Abstract
Hepatitis C infection produces chronic liver injury that is significantly exacerbated by alcohol consumption. While multiple mechanisms contribute to this synergy, a viral-induced loss of antioxidant responses has been shown to play an important role. This study examined the effects of HCV infection and alcohol on the regulation of the transcription factor FOXO3, an important regulator of SOD2 expression, a tumor suppressor, and a component of the hepatic antioxidant response system. FOXO3 was activated by either HCV or alcohol alone but suppressed by the combination. To understand this paradoxical result, we applied a capillary isoelectric focusing (IEF) method to determine the pattern of FOXO3 post-translational modifications (PTMs) induced by HCV and alcohol. We observed the presence of multiple different nuclear and cytosolic species of FOXO3 and used anti phosphoserine, acetyl-lysine, methylarginine and ubiquitin antibodies to identify the PTM patterns present in each species. HCV caused multiple changes including phosphorylation of FOXO3 at S-574, a novel JNK site, which promoted nuclear translocation and transcription. Ethanol suppressed arginine-methylation of FOXO3 promoting nuclear export and degradation of the JNK phosphorylated form. Human liver biopsy samples showed the presence of the HCV-specific form of FOXO3 in HCV-infected livers but not in normal liver or nonalcoholic steatohepatitis.
Conclusion
The development of this novel IEF method for the simultaneous quantification of differently modified FOXO3 species allowed us to demonstrate how HCV and alcohol combine to modify a complex pattern of FOXO3 PTMs that contribute to pathogenesis. This approach will allow further dissection of the role of protein PTMs in viral liver disease.
Keywords: Forkhead box transcription factors, arginine methylation, viral hepatitis, isoelectric focusing, c-Jun N-terminal kinase
Hepatitis C and alcohol each cause liver injury that results from a combination of immune-mediated cytotoxicity and alterations in adaptive signaling pathways within hepatocytes. While these two disease causing agents produce liver injury by themselves, there is considerable evidence that when present in combination, HCV and alcohol have effects that do not occur with either stimulus alone. In epidemiological studies, the alcohol-HCV combination results in rapid fibrosis progression, impaired viral clearance and enhanced carcinogenesis (1). In cell culture, synergistic effects include induction of cell death pathways, mitochondrial ROS production, and suppression of antioxidant protein expression (2).
Recent studies have shown that the function of FOXO transcription factors is altered as a consequence of HCV infection potentially contributing to insulin resistance and impaired activation of starvation-induced autophagy (3). FOXO transcription factors control expression of proteins responsible for longevity, antioxidant response, cell cycle arrest, insulin sensitivity, apoptosis, and autophagy (4, 5). FOXO3 is also a tumor suppressor (4, 6). FOXO proteins are regulated by a complex series of post-translational modifications (PTMs) that have collectively been suggested to constitute a “FOXO code” (4). Among the best understood of these PTMs are three Akt phosphorylations which cause nuclear export of the protein, but additional PTMs such as alternative phosphorylations, acetylation, ubiquitination, and methylation have all been shown to alter the stability of the protein in the nucleus, its transcriptional profile, or both (4, 7).
Preliminary studies from our lab have recently shown that HCV and alcohol each increased FOXO3 transcriptional activity but the combination of HCV and alcohol together suppressed FOXO3-dependent gene expression. In addition, the loss of FOXO3 activity was associated with liver injury (8, 9). We reasoned that changes in FOXO3 activity likely resulted from specific FOXO3 posttranslational modifications (PTMs) induced under these conditions. Identification of FOXO3 PTMs has been achieved primarily with PTM-epitope specific antibodies and mass spectroscopy but these approaches are not well suited to assess the diversity of modified species or multiple modifications present on the same molecule.
In the present study we have applied a novel capillary isoelectric focusing method (cIEF) to distinguish different species of FOXO3. We used this along with mutation analysis to compare HCV- and alcohol-induced functional changes in FOXO3 with PTM molecular signatures. We observed that HCV’s activating effect on FOXO3 resulted from JNK-dependent phosphorylation at a previously unrecognized site. JNK activation and JNK-dependent FOXO3 species were also observed in the majority of analyzed tissue samples from HCV positive patients. Suppression of FOXO3 activity by alcohol resulted from a loss of FOXO3 arginine methylation that occurred in the combined HCV-alcohol condition. This decreased the half-life of the protein and reduced its accumulation in the nucleus. The application of cIEF combined with more conventional mutation analysis thus shows the complexity of disease-related FOXO3 modifications and how this pattern of interactive modifications contributes to viral-host-environment interactions that determine disease outcomes.
Experimental Procedures
Cell culture, transfection and viral infection
Huh7.5 cells and Huh7.5 reporter cells (10) expressing an RFP reporter protein (obtained from Dr. Charles Rice) were maintained in Dulbecco’s Modified Eagle’s Medium (Invitrogen, Carlsbad, CA) containing 10% FBS, 50U mL−1 penicillin and 50 mg mL−1 streptomycin. Cells were treated where indicated with 50mM ethanol for 48 hours, 2.5 mM betaine, 0.1 mM S-adenosylmethionine or 20 µM JNK inhibitor (SP600125) for 16 h prior to harvest.
Cells were transfected using lipofectamine LTX transfection reagent (Invitrogen) according to the manufacturer’s protocol. pECE-HA-FOXO3, and pFHRE-luc (11) were provided by M. Greenberg via Addgene; pCDNA3 Flag MKK7B2Jnk1a1 (12) was provided by RJ Davis via Addgene. FOXO3 point mutations were generated by site directed mutagenesis (Quickchange kit, Stratagene).
DNA coding for the Japanese Fulminant Hepatitis 1 (JFH1) sequence of HCV was obtained from Dr. T. Wakita. Plasmid was propagated, reverse transcribed and the resulting RNA used to transfect (by electroporation) Huh7.5 cells for the production of intact viral particles as described (13). Huh7.5 cells (3×105) were seeded onto T-25 flasks and infected the following day with HCV at a multiplicity of infection of 0.5–1.0.
Luciferase reporter assays
Huh 7.5 cells were cultured either without infection or for 4 days after JFH1 infection. Cells were then seeded onto 24 well plates. After 24 h, cells were serum starved for 5–16 h and then cotransfected using lipofectamine–LTX (Invitrogen, Carlsbad, CA) with FHRE-luc reporter vector (11), pRL-tk vector (renilla luciferase reporter) and pECE-HA-FOXO3 vector (WT or mutants, 0.2 µg per well) and where indicated pFlagMKK7JNK1a1 (30 ng per well). Cells were subsequently incubated for 48 h prior to lysis and luciferase determination with the Dual luciferase assay kit (Promega) on a single tube Glomax 20/20 luminometer (Promega). Results were expressed as firefly luciferase/renilla luciferase activity.
Cell fractionation and Western blots
Whole cell lysates were prepared from cells that had been washed and harvested by centrifugation in phosphate buffered saline (PBS) pH 7.5. Cell pellets were resuspended in RIPA buffer that contained 50 mM Tris, pH 7.5, 150 mM sodium chloride, 1% NP-40, 0.5% sodium deoxycholate, 0.1 mM EDTA, and 1% protease and phosphatase inhibitors (Sigma-Aldrich, Saint Louis, MO). Lysates were centrifuged at 14,000 rpm for 15 min; supernatants were collected and protein concentration was measured using the Bio-Rad protein assay kit (Bio-Rad Laboratories, Inc. Hercules, CA).
Protein extracts (15 µg) were subjected to 10% SDS-polyacrylamide gel electrophoresis (SDS-PAGE), electrophoretically transferred to nitrocellulose membranes (Amercham Hybond ECL, GE Healthcare), and blocked in 3% BSA/PBS at RT for 1 hour. Primary antibodies were incubated overnight at manufacturer recommended concentrations. The antibodies used are detailed in supplementary methods. Immunoblots were detected with the ECL Plus Western Blotting Detection System (Amersham Biosciences, Piscataway, NJ) or using near-infrared fluorescence with the ODYSSEY Fc, Dual-Mode Imaging system (Li-COR). Expression levels were evaluated by quantification of relative density of each band normalized to that of the corresponding β-actin or GAPDH band density.
Capillary isoelectric focusing
cIEF analysis was performed using a Nanopro-1000 instrument (ProteinSimple, Santa Clara, CA). Protein samples were diluted with sample diluent (Bicine/CHAPS, protease and phosphatase inhibitors) to 1.6 mg/ml, treated with 12M urea/40 mM DTT (1:1 for a final urea concentration of 6M) for 5 min at RT, then supplemented with equal volume of Bicine/CHAPS buffer, 75 % v/v of G2 ampholyte premix (ProteinSimple), containing 8% v/v ampholyte mixture. For FOXO3 analysis the mixture contained 50% pH 2.5–5, 33% pH 5–8, and 17% pH 3–10 (GE Healthcare) for a final concentration of 6% v/v of ampholytes in the capillary. The mixture was supplemented with pI Standard Ladder 1 (ProteinSimple, p/n 040–644). When immunoprecipitated proteins were analyzed, 12M urea/40 mM DTT was added directly to the beads, and eluate was used for analysis. The urea concentration in the capillary was not allowed to exceed 2M and total salt concentration did not exceed 50mM.
Proteins were focused for 40 minutes at 60 mW, crosslinked to the capillary for 90 sec and incubated with primary (1:50) and secondary (1:100) antibodies for 120 and 60 min respectively. Data is presented as relative chemiluminescence intensity vs. distance along the capillary and these distances were transformed into pI values with internal fluorescent pI standards. This method led to an average variation in pI values for replicates of less than 0.05 pH units.
Human Liver specimens
De-identified human liver specimens from patients with Hepatitis C cirrhosis, nonalcoholic steatohepatitis cirrhosis, and normal liver (transplant donors) were obtained from the Liver Center Tissue Bank at the University of Kansas Medical Center. All studies using human tissue samples were approved by the Human Subjects Committee of the University of Kansas Medical Center. Subcellular fractions were isolated from frozen specimens by homogenization, passing the sample through a cell strainer (BD Falcon-40µm), and further fractionation as described for the cell culture specimens.
Statistics
Results are expressed as mean ± SD. The Student t test, paired t test, or one-way ANOVA with Bonferroni post hoc test was used for statistical analyses. P < 0.05 was considered significant.
Results
Effects of HCV and alcohol on FOXO3
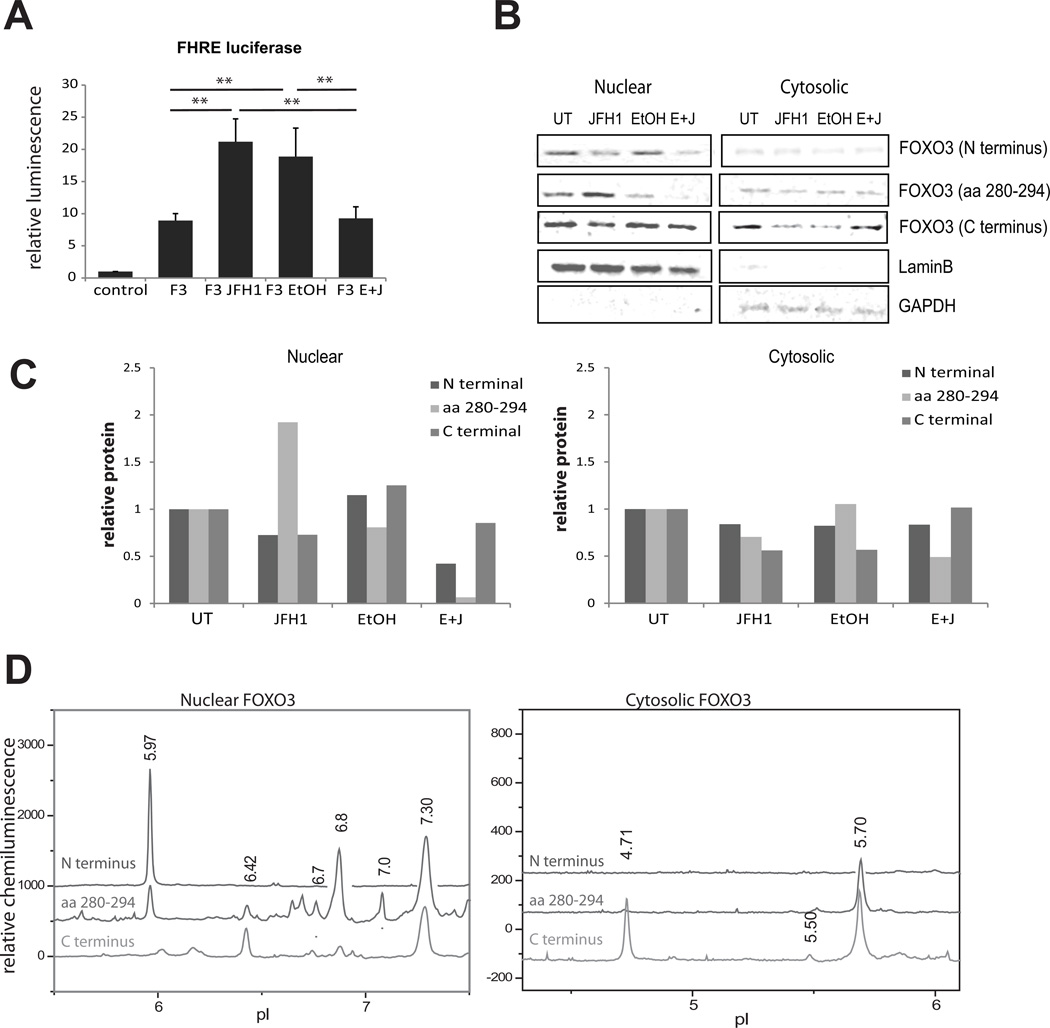
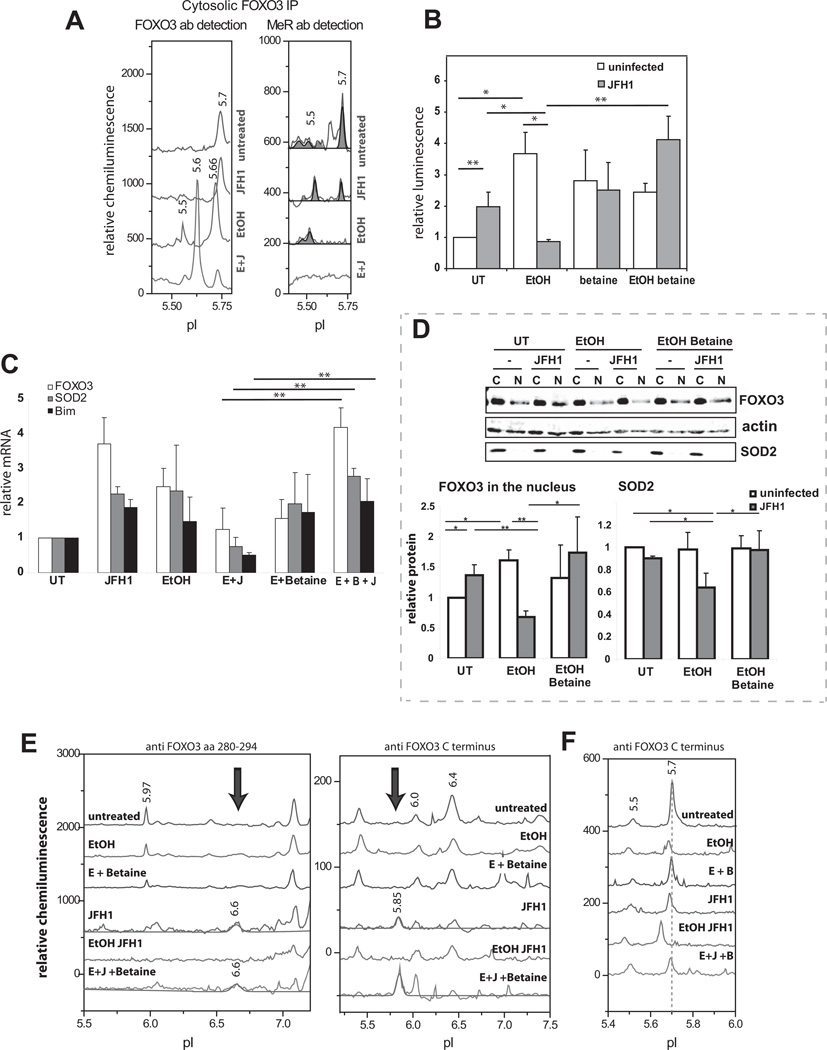
Huh 7.5 cells were infected with the JFH1 strain of HCV and/or treated with 50 mM ethanol for 48 h. This cell line was chosen because it is permissive for the entire HCV lifecycle, and it expresses the alcohol metabolizing enzyme, ADH. Fig. 1A demonstrates that overexpression of FOXO3 led to a 10-fold increase in FHRE-luciferase reporter activity. HCV infection and ethanol treatment each caused a further 2-fold increase in activity but this was absent when the two stimuli were combined. We immunoblotted nuclear and cytosolic fractions with three different FOXO3 antibodies (Fig 1B). HCV increased the amount of nuclear FOXO3 detected with an antibody directed against aa 280–294, but not with two other antibodies. Ethanol failed to increase nuclear FOXO3 detected by any of the antibodies, and the HCV/ethanol combination decreased nuclear FOXO3 detected with two of the antibodies but not the third. These results suggested that changes in FOXO3 transcriptional activity were not explained solely by alterations in the cytosol-nuclear translocation of the protein, and that the different conditions generated antigenically different forms of FOXO3.
Figure 1. HCV infection and alcohol exposure alter FOXO3 transcriptional activity and localization in Huh7.5 cells.
A. FHRE-luciferase reporter activity was measured for control Huh7.5 cells, Huh7.5 cells transfected with wild-type FOXO3 (F3), and FOXO3 overexpressing cells that were HCV-infected (F3 JFH1), treated with 50 mM ethanol for 48h (F3 EtOH) or both infected and ethanol treated (F3 E+J). Data are presented as mean ± SD.*p < 0.05, **p < 0.01. n=3. B. Western blot analysis of FOXO3 in nuclear and cytosolic fractions isolated from Huh7.5 cells treated as in (A). Results using three different FOXO3 antibodies are shown. C. Densitometry analysis of the western blot data from panel B. D. Nuclear and cytosolic extracts of Huh7.5 cells were analyzed by cIEF followed by immunodetection using antibodies against N terminus, central region (aa 280–294) and C terminus of FOXO3. Absolute values of the baselines of the luminescence traces have been shifted arbitrarily to allow placement of all three traces on the same axis. The scaling for each trace was not changed and is identical.
Since most of the described FOXO3 PTMs produce a change in net charge of the molecule, we developed a capillary isoelectric focusing method (cIEF) to resolve different FOXO3 species. A similar method has been previously adapted for several signaling kinases (14, 15). Following liquid phase capillary isoelectric focusing, proteins were crosslinked to the capillary and FOXO3 was detected using one of 3 different FOXO3 antibodies recognizing epitopes at the N-terminal, central region, or C-terminus (Fig 1D). The identity of the FOXO3 peaks was confirmed by recognition by at least 2 of the 3 antibodies and by analysis of overexpressed HA-FOXO3 (Fig S1). In cytosolic extracts, two major peaks were observed with pI 4.7 and 5.7. For nuclear extracts, a set of at least 5 FOXO3 species was recognized with overlapping specificity of each of the 3 antibodies.
Effects of HCV and ethanol on FOXO3 posttranslational modifications
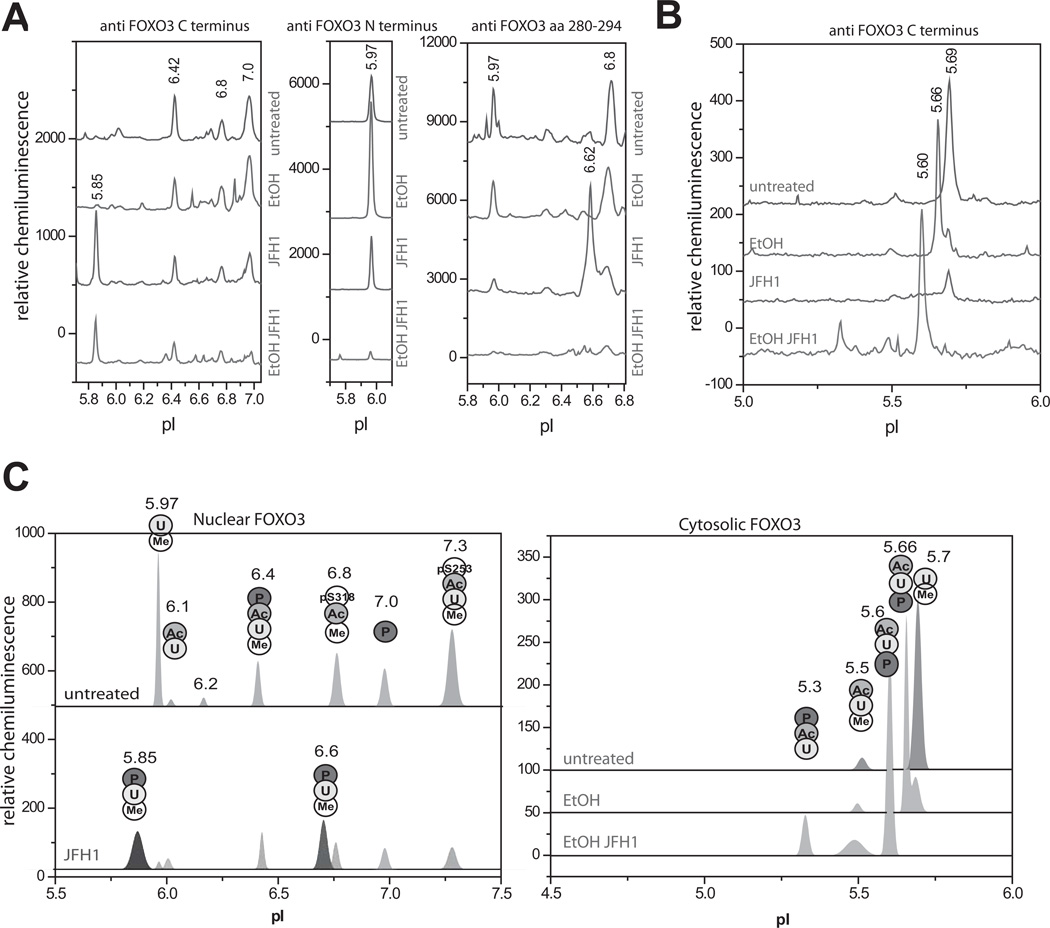
We next studied the effects of HCV and alcohol on the FOXO3 nuclear and cytosolic species. Ethanol had no effect on FOXO3 nuclear peak pIs, but changed the proportions between them with an increase in the 5.97 species and decreases in 6.42 and 6.8 species (Fig 2A). Ethanol also created a new peak with pI 5.66 in the cytosol (Fig 2B). HCV decreased cytosolic FOXO3, decreased all nuclear species present in uninfected cells, and caused the appearance of two new nuclear peaks at pI 6.62 (seen best with the 280–294 antibody) and at pI 5.85 (seen best with the C-term antibody) (Fig 2A,B). The combination of HCV and ethanol reduced the magnitude of all nuclear peaks, including both of the HCV specific peaks and caused the appearance of a new cytosolic peak at pI 5.60 (Fig 2A,B).
Figure 2. Effects of HCV on FOXO3 posttranslational modifications.
A, B. HCV-infected and uninfected Huh7.5 cells were treated with 50 mM ethanol for 48h as described in the text. Nuclear (A) and cytosolic (B) extracts were analyzed by cIEF using indicated antibodies. Baseline levels of each trace were shifted as for Fig. 1. C. Summary of confirmed FOXO3 posttranslational modifications for each nuclear and cytosolic FOXO3 peak found using immunoprecipitation and PTM specific antibodies. U – ubiquitination, Me – arginine methylation, Ac – lysine acetylation, P – serine/threonine phosphorylation, pS318 – phosphorylation on Ser318, pS253 – phosphorylation on Ser253.
In order to identify the molecular nature of the different nuclear peaks we performed a series of reciprocal immunoprecipitations followed by cIEF analysis using either a FOXO3 antibody for IP and a PTM specific antibody for detection, or the reverse. When site specific phosphoantibodies were available these were used as well. For nuclear peaks we were generally able to confirm the presence of a PTM by its appearance in both IPs. It was not always possible to perform IP with the PTM specific antibodies in cytosolic extracts so the identity of cytosolic peaks was determined by immunoprecipitation with FOXO3 antibody and detection with both antibodies. A summary of the results is shown in Fig. 2C. The individual IP analysis supporting these assignments is shown in Figs S2 and S3. Most of the nuclear species were ubiquitinated and methylated. We also detected acetylated species (mainly the pI 6.42 peak) and species phosphorylated on the Akt sites (S253 and S318). Both novel HCV nuclear species were also phosphorylated as they were recognized by pSer/Thr antibody.
HCV induces JNK phosphorylation of FOXO3 at a novel site
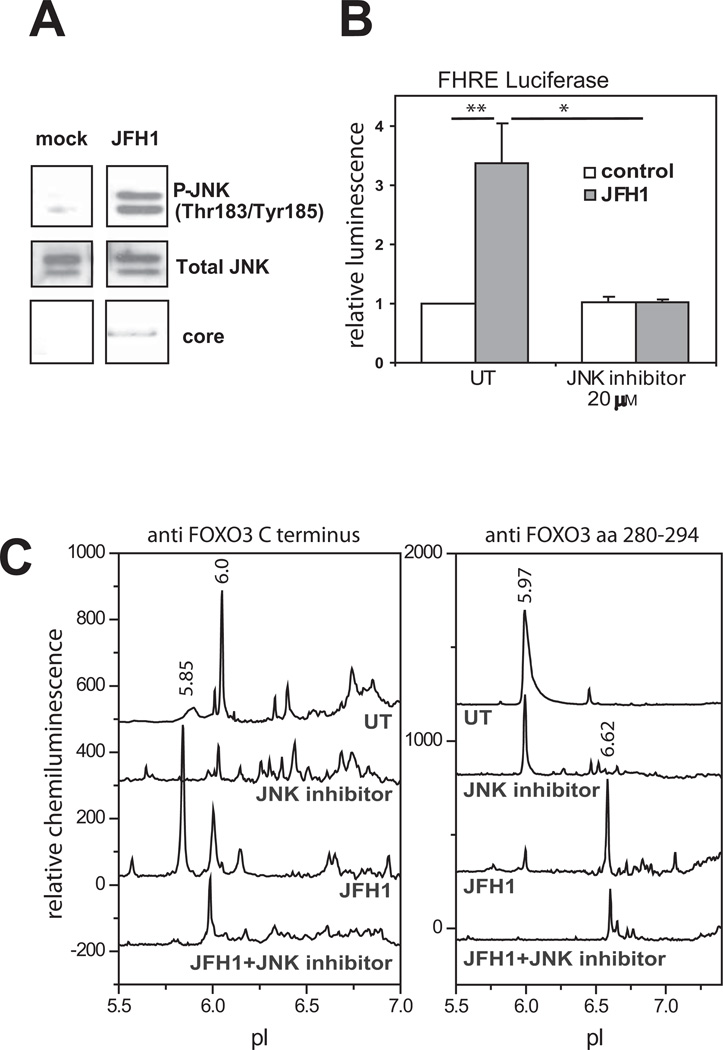
HCV has been reported to increase phosphorylation of the MAPkinases JNK, ERK and p38 (16). We also observed JNK activation (Fig. 3A). Since JNK phosphorylation has been reported to activate FOXO4 and FOXO1 we tested the effect of a JNK inhibitor on the process. As shown in Fig, 3B, the JNK inhibitor, SP600125, prevented the HCV induced increase in FHRE-reporter activity. Formation of the HCV-induced pI 5.85 FOXO3 peak was similarly prevented by JNK inhibitor (Fig 3C). A p38 inhibitor did not affect the ability of HCV to induce FOXO activity (Fig S4A).
Figure 3. JNK-dependence of HCV-induced effects on FOXO3.
A. Western bot analysis of phosphorylated forms of JNK (pJNKT183/Y185) and total JNK in infected and uninfected Huh7.5 cells. B. FHRE-luciferase reporter assays in control and HCV-infected Huh7.5 cells in the presence or absence of JNK inhibitor (SP600125). C. cIEF analysis of FOXO3 species present in nuclear extracts from Huh7.5 cells treated as above. Extracts were probed with either C-terminus (left panel) or aa 280–295 (right panel) FOXO3 primary antibodies. Baseline levels of each trace were shifted as for Fig. 1.
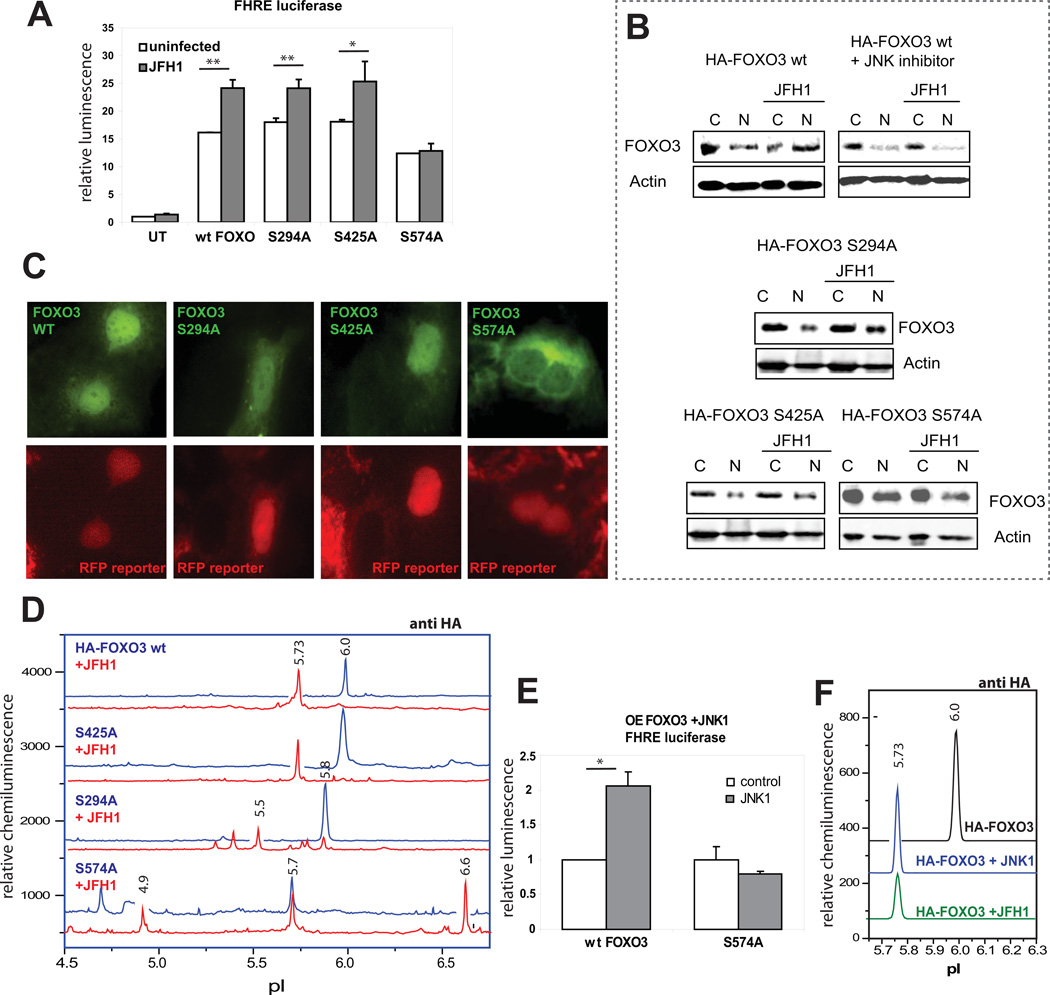
To identify possible sites of HCV stimulated JNK phosphorylation of FOXO3 we prepared a series of mutants at potential JNK phosphorylation sites (Fig S4B). HA-tagged wt FOXO3, FOXO3 S294A, S425A and S574A were transfected into Huh 7.5 cells which were then infected with JFH1. Overexpression of FOXO3 increased FHRE-reporter activity at least 10 fold. JFH1 further stimulated FHRE-luciferase reporter activity of all constructs except S574A (Fig 4A). In addition, HCV caused nuclear translocation of the wt, S294A, and S425A mutants but not the S574A mutant as assessed by either fractionation and western blotting (Fig 4B, densitometry analysis in Fig. S4C) or immunofluorescence (Fig. 4C).
Figure 4. HCV induces JNK-mediated phosphorylation of FOXO3 at serine 574.
A. FHRE-luciferase reporter assays in control and HCV-infected Huh7.5 cells that were either untransfected (UT), or overexpressing HA-FOXO3 wt, S294A, S425A or S574A mutants of FOXO3. *p < 0.05, **p < 0.01. B. Western blot analysis of overexpressed HA-FOXO3 wt, S294A, S425A and S574A in nuclear and cytosolic fractions of infected and control Huh7.5 cells. 20 µM JNK inhibitor (SP600125) was added as indicated. Numbers indicate the ratio of the density of the nuclear FOXO3 band in HCV infected/control cells. C. Immunofluorescence images of HCV-infected Huh7.5 reporter cells overexpressing HA-tagged wt or mutant FOXO3 constructs two days after infection. FOXO3 is in green (anti HA antibody), and RFP reporter protein is in red (infected cells have a nuclear distribution of the RFP reporter protein). D. cIEF analysis of nuclear FOXO3 species with anti-HA antibody in control and HCV-infected Huh7.5 cells overexpressing HA-tagged constructs of WT and mutant FOXO3. Baseline levels of each trace were shifted as for Fig. 1. E. FHRE-luciferase reporter assays in cells overexpressing WT or mutant FOXO3 S574A with or without co-transfection with Flag-MKK7-JNK1 fusion protein as indicated. F. cIEF analysis with anti-HA antibody of nuclear FOXO3 species in cells expressing wt HA-FOXO3 under control conditions, after cotransfection with the Flag-MKK7-JNK1 fusion protein, and after HCV infection.
We further examined the effect of HCV on FOXO3 mutants by cIEF (Fig 4D). As seen previously, HCV caused an acidic shift of the dominant nuclear FOXO3 peak from pI 6.0 to pI 5.7 (Fig 4E) and this was blocked by JNK inhibitor (Fig S4E). The S425A substitution had no effect on this shift, but the S574A mutation completely abolished the formation of the acidic shift species (Fig 4D). The effect of HCV on the S294 mutant was more complex and infection resulted in loss of the single dominant species and its replacement with multiple more acidic forms. To confirm that S574 is phosphorylated by JNK, we overexpressed a constitutively active form of JNK1 in cells transfected with either wild-type or S574A FOXO3. Fig. 4E shows that like HCV, JNK1 stimulates FHRE-luciferase activity of wild-type, but not S574A FOXO3. Fig. 4F also shows that JNK1 also generated a novel FOXO3 peak with identical pI to that produced by HCV. Finally, we used LC MS to analyze FOXO3 from cells infected with HCV. A peptide-ion corresponding to the residues 570–606 was observed with phosphorylation on S574 (Fig. S5). These results demonstrate that S574 is a previously unrecognized site that is necessary for HCV to cause the JNK-dependent alteration in protein pI, nuclear localization, and transcriptional activity.
HCV-Ethanol effects on FOXO3 are mediated by decreased methylation
Arginine methylation has been shown to regulate the stability and nuclear localization of FOXO1(17) and since ethanol is known to alter cellular methylation potential (18), we examined whether changes in methylation could be responsible for ethanol effects on FOXO3. We addressed this question using cIEF of immunoprecipitated FOXO3. Fig 5A shows that cytosolic FOXO3 from untreated cells was methylated but the novel ethanol induced cytosolic species at pI 5.66 was not. Functional consequences of FOXO3 methylation defects were tested using the methyl donor, betaine (19). Addition of betaine completely prevented the HCV/ethanol-induced inhibition of FHRE reporter activity (Fig. 5B) and decrease in FOXO3 target gene mRNA expression (Fig. 5C). Betaine also restored HCV-induced nuclear translocation of FOXO3 in the presence of ethanol and prevented the decrease in steady-state levels of SOD2 protein (Fig. 5D). Fig. 5E demonstrates that betaine also restored both of the HCV-induced nuclear species of FOXO3 (pI 5.85 and 6.62) that are decreased or eliminated by the HCV/ethanol combination. Betaine also prevented the acidic shift of cytosolic FOXO3 caused by ethanol (Fig 5F). Nearly identical results were obtained using SAM as a methyl donor instead of betaine (Fig. S6A–D).
Figure 5. HCV/Ethanol suppresses arginine methylation of FOXO3.
HCV-infected and uninfected Huh7.5 cells were treated with 50 mM ethanol for 48h where indicated. A. cIEF analysis of cytosolic FOXO3 species immunoprecipitated with anti-FOXO3 antibody (C terminal epitope) and detected using either FOXO3 specific or methyl-arginine specific antibody. Baseline levels of each trace were shifted as for Fig. 1. B. FHRE-luciferase reporter assays in control and HCV-infected Huh7.5 cells in the presence of ethanol and 2.5 mM betaine as indicated. C. mRNA levels for FOXO3, SOD2 and Bim determined by real-time RT-PCR. D. Western blot of FOXO3 and SOD2 protein levels in cytosolic and nuclear fractions from cells treated as above. Lower panel shows the densitometry quantification of nuclear FOXO3/actin and SOD2/actin signal ratios of three independent experiments. *p < 0.05, **p < 0.01, n≥3 in all experiments. E, F. cIEF analysis of nuclear (E) and cytosolic (F) FOXO3 species in control and HCV-infected Huh7.5 cells in the presence of ethanol and betaine as indicated. Arrows indicate the novel nuclear HCV-specific FOXO3 species. Baseline levels of each trace were shifted as for Fig. 1.
Properties of FOXO3 methylation resistant mutants
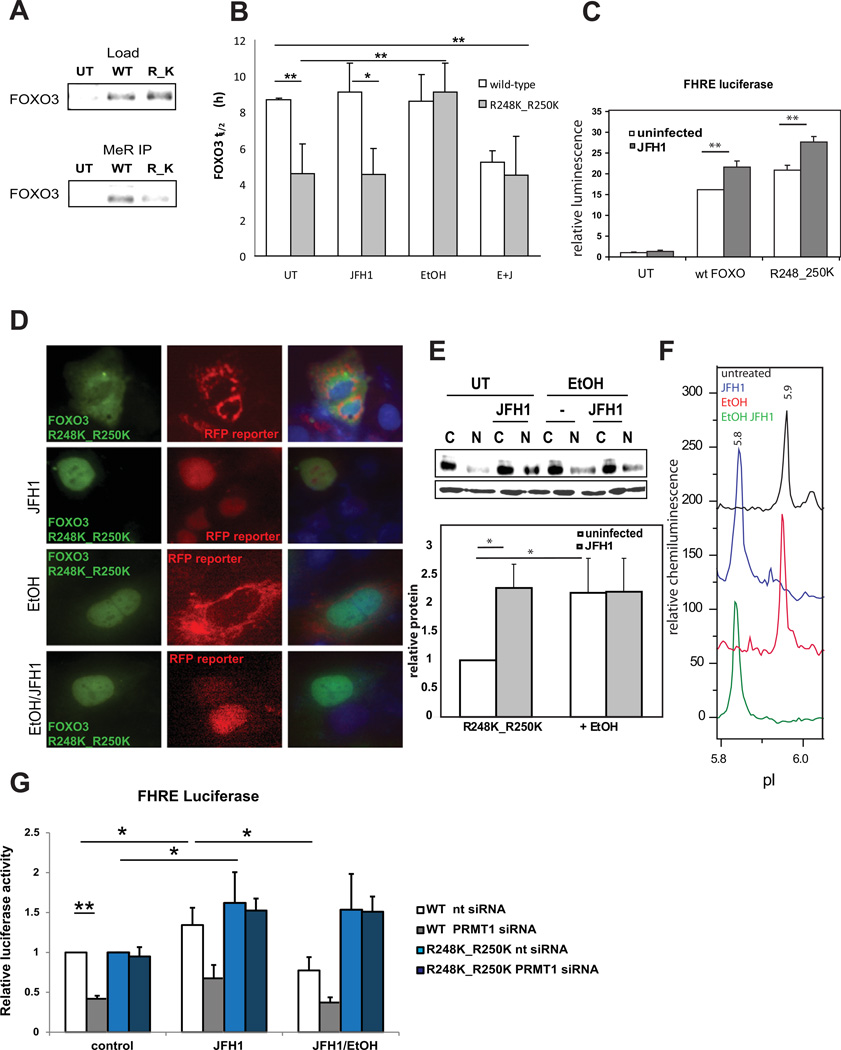
To further test the hypothesis that the combined HCV-ethanol effect results from FOXO3 demethylation, we generated a FOXO3 R248K_R250K double mutant missing the arginines methylated in FOXO1 (17). This mutant demonstrated less total protein methylation than the wt-FOXO3 (Fig. 6A) and its half-life was significantly reduced (Fig. 6B). The methylation-defective mutant was similar to the wild-type protein in that it was still transcriptionally active (Fig. 6C), it was primarily cytosolic in uninfected cells (Fig. 6D,E) and it translocated to the nucleus in response to HCV infection. The FOXO3 R248K_R250K double mutant differed from the wild-type protein in two respects. Its nuclear translocation was not inhibited by the combination of HCV and ethanol (Fig. 6D,E), and formation of the HCV-specific pI 5.85 peak was not inhibited by ethanol (Fig. 6F, compare to Fig 2A or S7B). Finally, we used siRNA to knock down expression of PRMT1, the methyltransferase responsible for the majority of arginine methylation (17). This knock down reduced nuclear levels and decreased transcriptional activity of wild-type but not mutant FOXO3 (Figs. 6G, S7C).
Figure 6. Mutation of R248 and R250 results in decreased methylation of FOXO3, nuclear exclusion and shorter half-life.
A. Western blot analysis of HA-FOXO3 WT and R248K_R250K (R_K) in whole cell extracts and after immunoprecipitation using methyl-arginine-specific or anti-HA (FOXO3) antibodies. B. Half-life time (h) of HA-FOXO3 WT and mutant in control and HCV-infected Huh7.5 cells in the presence of 50 mM ethanol for 48h. *p < 0.05, **p < 0.01. n≥3. Data are presented as mean ± SD. C. FHRE-luciferase reporter assays in control and HCV-infected Huh7.5 cells overexpressing WT and mutant FOXO3 R248K_R250K. **p < 0.01 n=3. D. Immunofluorescence images of Huh7.5 cells overexpressing HA-FOXO3 R248K_R250K two days after infection in the presence or absence of 50 mM ethanol for 48h as indicated. FOXO3 is in green (anti HA antibody), RFP reporter protein is in red (infected cells have a nuclear distribution of the RFP reporter protein) and DAPI is in blue. E. FOXO3 western blot analysis of nuclear and cytosolic fractions of cells treated as above. The diagram represents densitometry quantification of nuclear FOXO3/actin signal ratio of three independent experiments. F. cIEF analysis of nuclear FOXO3 R248K_R250K species detected with anti-HA antibody in control and HCV-infected Huh7.5 cells in the presence or absence of ethanol as indicated. Baseline levels of each trace were shifted as for Fig. 1. G. FHRE-luciferase reporter assays in control and HCV-infected Huh7.5 cells overexpressing WT and mutant FOXO3 R248K_R250K and co-expressing PRMT1 specific siRNA or scrambled siRNA as a negative control. Cells were treated with ethanol (50 mM for 48 h) where indicated. *p < 0.05, **p < 0.01 n=3.
FOXO3 modifications in human Hepatitis C infection
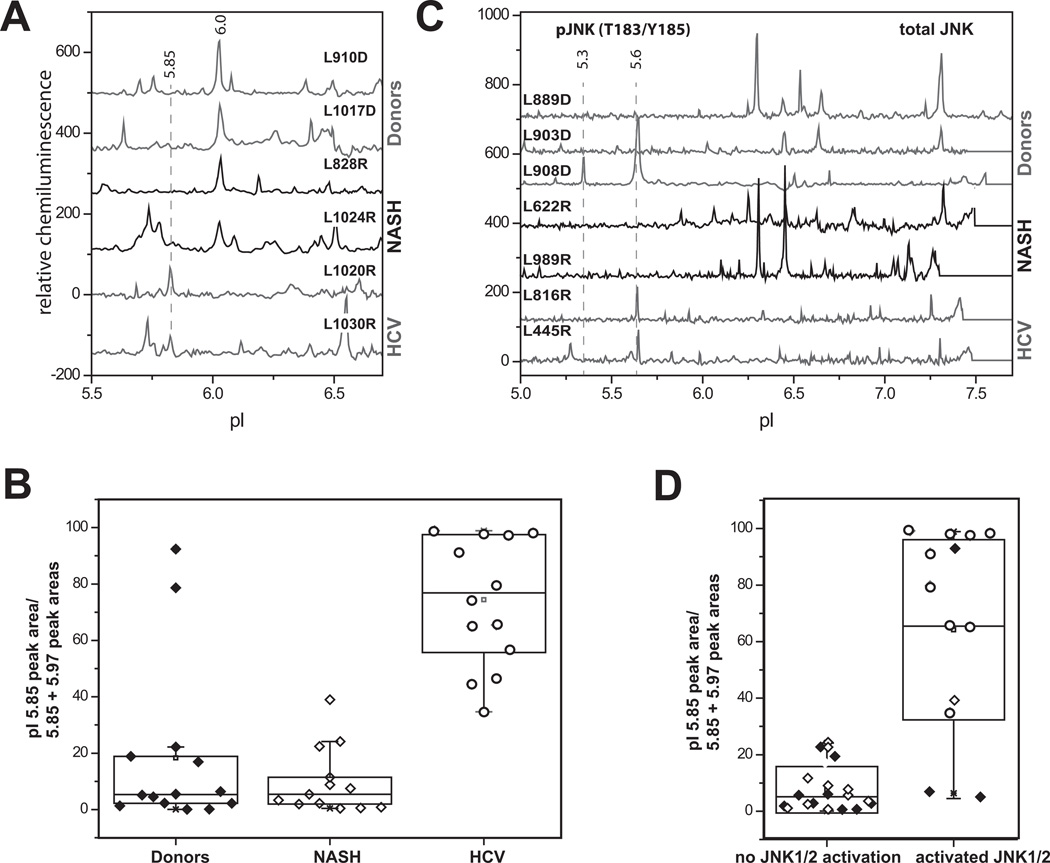
We compared the pattern of FOXO3 species present in human liver nuclear extracts from normal donor livers, HCV cirrhosis, and NASH cirrhosis. We observed the presence of a peak in the region of pI 6.0 for majority of the samples and a peak in the region of pI 5.85 in all HCV positive and some HCV negative samples (Fig 7A). We quantitated the presence of an HCV-like effect as the 5.85 peak area divided by the sum of the areas of the 5.85 + 6.0 peaks. As shown in Fig. 7B, the 5.85 peak accounted for approximately 60–90% of total in HCV vs. only approximately 10% in NASH or normal liver (P<0.01). To determine if this peak was related to JNK activation we simultaneously analyzed the samples for the phosphorylated JNK species (pI 5.3 and 5.6) in our samples by cIEF (Fig. 7C). Fig 7D shows that regardless of disease condition, there was a strong correlation between detection of pJNK species and the proportion of the pI 5.85 form of FOXO3.
Figure 7. FOXO3 modifications in human Hepatitis C infection.
A. Examples of cIEF analysis of FOXO3 nuclear species in donor livers, NASH and livers of HCV-positive patients. Baseline levels of each trace were shifted as for Fig. 1. B. Quantification of the pI 5.85 peak area divided by the total area under pI 5.85 and pI 5.97 peaks in three groups as indicated. C. Examples of cIEF analysis of JNK species detected by anti-total JNK ab in donor livers, NASH and livers of HCV-positive patients. pJNKT183/Y185 species are indicated by dashed lines. Baseline levels of each trace were shifted as for Fig. 1. D. pI 5.85/[pI 5.85 + pI 5.97] values as in B in the samples of patients with pJNK species present (right) or no pJNK species present (left). Donor samples are shown in black diamonds, NASH – in white diamonds and HCV-positive – in circles.
Discussion
FOXO transcription factors regulate hepatic growth and metabolism and respond to stress conditions (20–22). FOXO1 is activated by HCV infection contributing to insulin resistance (23, 24), and FOXO3 activity was noted to be increased in HCV infection where it modulated innate immune signaling (25). The molecular mechanisms of FOXO3 regulation by HCV and how alcohol modifies HCV’s FOXO3 effects in the liver has not been determined. In the present study we have shown that HCV and ethanol induce specific and interactive patterns of FOXO3 post-translational modifications that alter the function of this transcription factor. We separated FOXO3 species by cIEF and determined that HCV, alcohol, and the combination of the two each produced a unique pattern of FOXO3 PTMs that can explain the observed changes in transcriptional activity.
The cIEF method was particularly useful for this study since it is able to separate multiple forms of the same protein provided they have altered pIs. There are, however, some limitations to this technique. The IEF step is very sensitive to detergents and salts and this limits the types of homoginization buffers that can be used. The antibodies used for detection must be capable of detecting either native or urea-denatured forms of the protein and not all antibodies that work for western blot work for cIEF. Furthermore, the factors that promote efficient crosslinking of the protein to the capillaries are only poorly understood, and it is possible that some species are missed entirely. Finally, at the present time the technique is not preparative and direct analysis of the peaks, for example by MS, is not possible. Nonetheless, cIEF is well suited to detect many of the common protein PTMs involved in regulation of protein function.
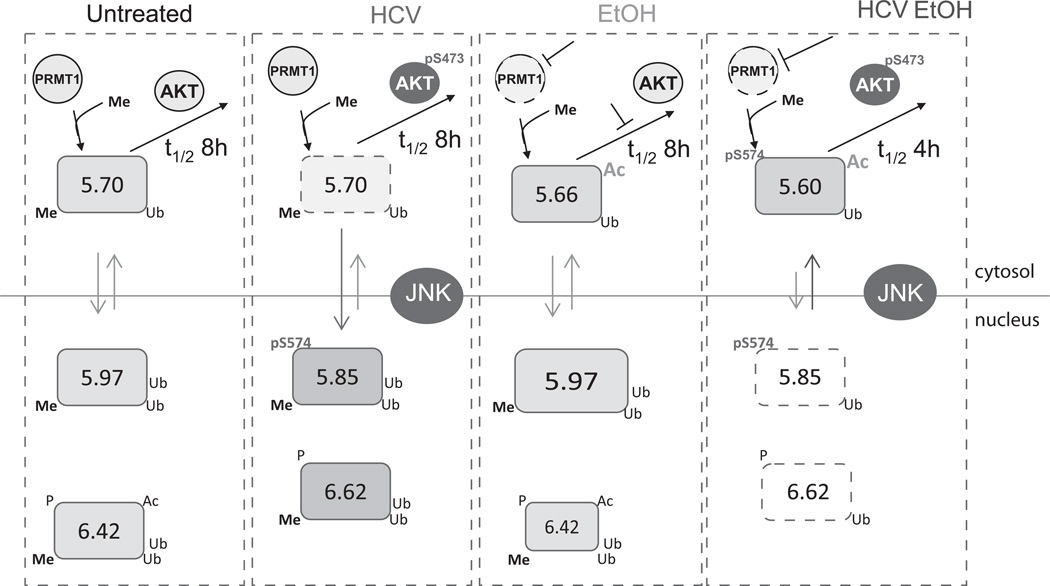
A schematic representation of the effects of HCV and ethanol on FOXO3 species is illustrated in Fig. 8. Under normal conditions, the majority of FOXO3 is in the nucleus forming several species that differ in the presence of PTMs including phosphorylation, acetylation and ubiquitination. In the cytosol, FOXO3 formed more acidic species. Under control conditions, all FOXO3 species were arginine methylated.
Figure 8. Proposed model of HCV and ethanol induced FOXO3 posttranslational modifications.
FOXO3 species are represented by boxes with indicated pI value and PTMs. HCV has an activating effect on FOXO3 that results from JNK-dependent phosphorylation at S574A. Alcohol treatment alone creates no novel species of FOXO3 in the nucleus but increases the proportion of pI 5.97 nuclear form. In the cytosol alcohol decreases arginine methylation by inhibiting PRMT1 activity. It maintains half-life by reducing the degradation rate of demethylated FOXO3. The combination of HCV and alcohol severely impairs the methylation of FOXO3. In combination with additional AKT activation this decreases the levels of FOXO3 in the nucleus and decreases the half-life of the protein.
The effect of HCV infection was to translocate FOXO3 to the nucleus and activate its transcriptional activity. In the nucleus, HCV-activated JNK phosphorylation of FOXO3 on S-574, and possibly other residues, and formed a novel FOXO3 species with a pI of 5.85. Serine-574 was absolutely necessary for HCV or JNK mediated FOXO3 activation and its phosphorylation resulted in the conversion of the pI 5.97 FOXO3 nuclear species to a more acidic one.
While JNK-induced S-574 phosphorylation was necessary, it was probably not sufficient for the all the HCV-induced changes. We were able to duplicate the formation of the 5.85 FOXO3 nuclear peak with active JNK1 expression but not the other HCV induced modifications that affect FOXO3 and produce a pI 6.62 peak. Furthermore, the addition of a single phosphate by itself should only shift FOXO3 pI by approximately 0.04 pH units. The generation of the 5.85 species with its acidic shift of 0.15 pH units thus likely involves either significant conformational changes or other modifications such as changes in ubiquitination. The importance of JNK, however, is consistent with the literature on other FOXOs as JNK plays a role in the oxidative stress dependent activation of FOXO4 by phosphorylation of T447 and T551 (26). Human liver specimens from HCV-infected patients similarly showed the presence of the HCV-specific 5.85 species of FOXO3. This species was not present in either normal livers or livers from patients with NASH. The HCV associated FOXO3 peak was also strongly associated with JNK activation in human livers demonstrating that the mechanisms involved in PTM formation are similar in vivo to those in our cellular model.
Ethanol treatment also increased FOXO3 transcriptional activity, but by different mechanisms. Ethanol did not cause FOXO3 to redistribute from cytosol to nucleus, and no novel nuclear species were observed after ethanol treatment. Ethanol did change the proportions of nuclear species, increasing the amount of a pI 5.97 deacetylated form and decreasing a 6.42 acetylated form. Acetylated FOXO3 has decreased DNA binding and activity (27, 28) and this shift away from the acetylated form may therefore contribute to an increase in transcriptional activity. Ethanol also generated a novel cytosolic FOXO3 species that was acetylated and demethylated. This resulted in an overall effect of increasing FOXO3 acetylation in the cytosol while decreasing it in the nucleus (Fig 8). Whether the stimulatory effects of ethanol result primarily from changes in FOXO3 acetylation remains to be determined.
The effects of HCV and ethanol in combination differed strikingly from those of each alone. The combination severely impaired arginine methylation of FOXO3, reduced its half-life and decreased both nuclear content and transcriptional activity. The role of demethylation in these effects is supported by the observations that a methylation deficient mutant of FOXO3 has a reduced half-life nearly identical to that produced by HCV and ethanol, and when demethylation was prevented by addition of betaine or SAM, the changes in FOXO3 no longer occurred. Thus ethanol mediated inhibition FOXO3 activity in the context of HCV infection is secondary to methylation changes. This is consistent with prior studies of FOXO1 where arginine methylation has been shown to prevent access of Akt to its phosphorylation site thus stabilizing the protein (17).
The combination of HCV-induced AKT activation (29) and ethanol-induced loss of FOXO3 methylation can explain most of the observed FOXO3 changes, but other effects probably occur as well. As shown in Fig. 6B, the methylation deficient FOXO3 mutant has a shorter half-life than wild-type FOXO3. It binds more strongly to the degradation promoting chaperone 14-3-3 (Fig. S7A), and its half-life is not further reduced by the HCV/ethanol combination. However, ethanol alone curiously increased the half-life of the mutant FOXO3 protein back to that seen for the wild-type protein (Fig. 6B). This could be a result of additional ethanol-induced modifications, such as increased acetylation that prevents degradation. A longer half-life of acetylated FOXO3 has been previously observed (30), and we observed acetylation of the demethylated cytosolic forms of FOXO3 (Fig. 2C). HCV infection was able to override this alcohol effect and shorten the half-life of demethylated FOXO3. The methylation status of the RRR motif at amino acids 248–250 is therefore likely a trigger regulating other FOXO3 PTMs during pathological states.
The reason that the HCV/ethanol combination suppresses protein arginine methylation is likely due to changes in PRMT1 activity. HCV has been shown to decrease activity of PRMT1 by activation of the phosphatase PP2A (31). We observed that the HCV/ethanol combination caused a 60% decrease in total protein arginine methylation (Fig. S8A). Ethanol slightly increased PP2A protein level, either with or without HCV infection (Fig. S8C), but did not change PRMT1 protein level (Fig. S8B), or SAM/SAH ratio (data not shown). Preliminary studies indicate that the additional effect of HCV/ethanol may be due to other modifications of the PRMT1 protein itself.
In conclusion, this study has examined the mechanisms by which HCV and alcohol modify the multifunctional transcription factor, FOXO3. FOXO3 is a tumor suppressor and is specifically involved in transcription of genes regulating cell cycle inhibition, apoptosis, and defense against oxidative stress. The use of a novel cIEF method has shown that HCV and ethanol have different molecular effects on FOXO3 when present in combination than they do when each is present alone. HCV effects primarily result from JNK activation and FOXO3 phosphorylation at a previously unrecognized site. Ethanol by itself affects FOXO3 primarily by changes in its acetylation. The combination results in FOXO3 arginine demethylation, and loss of FOXO3 nuclear localization and degradation. The ability of exogenous methyl donors to reverse the HCV/ethanol effects on FOXO3 could offer potential therapeutic utility. The methods developed in this study provide new insight into the molecular events modulating synergistic viral and environmental effects on the liver.
Supplementary Material
Acknowledgements
The human liver specimens used in this study were derived from samples provided by the University of Kansas Liver Center Tissue Bank. We thank Dr. Charles Rice for providing Huh7.5 cells and Dr. T. Wakita for providing JFH1 virus.
Grant support: This study was supported by grant AA012863 from the National Institute on Alcoholism and Alcohol Abuse, grant CA122499 from the National Cancer Institute and by a grant from the Hubert and Richard Hanlon Trust.
Abbreviations
- PTM
post-translational modification
- cIEF
capillary isoelectric focusing
- HCV
Hepatitis C virus
- JNK
c-Jun N-terminal kinase
- JFH1
Japanese Fulminant Hepatitis Virus-1
- ADH
alcohol dehydrogenase
- FHRE
forkhead box response element
- IP
immunoprecipitation
- LC-MS
liquid chromatography – mass spectroscopy
- SOD2
Mn-superoxide dismutase
- SAM
S-adenosylmethionine
- PRMT1
protein arginine methyl transferase 1
- NASH
nonalcoholic steatohepatitis
Footnotes
Disclosures: The authors have no financial, professional or personal conflicts of interest to disclose.
References
- 1.Safdar K, Schiff ER. Alcohol and hepatitis C. Semin Liver Dis. 2004;24:305–315. doi: 10.1055/s-2004-832942. [DOI] [PubMed] [Google Scholar]
- 2.Choi J. Oxidative stress, endogenous antioxidants, alcohol, and hepatitis C: pathogenic interactions and therapeutic considerations. Free Radic Biol Med. 2012;52:1135–1150. doi: 10.1016/j.freeradbiomed.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 3.Tikhanovich I, Cox J, Weinman S. FOXO Transcription Factors in Liver Function and Disease. J. Gastroenterol. Hepatol. 2013 doi: 10.1111/jgh.12021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calnan DR, Brunet A. The FoxO code. Oncogene. 2008;27:2276–2288. doi: 10.1038/onc.2008.21. [DOI] [PubMed] [Google Scholar]
- 5.Burgering BM. A brief introduction to FOXOlogy. Oncogene. 2008;27:2258–2262. doi: 10.1038/onc.2008.29. [DOI] [PubMed] [Google Scholar]
- 6.van der Horst A, Burgering BM. Stressing the role of FoxO proteins in lifespan and disease. Nat Rev Mol Cell Biol. 2007;8:440–450. doi: 10.1038/nrm2190. [DOI] [PubMed] [Google Scholar]
- 7.Zhao Y, Wang Y, Zhu WG. Applications of post-translational modifications of FoxO family proteins in biological functions. J Mol Cell Biol. 2011 doi: 10.1093/jmcb/mjr013. [DOI] [PubMed] [Google Scholar]
- 8.Tikhanovich I, Zhao J, Kuravi S, Weinman S. FOXO3 is a novel cytoprotection factor for alcoholic liver injury that is disrupted by HCV-specific posttranslational modifications. Hepatology. 2012;56:293A. [Google Scholar]
- 9.Ren JY, Zeng Z, Tumurbaatar B, Campbell RV, Chaturvedi G, Zhao J, Weinman SA. HCV induces FOXO3a translocation to cytosol during alcohol-associated inflammation and hepatocellular carcinoma. Hepatology. 2011;54:1312A–1312A. [Google Scholar]
- 10.Jones CT, Catanese MT, Law LM, Khetani SR, Syder AJ, Ploss A, Oh TS, et al. Real-time imaging of hepatitis C virus infection using a fluorescent cell-based reporter system. Nat Biotechnol. 2010;28:167–171. doi: 10.1038/nbt.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 12.Lei K, Nimnual A, Zong WX, Kennedy NJ, Flavell RA, Thompson CB, Bar-Sagi D, et al. The Bax subfamily of Bcl2-related proteins is essential for apoptotic signal transduction by c-Jun NH(2)-terminal kinase. Mol Cell Biol. 2002;22:4929–4942. doi: 10.1128/MCB.22.13.4929-4942.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kato T, Date T, Murayama A, Morikawa K, Akazawa D, Wakita T. Cell culture and infection system for hepatitis C virus. Nat Protoc. 2006;1:2334–2339. doi: 10.1038/nprot.2006.395. [DOI] [PubMed] [Google Scholar]
- 14.Fan AC, Deb-Basu D, Orban MW, Gotlib JR, Natkunam Y, O'Neill R, Padua RA, et al. Nanofluidic proteomic assay for serial analysis of oncoprotein activation in clinical specimens. Nat Med. 2009;15:566–571. doi: 10.1038/nm.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Neill RA, Bhamidipati A, Bi X, Deb-Basu D, Cahill L, Ferrante J, Gentalen E, et al. Isoelectric focusing technology quantifies protein signaling in 25 cells. Proc Natl Acad Sci U S A. 2006;103:16153–16158. doi: 10.1073/pnas.0607973103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qadri I, Iwahashi M, Capasso JM, Hopken MW, Flores S, Schaack J, Simon FR. Induced oxidative stress and activated expression of manganese superoxide dismutase during hepatitis C virus replication: role of JNK, p38 MAPK and AP-1. Biochem J. 2004;378:919–928. doi: 10.1042/BJ20031587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamagata K, Daitoku H, Takahashi Y, Namiki K, Hisatake K, Kako K, Mukai H, et al. Arginine methylation of FOXO transcription factors inhibits their phosphorylation by Akt. Mol Cell. 2008;32:221–231. doi: 10.1016/j.molcel.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 18.Kharbanda KK. Alcoholic liver disease and methionine metabolism. Semin Liver Dis. 2009;29:155–165. doi: 10.1055/s-0029-1214371. [DOI] [PubMed] [Google Scholar]
- 19.Fowler B. Homocysteine: overview of biochemistry, molecular biology, and role in disease processes. Semin Vasc Med. 2005;5:77–86. doi: 10.1055/s-2005-872394. [DOI] [PubMed] [Google Scholar]
- 20.Haeusler RA, Kaestner KH, Accili D. FoxOs function synergistically to promote glucose production. J Biol Chem. 2010;285:35245–35248. doi: 10.1074/jbc.C110.175851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leyendecker M, Korsten P, Reinehr R, Speckmann B, Schmoll D, Scherbaum WA, Bornstein SR, et al. Ceruloplasmin expression in rat liver cells is attenuated by insulin: role of FoxO transcription factors. Horm Metab Res. 2011;43:268–274. doi: 10.1055/s-0031-1271692. [DOI] [PubMed] [Google Scholar]
- 22.Tao R, Wei D, Gao H, Liu Y, DePinho RA, Dong XC. Hepatic FoxOs regulate lipid metabolism via modulation of expression of the nicotinamide phosphoribosyltransferase gene. J Biol Chem. 2011;286:14681–14690. doi: 10.1074/jbc.M110.201061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Banerjee A, Meyer K, Mazumdar B, Ray RB, Ray R. Hepatitis C virus differentially modulates activation of forkhead transcription factors and insulin-induced metabolic gene expression. J Virol. 2010;84:5936–5946. doi: 10.1128/JVI.02344-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deng L, Shoji I, Ogawa W, Kaneda S, Soga T, Jiang DP, Ide YH, et al. Hepatitis C Virus Infection Promotes Hepatic Gluconeogenesis through an NS5A-Mediated, FoxO1-Dependent Pathway. J Virol. 2011;85:8556–8568. doi: 10.1128/JVI.00146-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Honda M, Takehana K, Sakai A, Tagata Y, Shirasaki T, Nishitani S, Muramatsu T, et al. Malnutrition impairs interferon signaling through mTOR and FoxO pathways in patients with chronic hepatitis C. Gastroenterology. 2011;141:128–140. 140, e121–e122. doi: 10.1053/j.gastro.2011.03.051. [DOI] [PubMed] [Google Scholar]
- 26.Essers MA, Weijzen S, de Vries-Smits AM, Saarloos I, de Ruiter ND, Bos JL, Burgering BM. FOXO transcription factor activation by oxidative stress mediated by the small GTPase Ral and JNK. EMBO J. 2004;23:4802–4812. doi: 10.1038/sj.emboj.7600476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 28.Wang F, Nguyen M, Qin FX, Tong Q. SIRT2 deacetylates FOXO3a in response to oxidative stress and caloric restriction. Aging Cell. 2007;6:505–514. doi: 10.1111/j.1474-9726.2007.00304.x. [DOI] [PubMed] [Google Scholar]
- 29.Parvaiz F, Manzoor S, Tariq H, Javed F, Fatima K, Qadri I. Hepatitis C virus infection: molecular pathways to insulin resistance. Virol J. 2011;8:474. doi: 10.1186/1743-422X-8-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang F, Chan CH, Chen K, Guan X, Lin HK, Tong Q. Deacetylation of FOXO3 by SIRT1 or SIRT2 leads to Skp2-mediated FOXO3 ubiquitination and degradation. Oncogene. 2012;31:1546–1557. doi: 10.1038/onc.2011.347. [DOI] [PubMed] [Google Scholar]
- 31.Duong FH, Christen V, Berke JM, Penna SH, Moradpour D, Heim MH. Upregulation of protein phosphatase 2Ac by hepatitis C virus modulates NS3 helicase activity through inhibition of protein arginine methyltransferase 1. J Virol. 2005;79:15342–15350. doi: 10.1128/JVI.79.24.15342-15350.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.