Parsons et al., 2008; Parsons et al., 2011; Sjoblom et al., 2006). Transcriptional repression of TSG through DNA methylation and histone modifications is a common mechanism of gene silencing in numerous types of cancer. Inhibition of epigenetic suppression in vitro using specific inhibitors to block DNA methyltransferase and/or histone deacetylase can reactivate expression of TSG silenced in cancer. Downregulation of dNTP catabolic enzymes such as SAMHD1 may lead to imbalances in the intracellular dNTP pool, which can induce mutations and genomic instability as key features of CTCL.
Supplementary Material
Acknowledgments
We thank Dr. Olivier Schwartz (Institut Pasteur) for the kind gift of SAMHD1 antibody (clone I19-18) and Heather Hoy for her excellent technical assistance. This work was supported in part by grants AI098524 and AI102822 to LW, and CA164911 to HW and PP from the National Institutes of Health. LW is supported in part by the Public Health Preparedness for Infectious Diseases Program of The Ohio State University.
Footnotes
Conflict of interest
The authors declare that they have no conflict of interests.
References
- Baldauf HM, Pan X, Erikson E, Schmidt S, Daddacha W, Burggraf M, et al. SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells. Nat Med. 2012;18:1682–9. doi: 10.1038/nm.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11:726–34. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Silva S, Hoy H, Hake TS, Wong HK, Porcu P, Wu L. Promoter methylation regulates SAMHD1 gene expression in human CD4+ T cells. J Biol Chem. 2013;288:9284–92. doi: 10.1074/jbc.M112.447201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Descours B, Cribier A, Chable-Bessia C, Ayinde D, Rice G, Crow Y, et al. SAMHD1 restricts HIV-1 reverse transcription in quiescent CD4+ T-cells. Retrovirology. 2012;9:87. doi: 10.1186/1742-4690-9-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstone DC, Ennis-Adeniran V, Hedden JJ, Groom HC, Rice GI, Christodoulou E, et al. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature. 2011;480:379–82. doi: 10.1038/nature10623. [DOI] [PubMed] [Google Scholar]
- Hrecka K, Hao C, Gierszewska M, Swanson SK, Kesik-Brodacka M, Srivastava S, et al. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature. 2011;474:658–61. doi: 10.1038/nature10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–20. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–6. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, Segeral E, et al. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature. 2011;474:654–7. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahouassa H, Daddacha W, Hofmann H, Ayinde D, Logue EC, Dragin L, et al. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat Immunol. 2012;13:223–8. doi: 10.1038/ni.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–12. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons DW, Li M, Zhang X, Jones S, Leary RJ, Lin JC, et al. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331:435–9. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–74. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- van Doorn R, Zoutman WH, Dijkman R, de Menezes RX, Commandeur S, Mulder AA, et al. Epigenetic profiling of cutaneous T-cell lymphoma: promoter hypermethylation of multiple tumor suppressor genes including BCL7a, PTPRG, and p73. J Clin Oncol. 2005;23:3886–96. doi: 10.1200/JCO.2005.11.353. [DOI] [PubMed] [Google Scholar]
- Wong HK, Mishra A, Hake T, Porcu P. Evolving insights in the pathogenesis and therapy of cutaneous T-cell lymphoma (mycosis fungoides and Sezary syndrome) Br J Haematol. 2011;155:150–66. doi: 10.1111/j.1365-2141.2011.08852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.