Abstract
Proline, glutamic acid, and leucine rich protein 1 (PELP1) is a large multi-domain protein that has been shown to modulate an increasing number of pathways and biological processes. The first reports describing the cloning and characterization of PELP1 showed that it was an estrogen receptor coactivator. PELP1 has now been shown to be a coregulator for a growing number of transcription factors. Furthermore, recent reports have shown that PELP1 is a member of chromatin remodeling complexes. In addition to PELP1 nuclear functions, it has been shown to have cytoplasmic signaling functions as well. In the cytoplasm PELP1 acts as a scaffold molecule and mediates rapid signaling from growth factor and hormone receptors. PELP1 signaling ultimately plays a role in cancer biology by increasing proliferation and metastasis, among other cellular processes. Here we will review 1) the cloning and characterization of PELP1 expression, 2) interacting proteins, 3) PELP1 signaling, and 4) PELP1-mediated biology.
Keywords: PELP1, Coactivator, Estrogen signaling, Hormone Resistance, extra-nuclear signaling
1. Identification
PELP1 was first identified as a 160kDa protein in a screen for Src homology 2 (SH2) domain-binding proteins (Park et al, 1995). In a subsequent study, a HeLa cell library was screened based on the peptide sequence of p160, and a cDNA clone that contained an open reading frame of 3,846 bp was obtained. p160 was named proline, glutamic acid, and leucine rich protein 1 (PELP1), based on the high abundance of these amino acids in the protein. While PELP1 contains 10 LXXLL motifs and several other motifs common to transcriptional regulators, the overall protein structure is not homologous to other known proteins (Vadlamudi et al, 2001).
Shortly after the initial report describing the cloning and characterization of PELP1 (Vadlamudi et al, 2001), Wong and colleagues reported the cloning of Modulator of Nongenomic Activity of ER (MNAR). MNAR was identified by a GST-pull down approach using the ERβ ligand-binding domain (ERβ-LBD) as bait. In this study, whole cell extracts from MCF-7 cells, untreated or stimulated with estrogen, were incubated with GST- ERβ-LBD, and proteins that bound to ERβ in a ligand dependent manner were identified by mass spectrometry-based peptide microsequencing. This manuscript was later retracted due to irreproducibility of some of the signaling data (Wong et al, 2009). Peptide sequences isolated from a 120 kDa protein were homologous to PELP1, but the cloned gene was not 100% identical. The published PELP1 sequence contained an additional 435-bp region and a number of amino acid substitutions within highly GC-rich and repetitive amino acid regions (Balasenthil & Vadlamudi, 2003). To determine if PELP1 and MNAR were similar proteins encoded from the same gene or actually the same protein, the PELP1 cDNA was resequenced and it was found that the amino acid differences between MNAR and PELP1 were the result of sequencing errors. It was also determined that the extra 435-bp present in the PELP1 cDNA was the result of cloning an immature transcript containing an unspliced intron, and therefore PELP1 and MNAR are in fact the same protein (Balasenthil & Vadlamudi, 2003).
2. PELP1 Expression
2.1 Normal tissue and development
PELP1 expression has been examined in normal tissues, during central nervous system and mammary gland development, and in cancer. The initial report describing the cloning of PELP1 found that it was differentially expressed in human tissues with the highest expression observed in testis, mammary gland, brain, skeletal muscle and lung tissue (Vadlamudi et al, 2001). In the mouse, PELP1 expression was observed in the hypothalamus, testis, ovary, uterus, adrenal gland, lung, pituitary, mammary gland, cerebellum and spleen, and immunohistochemistry showed that cytoplasmic and nuclear subcellular localization varied depending on the tissue type. During murine mammary gland development, PELP1 expression was found to be elevated during pregnancy and then reduced during lactation (Vadlamudi et al, 2001).
Estrogen and ERs have been shown to play a significant role in the development of the central nervous system (Beyer, 1999; McEwen & Alves, 1999). Since PELP1 was first identified as an ER coactivator there have been a number of studies specifically examining PELP1 expression in the mouse, rat, and monkey brain (Khan et al, 2006; Khan et al, 2005; Pawlak & Beyer, 2005). In BALB/c mice, PELP1 expression gradually declined in the midbrain and hypothalamus between embryonic day 15 and postnatal day 15, while remaining fairly constant in the cortex (Pawlak & Beyer, 2005). PELP1 expression was also found in primary neuronal and astroglial cell cultures from the mouse striatum and midbrain (Pawlak & Beyer, 2005). In the rat, PELP1 was cloned from the hypothalamus and found to be 86% homologous to human PELP1, containing a glutamic acid rich region, and LXXLL and PXXP motifs. PELP1 was found expressed in many regions of the rat brain with intense IHC staining in the hypothalamus, cerebral cortex, hippocampus, amygdala, and cerebellum (Khan et al, 2005). Similar PELP1 expression patterns were observed in the monkey brain (Khan et al, 2006). Interestingly, PELP1 and ER colocalized in many, but not all, rat neuronal tissues (Khan et al, 2005). However, PELP1 and the glucocorticoid receptor (GR) showed an absolute colocalization in the monkey and rat brain, suggesting that PELP1 may modulate GR activity in the brain (Khan et al, 2006).
While PELP1 does appear to play a role in mammary gland and brain development, it is not known if PELP1 expression is essential for the development of these organs or embryonic development in general. A PELP1 knockout mouse would provide additional information on the role of PELP1 in development, and tissue specific knockout of PELP1 (mammary epithelial cells, neuronal cells, etc.) would also help delineate the role of PELP1 in disease progression. To our knowledge these mouse models have not been developed.
2.2 Cancer
PELP1 expression has been shown to be dysregulated in many different types of cancer, with PELP1 expression in breast cancer being the most frequently reported. The first report describing cloning and characterization found that PELP1 was overexpressed in a handful of breast tumors compared to the normal adjacent tissue (Vadlamudi et al, 2001). This report also showed differential expression of PELP1 protein in a series of 9 breast cancer cell lines. MCF-7 cells appeared to have the highest expression, while T47D and MDA-453 cells had low to no expression. Subsequent studies in breast tumor samples have shown that PELP1 is overexpressed in 60–80% of breast tumors (Habashy et al, 2010; Vadlamudi et al, 2005b). High PELP1 expression has been shown to be associated with tumor grade (Habashy et al, 2010; Kumar et al, 2009; Rajhans et al, 2007), proliferation (Habashy et al, 2010; Kumar et al, 2009), node positive invasive breast cancer and distant metastasis (Habashy et al, 2010; Rajhans et al, 2007), and a decrease in breast cancer specific survival and disease free survival (Habashy et al, 2010). Interestingly, in the study with the largest number invasive breast cancers high PELP1 expression was inversely associated with ER, PR, AR and luminal cytokeratins, and positively associated with basal cytokeratins and p53 expression (Habashy et al, 2010). PELP1 has also been reported to display aberrant localization in some breast tumors; while PELP1 is found localized to the nucleus in normal breast tissue, cytoplasmic localization has been observed in a significant number of invasive breast cancers (Kumar et al, 2009; Vadlamudi et al, 2005b). Analysis of recurrence free survival in ER-positive patients indicates that patients with high cytoplasmic PELP1 responded poorly to tamoxifen treatment, while those with low levels of cytoplasmic PELP1 responded well (Kumar et al, 2009). Together, these reports suggest that PELP1 expression may play a significant role in both ER-positive and ER-negative breast cancer. It is likely that in ER-positive breast cancer the predominate role of PELP1 is enhancing ER extra-nuclear and nuclear actions, while in ER-negative breast cancer PELP1 is enhancing growth factor signaling pathways and promoting permissive transcriptional regulation through chromatin remodeling (see section 4. PELP1 signaling).
Increased PELP1 expression has also been reported in female cancers of the reproductive tract, in particular endometrial and ovarian. In endometrial cancer PELP1 was expressed in all 60 endometrial tumors tested, and alterations in subcellular localization were observed. Strong cytoplasmic staining was found in over 30% of the endometrial tumors, with PELP1 and ERβ colocalized to the cytoplasm of high grade endometrial tumors. This study also examined PELP1 expression in benign endometrial tissue from proliferative, secretory and postmenopausal stages. Moderate to strong expression of PELP1 was observed in all stages, but the nuclear and cytoplasmic localization varied. While both nuclear and cytoplasmic staining was observed in proliferative and secretory samples, postmenopausal samples exhibited predominately cytoplasmic staining (Vadlamudi et al, 2004). In ovarian cancer PELP1 expression has been examined by two independent groups. Dimple et al. found PELP1 to be expressed in 85% of samples, with high staining in 60% of tumors. PELP1 expression was mostly cytoplasmic when compared to normal ovary, and all subtypes of ovarian cancer (serous, endometrioid, clear cell carcinoma, and mucinous) showed elevated PELP1 staining (Dimple et al, 2008). More recently, Aust et al found PELP1 expression in 76.2% of ovarian cancer samples, and the prevalence of PELP1 expression was lower in mucinous tumors compared to serous and endometriod tumors. This study also found that PELP1 expression was associated with better disease-free survival and overall survival (Aust et al, 2013).
Similar to breast cancer, PELP1 expression in prostate cancer and astrocytic brain tumors was found to be associated with high tumor grade and poor prognosis (Kefalopoulou et al, 2011; Nair et al, 2007; Yang et al, 2012). Two separate studies examined PELP1 expression in prostate tumors. In a recent study of 35 prostate tumors PELP1 was expressed in all tumors and co-expression of PELP1, AR, and ERβ was observed in all Gleason Grade 8–9 tumors (Yang et al, 2012). An earlier study reported similar results, with PELP1 expression observed in all 40 tumors tested, with 2–3 fold higher PELP1 expression in high grade tumors compared to low grade tumors (Nair et al, 2007). In astrocytic brain tumors PELP1 expression, along with the coactivators AIB1 and TIF2, was found to increase in high grade tumors compared to low grade tumors. Additionally, all three coactivators were associated with worse prognosis (Kefalopoulou et al, 2011).
While PELP1 expression has been associated with a higher tumor grade and poor prognosis in a number of cancer types, this is not the case in colon and salivary gland tumors. In colon carcinoma samples, PELP1 expression was significantly increased in the epithelial cells of carcinoma samples compared to normal mucosa, and was found to be an independent favorable prognostic factor (Grivas et al, 2009). A separate study by the same group showed that PELP1 was expressed in a dot-like pattern in the nuclei of epithelial and stromal cells. Statistical analysis revealed an increase in PELP1 expression in myofibroblasts from normal mucosa through adenomas to carcinomas, but this study did not look for associations with tumor grade or prognosis (Tzelepi et al, 2009). Two studies of salivary gland tumors found that PELP1 was expressed in a majority of tumors, 94% (Vadlamudi et al, 2005a) or 73% (Williams et al, 2007), but expression was not associated with any markers of disease progression (Vadlamudi et al, 2005a), suggesting that perhaps PELP1 upregulation may be early event in the development of these tumor types.
Collectively, PELP1 has been found overexpressed in a number of different tumor types and frequently PELP1 expression is associated with higher tumor grade and poor prognosis (Habashy et al, 2010; Kefalopoulou et al, 2011; Kumar et al, 2009; Nair et al, 2007; Rajhans et al, 2007; Yang et al, 2012), but in a few tumor types PELP1 expression was not an indicator of an aggressive phenotype. These differences may be related to expression of PELP1-interacting proteins that promote cell growth pathways, or at what point in tumor development PELP1 expression becomes dysregulated.
In a number of tumor types PELP1 was found localized to the cytoplasm or both the nucleus and cytoplasm (Dimple et al, 2008; Kumar et al, 2009; Vadlamudi et al, 2004; Vadlamudi et al, 2005b), suggesting that extra-nuclear functions of PELP1 may play a role in disease progression. Of note, while cytoplasmic PELP1 localization has been described, some reports have indicated that they do not observe cytoplasmic PELP1 staining (Habashy et al, 2010; Yang et al, 2012). The conflicting reports on PELP1 localization may be related to the PELP1 antibodies used for IHC. A number of publications that do not report cytoplasmic PELP1 staining used a Novous Biological PELP1 antibody, while publications that report cytoplasmic PELP1 staining primarily used a non-commercial laboratory developed antibody. The peptide sequence of PELP1 that was used to generate PELP1-specific antibodies may also play a role in whether the antibody will recognize cytoplasmic PELP1. It is possible that PELP1-associated proteins compete with antibody binding in a cell-compartment-specific manner.
Many of the studies reviewed above have examined co-expression of PELP1 with ERα and ERβ. In tissue types where very little ERα protein expression is observed (i.e. colorectal, salivary gland, and astrocytic brain tumors), co-expression of PELP1 and ERβ is observed, often in the cytoplasm (Grivas et al, 2009; Kefalopoulou et al, 2011; Vadlamudi et al, 2005a; Yang et al, 2012). Considering that PELP1 was initially identified in a screen for ERβ interacting proteins it is likely that ERβ-PELP1 interactions may play a mechanistic role in cancer progression.
3. Interacting proteins
The overall domain structure of PELP1 is unique, but there are a number of motifs and domains common to transcriptional regulators. The N-terminus contains 10 LXXLL nuclear receptor interaction motifs, three PxxP motifs for interacting with proteins containing SH3 domains, and a nuclear localization signal. PELP1 also contains a cysteine rich region that may form a zinc finger, and a glutamic acid-rich region that is located between two proline-rich regions in the C-terminus (Figure 1). Although PELP1 has been associated with chromatin remodeling, it does not possess any known enzymatic activity. Based on the size (1130 amino acids) and unique structure of PELP1, it is not surprising that a large number of interacting proteins have been identified and that PELP1 has frequently been shown to function as a scaffolding molecule. This section will review PELP1 interacting proteins that have been identified by a GST-pull down approach or co-immunoprecipitation, and has been divided into nuclear and cytoplasmic interactions. Table 1 is a summary of known PELP1 interacting proteins.
Figure 1.
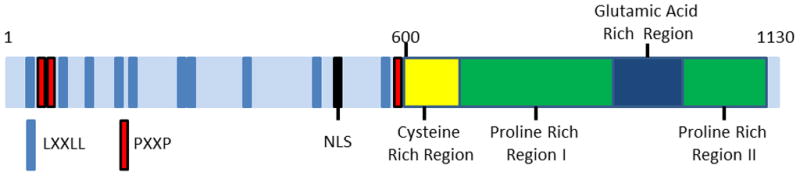
Schematic representation of PELP1 domain structure.
Table 1.
Summary of PELP1 interacting proteins. Includes PELP1 region required for interaction, if known, and the functional significance of the interaction.
| PELP1 interacting protein | PELP1 interacting region | Function | References |
|---|---|---|---|
| ERα/β | Amino acids 1–400; LXXLL motifs #4 and #5 | Extra-nuclear signaling and co-activator of ER | (Barletta et al, 2004; Vadlamudi et al, 2004; Vadlamudi et al, 2001) |
| AR | Amino acids 1–400 | Extra-nuclear signaling and co-activator of AR | (Nair et al, 2007; Unni et al, 2004; Yang et al, 2012) |
| GR | Glutamic acid rich region and amino acids 1–400 | Context dependent GR coactivator and corepressor | (Kayahara et al, 2008) |
| ERRα | Not determined | Increase expression of aromatase | (Rajhans et al, 2008) |
| RXR | N-terminal LXXLL | RXR coactivator | (Singh et al, 2006) |
| Nur77 | Not determined | Chromatin remodeling complex | (Rambaud et al, 2009) |
| STAT3 | Amino acids 1–600 | Growth factor induced phosphorylation and activation of STAT3 | (Manavathi et al, 2005) |
| SRF | Not determined | Inhibition of SRF-dependent genes | (Choi et al, 2004) |
| BCAS3 | Amino acids 1–400, 600–866 | Enhance coactivator function of BCAS | (Gururaj et al, 2007) |
| FHL2 | N-terminal LXXLL (aa 1–600) | Increase FHL2 transcriptional activity | (Nair et al, 2007) |
| PNRC2 | Proline rich region II (aa 966–1130) | Cooperate with ERRα to promote aromatase expression. | (Rajhans et al, 2008) |
| CBP/p300 | Not determined | Activate gene expression | (Vadlamudi et al, 2001) |
| HDAC2 | Glutamic acid rich region | Repress gene expression | (Choi et al, 2004) |
| Histone H1 | Glutamic acid-rich region and proline-rich region I | Acetylation and displacement of histone H1 | (Nair et al, 2004) |
| Histone H3 | Glutamic acid-rich region and proline-rich region I | Modulation of histone H3 methylation | (Nair et al, 2004; Nair et al, 2010b) |
| KDM1 | Amino acids 400–600 | Modulation of histone H3 methylation | (Fanis et al, 2012; Nair et al, 2010b; Rosendorff et al, 2006) |
| CARM1 | Amino acids 400–600 | Activation of E2-dependent genes | (Mann et al, 2013) |
| TTLL4 | Not determined | Polygutamylation of PELP glutamic acid rich region and chromatin remodeling | (Kashiwaya et al, 2010) |
| LAS1L | Not determined | Chromatin remodeling | (Fanis et al, 2012; Finkbeiner et al, 2011; Kashiwaya et al, 2010) |
| SENP3 | Not determined | Desumoylation; associated with chromatin remodeling and ribosome biogenesis | (Fanis et al, 2012; Finkbeiner et al, 2011; Kashiwaya et al, 2010) |
| CDK2/4 | Amino acids 200–350 | Phosphorylation of PELP1, transactivation of E2F target genes and cell cycle progression | (Nair et al, 2010a) |
| pRB | Amino acids 1–330 | Enhance phosphorylation of pRB; cell cycle progression | (Balasenthil & Vadlamudi, 2003; Nair et al, 2010a) |
| EGFR | Not determined | Enhance growth factor signaling | (Vadlamudi et al, 2005b) |
| PI3K/p85 | Not determined | Activation of PI3K/Akt pathway | (Vadlamudi et al, 2005b) |
| c-Src | N-terminal PxxP, C-terminal amino acids 887–962 | Enhance extra-nuclear signaling and phosphorylation of ER | (Barletta et al, 2004; Cheskis et al, 2008) |
| ILK1 | Amino acids 601–886 | Promote migration and invasion | (Chakravarty et al, 2010) |
| HRS | Amino acids 1–400 | Sequester PELP1 in the cytoplasm and enhance MAPK activation | (Rayala et al, 2006a) |
| Gβ | N-terminal region | G-protein regulated meiosis in response to androgens | (Haas et al, 2005) |
| SUMO-1/2 | Not determined | Transcriptional regulation and ribosome biogenesis | (Fanis et al, 2012; Finkbeiner et al, 2011; Rosendorff et al, 2006) |
| Chtop/5FMC | Not determined | Transcriptional regulation | (Fanis et al, 2012) |
| TEX10-WDR18 complex | Not determined | Ribosome biogenesis | (Finkbeiner et al, 2011) |
3.1 Nuclear interactions
The initial reports describing the cloning of PELP1/MNAR found that it interacted with ER and played a role in increasing ER transcriptional activity (Vadlamudi et al, 2001). The N-terminal domain of PELP1 containing LXXLL motifs #4 and #5 were found to interact with the AF2 domain of ERα in a ligand dependent manner (Barletta et al, 2004). Since the initial cloning and characterization of PELP1, it has been found to interact with a number of additional nuclear receptors including GR (Kayahara et al, 2008), AR (Nair et al, 2007; Unni et al, 2004; Yang et al, 2012), ERRα (Rajhans et al, 2008), and RXR (Singh et al, 2006), and the transcription factor STAT3 (Manavathi et al, 2005). While PELP1 presumably interacts with nuclear receptors via one or more of its LXXLL motifs, as it has been shown with ER (described above) and RXR (Singh et al, 2006), this is not a universal mechanism. GR was shown to interact with the C-terminal region of PELP1 that contains the glutamic acid rich region. Although, PELP1 may also interact with GR via the N-terminal 400 amino acids that contain the majority of the LXXLL motifs, as expression of this fragment of PELP1 was able to inhibit GR transcriptional activity (Kayahara et al, 2008). Similar to ER and RXR, STAT3 was shown to interact with the N-terminal 400 amino acids of PELP1 (Manavathi et al, 2005). PELP1 has also been found in complexes with Nur77 (Rambaud et al, 2009) and SRF (Choi et al, 2004), but the domains involved were not determined.
In addition to transcription factors, PELP1 has been shown to interact with other transcriptional coregulators and components of chromatin. BCAS3, an ER coactivator, interacts with PELP1 through the PELP1 N-terminal 400 amino acids, and the region that contains 2 additional LXXLL motifs (amino acids 600–866). Additionally, it was determined that the BCAS3 bromodomain was required for the BCAS3-PELP1 interaction (Gururaj et al, 2007). PELP1 binds to the LIM3 and LIM4 regions of the coactivator FHL2. Similar to BCAS3, FLH2 was shown to interact with PELP1 amino acids 1–400 and 400–600. Interestingly, although the C-terminal region of PELP1 did not associate with FHL2, it was found to be necessary to promote FLH2 transcriptional activation (Nair et al, 2007). One coactivator, PNRC2, was found to interact with C-terminal 966–1130 amino acids of PELP1, which contains proline-rich region II (Rajhans et al, 2008). Finally, while the domains involved were not determined, co-immunoprecipitation studies indicated that PELP1 also binds the transcriptional co-activators CBP and p300 (Vadlamudi et al, 2001).
Recent reports indicate that PELP1 protein interactions play a significant role in chromatin remodeling at target gene promoters. Choi et al reported that the PELP1 N-terminal domain interacts with HDAC2 while the C-terminal glutamic acid rich region preferentially associates with unacetylated histone H3. The result of these interactions was the inhibition of SRF-mediated gene expression (Choi et al, 2004). Interestingly, Nair and colleagues found by ChIP/re-ChIP that PELP1 and acetylated histone H3 were associated following estrogen treatment. Furthermore, PELP1 interacts with both histone H1 and H3, with higher affinity for H1. The regions required for binding were the C-terminal glutamic acid-rich region and the proximal proline-rich region. Additionally, both of these regions were required for efficient transactivation of estrogen-induced genes (Nair et al, 2004). While these results are contradictory, it is possible that PELP1 actions are context dependent and it can act as both a co-repressor by recruiting HDAC2 at SRF-dependent genes, and a co-activator on estrogen-induced genes by displacing H1 and allowing histone acetyl transferases to modify chromatin structure and promote gene expression.
In a separate report, Nair and colleagues also found that PELP1 specifically recognizes di-methylated histone H3K4 and H3K9 through the N-terminal glutamic acid-rich region (amino acids 886–990). Interestingly, in the absence of ERα, PELP1 preferentially interacts with di-methyl H3K9 a marker of transcriptional repression. Addition of ERα decreased the PELP1/H3K9 interaction, and the addition of KDM1, a lysine demethylase, lead to PELP1 specific binding to di-methyl H3K4, a marker of transcriptional activation. This same study mapped the interaction between PELP1 and KDM1 to amino acids 400–600 of PELP1. Overall, the results of this elegant study suggest that PELP1 alters the substrate specificity of KDM1 from H3K4 to H3K9 and that demethylation of H3K9 by KDM1 requires a functional complex composed of KDM1-ERα-and PELP1 (Nair et al, 2010b). Importantly, two additional reports have identified PELP1 and KDM1 in nuclear multiprotein complexes (Fanis et al, 2012; Rosendorff et al, 2006). In support of the above studies, Mann et al. recently showed that PELP1 specifically interacted with histones modified by arginine dimethylation and citrullination and lysine dimethylation. Additionally, they found that PELP1 interacts with the arginine methyltransferase CARM1. The CARM1/PELP1 interaction was mapped to amino acides 400–600 of PELP1 and resulted in an increase in the transcription of ER target genes (Mann et al, 2013).
Posttranslational modifications of PELP1 have also been shown to alter protein-protein interactions. Expression of TTLL4, a tubulin polyglutamylase previously shown to have non-tubulin protein targets, was shown to promote polyglutamylation of PELP1. Polyglutamylation of PELP1 enhanced the interaction of PELP1 with histone H3 and LAS1L, but inhibited PELP1-SENP3 binding (Kashiwaya et al, 2010). Sumoylation likely impacts PELP1 protein interactions as well. PELP1 was identified in screens for both SUMO-1 and SUMO-2 interacting proteins (Matafora et al, 2009; Rosendorff et al, 2006), and is both a non-covalent binding partner of SUMO-2 (Rosendorff et al, 2006), and covalently modified by SUMO-1/2 at K826 (Finkbeiner et al, 2011). Phosphorylation of PELP1 may also impact protein complex formation. CDK/cyclin complexes have been shown to bind and phosphorylate PELP1, which results in enhanced coactivator function, but alterations in protein complexes resulting form phosphorylation as not been demonstrated experimentally (Nair et al, 2010a).
The described experimental data supports the hypothesis that PELP1 is acting as a scaffolding molecule that facilitates assembly of complexes involved in gene repression and activation, likely through chromatin remodeling. In addition, the number of LXXLL motifs and binding proteins identified suggests that PELP1 could be acting as a scaffolding molecule that facilitates cross-talk between NR family members and other transcriptional regulators. Taken together these data demonstrate PELP1 promiscuity in facilitating a variety of cellular signaling and transcriptional activities. Perhaps PELP1 specializes in coordinating the transition from signaling to transcriptional (gene regulation) responses.
3.2 Cytoplasmic Interactions
PELP1 has predominately been shown to interact with nuclear proteins, but there are a significant number of reports indicating that PELP1 functions as a scaffolding molecule in the cytoplasm as well. Expression of the NLS (nuclear localization signal) mutant PELP1 (PELP1-Cyto) was shown to interact with the p85 subunit of PI3K and EGFR in breast cancer cell line models (Vadlamudi et al, 2005b). Expression of PELP1-Cyto was also shown to increase c-Src activity (Vadlamudi et al, 2005b). Not surprisingly, PELP1 and Src interact, and this was shown to occur via the first N-terminal PxxP domain of PELP1 and the c-Src SH3 domain. Additionally, PELP1 C-terminal amino acids 887–962 interacted with the Src SH2 domain (Barletta et al, 2004). Integrin linked kinase 1 (ILK1) has also been shown to interact with PELP1 in the cytoplasm via PELP1 amino acids 601–886 (Chakravarty et al, 2010). In breast cancer cells estrogen stimulated PELP1-ILK1 interaction involved activation of a pathway that also included ER, Src, and PI3K, all of which have been shown to interact with PELP1. Thus, PELP1 cytoplasmic interactions likely promote extra-nuclear ER signaling and also growth factor signaling pathways in ER-negative cells, both of which contribute to a more aggressive cancer phenotype.
Although differential localization of PELP1 has been observed in numerous tissue and tumor types (see above), the mechanisms governing PELP1 trafficking between intracellular compartments is not well understood. One hint was provided by studies identifying hepatocyte growth factor-regulated tyrosine kinase substrate (HRS) as a PELP1 interacting protein in a yeast two-hybrid screen. HRS amino acids 391–600 were found to be required to interact with amino acids 1–400 of PELP1. Interestingly, overexpression of HRS resulted in translocation of PELP1 to the cytoplasm (Rayala et al, 2006a), and suggesting it serves to sequester PELP1 outside of the nucleus.
4. PELP1 signaling
PELP1 is involved in a multitude of biological processes within the cell via its diverse signaling capacity and cellular localization profile. Due to its modular domain structure and the possession of several protein binding motifs (described above), PELP1 functions chiefly to facilitate multi-protein complexes that increase the cellular signaling efficiency and genomic responsiveness to external stimuli. For this reason, understanding the mechanisms by which PELP1 functions and is dysregulated in disease states, such as cancer, is becoming increasingly important. In this section we will review critical PELP1 signaling/genomic findings that may lead to a better understanding of how therapies targeting PELP1 pathways may be developed or optimized.
4.1 Estrogen signaling
PELP1 participates in cellular responses to estrogen by two mechanisms; it scaffolds ER containing signaling complexes in the cytoplasm to activate kinase cascades and participates in ER transcriptional complexes as a coactivator. While these functions are often referred to separately, reports suggests that hormone induced cytoplasmic signaling is well integrated with genomic responses to hormone (Dressing et al, 2009). There is increasing evidence that hormone bound receptors form complexes in the cytoplasm, which function to initiate kinase signaling and phosphorylation events that subsequently facilitate compartmental shuttling and chromatin actions of these same complexes in the nucleus (Beguelin et al, 2010). PELP1 may serve to scaffold estrogen-induced cytoplasmic signaling complexes that are ultimately reorganized in the nucleus to activate transcription of ER gene targets.
Upon estrogen stimulation of breast cancer cells, ER induces rapid (5–10 min) activation of MAPK in a c-Src dependent manner (Boonyaratanakornkit, 2011). PELP1 has been shown to be a critical mediator of estrogen-induced MAPK activation via c-Src. Mutation of the PELP1 PXXP c-Src interaction site abolished estrogen-induced MAPK activation and ER transcriptional activity. ER is also able to interact with c-Src, further increasing signaling efficiency to MAPK (Barletta et al, 2004). The PELP/ER/c-Src signaling axis has also been shown to include growth factor receptors (EGFR, HER2) and the stimulation of the PI3K/AKT and integrin-linked kinase 1 (ILK1) pathway (Chakravarty et al, 2010; Vadlamudi et al, 2005b; Vallabhaneni et al, 2010). Here, PELP1 serves to concentrate low abundant signaling molecules in order to efficiently activate downstream signaling upon hormone exposure.
Recent studies have demonstrated that PELP1 is localized to the cytoplasm in a large cohort of breast tumors (Vadlamudi et al, 2005b). Mechanistic studies demonstrated that overexpression of PELP1-Cyto in unstimulated breast cancer cells was able to drive MAPK signaling and constitutive AKT activation resulting in increased phosphorylation of ER at Serine 118 and Serine 167 (Vadlamudi et al, 2005b). Additionally, HRS, which functions to regulate trafficking of growth factor-receptor complexes through early endosomes, has been shown to bind PELP1 and sequester it in the cytoplasm, thus increasing MAPK activation while decreasing ER genomic actions (Rayala et al, 2006a). Interestingly, while cytoplasmic PELP1 induces increased mitogenic cell signaling, it also sensitizes the cell to TNF-alpha-induced apoptosis, a mechanism that could potentially be exploited therapeutically for breast tumors with altered PELP1 localization (Rayala et al, 2006b).
Not surprisingly, due to the participation of PELP1 in multiple growth factor and hormone induced signaling complexes with kinases, PELP1 is phosphorylated on many sites, which serve primarily to enhance its coactivation of transcription factor complexes. Growth factor stimulation of PELP1 expressing breast cancer cells induces PELP1 phosphorylation in a PKA dependent manner, leading to increased ER transcriptional activity (Nagpal et al, 2008). Similarly, cyclin-dependent kinase (CDK) activation during cell cycle progression induces PELP1 phosphorylation on Ser477 and Ser991 leading to increased transactivation of E2F1 target genes and ER signaling (Nair et al, 2010a). CDK activation during S and G2 phases has also been shown to induce PELP1 nucleolar localization and activation of ribosomal RNA transcription (Gonugunta et al, 2011). Collectively, these data support a model where PELP1 cytoplasmic signaling contributes to increased coregulator and transcription factor phosphorylation events which serve to integrate the cytoplasmic and genomic actions of hormone signaling.
In addition to its cytoplasmic actions, PELP1 functions in the nucleus to coactivate multiple transcription factors, of which the most studied is ER. PELP1 interacts with ER and p300/CBP to enhance estrogen mediated transcriptional activation in a dose-dependent manner (Vadlamudi et al, 2001). PELP1 has been shown to function in a variety of capacities to increase ER transcriptional activity, for example it displaces histone H1 to activate transcription of pS2, PR and IGF, and interacts with pRB to enhance cyclin D1 expression (Balasenthil & Vadlamudi, 2003; Nair et al, 2004). Additionally, KDM1 interacts with PELP1 to promote estrogen mediated transcription of GREB1C, CBX5, myc, and max (Nair et al, 2010b). ER and PELP1 complexes also function to induce estrogen target genes MTA3 and BCAS3 (Gururaj et al, 2007; Mishra et al, 2004). Collectively, these data illustrate that PELP1 is intimately linked to ER and serves to scaffold critical protein complexes necessary for estrogen mediated signaling and gene regulation that comprise the estrogen responsive biological outcomes.
4.2 Other Transcription Factors and signaling molecules
Multiple nuclear receptors and other transcription factors rely on PELP1 scaffolding functions to elicit efficient responses to stimuli. In prostate cell models PELP1 binds AR and c-Src to facilitate rapid androgen initiated cytoplasmic signaling that culminates in CREB mediated cfos promoter activation (Unni et al, 2004). In the nucleus, PELP1 binds FHL2 in a c-Src dependent manner to enhance FHL2 transcriptional activity. Furthermore, this complex binds AR to increase androgen mediated transcriptional events (Nair et al, 2007). Estrogen bound ERβ and AR complexes are scaffolded by PELP1 on androgen responsive elements (AREs) in the chromatin to activate genes and this mechanism allows cells to bypass the need for androgens to activate AR target genes (Yang et al, 2012). PELP1 and AR also interact in response to androgens in a Xenopus laevis oocyte model; PELP1 complexes to the AR ligand binding domain via its LXXLL containing region to augment c-Src dependent transcriptional activation. Xenopus PELP1 also binds Gβ and is involved in G-protein regulated meiosis in response to androgens (Haas et al, 2005).
In addition to ER and AR, PELP1 functions as a coactivator for variety of other nuclear transcription factors including glucocorticoid receptors (GR) (Kayahara et al, 2008), ERβ (Vadlamudi et al, 2004; Yang et al, 2012), RXRα (Singh et al, 2006), E2F1 (Nair et al, 2010a), and ERRα/PCRC2 (Rajhans et al, 2008). Upon growth factor stimulation PELP1/STAT3 complexes enhance transcription of cyclin D1, c-myc, and c-fos in a c-Src and MAPK dependent manner (Manavathi et al, 2005). On the DNA, PELP1 participates in chromatin remodeling complexes such as Five Friends of Methylated Chtop (Fanis et al, 2012) and MLL1-WDR5 (Kashiwaya et al, 2010). Interestingly, PELP1 has also been observed to interact with HDAC2 and, in certain circumstances, serves as a corepressor for GR, Nur77, AP1, NF-κB, TCF/SRF (Choi et al, 2004). PELP1 interacts with a diverse array of partners and is involved in a variety of signaling pathways and cellular processes. A greater understanding of the mechanisms by which PELP1 contributes to cancer may lead to ways we can exploit these pathways therapeutically.
5. PELP1- mediated Biology
PELP1 has been shown to regulate a diverse number of cellular processes. Here we will review how PELP1 expression impacts 1) proliferation and tumorigenesis, 2) apoptosis and autophagy, 3) migration, invasion, and metastasis, and 4) hormone resistance.
5.1 Proliferation/Tumorigenesis
Numerous studies have shown that PELP1 is a driver of proliferation in hormonal tissues and cancer. A transgenic mouse model expressing PELP1-Cyto under an MMTV promoter displayed increased proliferation, as measured by BrdU incorporation in mammary epithelial cells, and mammary gland hyperplasia (Kumar et al, 2009). Conversely, in mice receiving MCF-7 xenografts, in vivo knockdown of PELP1 using nanoliposomal siRNA showed decreased tumor volume in part due to decreased proliferation, as measured by Ki-67 staining (Cortez et al, 2012). Similar results have been reported in ovarian cancer cell lines, where stable shRNA knockdown of PELP1 caused decreased proliferation under both low and high serum conditions and a reduction in soft-agar colony growth (Dimple et al, 2008). This decreased proliferation was in part due to attenuated cyclin D1 promoter activation. Moreover, PELP1 is a known regulator of the G1/S-phase transition through upregulation of cyclin D1 and phosphorylation of pRb (Balasenthil & Vadlamudi, 2003). Initial reports suggested that PELP1 specifically promoted estrogen-induced cell growth; however, more recent reports point to an additional role for PELP1 regulated proliferation independent of estrogen. For example, PELP1 has been shown to have proliferative role in ER-negative breast cancer cell lines and knockdown resulted in a variety of phenotypic changes, which included decreased proliferation (Roy et al, 2012).
In addition to proliferation, PELP1 overexpression was sufficient to induce cellular transformation. Expression of PELP1 in rat kidney epithelial cells and mouse fibroblasts resulted in increased focus formation and soft agar growth compared to controls (Rajhans et al, 2007). Upon co-transfection with c-Src, transformation potential was enhanced compared to PELP1 or c-Src transfection alone, indicating that these proto-oncogenes can cooperate to induce cellular transformation (Rajhans et al, 2007). Rajhans et al also showed MCF-7 breast cancer cells overexpressing PELP1 substantially increased estrogen-driven, anchorage-independent growth. PELP1-Cyto expression in MCF-7s showed similar results in potentiating estrogen-driven, anchorage independent growth. Overexpression of PELP1 in ER-positive cells has also been shown to promote estrogen-independent growth. In the absence of exogenous estrogen, MCF7-PELP1 xenografts displayed tumor formation in 50% of mice, while mice receiving MCF7-vector control cells showed no tumor formation (Rajhans et al, 2007).
Collectively, these reports demonstrate that PELP1 is a proto-oncogene and plays an important role in tumorigenesis and cellular proliferation in a variety of cell lines and tissues. Clinical data also support a role for PELP1 in proliferation and tumorigenesis, as numerous studies have shown that PELP1 expression is linked to larger and higher grade tumors, and acts as a marker of poor prognosis in multiple cancer types (Chakravarty et al, 2011; Habashy et al, 2010; Kefalopoulou et al, 2011; Rajhans et al, 2007). While PELP1 can enhance estrogen-driven effects in cells, its role has been expanded to include estrogen independent events.
5.2 Autophagy and Apoptosis
Though understudied, PELP1 has been implicated in autophagy of cancer cell lines. Data from Kumar et al showed that upon treatment with the phytoestrogen resveratrol, PELP1 co-localized with GFP-LC3, a marker of autophagosome formation (Ohshiro et al, 2007). This interaction was abolished by the autophagy-specific inhibitor 3-methyladenine. PELP1 was also co-recruited to autophagosomes with HRS, a known trafficking molecule. These data indicate that PELP1 may function to modulate cellular autophagy, but further study is necessary to determine its mechanistic role.
Similarly, the mechanisms by which PELP1 expression alters apoptosis pathways understudied. However, (as mentioned above), PELP1-Cyto has been shown to sensitize cells to tumor necrosis factor -α induced, caspase-dependent apoptosis, whereas wild-type PELP1 expressing cells were not sensitive (Rayala et al, 2006b). PELP1-cyto cells also showed less Bcl-2 protein expression. Interestingly, expression of wild-type PELP1 enhanced 9-cis retinoic acid induced cell death in MCF-7 cells (Singh et al, 2006). Together these reports suggest that PELP1 expression can sensitize cells to apoptosis-inducing stimuli, a phenomenon that deserves further attention.
5.3 Migration, Invasion, and Metastasis
A significant number of reports provide evidence that PELP1 expression contributes to cytoskeletal changes that contribute to a more aggressive cancer phenotype. In MCF-7 cells overexpressing PELP1, E2 stimulation increased motility compared to control, and PELP1 siRNA knockdown caused significant reduction of this phenotype. PELP1 was also found to be necessary for optimal in vitro estrogen-induced migration and cytoskeletal reorganization. This report also showed a significant increase in metastases of ZR75-PELP1 and MCF7-PELP1-Cyto overexpressing cells compared to control in tail vein and cardiac injection models (Chakravarty et al, 2010). Additionally, in series of MCF10A cells ranging from wild-type cells (normal mammary epithelial cells) to highly tumorigenic and metastatic variant cell lines, PELP1 expression was also positively correlated with MCF10A variants that exhibit higher metastatic potential (Rajhans et al, 2007). PELP1 expression regulates several proteins that are associated with metastasis, MTA1 and MTA3 (Mishra et al, 2003; Mishra et al, 2004). Clinical data showing that PELP1 expression is associated with node-positive and metastatic breast tumors further supports a role for PELP1 in invasion and metastasis (Rajhans et al, 2007).
Though PELP1 was initially discovered as an ER co-activator, PELP1 has recently been shown to promote metastasis in ER-negative breast cancer cells. In ER-negative MDA-MB-231 cells, PELP1 knockdown was shown to decrease the ability of these cells to proliferate, migrate, and invade in vitro. This was verified in vivo, using MDA-MB-231 xenografts with PELP1 knockdown, which exhibited decreased tumor burden compared to control cells. This study also showed that PELP1 expression may promote migration and invasion through its regulation of EMT genes. PELP1 knockdown led to decreased expression of classical EMT genes including MMPs, ZEB1/2, SNAI1, and TWIST1 (Roy et al, 2012).
PELP1-mediated regulation of metastasis has also been examined in an ovarian cancer model, where it was shown to be a strong regulator of the EMT phenotype. Overexpression of PELP1 in ovarian cancer cells led to increased migratory potential in boyden chamber assay, and PELP1 knockdown blocked this migration (Chakravarty et al, 2011). PELP1 knockdown cells also inhibited EGF-induced cytoskeletal changes, including formation of filopodia and ruffles. Similar to breast cancer cells, knockdown of PELP1 also resulted in a decreased expression of many EMT genes including MMP2, MMP9, MTA1, Myc, and SMAD2. In an ovarian xenograft model, PELP1 siRNA delivery to SKOV3ip1 tumors resulted in a reduction in metastatic tumor burden compared to control siRNA (Chakravarty et al, 2011).
5.4 Hormone Resistance
Endocrine therapy is highly effective for the treatment ER-positive breast cancer. The selective estrogen receptor modulator, tamoxifen, is the most studied and longest used endocrine therapy available. However, intrinsic and acquired resistance to therapies targeting ER occurs in many women, resulting in disease progression. Aberrant genomic and extra-nuclear ER signaling has been shown to contribute to endocrine-therapy resistance, including loss of ER, dysregulation of co-activators, altered receptor tyrosine kinase signaling, disruption of cell cycle regulators, and increased cell survival signaling (Musgrove & Sutherland, 2009).
Extra-nuclear signaling of PELP1 has been shown to modulate several pathways implicated in hormone resistance. MCF-7 cells expressing PELP1-Cyto were found to display tamoxifen resistance, as measured by proliferation assay. This work was verified in vivo by showing that mice receiving MCF7/PELP1-cyto xenografts did not respond to tamoxifen treatment. Additionally, in women treated with tamoxifen, high cytoplasmic PELP1 was associated with a decreased recurrence free survival (Kumar et al, 2009). Subsequent work has highlighted the requirement for a functional Src axis in PELP1 driven tamoxifen resistance. Blocking Src with the inhibitor dastatinib in MCF-7-PELP1 xenografts decreased tumor volume, and co-treatment with tamoxifen showed a further decrease (Vallabhaneni et al, 2010).
In addition to cytoplasmic localization, recent work has shown that targeting of PELP1 nuclear complexes has promise for circumventing hormone resistance. Cortez et al, showed that PELP1 interaction with KDM1 potentiates ERα target gene transcription by regulating the substrate specificity of KDM1 towards activating epigenetic marks (Cortez et al, 2012). KDM1 inhibition with pargyline, a clinically available monoamine oxidase B inhibitor, sensitized resistant cell lines to tamoxifen. Additionally, the PELP1-KDM1 axis also regulates the HER2- aromatase pathway leading to local estrogen synthesis (Vadlamudi et al, 2010). This pathway is a known contributor to hormone independence and may play a role in aromatase inhibitor resistance as well. PELP1 likely also contributes to tamoxifen resistance through its cell cycle regulation of cyclin D1 and Rb activation, which are known to contribute to resistance (Musgrove & Sutherland, 2009).
Collectively, these data indicate that PELP1 contributes to hormone therapy resistance via several mechanisms. Thus, therapies that target PELP1 mediated signaling may be a viable option to treat endocrine-therapy resistant tumors. Furthermore, targeting PELP1 prior to the development of resistance may lead to better overall survival for women diagnosed with breast cancer.
6. Therapeutic Targeting of the PELP1
Currently there are no PELP1 specific inhibitors and the lack of enzymatic activity makes the development of PELP1 inhibitors more challenging. Therefore, targeting PELP1 interacting molecules, or downstream pathways is attractive option for PELP1 expressing tumors. This approach has yielded promising results. For example, the Src inhibitor dastatanib and the KDM1 blocker pargyline were both effective in reducing PELP1 driven tumors in mice receiving MCF7-PELP1 xenografts (Nair et al, 2010b; Vallabhaneni et al, 2010). In addition, roscovitine, a CDK inhibitor was also shown to downregulate PELP1 and suppress endocrine-resistant tumor growth in mouse xenografts by arresting cell cycle progression through decreased cyclin D1 expression, and reduced phosphorylation of pRb and CDK2 (Nair et al, 2011). Most recently Rivindranathon and colleagues developed a small molecule peptidomimetic (D2) that targets the LXXLL motif of the AR. D2 was able to block the AR/PELP1 interaction and inhibit DHT-induced gene expression and proliferation of prostate cancer cell lines. Additionally, D2 was found to inhibit growth of C4-2 subcutaneous tumors in vivo (Ravindranathan et al, 2013). The development of D2 also suggests that it may be possible to specifically target PELP1 LXXLL motifs to inhibit PELP1/NR interactions in general.
Other therapeutic PELP1 targets that already have pharmacological inhibitors in clinical trials include PI3K, MAPK (through MEK inhibition), HER2, EGFR, and STAT3. Figure 2 is a summary of pharmacological inhibitors that may be used to target PELP1 signaling pathways. Future studies examining the response to targeted therapies in PELP1 expressing tumors has the potential to identify targets that could further enhance response to treatment.
Figure 2.
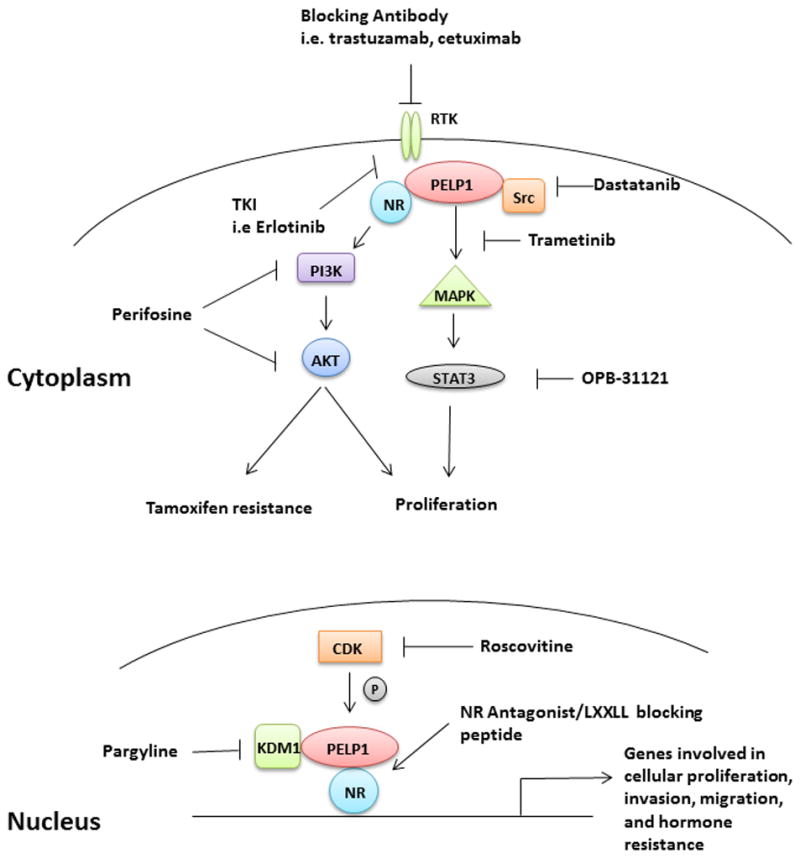
Targeting PELP1 pathways during tumorigenesis. Diagram of PELP1 signaling complexes and pathways in the cytoplasm and nucleus that could be targeted with pre-clinical and clinical drugs. At the membrane, PELP1 interacting RTKs could be targeted through blocking antibodies such as cetuzimab or kinase inhibitors such as erlotinib (both targeted to EGFR). In the cytoplasm, PELP1 interacting molecules such as Src or PI3K could be inhibited using the small molecule inhibitors dasatanib or perifosine, respectively. In the nuclear compartment, PELP1 interacts with different proteins including KDM1A and CDKs, both of which have candidate therapies available. Finally, exciting pre-clinical data suggest that targeting PELP1 protein-protein interactions may be a viable treatment option, as interfering with the LXXLL motif between PELP1 and AR successfully blocked tumor growth.
Highlights.
PELP1 expression is dysregulated in a significant number of cancer types.
Overexpression of PELP1 has been correlated with higher tumor grade and poor prognosis is some tumor types.
PELP1 interacts with numerous proteins and functions as a scaffolding protein in the cytoplasm and nucleus.
PELP1 signaling pathways modulate cellular proliferation, migration and invasion, and hormone resistance.
Acknowledgments
The authors would like to recognize their funding sources: NIH/NCI R01 CA159712 (CAL), National Center for Advancing Translational Sciences of the National Institutes of Health Award UL1TR000114 (JHO), and NIH/NCI K07CA131501-02 (JHO).
Abbreviations
- AIB1
amplified in breast cancer 1
- AR
androgen receptor
- BCAS3
breast carcinoma amplified sequence 3
- CBP
CREB binding protein
- ChIP
chromatin immunoprecipitation
- E2
estradiol
- ER
estrogen receptor
- ERRα
estrogen-related receptor alpha
- FHL2
four and a half LIM domains 2
- GR
glucocorticoid receptor
- HDAC
histone deacetylase
- IHC
immunohistochemistry
- ILK1
integrin-linked kinase 1
- KDM1
lysine-specific histone demethylase 1
- LIM domain
Lin-1, Isl-1, and Mec-3 domain
- MNAR
Modulator of Nongenomic Activity of ER
- PELP1
Proline, glutamic acid and leucine rich protein 1
- RXR
retinoid X receptor
- SH2
c-Src homology domain 2
- SH3
c-Src homology domain 3
- STAT3
signal transducer and activator or transcription 3
- TIF2
transcriptional mediators/intermediary factor 2
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aust S, Horak P, Pils D, Pils S, Grimm C, Horvat R, Tong D, Schmid B, Speiser P, Reinthaller A, Polterauer S. The prognostic value of estrogen receptor beta and proline-, glutamic acid- and leucine-rich protein 1 (PELP1) expression in ovarian cancer. BMC Cancer. 2013;13:115. doi: 10.1186/1471-2407-13-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasenthil S, Vadlamudi RK. Functional interactions between the estrogen receptor coactivator PELP1/MNAR and retinoblastoma protein. J Biol Chem. 2003;278:22119–22127. doi: 10.1074/jbc.M212822200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barletta F, Wong CW, McNally C, Komm BS, Katzenellenbogen B, Cheskis BJ. Characterization of the interactions of estrogen receptor and MNAR in the activation of cSrc. Mol Endocrinol. 2004;18:1096–1108. doi: 10.1210/me.2003-0335. [DOI] [PubMed] [Google Scholar]
- Beguelin W, Diaz Flaque MC, Proietti CJ, Cayrol F, Rivas MA, Tkach M, Rosemblit C, Tocci JM, Charreau EH, Schillaci R, Elizalde PV. Progesterone receptor induces ErbB-2 nuclear translocation to promote breast cancer growth via a novel transcriptional effect: ErbB-2 function as a coactivator of Stat3. Mol Cell Biol. 2010;30:5456–5472. doi: 10.1128/MCB.00012-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer C. Estrogen and the developing mammalian brain. Anatomy and embryology. 1999;199:379–390. doi: 10.1007/s004290050236. [DOI] [PubMed] [Google Scholar]
- Boonyaratanakornkit V. Scaffolding proteins mediating membrane-initiated extra-nuclear actions of estrogen receptor. Steroids. 2011;76:877–884. doi: 10.1016/j.steroids.2011.02.017. [DOI] [PubMed] [Google Scholar]
- Chakravarty D, Nair SS, Santhamma B, Nair BC, Wang L, Bandyopadhyay A, Agyin JK, Brann D, Sun LZ, Yeh IT, Lee FY, Tekmal RR, Kumar R, Vadlamudi RK. Extranuclear functions of ER impact invasive migration and metastasis by breast cancer cells. Cancer Res. 2010;70:4092–4101. doi: 10.1158/0008-5472.CAN-09-3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarty D, Roy SS, Babu CR, Dandamudi R, Curiel TJ, Vivas-Mejia P, Lopez-Berestein G, Sood AK, Vadlamudi RK. Therapeutic targeting of PELP1 prevents ovarian cancer growth and metastasis. Clin Cancer Res. 2011;17:2250–2259. doi: 10.1158/1078-0432.CCR-10-2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheskis BJ, Greger J, Cooch N, McNally C, McLarney S, Lam HS, Rutledge S, Mekonnen B, Hauze D, Nagpal S, Freedman LP. MNAR plays an important role in ERa activation of Src/MAPK and PI3K/Akt signaling pathways. Steroids. 2008;73:901–905. doi: 10.1016/j.steroids.2007.12.028. [DOI] [PubMed] [Google Scholar]
- Choi YB, Ko JK, Shin J. The transcriptional corepressor, PELP1, recruits HDAC2 and masks histones using two separate domains. J Biol Chem. 2004;279:50930–50941. doi: 10.1074/jbc.M406831200. [DOI] [PubMed] [Google Scholar]
- Cortez V, Mann M, Tekmal S, Suzuki T, Miyata N, Rodriguez-Aguayo C, Lopez-Berestein G, Sood AK, Vadlamudi RK. Targeting the PELP1-KDM1 axis as a potential therapeutic strategy for breast cancer. Breast Cancer Res. 2012;14:R108. doi: 10.1186/bcr3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimple C, Nair SS, Rajhans R, Pitcheswara PR, Liu J, Balasenthil S, Le XF, Burow ME, Auersperg N, Tekmal RR, Broaddus RR, Vadlamudi RK. Role of PELP1/MNAR signaling in ovarian tumorigenesis. Cancer Res. 2008;68:4902–4909. doi: 10.1158/0008-5472.CAN-07-5698. [DOI] [PubMed] [Google Scholar]
- Dressing GE, Hagan CR, Knutson TP, Daniel AR, Lange CA. Progesterone receptors act as sensors for mitogenic protein kinases in breast cancer models. Endocr Relat Cancer. 2009;16:351–361. doi: 10.1677/ERC-08-0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanis P, Gillemans N, Aghajanirefah A, Pourfarzad F, Demmers J, Esteghamat F, Vadlamudi RK, Grosveld F, Philipsen S, van Dijk TB. Five friends of methylated chromatin target of protein-arginine-methyltransferase[prmt]-1 (chtop), a complex linking arginine methylation to desumoylation. Molecular & cellular proteomics: MCP. 2012;11:1263–1273. doi: 10.1074/mcp.M112.017194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkbeiner E, Haindl M, Muller S. The SUMO system controls nucleolar partitioning of a novel mammalian ribosome biogenesis complex. Embo J. 2011;30:1067–1078. doi: 10.1038/emboj.2011.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonugunta VK, Nair BC, Rajhans R, Sareddy GR, Nair SS, Vadlamudi RK. Regulation of rDNA transcription by proto-oncogene PELP1. PLoS One. 2011;6:e21095. doi: 10.1371/journal.pone.0021095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivas PD, Tzelepi V, Sotiropoulou-Bonikou G, Kefalopoulou Z, Papavassiliou AG, Kalofonos H. Expression of ERalpha, ERbeta and co-regulator PELP1/MNAR in colorectal cancer: prognostic significance and clinicopathologic correlations. Cell Oncol. 2009;31:235–247. doi: 10.3233/CLO-2009-0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gururaj AE, Peng S, Vadlamudi RK, Kumar R. Estrogen induces expression of BCAS3, a novel estrogen receptor-alpha coactivator, through proline-, glutamic acid-, and leucine-rich protein-1 (PELP1) Mol Endocrinol. 2007;21:1847–1860. doi: 10.1210/me.2006-0514. [DOI] [PubMed] [Google Scholar]
- Haas D, White SN, Lutz LB, Rasar M, Hammes SR. The modulator of nongenomic actions of the estrogen receptor (MNAR) regulates transcription-independent androgen receptor-mediated signaling: evidence that MNAR participates in G protein-regulated meiosis in Xenopus laevis oocytes. Mol Endocrinol. 2005;19:2035–2046. doi: 10.1210/me.2004-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habashy HO, Powe DG, Rakha EA, Ball G, Macmillan RD, Green AR, Ellis IO. The prognostic significance of PELP1 expression in invasive breast cancer with emphasis on the ER-positive luminal-like subtype. Breast Cancer Res Treat. 2010;120:603–612. doi: 10.1007/s10549-009-0419-9. [DOI] [PubMed] [Google Scholar]
- Kashiwaya K, Nakagawa H, Hosokawa M, Mochizuki Y, Ueda K, Piao L, Chung S, Hamamoto R, Eguchi H, Ohigashi H, Ishikawa O, Janke C, Shinomura Y, Nakamura Y. Involvement of the tubulin tyrosine ligase-like family member 4 polyglutamylase in PELP1 polyglutamylation and chromatin remodeling in pancreatic cancer cells. Cancer Res. 2010;70:4024–4033. doi: 10.1158/0008-5472.CAN-09-4444. [DOI] [PubMed] [Google Scholar]
- Kayahara M, Ohanian J, Ohanian V, Berry A, Vadlamudi R, Ray DW. MNAR functionally interacts with both NH2- and COOH-terminal GR domains to modulate transactivation. Am J Physiol Endocrinol Metab. 2008;295:E1047–1055. doi: 10.1152/ajpendo.90429.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kefalopoulou Z, Tzelepi V, Zolota V, Grivas PD, Christopoulos C, Kalofonos H, Maraziotis T, Sotiropoulou-Bonikou G. Prognostic value of novel biomarkers in astrocytic brain tumors: nuclear receptor co-regulators AIB1, TIF2, and PELP1 are associated with high tumor grade and worse patient prognosis. J Neurooncol. 2011 doi: 10.1007/s11060-011-0637-y. [DOI] [PubMed] [Google Scholar]
- Khan MM, Hadman M, De Sevilla LM, Mahesh VB, Buccafusco J, Hill WD, Brann DW. Cloning, distribution, and colocalization of MNAR/PELP1 with glucocorticoid receptors in primate and nonprimate brain. Neuroendocrinology. 2006;84:317–329. doi: 10.1159/000097746. [DOI] [PubMed] [Google Scholar]
- Khan MM, Hadman M, Wakade C, De Sevilla LM, Dhandapani KM, Mahesh VB, Vadlamudi RK, Brann DW. Cloning, expression, and localization of MNAR/PELP1 in rodent brain: colocalization in estrogen receptor-alpha- but not in gonadotropin-releasing hormone-positive neurons. Endocrinology. 2005;146:5215–5227. doi: 10.1210/en.2005-0276. [DOI] [PubMed] [Google Scholar]
- Kumar R, Zhang H, Holm C, Vadlamudi RK, Landberg G, Rayala SK. Extranuclear coactivator signaling confers insensitivity to tamoxifen. Clin Cancer Res. 2009;15:4123–4130. doi: 10.1158/1078-0432.CCR-08-2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavathi B, Nair SS, Wang RA, Kumar R, Vadlamudi RK. Proline-, glutamic acid-, and leucine-rich protein-1 is essential in growth factor regulation of signal transducers and activators of transcription 3 activation. Cancer Res. 2005;65:5571–5577. doi: 10.1158/0008-5472.CAN-04-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann M, Cortez V, Vadlamudi R. PELP1 oncogenic functions involve CARM1 regulation. Carcinogenesis. 2013 doi: 10.1093/carcin/bgt091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matafora V, D’Amato A, Mori S, Blasi F, Bachi A. Proteomics analysis of nucleolar SUMO-1 target proteins upon proteasome inhibition. Molecular & cellular proteomics: MCP. 2009;8:2243–2255. doi: 10.1074/mcp.M900079-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen BS, Alves SE. Estrogen actions in the central nervous system. Endocrine reviews. 1999;20:279–307. doi: 10.1210/edrv.20.3.0365. [DOI] [PubMed] [Google Scholar]
- Mishra SK, Mazumdar A, Vadlamudi RK, Li F, Wang RA, Yu W, Jordan VC, Santen RJ, Kumar R. MICoA, a novel metastasis-associated protein 1 (MTA1) interacting protein coactivator, regulates estrogen receptor-alpha transactivation functions. J Biol Chem. 2003;278:19209–19219. doi: 10.1074/jbc.M301968200. [DOI] [PubMed] [Google Scholar]
- Mishra SK, Talukder AH, Gururaj AE, Yang Z, Singh RR, Mahoney MG, Franci C, Vadlamudi RK, Kumar R. Upstream determinants of estrogen receptor-alpha regulation of metastatic tumor antigen 3 pathway. J Biol Chem. 2004;279:32709–32715. doi: 10.1074/jbc.M402942200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musgrove EA, Sutherland RL. Biological determinants of endocrine resistance in breast cancer. Nat Rev Cancer. 2009;9:631–643. doi: 10.1038/nrc2713. [DOI] [PubMed] [Google Scholar]
- Nagpal JK, Nair S, Chakravarty D, Rajhans R, Pothana S, Brann DW, Tekmal RR, Vadlamudi RK. Growth factor regulation of estrogen receptor coregulator PELP1 functions via Protein Kinase A pathway. Mol Cancer Res. 2008;6:851–861. doi: 10.1158/1541-7786.MCR-07-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair BC, Nair SS, Chakravarty D, Challa R, Manavathi B, Yew PR, Kumar R, Tekmal RR, Vadlamudi RK. Cyclin-dependent kinase-mediated phosphorylation plays a critical role in the oncogenic functions of PELP1. Cancer Res. 2010a;70:7166–7175. doi: 10.1158/0008-5472.CAN-10-0628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair BC, Vallabhaneni S, Tekmal RR, Vadlamudi RK. Roscovitine confers tumor suppressive effect on therapy-resistant breast tumor cells. Breast Cancer Res. 2011;13:R80. doi: 10.1186/bcr2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair SS, Guo Z, Mueller JM, Koochekpour S, Qiu Y, Tekmal RR, Schule R, Kung HJ, Kumar R, Vadlamudi RK. Proline-, glutamic acid-, and leucine-rich protein-1/modulator of nongenomic activity of estrogen receptor enhances androgen receptor functions through LIM-only coactivator, four-and-a-half LIM-only protein 2. Mol Endocrinol. 2007;21:613–624. doi: 10.1210/me.2006-0269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair SS, Mishra SK, Yang Z, Balasenthil S, Kumar R, Vadlamudi RK. Potential role of a novel transcriptional coactivator PELP1 in histone H1 displacement in cancer cells. Cancer Res. 2004;64:6416–6423. doi: 10.1158/0008-5472.CAN-04-1786. [DOI] [PubMed] [Google Scholar]
- Nair SS, Nair BC, Cortez V, Chakravarty D, Metzger E, Schule R, Brann DW, Tekmal RR, Vadlamudi RK. PELP1 is a reader of histone H3 methylation that facilitates oestrogen receptor-alpha target gene activation by regulating lysine demethylase 1 specificity. EMBO Rep. 2010b;11:438–444. doi: 10.1038/embor.2010.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshiro K, Rayala SK, Kondo S, Gaur A, Vadlamudi RK, El-Naggar AK, Kumar R. Identifying the estrogen receptor coactivator PELP1 in autophagosomes. Cancer Res. 2007;67:8164–8171. doi: 10.1158/0008-5472.CAN-07-0038. [DOI] [PubMed] [Google Scholar]
- Park I, Chung J, Walsh CT, Yun Y, Strominger JL, Shin J. Phosphotyrosine-independent binding of a 62-kDa protein to the src homology 2 (SH2) domain of p56lck and its regulation by phosphorylation of Ser-59 in the lck unique N-terminal region. Proc Natl Acad Sci U S A. 1995;92:12338–12342. doi: 10.1073/pnas.92.26.12338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlak J, Beyer C. Developmental expression of MNAR mRNA in the mouse brain. Cell Tissue Res. 2005;320:545–549. doi: 10.1007/s00441-005-1090-z. [DOI] [PubMed] [Google Scholar]
- Rajhans R, Nair HB, Nair SS, Cortez V, Ikuko K, Kirma NB, Zhou D, Holden AE, Brann DW, Chen S, Tekmal RR, Vadlamudi RK. Modulation of in situ estrogen synthesis by proline-, glutamic acid-, and leucine-rich protein-1: potential estrogen receptor autocrine signaling loop in breast cancer cells. Mol Endocrinol. 2008;22:649–664. doi: 10.1210/me.2007-0350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajhans R, Nair S, Holden AH, Kumar R, Tekmal RR, Vadlamudi RK. Oncogenic potential of the nuclear receptor coregulator proline-, glutamic acid-, leucine-rich protein 1/modulator of the nongenomic actions of the estrogen receptor. Cancer Res. 2007;67:5505–5512. doi: 10.1158/0008-5472.CAN-06-3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaud J, Desroches J, Balsalobre A, Drouin J. TIF1beta/KAP-1 is a coactivator of the orphan nuclear receptor NGFI-B/Nur77. J Biol Chem. 2009;284:14147–14156. doi: 10.1074/jbc.M809023200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindranathan P, Lee TK, Yang L, Centenera MM, Butler L, Tilley WD, Hsieh JT, Ahn JM, Raj GV. Peptidomimetic targeting of critical androgen receptor-coregulator interactions in prostate cancer. Nature communications. 2013;4:1923. doi: 10.1038/ncomms2912. [DOI] [PubMed] [Google Scholar]
- Rayala SK, Hollander P, Balasenthil S, Molli PR, Bean AJ, Vadlamudi RK, Wang RA, Kumar R. Hepatocyte growth factor-regulated tyrosine kinase substrate (HRS) interacts with PELP1 and activates MAPK. J Biol Chem. 2006a;281:4395–4403. doi: 10.1074/jbc.M510368200. [DOI] [PubMed] [Google Scholar]
- Rayala SK, Mascarenhas J, Vadlamudi RK, Kumar R. Altered localization of a coactivator sensitizes breast cancer cells to tumor necrosis factor-induced apoptosis. Mol Cancer Ther. 2006b;5:230–237. doi: 10.1158/1535-7163.MCT-05-0276. [DOI] [PubMed] [Google Scholar]
- Rosendorff A, Sakakibara S, Lu S, Kieff E, Xuan Y, DiBacco A, Shi Y, Gill G. NXP-2 association with SUMO-2 depends on lysines required for transcriptional repression. Proc Natl Acad Sci U S A. 2006;103:5308–5313. doi: 10.1073/pnas.0601066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Chakravarty D, Cortez V, De Mukhopadhyay K, Bandyopadhyay A, Ahn JM, Raj GV, Tekmal RR, Sun L, Vadlamudi RK. Significance of PELP1 in ER-negative breast cancer metastasis. Mol Cancer Res. 2012;10:25–33. doi: 10.1158/1541-7786.MCR-11-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RR, Gururaj AE, Vadlamudi RK, Kumar R. 9-cis-retinoic acid up-regulates expression of transcriptional coregulator PELP1, a novel coactivator of the retinoid X receptor alpha pathway. J Biol Chem. 2006;281:15394–15404. doi: 10.1074/jbc.M601593200. [DOI] [PubMed] [Google Scholar]
- Tzelepi V, Grivas P, Kefalopoulou Z, Kalofonos H, Varakis JN, Sotiropoulou-Bonikou G. Expression of estrogen receptor co-regulators NCoR and PELP1 in epithelial cells and myofibroblasts of colorectal carcinomas: cytoplasmic translocation of NCoR in epithelial cells correlates with better [corrected] prognosis. Virchows Arch. 2009;454:41–53. doi: 10.1007/s00428-008-0708-4. [DOI] [PubMed] [Google Scholar]
- Unni E, Sun S, Nan B, McPhaul MJ, Cheskis B, Mancini MA, Marcelli M. Changes in androgen receptor nongenotropic signaling correlate with transition of LNCaP cells to androgen independence. Cancer Res. 2004;64:7156–7168. doi: 10.1158/0008-5472.CAN-04-1121. [DOI] [PubMed] [Google Scholar]
- Vadlamudi RK, Balasenthil S, Broaddus RR, Gustafsson JA, Kumar R. Deregulation of estrogen receptor coactivator proline-, glutamic acid-, and leucine-rich protein-1/modulator of nongenomic activity of estrogen receptor in human endometrial tumors. J Clin Endocrinol Metab. 2004;89:6130–6138. doi: 10.1210/jc.2004-0909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadlamudi RK, Balasenthil S, Sahin AA, Kies M, Weber RS, Kumar R, El-Naggar AK. Novel estrogen receptor coactivator PELP1/MNAR gene and ERbeta expression in salivary duct adenocarcinoma: potential therapeutic targets. Hum Pathol. 2005a;36:670–675. doi: 10.1016/j.humpath.2005.03.016. [DOI] [PubMed] [Google Scholar]
- Vadlamudi RK, Manavathi B, Balasenthil S, Nair SS, Yang Z, Sahin AA, Kumar R. Functional implications of altered subcellular localization of PELP1 in breast cancer cells. Cancer Res. 2005b;65:7724–7732. doi: 10.1158/0008-5472.CAN-05-0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadlamudi RK, Rajhans R, Chakravarty D, Nair BC, Nair SS, Evans DB, Chen S, Tekmal RR. Regulation of aromatase induction by nuclear receptor coregulator PELP1. J Steroid Biochem Mol Biol. 2010;118:211–218. doi: 10.1016/j.jsbmb.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadlamudi RK, Wang RA, Mazumdar A, Kim Y, Shin J, Sahin A, Kumar R. Molecular cloning and characterization of PELP1, a novel human coregulator of estrogen receptor alpha. J Biol Chem. 2001;276:38272–38279. doi: 10.1074/jbc.M103783200. [DOI] [PubMed] [Google Scholar]
- Vallabhaneni S, Nair BC, Cortez V, Challa R, Chakravarty D, Tekmal RR, Vadlamudi RK. Significance of ER-Src axis in hormonal therapy resistance. Breast Cancer Res Treat. 2010 doi: 10.1007/s10549-010-1312-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams MD, Roberts D, Blumenschein GR, Jr, Temam S, Kies MS, Rosenthal DI, Weber RS, El-Naggar AK. Differential expression of hormonal and growth factor receptors in salivary duct carcinomas: biologic significance and potential role in therapeutic stratification of patients. Am J Surg Pathol. 2007;31:1645–1652. doi: 10.1097/PAS.0b013e3180caa099. [DOI] [PubMed] [Google Scholar]
- Wong CW, McNally C, Nickbarg E, Komm BS, Cheskis BJ. Retraction for Wong et al., “Estrogen receptor-interacting protein that modulates its nongenomic activity-crosstalk with Src/Erk phosphorylation cascade”. Proc Natl Acad Sci U S A. 2009;106:14180. doi: 10.1073/pnas.0907607106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Ravindranathan P, Ramanan M, Kapur P, Hammes SR, Hsieh JT, Raj GV. Central role for PELP1 in nonandrogenic activation of the androgen receptor in prostate cancer. Mol Endocrinol. 2012;26:550–561. doi: 10.1210/me.2011-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]


