Abstract
Objectives
The role of social media in biomedical knowledge mining, including clinical, medical and healthcare informatics, prescription drug abuse epidemiology and drug pharmacology, has become increasingly significant in recent years. Social media offers opportunities for people to share opinions and experiences freely in online communities, which may contribute information beyond the knowledge of domain professionals. This paper describes the development of a novel Semantic Web platform called PREDOSE (PREscription Drug abuse Online Surveillance and Epidemiology), which is designed to facilitate the epidemiologic study of prescription (and related) drug abuse practices using social media. PREDOSE uses web forum posts and domain knowledge, modeled in a manually created Drug Abuse Ontology (DAO) (pronounced dow), to facilitate the extraction of semantic information from User Generated Content (UGC). A combination of lexical, pattern-based and semantics-based techniques is used together with the domain knowledge to extract fine-grained semantic information from UGC. In a previous study, PREDOSE was used to obtain the datasets from which new knowledge in drug abuse research was derived. Here, we report on various platform enhancements, including an updated DAO, new components for relationship and triple extraction, and tools for content analysis, trend detection and emerging patterns exploration, which enhance the capabilities of the PREDOSE platform. Given these enhancements, PREDOSE is now more equipped to impact drug abuse research by alleviating traditional labor-intensive content analysis tasks.
Methods
Using custom web crawlers that scrape UGC from publicly available web forums, PREDOSE first automates the collection of web-based social media content for subsequent semantic annotation. The annotation scheme is modeled in the DAO, and includes domain specific knowledge such as prescription (and related) drugs, methods of preparation, side effects, routes of administration, etc. The DAO is also used to help recognize three types of data, namely: 1) entities, 2) relationships and 3) triples. PREDOSE then uses a combination of lexical and semantic-based techniques to extract entities and relationships from the scraped content, and a top-down approach for triple extraction that uses patterns expressed in the DAO. In addition, PREDOSE uses publicly available lexicons to identify initial sentiment expressions in text, and then a probabilistic optimization algorithm (from related research) to extract the final sentiment expressions. Together, these techniques enable the capture of fine-grained semantic information from UGC, and querying, search, trend analysis and overall content analysis of social media related to prescription drug abuse. Moreover, extracted data are also made available to domain experts for the creation of training and test sets for use in evaluation and refinements in information extraction techniques.
Results
A recent evaluation of the information extraction techniques applied in the PREDOSE platform indicates 85% precision and 72% recall in entity identification, on a manually created gold standard dataset. In another study, PREDOSE achieved 36% precision in relationship identification and 33% precision in triple extraction, through manual evaluation by domain experts. Given the complexity of the relationship and triple extraction tasks and the abstruse nature of social media texts, we interpret these as favorable initial results. Extracted semantic information is currently in use in an online discovery support system, by prescription drug abuse researchers at the Center for Interventions, Treatment and Addictions Research (CITAR) at Wright State University.
Conclusion
A comprehensive platform for entity, relationship, triple and sentiment extraction from such abstruse texts has never been developed for drug abuse research. PREDOSE has already demonstrated the importance of mining social media by providing data from which new findings in drug abuse research were uncovered. Given the recent platform enhancements, including the refined DAO, components for relationship and triple extraction, and tools for content, trend and emerging pattern analysis, it is expected that PREDOSE will play a significant role in advancing drug abuse epidemiology in future.
Keywords: Entity Identification, Relationship Extraction, Triple Extraction, Sentiment Extraction, Semantic Web, Drug Abuse Ontology, Prescription Drug Abuse, Epidemiology
1. Introduction
Over the past decade, the illicit use of pharmaceutical opioids has emerged as a major public health problem in the United States [18], [24], [40], [50], [51], [52], [61], [72]. In 2010, the lifetime, non-medical use of pharmaceutical opioids was reported by nearly 14% of the US population aged 12 years and older [58]. Increases in illicit pharmaceutical opioid use resulted in escalating accidental overdose death rates [51], expanded pathways to heroin addiction [18] and significant increases in opioid use disorders [41]. The development of effective prevention and policy measures requires timely and reliable information on new and emerging drug trends. Although existing epidemiological data systems, such as the National Survey on Drug Use and Health (NSDUH), the Community Epidemiology Work Group (CEWG), and the Drug Abuse Warning Network (DAWN), provide critically important data about drug abuse trends, they lag in time. Additional methods are needed to enhance early identification of emerging trends and expand access to hard-to-reach populations.
The World Wide Web has been identified as a very useful, but underutilized tool for reaching hidden populations of illicit drug users, as well as detecting patterns and changes in drug abuse trends of pharmaceutical opioids and other drugs [32], [44], [49], [60]. Many Web 2.0 empowered social platforms, including web forums, provide a venue for illicit drug users to freely share their experiences, post questions, comments, and opinions about different drugs. Such UGC can be used as a very rich source of unsolicited self-disclosures of drug use behaviors [8], [9], [10], [11], [33].
Web-based data have been used in drug abuse research to examine a variety of topics, including tampering methods for selected pharmaceutical products [25] recreational use of Salvia divinorum [5], [39] and to explore user endorsement of the illicit use of selected pharmaceutical opioids [15]. The utility of web-based data has been recognized by the post-marketing surveillance systems designed to monitor illicit use of pharmaceutical drugs [14], [21]. However, most prior web-based studies of pharmaceutical opioids and other drugs relied on web-based surveys and manual searches to obtain relevant information. Overall, very few attempts have been made to (semi) automatically analyze UGC web forums or other social media sites to explore drug use phenomena. More importantly, prior studies of web-based data were limited in their scope, lacked methodological rigor and relied on manual coding to perform content analysis [7], [9], [15], [21], [25]. Manual coding in qualitative research as well as in content analysis studies of media communications requires that researchers perform the following steps: 1) “read” a text document; 2) “break” it into manageable segments; 3) attach labels (or codes) to those segments, and 4) subsequently interpret and analyze them [43], [63]. Manual coding is therefore a labor intensive and time consuming process, and its wider application to web-based data is impractical [12]. Further, existing content analysis programs are not able to automatically capture complex semantic relationships between concepts, nor process information expressed in colloquial language often found in web forums [2], [6]. Hence, the development of automated methods to aid the collection, extraction and coding of UGC related to illicit drug use will result in harnessing the full potential of the Web for drug abuse epidemiology research. To accomplish these tasks, we created a Semantic Web application called PREDOSE, which is capable of extracting semantic information from social media and providing levels of content analysis that support more effective epidemiologic description of prescription drug abuse. The overall goal of PREDOSE is therefore to help researchers gain knowledge of the attitudes and behaviors of drug abusers related to the illicit use of pharmaceutical opioids such as buprenorphine. The specific contributions of this research are as follows:
Use of a novel semantic web platform to extract semantic information from social media for drug abuse epidemiology
Evaluation of techniques for extracting entities, relationships and triples implemented in the PREDOSE platform
In-use components for data analysis and interpretation, available in the PREDOSE platform that support content exploration, trend detection and emerging pattern analysis
The rest of the paper is organized as follows: the overall Approach is discussed in Section 2, including Data Collection (2.1), Automatic Coding (2.2) and Data Analysis and Interpretation (2.3). Section 2.2 discusses the approach to Automatic Coding, including the Drug Abuse Ontology (DAO) (2.2.1), Entity Identification (2.2.2), Relationship Extraction (2.2.3), Triple Extraction (2.3.4) and Sentiment Identification (2.3.5). Section 3 discusses components developed for Data Analysis and Interpretation, including the Content Explorer (3.1), Trend Explorer (3.2) and Emerging Pattern Explorer (3.3). Finally, Section 4 discusses the implications of PREDOSE to prescription drug abuse research in general. Section 5 presents the Conclusion, followed by Acknowledgement in Section 6. Supplementary materials are presented in the Appendices.
2. Approach
The PREDOSE platform consists of three distinct modules: 1) a Data Collection module; 2) an Automatic Coding module and 3) a Data Analysis and Interpretation module (all shown in Figure 1).
Figure1.
The PREDOSE Platform
2.1. Data Collection Module
The data collection module enables collection of User Generated Content (UGC) for analysis in PREDOSE from three online web forums. Each site was selected specifically because it: 1) allows free discussion of psychoactive drug use; 2) contains information on illicit pharmaceutical drug use and 3) is publicly accessible on the web (please note that in compliance with Institutional Review Board (IRB) guidelines at Wright State University, the names of the selected web sites have not been disclosed in this manuscript).
The selected sites were scraped using a suite of custom web crawlers1 (Figure 1, Stage 1 Step 2) developed to fetch data from the web and store it locally in an informal text database (Figure 1, Stage 1 Step 4). The crawlers collect a variety of data, including actual post content and a variety of metadata, including unique post identifiers (post_id), unique user identifiers (user_id) and web site source identifiers (source_id). Table 1 shows that approximately 1 million posts (1,066,502) were collected from 35,974 users – as of the most recent crawl (May 2012). SiteX has the greatest number of posts and users, as well as the most expansive date range (starting October 1999). SiteY and SiteZ are more recent (from October 2004 and February 2004 respectively). SiteZ appears to be the most interactive, averaging approximately 73 posts per user, among 5,449 users.
Table 1.
Crawler Statistics
| SiteX | SiteY | SiteZ | TOTAL | |
|---|---|---|---|---|
| Number of posts | 446,070 | 220,698 | 399,734 | 1,066,502 |
| Number of users | 27,420 | 3,105 | 5,449 | 35,974 |
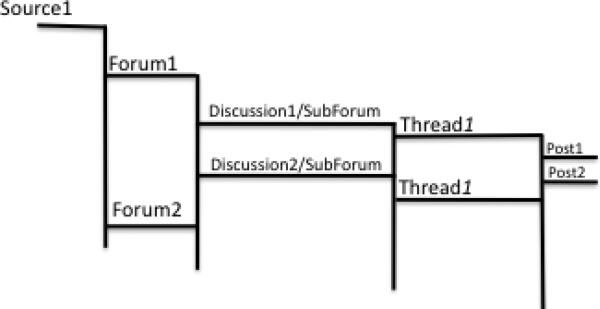
Table 2 depicts the schema of the post table in the informal text database. In this database, raw post content is stored using the content field. Each post also contains a timestamp (post_datetime), to facilitate longitudinal data display and analysis. The post type (post_type) field indicates whether a post is an original or reply to an earlier post. Knowledge of such distinctions is important in distinguishing users that initiate discussions on topics that subsequently become trends. The post thread id (post_thread_id) is also maintained in the database to capture provenance, i.e., link posts to their parent thread. Furthermore, each thread belongs to a unique discussion or sub-forum, which in turn belongs to a unique forum on a given site. Figure 2 shows this hierarchy of post membership.
Table 2.
Post Table Schema
| Field | Description |
|---|---|
| post_id | unique auto generated post id |
| post_type | whether original or reply post |
| post_thread_id | thread id of thread in which post appears |
| user_id | website user id of user submitting post |
| content | actual text content of post |
| source_id | id of website in which post appears |
| post_datetime | timestamp of post |
| post_url | relative url of post |
| post_seq_num | post position in thread |
| entity_annotated | whether all entities verified by domain experts |
| drugs | list of all drugs spotted in post |
| sentiment_annotated | whether all sentiments verified by domain experts |
| trip_patterns_annotated | whether all triple patterns verified by domain experts |
| tag | whether training set post, test set post or neither |
Figure 2.
Generic Web Forum Structure
In the early phase of the study, geographic locations were also collected whenever provided by users. Unfortunately, among the 35,974 users, a little less than half (~15,000) provided no location information in their profiles. Among those who did provide locations, many were abstruse in nature, including a proliferation of slang and colloquial references to known geographic locations. Many included cryptic expressions such as “Frisco,” “earth,” “The YAY” and “HELLAWARE,” which may be difficult to disambiguate programmatically. Hence, we postponed the issue of location disambiguation from such texts for future research. Notably, a lack of geographic and demographic information in social media platforms such as web forums is a known limitation of web-based studies. Unlike social media sources such as Twitter, location disambiguation problems are compounded by privacy restrictions against public access to user profiles. Such restrictions preclude IP address-based resolution of geographic locations. In this work therefore, we focus on extraction and analysis of fine-grained information from web posts, for which much of the metadata is stored using Lucene2 for fast retrieval. In the next section we discuss approaches to semantic information extraction from web forum posts.
2.2. Automatic Qualitative Coding Module
In PREDOSE semantic information extraction is achieved using the following components: 1) the Drug Abuse Ontology (DAO) creation; 2) an Entity Identification component; 3) a Relationship Extraction component; 4) a Triple Extraction component and 5) techniques for Sentiment Extraction. To illustrate the role of each component, consider the following motivating scenario.
Motivating Scenario
Consider a manually annotated snippet from a post, written by a user, which contains multiple drug-mentions (in boldface), sentiment clues (underlined), relationships (enclosed with double squared braces) and relations - which indicate a relationship between drug-mentions (enclosed with angle braces), dosages (enclosed with double curly braces) and time intervals (enclosed with double parentheses):
So I was recently discharged from a detox facility . . . I was in detox for ((5 days)), and they [[gave me]] a tapering Suboxone dose ((every day)) . . . I was sent home with {{5 × 2 mg}} Suboxones. I also got a bunch of phenobarbital. <I took all {{180 mg}} and it didn't do EXPLETIVE except [[make me]] a walking zombie for {{2 days}}>. I waited ((24 hours)) after my last {{2 mg}} dose of Suboxone and <[[tried injecting]] {{4 mg}} of the bupe>. <It [[gave me]] a bad headache>, ((for hours)), and I almost vomited. I could [[feel]] the bupeworking but overall the experience sucked.
The CITAR research team inspected this snippet and agreed that the code: “Suboxone used by injection, negative experience” could be derived from the two relations: “<[[tried injecting]] {{4 mg}} of the bupe>” and “<It [[gave me a]] bad headache>.” In these two relations, the two surface forms ‘bupe,’ and ‘bad headache’ are concepts or entities, where the surface form ‘bupe’ is a reference to the brand name Suboxone (for the standard drug Buprenorphine), and ‘bad headache’ is a reference to the concept Cephalalgia. (Please note that reference ‘bad headache’ is also a sentiment expression as well as a known Side Effect). Additionally, the surface form ‘injecting’ is both a method of administration (Intravenous) and an entity modifier – that modifies the concept ‘bupe’ (i.e., Suboxone) – that refers to the combined concept 'Suboxone Injection'. Furthermore, note that the phrase [[gave me]] is a causality association in reference to the relationship CAUSES. The snippet also contains additional references to known constructs such as time intervals and dosage.
It is an unreasonable expectation that a human coder should manually identify the relevant annotations in this snippet, or using texts on a web scale. Although human involvement in the coding process cannot be completely replaced, manual coding is impractical for scenarios involving massive amounts of heterogeneous data. Therefore, to effectively analyze web based social media, a coding module must be able to (semi) automatically identify, extract and annotate documents with entities, relationships, triples and domain specific constructs, needed for content analysis. Moreover, the provision of tools that facilitate interpretation of the annotated data is important in alleviating the overall manual effort required to perform such analysis.
For instance, in the above snippet, the qualitative code “Suboxone used by injection, negative experience” can be interpreted as semantically equivalent to the two relations: <tried injecting 4 mg of the bupe> and <It gave me a bad headache> as well as the more concise relation which states that “<Suboxone_Injection CAUSES bad headache>.” Such relations are called triples. A triple is an association between two concepts, in the form subject ⇒ predicate ⇒ object, in which the predicate conveys an association between the subject and object. In the assertion which states that <Suboxone_Injection CAUSES bad headache> the predicate (or relationship) CAUSES expresses the causality association between the subject, ‘Suboxone Injection’ and the object, ‘bad headache’. The concept ‘bad headache’ is of type Cephalalgia, where the type of the concept is a generalization of a class of things to which it belongs. Such type and class definitions are maintained in the DAO schema.
The information extraction layer of the PREDOSE platform (Figure 3, also shown in Figure 1, Stage 2) therefore utilizes the DAO to extract entities, relationships and triples. For sentiments, an adaptation of the technique originally developed in [19] by Chen et. al (a co-author in this work) is made for web forum texts. The Automatic Qualitative Coding Module of PREDOSE therefore consists of the following five components: 1) the Drug Abuse Ontology; 2) an entity identification component; 3) a relationship identification component 4) a triple extraction component and 5) a sentiment extraction component. We begin with the Drug Abuse Ontology (DAO) in the next section.
Figure 3.
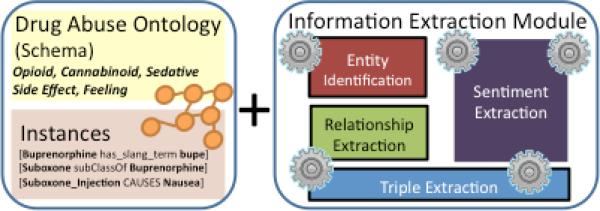
Information Extraction Layer
2.2.1. Drug Abuse Ontology
The Drug Abuse Ontology (DAO) is a formal representation of concepts and relationships between them for the prescription drug abuse domain. Figure 3 (left) shows that the DAO consists of a: 1) schema and 2) an instance base of assertions. The schema contains classes, type definitions and relationships permissible between them, defined manually during the ontology creation process. Such definitions serve as the basis for the annotation scheme in PREDOSE, and include both hierarchical and associative relationships. A hierarchical relationship (property) between two classes, expresses membership between them. For example, Drugs occur in different Classes such as Cannabinoids, Buprenorphine, Opioids, Sedatives and Stimulants. Hence, a Cannabinoid is a type of Drug, and the isA hierarchical property expresses the relationship between them. Associative relationships are non-hierarchical relationships between classes. For instance, the statement that “Suboxone_Injection CAUSES bad headache” expresses a causality association between a Drug and a Side Effect and does not imply membership of one concept to the other.
The current DAO contains 43 classes and 20 properties. Although it is a relatively shallow representation, the DAO is very precise, given its creation by domain experts (CITAR researchers). The DAO is also enriched with links to concepts in external ontologies, through a very careful manually supervised process. Among the 43 DAO classes, 11 classes have been mapped to URIs in DrugBank3, Freebase4, DBpedia5 and the Cyc6 ontologies, using the sameAs property. The DAO also contains 16 object properties, which are properties that associate class instances amongst each another. For example, in the relation <‘bupe’ has_side_effect ‘headache’>, the object property has_side_effect links the surface form ‘bupe’ of the class Suboxone to the surface form ‘headache’ of the class, Side Effect (i.e., the schema level assertion that Suboxone has_side_effect Side Effect). Other object properties modeled in the DAO include, has_preparation_method, induces_feeling, is_treated_with etc.
In addition to object properties, the DAO models two data type properties (has_value and has_slang_term). Data type properties associate a concept with a literal value. For example, the property has_slang_term associates a drug with a slang term (e.g. <Buprenorphine has_slang_term bupe>). The ability to accurately and formally represent slang term-to-drug associations is important for two reasons. The first is that accurate slang term mappings will positively impact search and retrieval of relevant documents when performing content analysis. Search methods devoid of such domain knowledge are at a disadvantage, and may be unable to retrieve relevant documents containing only slang term mentions but no standard references to a given drug. This problem is reflected in a gold standard dataset of 601 web forum posts, in which a ratio of slang term mentions to the standard drug label for Buprenorphine of 33:1 was observed. For the drug Loperamide, the ratio was 24:1 in this gold standard. An inability to capture such associations, not only affects search, but also the quality and granularity of semantic information that can be extracted from the corpus. To facilitate these mappings, we utilized various slang to drug mapping sources, including numerous online dictionaries and resources, such as DrugSlang7, NIDA, NDCP8 and www.erowid.com. Altogether, 307 slang terms were collected, curated and added to the DAO. A total of 193 concepts contained slang term mappings.
The DAO is the result of a joint manual effort, between domain experts at CITAR and computer scientists at Kno.e.sis. It was created and edited using the Protégé Ontology Editor9. A web-based version is available for browsing (http://knoesis-hpco.cs.wright.edu/drug-abuse-ontology/) using the Ontology Browser software. The raw ontology data file is also available online (http://knoesishpco.cs.wright.edu/predose/ontologies/DAO.owl). In the next section, we discuss the use of the DAO for entity identification.
2.2.2. Entity Identification
The DAO is used together with a lexical entity spotter (hereafter spotter) for entity identification. The choice of the DAO and spotter combination is appealing, since the DAO is the first ontology for prescription drug abuse and hence, is expected to be most reliable in capturing formal representations of slang references to standard drugs. Note that a survey on techniques for named entity identification (NEI) for a variety of texts is covered in [71]. Specifically, the spotter is a customized implementation of a data structured called a Prefix Trie [17], developed at Kno.e.sis for text annotation. A Prefix Trie is a special modification of a tree data structure, in which child nodes have the same prefix as their parent. The trie enables fast lookup and annotation of strings of arbitrary length. Hence, the spotter relies on term mappings, which have been modeled in the DAO for entity spotting (e.g. ‘bupe’ to Buprenorphine). The spotter is therefore merely a means to an end. Prior to spotting, candidate entities from the DAO, as well as web post content are lowercased and normalized by removing punctuation. The snippet below shows an anecdotal output of annotations obtained using the spotter, from a segment of text in our running example.
![]()
The snippet shows that the DAO is currently capable of mapping units (e.g. mg -> MILLIGRAM) and slang terms (e.g. bupe – Buprenorphine), based on a lexical lookup in the ontology.
The spotter is therefore most effective when one-to-one mappings exist between slang terms and ontological concepts. It is most ineffective when multiple candidate matches exists (e.g. hydro for both Marijuana and Hydrocodone). Entity disambiguation is required to determine the correct match in such scenarios. While entity disambiguation is crucial, it was observed that the pervasive use of slang, and the domain specific nature of such texts, mere entity identification without disambiguation still yields useful annotations. In particular, in an evaluation on a gold standard dataset of 601 web forum posts, created by researchers at CITAR, the spotter had 84.9% precision and 72.5% recall in spotting entities. More specifically, out of a total of 3639 manually observed annotations from the 601 posts, 2640 annotations were correctly predicted by the Prefix Trie Spotter as true positives, while 683 of the spotter-predicted slang term to drug mappings were incorrect (false positives). For recall, only 999 out of 3639 annotations were not predicated or missed (false negatives) altogether. These are favorable early results for entity identification on such texts.
To facilitate the creation of the gold standard, both the Prefix Trie Spotter and the DAO, were used to initially spot the entire corpus. The spotted corpus was then presented to the domain experts as a starting point for the annotation process. The Content Explorer (discussed in Section 3.1) in the PREDOSE platform provides utilities that facilitate annotation, including features for adding, accepting, editing and deleting pre-annotations. The DAO therefore used both for entity identification as well as the creation of training and test sets within PREDOSE. The aforementioned gold standard dataset is available online for review in the Content Explorer of the PREDOSE system (http://knoesis-hpco.cs.wright.edu/predose/username: guest, password: guest).
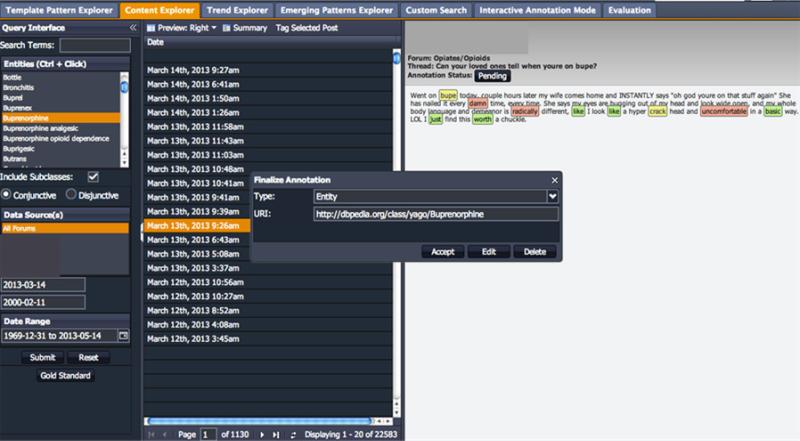
While the performance of our entity identification techniques is reasonable, the inability to disambiguate entities is a noted limitation. This limitation has far reaching consequences when attempting to obtain semantic information for data analysis and interpretation. Furthermore, such a large number of false negatives (999) are an indication that the DAO is likely incomplete. Fortunately, the Content Explorer provides functionality for on-the-spot entity adjudication. Annotations can be added, edited or deleted individually in the PREDOSE interface (shown in Figure 4). The context-aware entity disambiguation technique implemented by Mendes [42] in DBpediaSpotlight10 is under consideration for application to the PREDOSE texts in future. In the next section, we discuss techniques for relationship extraction, which is the precursor to triple extraction.
Figure 4.
Content Explorer
2.2.3. Relationship Extraction
The second type of semantic information extracted in the PREDOSE platform is relationships. Consider the relation “<It [[gave me]] a bad headache>” in which the relationship-conveying phrase (hereafter relphrase) “gave me” conveys the causality relationship (CAUSES) between the anaphora ‘It’ (for Buprenorphine) and the concept Cephalalgia (for headache). Our attitude towards relationship extraction in PREDOSE is to first map less complex relphrases to standard relationships and then systematically extend to more complex relphrase-to-relationship mappings. To achieve this we compiled a set of target relationships (called a target lexicon), using the 54 unique predicates from the Unified Medical Language System – UMLS (specifically, UMLS 2012AB), instead of the 20 DAO relationships. The use of UMLS predicates is a practical step since UMLS relations are formally specified and considered as representative of the more salient predicates across web forum posts. While the 20 relationships in the DAO are precise, they may be limited in coverage across this domain (as the DAO is an evolving ontology).
Given the target lexicon, the next step is to map relphrases that occur in text to appropriate relationships from this target. To achieve this, we selected a technique that is independent of the target lexicon, but rather focuses on synonymy among relationship-conveying expressions. For example, the relationship CAUSES is used interchangeably with ‘get,’ ‘have,’ ‘induce,’ ‘stimulate,’ ‘make.’ Hence, to capture the relationship context of a phrase in social media texts we adopt a two-step approach. We first use WordNet11 to obtain a word set that represents the lexical context of a UMLS predicate (from the target lexicon) based on WordNet SynSets. Then we obtain a word set that represents the lexical context of a relphrase also based on WordNet Synsets. Given these two word sets containing the contexts of UMLS predicates and relphrases, we then rely on the synonymy of the associated Synsets to uncover the semantic similarity between the UMLS predicate and the relphrase.
For example, consider a WordNet Synset for the WordNetKey (or headword) ‘cause’, which contains the following synonyms, cause → [cause, get, have, induce, stimulate, make]. Consider also another SynSet for the headword ‘give’ (which is lexically similar to ‘gave’) that contains the following synonyms: give → [throw, give, have, hold, make]. A mapping between the relphrase ‘gave me’ and ‘cause’ is possible based on the overlapping synonyms in the two SynSets sim(gave me, cause) → [have, make]. To obtain the relationship mapping for a relphrase to the appropriate UMLS predicate, we then compare all the mapped SynSets for the relphrase, including unigrams, bigrams and trigrams of the parsed relphrase (e.g. the SynSets for ‘gave,’ ‘me’ and ‘gave me’) with the mapped SynSets for all 54 UMLS predicates. Given the SynSet tokens for the relphrase as the query, the highest ranked Lucene document (i.e., the SynSets of a given UMLS predicate) is then taken as the predicate for the relphrase.
Hence, by using this largely lexical-based approach to relationship extraction, based on the premise that synonyms in WordNet Synsets, can provide enough semantics to capture overlapping similarity with unstructured relationship mentions in text (i.e. relphrases), a method for relationship identification in unstructured social media texts has been developed. We evaluated this lexical approach to relationship identification using three evaluators with domain expertise on prescription drug abuse. Each evaluator manually scored our predicated target predicates, using a dataset of 2736 unique relphrases, randomly selected from web forum posts, using a web interface in PREDOSE. The reader is encouraged to review the evaluation online: http://knoesis-hpco.cs.wright.edu/predose/, username: guest, password: guest – select the Evaluation Tab. Our evaluation showed that for a total of 183 scored relphrases, 66 were correct and 117 were incorrect, yielding 36% precision. This suggests that if the PREDOSE platform is used to search and visualize web forum content based on the extracted relationships, 1 in 3 such relationships will be correct on average. These results are significant, given the manual effort required for qualitative coding, and the semantic equivalence, between triples and codes.
One caveat is that this technique is only reliable when mapping relphrases and target predicates that share synonymy, homonymy and homophony, but not antonymy and negation. In cases where the same headword exists for a relphrase SynSet expansion and the UMLS SynSet expansion, a negation may lead to a false positive. For example, the relphrase “doesnt contain any” and the UMLS predicate “contains” bothshare the lemma – contain. Tokenizing the relphrase (doesnt contain any) into unigrams, bigrams and trigram, and then aggregating the SynSets for each token, our algorithm will incorrectly predict the target predicate as the UMLS predicate “contains” for the relphrase “doesnt contain any.” This is because of a high Lucene score between the SynSets for the relationship ‘contain’ in the relphrase and the SynSets for the UMLS predicate ‘contains.’ A similar scenario arises when a relphrase and the target predicate contain prepositions that imply opposite semantics. For example, the relphrase “leads to” and the target predicate “result of” both contain the prepositions “of” and “to,” which have different semantics. However, the base words “leads” and “results” are similar and will match based on our algorithm. In future we intend to explore a rule-based approach for relphrase mapping, which will prune target predicates based on permissible relationships in the ontology. We also project that the use of background knowledge will also resolve the relationship directionality issue, not addressed here. In the next section we discuss triple extraction from such social media texts.
2.2.4. Triple Extraction
Triple extraction from unstructured social media is a complex task. Social media content is known for poor grammar, slang and abbreviation and is fraught with misspelling and colloquialisms. Hence, it is difficult to predict the structure of social media texts. The linguistic structure of scientific literature, new articles and Wikipedia however, can be predicted and hence, machine learning, NLP and even semantic web techniques can be applied in these situations for triple extraction to some degree. Therefore, traditional approaches for triple extraction approaches are not directly applicable to web forum texts, and in PREDOSE, we made some adaptations to be able to extract triples from unstructured text. For completeness, various techniques [56], [66], [67], [68] that utilize rule-based and pattern-based approaches for triple extraction from (semi) structured text (an overview of triple extraction techniques is covered in Appendix A). Top-down approaches to triple extraction rely on domain knowledge to model triple patterns that occur in text. However, this approach has several limitations. The first is that associations between concepts do not always occur as infix, but may appear in prefix or postfix form. Another issue is that many associations that exist in text may not be covered in the ontology. Bottom-up approaches to triple extraction [66], [68] rely on the corpus itself to reveal patterns that could point to concept associations, such as <* ⇒ predicate ⇒ *>, <* ⇒ predicate ⇒ object> and <subject ⇒ * ⇒ *> also not present in the background knowledgebase. In PREDOSE, we used a top-down approach to triple extraction we call triple pattern extraction, due to the challenge of recognizing prefix and postfix relationships. The DAO schema is used to extract triple patterns from text within some variable window of concept pair mentions. Hence, a triple pattern is an association between two concepts in which the relationship appears as infix; that is a subject, followed by a predicate, followed by an object.
We first extracted triple pattern candidates from the corpus of 1 million web forum posts, by retrieving all concept pairs in the DAO within some variable window. Relationships were then extracted in a postprocessing step based on the method in Section 2.2.3. To obtain the triple patterns from text, we used a rule-based declarative information extraction framework, called SystemT [20], [38], which is designed for information extraction across heterogeneous texts. SystemT provides an Annotation Query Language (AQL)12 for pattern specification, which in previous research we [16] demonstrated has applicability to Smoker Semantic Type (SST) identification in doctor's progress notes. Details on AQL are covered in Appendix B.
We evaluated our top-down triple pattern extraction technique using the same dataset used for relphase evaluation discussed in Section 2.2.3. To create a triple, we replaced the relphrase with the mapped UMLS predicate in the candidate triple, and presented the modified triple for evaluation to the user. A screenshot of the evaluation interface is shown in Appendix C. Our results showed that across 196 evaluated triple patterns, 66 were correct and 131 were incorrect. This is a 33% precision in triple extraction. While seemingly low, this is a significant intermediate milestone in the broader context of automatic semantic information extraction from such unstructured text. The identification of triple patterns has significance in a variety of tasks including search, content analysis, trend detection and emerging pattern exploration. We discuss the role of entities, relationships and triples in this context, in Section 3. Next we briefly discuss the role of sentiment analysis in the PREDOSE platform.
2.2.5 Sentiment Identification
We use the state-of-the-art sentiment extraction technique implemented by Chen et al. [19] to identify sentiment clues in web-forum posts in the PREDOSE platform. This technique is capable of: 1) recognizing sentiment bearing expressions including both formal and slang words/phrases; and 2) assessing the topic-dependent polarity of each sentiment clue associated with a specific entity. For example, referring to the running example, the negative expression “didn't do EXPLETIVE” is in reference to the drug phenobarbital, while the expressions “bad headache,” “almost vomited,” and “sucked” are associated with the drug “bupe” (buprenorphine). In addition, sentiment clues can be very diverse, occurring in various forms, ranging from single words to multi-word phrases. The techniques used for sentiment identification are designed to identify on-target, multiple-word, standard as well as colloquial sentiment expressions. Using this approach, Chen et al., automatically identified sentiment clues in PREDOSE, given the target drug names identified by the Prefix Trie spotter, on the entire corpus. All posts have therefore been annotated with sentiment expressions and rendered in the Content Explorer (discussed in Section 3.1) in which they serve as visual clues during context exploration. In the Trend Explorer (discussed in Section 3.2) sentiment expressions are used to explore shifts in attitude towards drug-specific discussion. In the next section, we discuss the use of the DAO, extracted entities, relationships and triples, along with sentiments for data analysis and interpretation in the PREDOSE platform.
3. Data Analysis and Interpretation Module
An integral part of the PREDOSE platform is to provide domain scientists with tools to analyze extracted semantic information from the social media sources. Such information includes both raw statistical data as well as extracted content. We therefore provide three distinct components in PREDOSE for such data analysis and interpretation. These are: 1) a content analysis component; 2) a trend detection component and 3) an emerging patterns explorer. We discuss each in turn in the following subsections, and also allow access to them online (http://knoesis-hpco.cs.wright.edu/predose/, username: guest, password: guest).
3.1 Content Explorer
The Content Explorer (as shown in Figure 4) supports search and browsing of posts based on entities, triples and sentiment annotations. Entity annotations and sentiment expressions serve as visual clues for content exploration in the Content Explorer, while sentiment clues also enable the detection and tracking of peoples’ attitudes towards specific drugs.
To perform a search using the Content Explorer, a user must first select a drug name by label from the Query Interface (Figure 4, left – Buprenorphine selected) and then select a Data Source(s) (i.e., SiteX, SiteY and/or SiteZ – use Ctrl + Click to select multiple) from which data will be displayed. The user must then select a “Date Range” (leave blank – recommend). Then the user must click Submit.
The query processor expands the selected drug with lexical variants and slang terms using the DAO and retrieves relevant posts containing such terms. Posts are annotated in an offline-processing step a priori using the spotter. Color-coded annotations are then provided in the content of each post as shown more closely in Figure 5. The legend for the various types of annotations displayed in the Content Explorer is shown in Table 3.
Figure 5.

PREDOSE Post Annotations
Table 3.
Legend for PREDOSE Annotations
| Annotation Type | Color Codes | |
|---|---|---|
| Non-Confirmed (a priori) | Confirmed | |
| Entity | Yellow (Pale) | Yellow (Dark) |
| Predicate (Relationship) | Blue (Pale) | Blue (Dark) |
| Sentiment Clue (Positive) | Green (Pale) | Green (Dark) |
| Sentiment Clue (Negative) | Red (Pale) | Red (Dark) |
| Triple Pattern | Black (White font) | White (Black font) |
When the user clicks an annotation the system provides options to edit, delete or accept the selection (Figure 4, Finalize Annotation window for the annotation ‘bupe’). These capabilities exist primarily to support building training and test sets for extraction and evaluation of our information extraction techniques. For completeness, annotations not present in the DAO can be added in the web interface. To do this, users can select any “non-highlighted” text in a post and choose the appropriate concept from a list of external sources (including drugbank, dbpedia, freebase and cyc) populated on the fly. Selected external concepts will then be indexed and become candidates for addition to the DAO – but not without manual adjudication.
The Content Explorer therefore facilitates data analysis by enabling filtering and browsing of post content based on the presence of semantic annotations in them. These annotations include entities, positive and negative sentiment expressions and triples. Next, we discuss the Trend Explorer for longitudinal trend visualization and exploration in the PREDOSE platform.
3.2 Trend Explorer
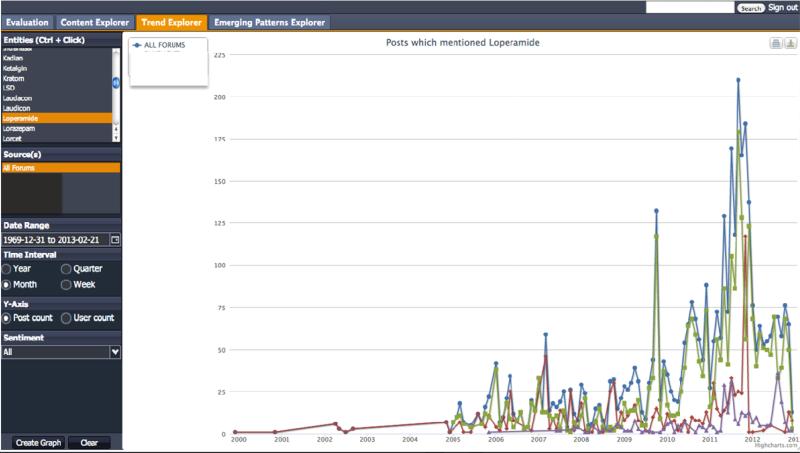
To deal with the information overload likely to occur when unconstrained temporal queries are executed in the Content Explorer, we implemented the Trend Explorer to perform statistical data charting. The longitudinal view of a contrived example is shown in Figure 6. To generate and visualize such data, a user must first select a drug name(s) (Loperamide selected), then a “Source(s),” then a “Date Range” and “Time Interval” (display by Month selected). Longitudinal data is available for display either by user or post frequency (Post frequency selected).
Figure 6.
Trend Explorer for Loperamide posts
An option also exists to filter by posts by mentions of sentiment expression generally associated with a negative, neutral or positive context. No sentiment polarity has been selected (default). Selecting a data point on the graph will execute a query for posts based on the selected point. Such posts will be loaded in the Context Explorer, and the system will switch tabs (to the Content Explorer) after the query executes. In this way, the user can analyze trends in the Trend Explorer, then select posts for content analysis in the Content Explorer, based on fluctuations in chatter in the data graphs in the Trend Explorer. Additionally, each line in the Trend Explorer graph represents data from a different web forum (recall three web forums were selected for this study – sites X, Y and Z), while the line in blue (ALL FORUMS) is the aggregation of data from each web forum. The user may enable/disable a line graph from appearing in the interface graph by clicking a line color in the legend (immediately beneath the Trend Explorer Tab Title). Users may also print or download the visualized graph(s) by clicking the printer icon or download icon in the upper right of the Trend Explorer. We assert that the main benefit of the Trend Explorer is that it can help identify significant changes in drug abuse trends, through longitudinal data visualization on dimensions of entities and sentiment expressions. In the next section, we discuss the Emerging Pattern Explorer, which augments the Trend Explorer for further data analysis.
3.3 Emerging Patterns Explorer
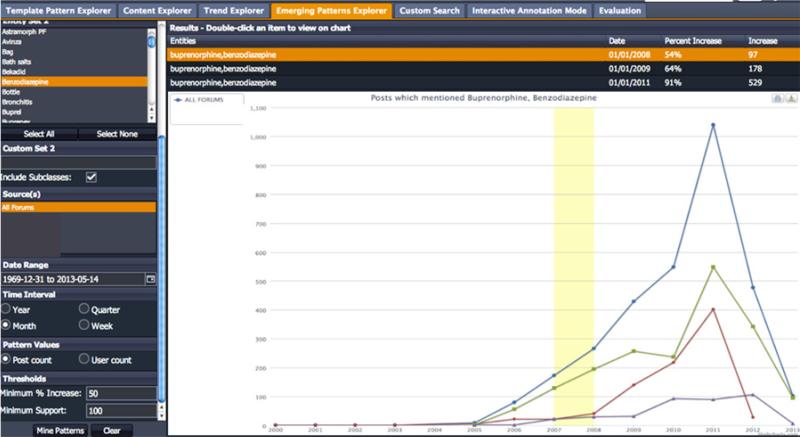
The Emerging Patterns Explorer (shown in Figure 7) complements the Trend Explorer by allowing further exploration of drug-drug discussions through co-occurrence relationships between drugs in posts. To access this feature, the user must first select a concept of interest from “Entity Set 1” (upper part of left scrollbar, too large to be shown – Buprenorphine selected) then select a second from “Entity Set 2” (shown in left panel – Benzodiazepine selected). Note that here also, multiple values from each set can be selected using Ctrl + Click action). Each entity set can also be expanded with custom search terms using the sets (“Custom Set 1” and “Custom Set 2”) in the query panel – Figure 7 (left). Similarly, the user must also select a “Date Range” and “Time Interval.” Then the user must select a threshold for a “Minimum % increase” in post counts, between two consecutive points data points (for example, – 200%). Finally, the user must indicate the Minimum Support, or minimum number of posts, which must contain the co-occurring pairs.
Figure 7.
Emerging Patterns Explorer
Figure 7 shows a snapshot of the variations in co-occurrence for Buprenorphine-Benzodiazepine with a 54% increase in co-occurrence mentions beginning with 173 posts in 2007 which increased to 266 posts in 2008 (yellow region in the graph for All Forums in blue). Such regions in the graph may reflect emerging patterns, which can be detected by browsing the co-occurrence relationships in the Content Explorer.
To summarize, we presented in this section various tools for data analysis and interpretation. In previous research, [27], [28], [73], based only on a functional Prefix Trie Spotter for entity identification, the PREDOSE platform was used to provide a dataset from which new information [27] about extra-medical use of loperamide for self-medication was discovered. Researchers coded and analyzed numerous mentions of extra-medical use of loperamide (e.g., Imodium® A-D), which is a piperidine derivative that acts on opioid receptors in the intestine and has been approved by the U.S. Food and Drug Administration for the control of diarrhea. Because of its general inability to cross the blood-brain barrier, loperamide is considered to have no abuse potential and is therefore available without a prescription [31].
Given the various enhancements, including an updated DAO, methods for entity, relationship and triple extraction, together with sentiment extraction and various tools for context exploration, trend detection and emerging patterns exploration, the PREDOSE platform is now more capable of supporting the needs of prescription drug abuse research. PREDOSE is therefore rapidly becoming an effective platform for automatic information filtering that can alleviate many laborious tasks currently undertaken manually in qualitative research.
4. Discussion
PREDOSE is a Semantic Web platform that aims to automate the extraction of semantic information from web-forum content to facilitate drug abuse research using social media. The system provided a dataset from which new knowledge regarding self-treatment of opioid withdrawal symptoms was obtained, after further application of traditional qualitative research methods on data initially extracted using PREDOSE. The platform is capable of extracting entities, relationships, triples and sentiments from such unstructured texts. A live online version of our system is available for guest access (http://knoesis-hpco.cs.wright.edu/predose/ - username: guest, password: guest) and the DAO is available for browsing online (http://knoesishpco.cs.wright.edu/drug-abuse-ontology, http://knoesis-hpco.cs.wright.edu/predose/ontologies/DAO.owl). Various components, including a Context Explorer, Trend Explorer and Emerging Patterns Explorer, have been developed and are in current use by prescription drug abuse researchers at CITAR to mine biomedical (more specifically prescription drug abuse) knowledge from social media. Two additional modules, namely: 1) a Template Pattern Explorer – to support exploration of ontology concepts and various other types of co-occurring data (http://knoesis-hpco.cs.wright.edu/knowledge-aware-search/) and 2) a component for Custom Search – that allows flexible search for entities and non-ontology keywords, are under development in the PREDOSE platform.
Given initial steps for entity identification, relationship extraction and triple extraction, there is considerable optimism about the implications of our platform for analysis of social media texts for prescription drug abuse research. In the early phase of this research, we investigated the use of Twitter data (i.e., tweets) as an alternative type of social media for content analysis. However, such efforts were met with limited success, given that only entities were used for the study. Given the various enhancements we consider twitter an appealing source of information for future research. Furthermore, we have given consideration for fact extraction from scientific literature to corroborate facts extracted from social media in the PREDOSE platform, to study the evolution of knowledge between online communities and the scientific community.
Nonetheless, in spite of promising intermediate milestones, the research objectives of the PREDOSE project are still subject to several limitations inherent to many web-based studies. The first issue is a lack of demographic indicators due to privacy restrictions from web-forum administrators. The unavailability of geographic information, whether not provided by social media participants or difficult to decipher due to the lack of an adopted structure by web forum users, limits the scope and granularity of longitudinal data analysis PREDOSE can perform. Moreover, opponents debate whether web-data can be taken as a representative sample of community behavior and practices in general. We believe that the loperamide-Withdrawal finding attests to the credibility of web-based sources as a viable source of community behavior and are optimistic about the use of social media for prescription drug abuse epidemiology. What is of considerable significance is that the PREDOSE platform is among the first to leverage semantic web resources in combination with lexical, pattern-based and rule-based approaches to facilitate drug abuse epidemiology using social media.
5. Conclusion
In this study, we described the development of PREDOSE, a platform to facilitate web-based research on the illicit use of pharmaceutical opioids, through the combination of lexical, pattern-based, rule-based and semantic web technologies using social media. PREDOSE is currently in use by prescription drug abuse researchers at CITAR, and has played a role in discovering new scientific finding in this area. Techniques for information extraction implemented in PREDOSE, including entity, relationship, triple and sentiment extraction offer to automate (or minimally supplement) the laborious manual coding currently undertaken by qualitative researchers in the domain. Additionally, various components for analyzing and interpreting extracted data have been implemented, including a Content Explorer, Trend Explorer and an Emerging Patterns Explorer. In future we plan to implement a module for entity disambiguation and enhance the existing modules for relationship, triple extraction and sentiment extraction. PREDOSE is the first such comprehensive platform for such biomedical (prescription drug abuse) knowledge mining using social media.
We present a Semantic Web platform for drug abuse research using Social Media.
Social Media texts are an important resource in identifying new epidemiological trends.
Extraction of appropriate semantic information is crucial to epidemiological research.
Acknowledgement
This research was conducted in close collaboration between Computer Scientists at the Ohio Center of Excellence in Knowledge-enabled Computing (Kno.e.sis; http://knoesis.org) who developed the computational components, while the epidemiologists at the Center for Interventions, Treatment and Addictions Research (CITAR; http://www.med.wright.edu/citar) specified application requirements and performed various evaluations. This project is sponsored by the National Institutes of Health (NIH) Grant No. R21 DA030571-01A1 (Amit P. Sheth and Raminta Daniulaityte, PIs). We wish to thank Pablo N. Mendes, Revathy Krishnamurthy, Nishita Jaykumar, Swapnil Soni, Mary Oberer, Michael Cooney, Matthan Sink and Sujan Perera for their involvement in the development of the PREDOSE project.
APPENDIX A
Relationship and Triple Extraction: A Brief Review
While our results for entity identification are reasonable, they do not guarantee that extracting relationships and triples from unstructured text will also have good outcomes. Relationship and triple extraction from such texts must also leverage context as well. In this appendix, we give a short review of existing techniques for relationship and triple extraction in the literature, to put our task in perspective.
Relationship Extraction
Ramakrishnan et. al. [54], developed a technique for relationship extraction that leverages background knowledge together with a rule-based method for identifying modified and complex entities (utilizing the MEdical Subject Headings - MESH), and then the UMLS to identify relationships between such entities using Medline abstracts. The approach first uses a linguistics-based parser to identify candidate entities in text that map to MESH terms, which in turn map to UMLS concepts. The relationships permissible between concepts are gleaned from the UMLS Semantic Network. While this technique performs reasonably well for structured scientific literature, its performance on the unstructured social media texts may not be equally successful. Consider the relation “<xanax [[made me have]] a headache>,” in which the relphrase “made me have” should map to the UMLS predicate “CAUSES.” Ramakrishnan's technique could possibly map the subject and object to the appropriate concepts in the UMLS. However, the relationship mapping for the relphrase “made me have” to the relationship CAUSES, which can be found by our technique may be difficult to map using this technique. The UMLS has no predicate containing any of the relationship tokens (made, me or have) expressed here.
In spite of this, the benefits of the UMLS for relationship mapping cannot be underestimated. We expect that the UMLS property hierarchy can be used to expand UMLS predicates with WordNet Synsets to capture a broader context for relationships. We intend to investigate the impact of this expansion in future work.
Triple Extraction
Ramakrishnan et. al. [55] implemented a method for detecting complex entities and subsequently triples, by leveraging the linguistic features of token sequences in text to help define dependency rules. The idea is that linguistic dependencies in natural language, abstract relationships between simple and compound entities. In [56] Ramakrishnan et. al. enhanced this triple extraction technique by applying point-wise mutual information to identify token sequences most likely to be complex entities based on co-occurrence statistics in the corpus. Given the focus on relationship extraction by Ramakrishnan et al. in [54] the research in [56] focused more on detecting complex entities for improvements in triple extraction.
Rindflesch et. al. [57] also implemented a method that combines the linguistics structure of token sequences with domain knowledge for extracting triples containing hypernymic associations expressed in taxonomic relations. Rindflesch utilizes an underspecified parser to resolve POS tags, and the UMLS Specialist Lexicon through MetaMap for entity identification. Similar to Ramakrishnan, Rindflesch uses the UMLS Semantic Network to find permissible relationships based on concept associations in the network.
Thomas et. al., in [68] describe a pattern-based method for domain model creation that includes a component for triple (fact) extraction using Wikipedia articles. In this approach a domain description is first created, by identifying initial ontology-based predicate patterns (e.g. X CAUSES Y) also in the corpus. Thomas uses these patterns to learn concepts that co-occur with predicates in text. Relationships are then modeled as a set of pattern generalizations, using evidence-based (vector) patterns, which can then be used to learn more facts from the corpus. An unsupervised algorithm based on statistical pertinence was developed for disambiguating overlapping relationship types (belonging to multiple patterns). Finally, a probabilistic multi-class classifier predicts the relationship for the candidate triple (or fact).
In [66] Suchanek et. al. presented a semi-supervised pattern-based technique for relation (triple) extraction from structured text. Similar to Thomas, the idea assumes that relationships (linguistic linkages) can be used to find patterns in text, which in turn can be used to find more linkages. In the first phase, patterns for a given linkage are assigned placeholders for subject and object, and designated as positive patterns. Given flexible bridges of linkages, new linkages (which are counter examples) called negative patterns were obtained by applying the newly learned positive patterns. Potentially valid relations containing matching subject-object pairs are those in a test set, for which among all positive patterns for a linkage were obtained from the corpus.
The above techniques have been developed for triple extraction from structured texts and may not work well in unstructured texts. For triple extraction, we intuitively rely on background knowledge for entity identification, similar to the methods by Ramakrishnan and Rindflesch. Specifically, we use a top-down approach for triple extraction using the DAO. And since DAO concepts do not map to UMLS concepts, we use WordNet13 to help with relationship extraction. The more generic bottom-up approaches by Thomas and Suchanek, which extract patterns from the corpus using the instances in text are under consideration for future work. Since PREDOSE is one of the first research projects to implement techniques for triple extraction from such social media texts, we approach this task incrementally.
APPENDIX B
AQL: Annotation Query Language
The Annotation Query Language (or AQL) is a rule-based SQL-like query language for pattern extraction from text. AQL is based on two primitives: 1) a dictionary primitive and 2) the view primitive. A dictionary is a collection of terms belonging to some arbitrary category. For example, the following listing shows that the Cannabinoid dictionary (Cannibinoid_dict) can be expressed a mapping between the ontology class Cannabinoid and a list of surface forms associated with this class, including slang terms from the DAO.
create dictionary Cannabinoid_dict as ( ‘Cannabinoids’, ‘Cannabis’, ‘K2’, ‘Spice’, ‘Synthetic cannabinoids’, ‘bud’, ‘dank’, ‘hash’, ‘herb’, ‘mids’, ‘mj’, ‘pot’, ‘schwag’, ‘weed’ );
The second AQL primitive is the view primitive. A view is a rule-based abstraction of a dictionary, much like SQL views are an abstraction over SQL query commands. The listing below shows the CannabinoidView.
create view CannabinoidView as extract dictionaries ‘Cannabinoid_dict’ on D.text as cannabinoids from Document D;
AQL also supports the composition of complex views built from existing dictionaries and views through nesting. For example, the CannabinoidDrugTriplePattern view is a view in that will annotate any span of text in which a Cannibaniod reference preceded a Drug reference (extracted using the DrugView), for up to four tokens.
create view CannabinoidDrugTriplePattern as select CombineSpans(LeftContextTok(D.match, 4), D.match) as match from DrugView D where ContainsDict(‘Cannabinoid_dict’, LeftContextTok(D.match, 4));
The CannabinoidDrugTriplePattern will therefore match patterns in the corpus in which users mention phrases such as “pot is the gateway drug,” “cannabis is comparable to cocaine,” “pot is made into hash” and “K2 is another name for spice.” The subject of each phrase after triple extraction is a Cannabinoid -“pot,” “cannibis” and “K2.” The Drug mentions in the object are the self-referential label “drug,” and “cocaine,” “hash” (for Hashish) and “spice” for Synthetic_Cannibis.
APPENDIX C
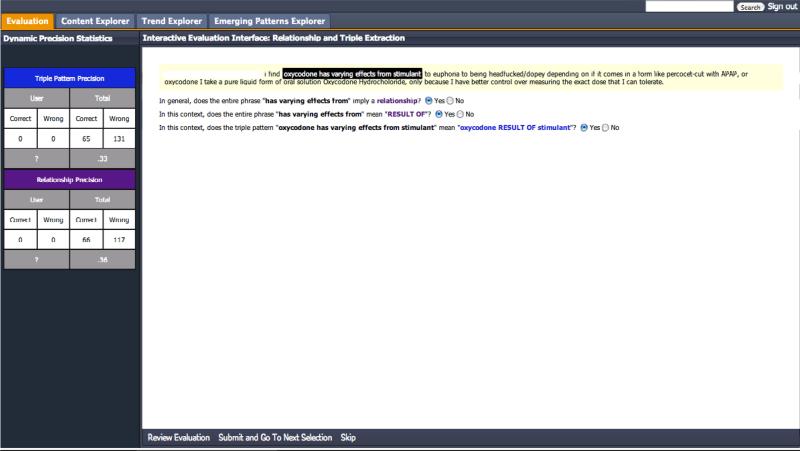
We automatically created 23 AQL query patterns based on permissible DAO schema triple patterns. Queries were then executed on the corpus (of 1 million posts) using Apache UIMA (Unstructured Information Management Architecture) [70]. To evaluate candidates, an Evaluation component was added to the PREDOSE web application. Hence, evaluation results can be reviewed online: http://knoesis-hpco.cs.wright.edu/predose/ (username: guest, password: guest). Figure 8 shows a snapshot from the live web application in which the triple pattern for evaluation is highlighted in black background and white font.
Figure 8.
Relationship and Triple Extraction Evaluation Interface
To review the evaluation, select the “Evaluation” Tab if not selected by default. Each evaluator was asked to determine first, whether the triple pattern in the text contains two ontology concepts and some perceivable relationship between them. That is:
Question 1: In general, does this triple pattern imply a valid relationship?
If no, then such triple patterns should be ignored and the evaluator should move on to the next item for evaluation by clicking “Skip” (on the bottom bar in Figure 8). Note that by clicking “Submit and Go To Next” the displayed triple pattern, will not be scored, since “No” would have been selected by default for Question 1. If the triple pattern does contain a valid relationship, then the evaluator should select “Yes” for this question. The second question in the evaluation is as follows:
Question 2: In this context, does the relphrase imply the suggested relationship by our system?
If yes, then our technique has correctly identified the (UMLS) relationship from the relphrase using the technique discussed in Section 2.2.3. Finally, the evaluator was asked:
Question 3: In this context, does the triple pattern imply the updated triple pattern based on the relationship suggested by our technique?
If yes, then our top-down pattern-based triple extraction technique, together with the mapped (UMLS) predicate for the relphrase was mapped correctly to a triple.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Web crawlers are software agents that recursively traverse hyperlinks of an arbitrary website, beginning at the homepage until all hyperlinks (on that site) have been traversed.
DrugBank (Drug and Drug target Database) - http://www.drugbank.ca/
Freebase Knowledge base - http://www.freebase.com/
DBpedia Knowledge base - http://dbpedia.org/About
Cyc ontology - http://en.wikipedia.org/wiki/Cyc
We collected 4,692 slang term mappings from the DrugSlang Dictionary available online at http://www.noslang.com/drugs/dictionary/
We collected 4,409 mappings from the Office of National Drug Control Policy (NDCP) available online at http://www.whitehousedrugpolicy.gov/streetterms/
Protégé Ontology Editor - http://protege.stanford.edu/
DBPediaSpotlight - http://dbpedia-spotlight.github.com/demo/
WordNet Lexical Database - http://wordnet.princeton.edu/
Annotation Query Language Reference Guide - http://bit.ly/WOHiJC
WordNet Lexical Database - http://wordnet.princeton.edu/
References
- 1.Aaronson A, Lang F. An overview of metamap: historical perspective and recent advances. Journal of the American Informatics Association. 2010:229–236. doi: 10.1136/jamia.2009.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alpers G, Winzelberg A, Classen C, Roberts H, Dev P, Koopman C. Evaluation of computerized text analysis in an internet breast cancer support group. Computers in Human Behavior. 2005;21:361–376. [Google Scholar]
- 3.ASI-MV Connect 2012 Retrieved 2012, from ASI-MV Connect: www.asi-mvconnect.com.
- 4.Bach N, Badaskar S. A Review of relation extraction. 2007 Retrieved from http://www.cs.cmu.edu/~nbach/papers/A-survey-on-Relation-Extraction.pwd. [Google Scholar]
- 5.Baggott M, Erowid E, Erowid F, Galloway G, Mendelson J. Use pat- terns and self-reported effects of salvia divinorum: An internet-based survey. Drug Alcohol Depend. 2010;83(111):250–256. doi: 10.1016/j.drugalcdep.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 6.Bantum E, Owen J. Evaluating the validity of computerized content analysis programs for identification of emotional expression in cancer narratives. Psychol Assess. 2009;21:79–88. doi: 10.1037/a0014643. [DOI] [PubMed] [Google Scholar]
- 7.Boyer E, Wines J. Impact of internet pharmacy regulation on opioid analgesic availability. J Stud Alcohol Drugs. 2008;69:703–708. doi: 10.15288/jsad.2008.69.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyer E, Babu K, Macalino G. Self-treatment of opioid withdrawal with a dietary supplement, kratom. Am J Addict. 2007;16:352–356. doi: 10.1080/10550490701525368. [DOI] [PubMed] [Google Scholar]
- 9.Boyer E, Lapen P, Macalino G, Hibberd P. Disseminaion of psychoactive substance information by innovative drug users. Cyberpsychol Behav. 2007;10:1–6. doi: 10.1089/cpb.2006.9999. [DOI] [PubMed] [Google Scholar]
- 10.Boyer E, Shannon M, Hibberd P. The internet and psychoactive substance use among innovative drug users. Pediatrics. 2005;115:302–305. doi: 10.1542/peds.2004-1199. [DOI] [PubMed] [Google Scholar]
- 11.Boyer E, Shannon M, Hibberd P. Web sites with misinformation about illicit drugs. N Engl J Med. 2001;345:469–471. doi: 10.1056/NEJM200108093450619. [DOI] [PubMed] [Google Scholar]
- 12.Brent E. Artificial Intelligence and the Internet. Sage; 2008. in the sage handbook of online research methods. [Google Scholar]
- 13.Broekstra J, Fluit C, Kampman A, Van Harmelen F, Stuckenshmidt H, Bhogal R, et al. The drug ontology project for elsevier - an rdf architecture enabling thesaurus-driven data integration. WWW Workshop on Application Design, Development and Implementation Issues in the Semantic Web. 2004 [Google Scholar]
- 14.Butler S, Budman S, Licari A, Cassidy T, Lioy K, Dickenson J, et al. National addictions vigilance intervention and prevention program (navippro): a real-time, product-specific, public health surveillance system for monitoring prescription drug abuse. Pharmacoepidemiol Drug Saf. 2008:1142–1145. doi: 10.1002/pds.1659. [DOI] [PubMed] [Google Scholar]
- 15.Butler S, Venuti S, Benoit C, Beaulaurier R, Houle B, Katz N. Internet surveillance: content analysis and monitoring of product-specific internet prescription opioid abuse-related postings. Clin J Pain. 2007;23:619–628. doi: 10.1097/AJP.0b013e318125c5cf. [DOI] [PubMed] [Google Scholar]
- 16.Cameron D, Bhagwan V, Sheth AP. 1st International Workshop on the role of semantic web in literature-based discovery (SWLBD) IEEE; Philadelphia, PA, USA: 2012. Towards Comprehensive longitudinal healthcare data capture. pp. 241–247. [Google Scholar]
- 17.Cameron D, Mendes PN, Sheth AP, Chan V. Semantics-empowered Text exploration for knowledge discovery. ACM Southeast Regional Conference. 2010:14. [Google Scholar]
- 18.Canfield M, Keller C, Frydrych L, Ashrafioun L, Purdy C, Blondell R. Prescription opioid use among patients seeking treatment for opioid dependence. J Addict Med. 2010;4:108–113. doi: 10.1097/ADM.0b013e3181b5a713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen L, Wang W, Nagarajan M, Sheth AP. Extracting diverse sentiment expressions with target-dependent polarity from twitter. ICWSM . 2012 [Google Scholar]
- 20.Chiticariu L, Krishnamurthy R, Li Y, Raghavan S, Reiss F, Vaithyanathan S. SystemT: An algebraic approach to declarative information extraction.. 48th Annual Meeting of the Association for Computation Linguistics; Stroudsburg, PA, USA. 2010. pp. 128–137. [Google Scholar]
- 21.Cicero T, Adams E, Geller A, Inciardi J, Munoz A, Schnoll SH, Senay E, Woody G. A postmarketing surveillance program to monitor ultram (tramadol hydrochloride) abuse in the united states. Drug Alcohol Depend. 1999;57:7–22. doi: 10.1016/s0376-8716(99)00041-1. [DOI] [PubMed] [Google Scholar]
- 22.Cicero T, Ellis M, Surratt H. Effect of abuse-deterrent formulation of oxycontin. N Engl J Med. 2012;367:187–189. doi: 10.1056/NEJMc1204141. [DOI] [PubMed] [Google Scholar]
- 23.Cicero T, RC R, Inciardi J, Woody G, Schnoll S, Munoz A. The development of a comprehensive risk-management program for prescription opioid analgesics: Research abuse, diversion and addiction-related surveillance (radars). Pain Medicine. 2007:157–170. doi: 10.1111/j.1526-4637.2006.00259.x. [DOI] [PubMed] [Google Scholar]
- 24.Compton W, Volkow N. Major increases in opioid analgesic abuse in the United States: concerns and strategies. Drug Alcohol Depend. 2006;81(2):103–107. doi: 10.1016/j.drugalcdep.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 25.Cone E. Ephemeral profiles of prescription drug and formulation tampering: evolving pseudoscience on the internet. Drug Alcohol Depend. 2006;83(1):S31–39. doi: 10.1016/j.drugalcdep.2005.11.027. [DOI] [PubMed] [Google Scholar]
- 26.Coplan P, Kale H, Sandstrom L, Chilicoat H. National changes in oxycontin, other oxycodone, and heroin exposures reported to poison centers with introduction of reformulated oxycontin. 2012.
- 27.Daniulaityte R, Carlson R, Falck R, Cameron D, Perera S, Chen L, et al. “i just wanted to tell you that loperamide will work”: A web based study of extra-medical use of loperamide. Drug Alcohol Depend. 2013;130(1-3):241–244. doi: 10.1016/j.drugalcdep.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Daniulaityte R, Carlson R, Falck R, Cameron D, Perera S, Chen L, et al. A web-based study of self-treatment of opioid withdrawal symptoms wih loperamide. College on Problems of Drug Dependence. 2012 [Google Scholar]
- 29.Daniulaityte R, Falck R, Carlson R. Illicit use of buprenorphine in a community sample of young adult non-medical users of pharmaceutical opioid. Drug Alcohol Depend. 2012;122:122. doi: 10.1016/j.drugalcdep.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DeVeaugh-Geiss A, Leukefeld C, Havens JC, Chilcoat H. Routes of administration frequency of abuse of oxycontin and immediate release oxycodone in a rural kentucky county following introduction of reformulated oxycontin. 2012.
- 31.Ericsson C, Johnson P. Safety and efficacy of loperamide. Am J Med. 1990;88:10S–4. doi: 10.1016/0002-9343(90)90269-j. [DOI] [PubMed] [Google Scholar]
- 32.F S, Ricciardi A, Corazza O, Deluca P, Davey Z, Rafanelli C. “psychonaut web mapping”: New drugs of abuse on the web: the role of the psychonaut web mapping project. Riv Psichiatr. 2010;45:88–93. [PubMed] [Google Scholar]
- 33.Falck R, Carlson R, Wang J, Siegal H. Sources of information about mdma (3,4- methylenedioxymethamphetamine): perceived accuracy, importance,. Alcohol Depend. 2004;74:45–54. doi: 10.1016/j.drugalcdep.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 34.Glen J, Widom J. SimRank: a measure of structural-context similarity. KDD. 2002:538–543. [Google Scholar]
- 35.Inflexxion Retrieved from Inflexxion. 2012 http://www.inflexxion.com.
- 36.Kaloyanides K, McCabe S, Cranford J, Teter C. Prevalence of illicit use and abuse of prescription stimulants, alcohol, and other drugs among college students: relationship with age at initiation of prescription stimulants. Pharmacotherapy. 2007;27(5):666–674. doi: 10.1592/phco.27.5.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kavuluru R, Thomas C, Sheth AP, Chan V, Wang W, Smith A. An up-to-date knowledge-based literature search and exploration framework for focused bioscience domains. International Health Informatics Symposium. 2012:275–284. [Google Scholar]
- 38.Krishnamurthy R, Li Y, Raghavan S, Reiss F, Vaithyanathan S, Zhu H. SystemT: A system for declarative information extraction. SIGMOD Record. 2008;37(4):7–13. [Google Scholar]
- 39.Lange J, Daniel J, Homer K, Reed M, Clapp J. Salvia divino- rum: effects and use among youtube users. Drug Alcohol Depend. 2010;83(108):138–140. doi: 10.1016/j.drugalcdep.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 40.Lankenau S, Teti M, Silva K, Bloom J, Harocopos A, Treese M. Initiation into prescription opioid misuse amongst young injection drug users. Int J Drug Policy. 2012;23:37–44. doi: 10.1016/j.drugpo.2011.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCabe S, Cranford J, West B. Trends in prescription drug abuse and dependence, co occurrence with other substance use disorders, and treatment utilization: Results from two national surveys. Addict Behav. 2008;33:1297–1305. doi: 10.1016/j.addbeh.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mendes PN, Jakob M, Garcia.-Silva A, Bizer C. 7th International Conference on Semantic Systems. I-Semantics. ACM; New York: 2011. DBpediaSpotlight: shedding light on the web of documents. pp. 1–8. [Google Scholar]
- 43.Miles M, Huberman A. Qualitative data analysis : an expanded sourcebook. Thousand Oaks: Sage Publications. 1994 [Google Scholar]
- 44.Miller P, Sonderlund A. Using the internet to research hidden populations. Addiction. 2010;105:1557–1567. doi: 10.1111/j.1360-0443.2010.02992.x. [DOI] [PubMed] [Google Scholar]
- 45.Monte A, Mandell T, Wilford B, Tennyson J, Boyer E. Diversion of byprenorphine/naloxone coformulated tablets in a region with high prescribing prevalence. AJ Addic Dis. 2009;16:226–231. doi: 10.1080/10550880903014767. [DOI] [PubMed] [Google Scholar]
- 46.Nadeau D, Sekine S. A survey of named entity recognition and classification. Lingvisticae Investigationes. 2007;30(1):3–26. [Google Scholar]
- 47.Navippro 2012 Retrieved from http://www.navippro.com/
- 48.NVivo 2012 Retrieved from www.qsrinternational.com.
- 49.P G, Vingoe L, Hunt N, Mounteney J, Hartnoll R. Drug information systems, early warning, and new drug trends: can drug monitoring systems become more sensitive to emerging trends in drug consumption? Subst Use Misuse. 2000;35:811–844. doi: 10.3109/10826080009148423. [DOI] [PubMed] [Google Scholar]
- 50.Paulozzi L, Xi Y. Recent changes in drug poisining mortality in the United States by urban-rural status and by drug type. Pharmacoepidemiol Drug Saf. 2008;17(10):997–1005. doi: 10.1002/pds.1626. [DOI] [PubMed] [Google Scholar]
- 51.Paulozzi L, Budnitz D, Xi Y. Increasing deaths from opioid analgesics in the United States. Pharmacoepidemiol Drug Saf. 2006;15(9):618–627. doi: 10.1002/pds.1276. [DOI] [PubMed] [Google Scholar]
- 52.Peavy K, Banta-Green C, Kingston S, Hanrahan M, Merill J, Coffin P. “hooked on” prescription-type opiates prior to using heroin: Results from a survey of syringe exchange clients. J Psychoactive Drugs. 2012;44:259–265. doi: 10.1080/02791072.2012.704591. [DOI] [PubMed] [Google Scholar]
- 53.Ramakrishnan C. Extracting, Representing and Mining Semantic Metadata from Text; Facilitating Knowledge Discovery in Biomedicine. Wright State University; Dayton, OH: 2008. [Google Scholar]
- 54.Ramakrishnan C, Kochut K, Sheth AP. A Framework for Schema-Driven Relationship Discovery from Unstructured Text.. International Semantic Web Conference; Athens, GA, USA. 2006. pp. 583–596. [Google Scholar]
- 55.Ramakrishnan C, Mendes PN, daGama RA. Joint Extraction of Compound Entities and Relationships for Biomedical Literature. Web Intelligence. 2008:398–401. [Google Scholar]
- 56.Ramakrishnan C, Mendes PN, Wang S, Sheth AP. Unsupervised Discovery of Compound Entities for Relationship Extraction. EKAW. 2008:146–155. [Google Scholar]
- 57.Rindflesch TF. The interaction of domain knowledge and linguistic structure in natural language processing: Interpreting hypernymic propositions in biomedical text. Journal of Biomedical Informatics. 2003:462–477. doi: 10.1016/j.jbi.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 58.SAMHSA Results from the 2010 National Survey on Drug Use and Health: Detailed Tables. 2011.
- 59.Sarawagi S. Infomation Extraction. Found Trends databases. 2008:261–377. [Google Scholar]
- 60.Schifano F, Deluca P. Psychonaut 2002 research group: Searching the internet for drug-related web sites: analysis of online available information on ecstasy (mdma). Am J Addict. 2007;16:479–83. doi: 10.1080/10550490701641181. [DOI] [PubMed] [Google Scholar]
- 61.Siegal H, Carlson R, Kenne D, Swora M. Probable relationship between opioid abuse and heroin use. Am Fam Physician. 2003;67:942–945. [PubMed] [Google Scholar]
- 62.Sloboda Z. Epidemiology of Drug Abuse. Springer; 2005. [Google Scholar]
- 63.Strauss A, Corbin J. Basics of qualitative research: grounded theory procedures and techniques. Sage Publications; 1990. [Google Scholar]
- 64.Substance Abuse and Mental Health Services Administration. Rockville, MD: 2009. [Google Scholar]
- 65.Substance Abuse and Mental Health Services Administration . Results from the 2008 National Survey on Drug Use and Health: National Findings. Office of Applied Studies, NSDUH Series H-36, HHS Publication No. SMA 09-4434; Rockville, MD: [Google Scholar]
- 66.Suchanek F, Ifrim GW. Combining Linguistic and Statistical Analysis to Extract Relations from Web Documents. KDD. 2006:712–717. [Google Scholar]
- 67.Sudweeks F, Simoff SJ. Doing Internet Research: Critical Issues and Methods for Examining the Net. Sage Publications; 1999. Complementary Explorative Data Analysis The Reconciliation of Quantitative and Qualitative Principles. [Google Scholar]
- 68.Thomas C. Knowledge Acquisition in a System. Wright State University; Dayton, OH: 2012. [Google Scholar]
- 69.Mircrowave op oxycontin pills. 2010 Topix.com Web Forum: Retrieved from Topix: www.topix.com/forum/drug/oxyconting/TSQ7F2SBHMJ1H2QUB.
- 70.UIMA 2010 Retrieved from Apache: http://uima.apache.org.
- 71.Wishart D, Knox C, Guo A, Cheng D, Shrivastava S, Tzur D, et al. Drugbank: a knowledgebase for drugs, drug actions, and drug targets. Nucleic Acid Research. 2008:901–906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zacny J, Bigelow GC, Foley K, Iguchi M, Sannerud C. College on problems of drug dependence task force on prescription opioid non-medical use and abuse: position statement. Drug Alcohol Depend. 2003;69:215–232. doi: 10.1016/s0376-8716(03)00003-6. [DOI] [PubMed] [Google Scholar]
- 73.Ziano J. Semantic app helps researchers understand prescription drug abuse. 2012 From semanticweb.com: http://tinyurl.com/9oeop2j.