Abstract
Flavoproteins catalyze a diversity of fundamental redox reactions and are one of the most studied enzyme families1,2. As monooxygenases, they are universally thought to control oxygenation by means of a peroxyflavin species that transfers a single atom of molecular oxygen to an organic substrate1,3,4. Here we report that the bacterial flavoenzyme EncM5,6 catalyzes the peroxyflavin-independent oxygenation-dehydrogenation dual oxidation of a highly reactive poly(β-carbonyl). The crystal structure of EncM with bound substrate mimics coupled with isotope labeling studies reveal previously unknown flavin redox biochemistry. We show that EncM maintains an unanticipated stable flavin oxygenating species, proposed to be a flavin-N5-oxide, to promote substrate oxidation and trigger a rare Favorskii-type rearrangement that is central to the biosynthesis of the antibiotic enterocin. This work provides new insight into the fine-tuning of the flavin cofactor in offsetting the innate reactivity of a polyketide substrate to direct its efficient electrocyclization.
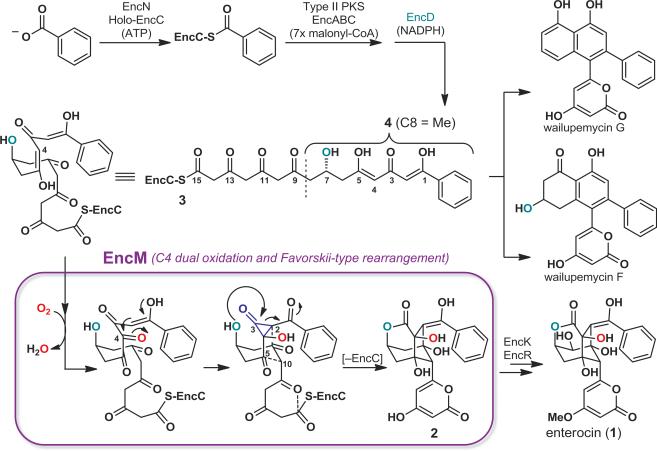
The antibiotic enterocin (compound 1, Fig. 1) is produced by various streptomycete bacteria7 and contains a unique, tricyclic caged core. Nearly 40 years ago, isotope labeling studies suggested the involvement of a rare oxidative Favorskii-type rearrangement during its biosynthesis8. More recently, discovery, expression, and biochemical analyses of the Streptomyces maritimus enterocin biosynthetic gene cluster including in vitro reconstitution of the metabolic pathway, demonstrated further involvement of the type II polyketide synthase, EncABC, and the NADPH-dependent reductase, EncD6,7,9 (Fig. 1). While type II polyketide synthase pathways typically yield polycyclic aromatic products like the antibiotic tetracycline and the anticancer agent doxorubicin10, aromatic polyketides called wailupemycins are formed only as minor products of the enterocin biosynthetic pathway7. Remarkably, the flavin adenine dinucleotide (FAD)-dependent “favorskiiase” EncM proved to be singly responsible for interruption of the more typical polycyclic aromatization of the poly(β-carbonyl) chain to direct generation of the rearranged desmethyl-5-deoxyenterocin (2)5,6. To date, detailed mechanistic studies of EncM have been hampered by the inherently high reactivity of the proposed EncM substrate, a putative acyl carrier protein (ACP)-bound C7,O4-dihydrooctaketide intermediate (EncC-octaketide) (3). To overcome this experimental limitation we employed synthetic substrate analogs (for synthesis see Supplementary Information), including the untethered C7,O4-dihydrotetraketide (4, Fig. 1), for structure-function analyses of recombinant EncM.
Figure 1. Overview of the Streptomyces maritimus enterocin biosynthetic pathway and proposed EncM catalysis.
The ACP EncC is primed with benzoate by ligase EncN, followed by seven iterative type II polyketide synthase (EncAB)-catalyzed elongation steps by decarboxylative Claisen condensations with malonyl-CoA. The ketoreductase EncD likely forms the (R)-7-hydroxyl during elongation. The linear (R)-C7,O4-dihydrooctaketide (3) can cyclize to various wailupemycins (e.g., G and F), while in the presence of EncM is preferentially converted into desmethyl-5-deoxyenterocin (2). Final pathway steps leading to enterocin (1) are catalyzed by EncR and EncK. EncM catalysis (blue box) involves C4 dual oxidation (see Fig. 3b) and a Favorskii-type rearrangement, followed by aldol-condensations and heterocycle formation (dashed lines). Functional studies of EncM were conducted with the substrate analog 4.
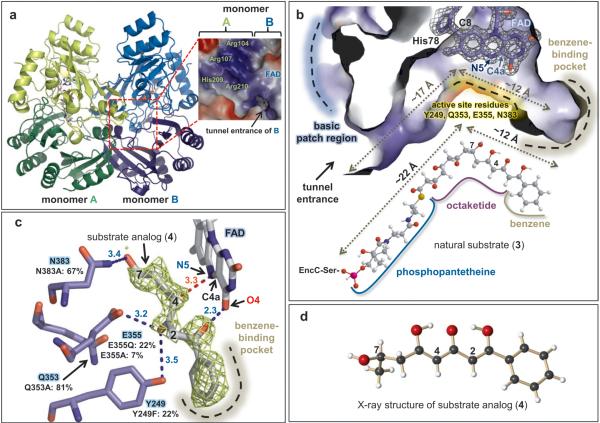
Several crystal structures of FAD-bound EncM were determined at resolutions up to 1.8 Å by molecular replacement against 6-hydroxy-D-nicotine oxidase (6HDNO) from Arthrobacter nicotinivorans11 (Fig.1, Supplementary Table 1). Structurally, EncM exhibits greater architectural similarity to flavin dehydrogenases rather than to oxygenases, such as 6HDNO (33% sequence identity for 444 equivalent amino acid residues, 2.2 Å root-mean-square-deviation (rmsd) for Cα-atoms, Z-score = 46.4), glucooligosaccharide oxidase12 (31% sequence identity for 415 equivalent residues, 2.3 Å rmsd, Z-score = 44.1) and aclacinomycin oxidoreductase13 (37% sequence identity for 316 equivalent residues, 2.5 Å rmsd, Z-score = 40.6). In contrast to these monomeric dehydrogenases, EncM exists as homodimer in crystal form and in solution (Fig. 2a, Supplementary Fig. 1). The monomeric subunits of the homodimer show high structural similarity (0.19 Å rmsd for Cα atoms), and each contains distinct domains for substrate-binding (residues 211-418) and FAD-binding (residues 2-210 and 419-461). The FAD-binding domain sequesters the ADP-ribosyl of the flavin cofactor, while the reactive isoalloxazine core resides at the substrate-cofactor domains’ interface (Figs 2a, b). As previously observed in 6HDNO, the flavin is covalently linked to EncM via the C8-methyl of the isoalloxazine ring system and a histidine residue (His78) (Fig. 2b).
Figure 2. Crystal structure of EncM.
a, Homodimeric EncM shown as a ribbon diagram (with flavin cofactors as color-coded stick model). Monomeric subunits are colored in green and blue with darker shades of each highlighting the substrate-binding domains and lighter shades emphasizing the flavin-binding domains. The basic patch abutting the active site tunnel entrance is magnified in the dashed red box (blue and red colors indicate positive and negative charges, respectively). b, Sliced-away interior view of the EncM substrate tunnel showing a covalent link between His78 and FAD (shown is the SIGMAA-weighted 2Fo - Fc electron density map contoured at 2.0 σ).The natural substrate 3 is shown below. Approximate lengths of the tunnel and substrate are indicated. c, SIGMAA-weighted Fo-Fc difference map calculated with the ligand omitted, contoured at 2.0 σ around modeled 4. Hydrogen-bonding interactions are indicated by blue dashed lines. The red dashed line shows the distance (Å) from the site of oxidation to the reactive N5 of FAD. Normalized activities of active site mutants are shown (native EncM = 100%). d, X-ray structure of chemically synthesized 4.
Structure comparisons with homologous flavin-dependent enzymes emphasized the unusually elongated L-shaped EncM ligand-binding tunnel that extends approximately 30 Å from the surface to a hydrophobic pocket at its base. This orthogonally arranged two-room tunnel is highly complementary to the shapes of the ACP-derived phosphopantetheine arm, the octaketide chain, and the terminal benzene moiety of 3 (Fig. 2b, Supplementary Fig. 2). The entrance of the tunnel of EncM sits near the dimer interface and adjacent to a surface exposed basic patch formed by a few positively charged residues, including Arg107 and Arg210, from the dyad related monomer (Fig. 2a). This positively charged region of EncM is highly complementary to the decidedly negative surface area of ACPs14, suggestive that EncC7 presents elongated polyketide intermediates to EncM through protein-protein interactions to limit deleterious side reactions of the highly reactive poly(β-carbonyl) chain. Support for the close association of EncM and EncC was obtained by protein-protein computational docking simulation using an EncC homology model (Supplementary Fig. 3). Moreover, disruption of the positive surface area of the EncM dimer with the EncM-R210E mutant, resulted in ~40% the relative activity as native EncM (Supplementary Fig. 4).
To explore the interaction of EncM with the polyketide reactant, we co-crystallized the enzyme with substrate analogs harboring the benzene moiety of 3 (Supplementary Table 1). The resulting SIGMAA-weighted Fo-Fc electron-density difference maps clearly indicated mimetic binding to the active site, although elevated B-factors and incomplete occupancy (e.g., ≈ 33 Å2 and 0.8, respectively for substrate 4) caused slightly disordered electron densities (Fig. 2c, Supplementary Fig. 5). Binding occurred with little overall structural perturbation to the EncM polypeptide backbone (e.g., 0.14 Å rmsd for 4) and no significant backbone or side-chain displacements in the binding region. The terminal benzene group sits at the end of a largely hydrophobic tunnel and forms aromatic-aromatic and van der Waals interactions with Tyr150, Trp152, and Leu357, respectively. Likely, the enol at C1 engages in hydrogen bonding with O4 of the flavin (2.3 Å), while the C3 ketone twists away from the flavin and may accept a hydrogen bond from the side-chain of Glu355 (3.2 Å), and possibly from Tyr249 (3.5 Å). Mutagenesis of these residues confirmed their importance for EncM activity (Fig. 2c). Notably, the putative C7-hydroxyl of 4 resides at the elbow of the L-shaped two-room tunnel and ostensibly serves as the pivot point in the natural substrate 3. The mutually orthogonal sections of the EncM ligand-binding pocket separate the C1–C6 triketide head from the C8–C15 pantothenate-linked tetraketide tail to uncouple the reactivity of the entire C1-C16 poly(β-carbonyl) chain. This chemical and structural disconnection prevents kinetically facile but unwanted cyclizationaromatization reactions, and instead favors the EncM-mediated oxidative Favorskii-type rearrangement (Fig. 2b).
We hypothesize that EncM performs a dual oxidation of 3 at C4 to effectively convert a 1,3-diketone to a 1,2,3-triketone. In this mechanistic scenario, C4 is now set up to undergo a facile electrophilic cyclization with C2 to trigger the proposed Favorskii-like rearrangement (Fig. 1). Typical flavin oxygenases are initially reduced with NAD(P)H to enable capture of O2 by reduced flavin (Flred) generating the flavin-C4a-peroxide oxygenating species4. EncM, however, lacks an NAD(P)H binding domain and functions in the absence of a flavin reductase6, raising questions surrounding the oxidative mechanism of EncM.
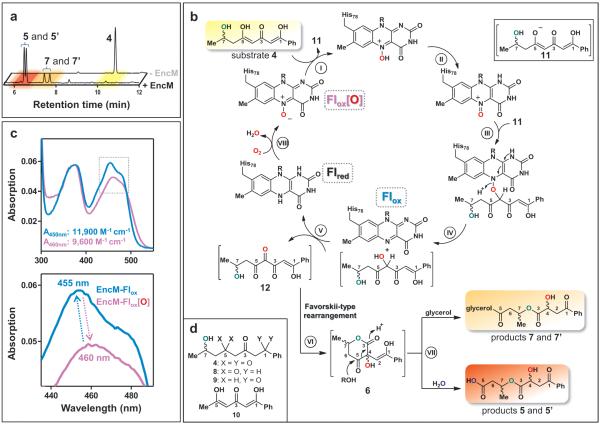
To gain further insight into the EncM chemical mechanism, we analyzed the in vitro reaction of EncM with either racemic or enantiopure 4 by reverse-phase HPLC and UV-Vis spectroscopy. Remarkably, 4 was converted in the absence of NAD(P)H into diastereomeric products 5 and 5’ without detectable intermediates (Fig. 3a). Through comprehensive NMR and MS analyses together with chemical synthesis (see Supplementary Information), we identified 5 and 5’ as ring-opened derivatives of the expected enterocin-like lactone 6 (Fig. 3b). Circular dichroism experiments proved that the configuration of 4 is maintained during the transformation (see Supplementary Information). We reasoned that a facile hydrolytic retro-Claisen ring cleavage15,16 of 6 occurs after an oxidative Favorskii-type rearrangement and lactonization (Fig. 3b, step VI) that is likely responsible for the racemization of C4. This proposed reaction was further substantiated by the observation that glycerol also effectuates the ring opening to form 7 and 7’ (Fig. 3a, Supplementary Figs 6, 7). During actual enterocin biosynthesis, this reaction is likely prevented through aldol condensations with the remainder of the ketide chain (Fig. 1). Notably, the C1 and C5 deoxo-substrate analogs 8 and 9, respectively, were not transformed by EncM, while the dehydroxy-substrate 10 (see Fig 3d or Supplementary Fig. 5 for compound structures) was converted into multiple unstable products that were not further characterized. This series of structure-activity relationships revealed that the triketone motif (C1–C6) is essential for catalysis and suggested that the C7-hydroxyl is critical for spatial and temporal control of the EncM catalyzed reaction.
Figure 3. Proposed EncM mechanism and spectral features of the flavin cofactor catalytic states.
a, Reverse-phase HPLC analysis (absorption detection at 254 nm) of enzymatic assays showing substrate analog 4 (upper lane; control assay without EncM) and diastereomeric product pairs 5/5’ and 7/7’ (lower lane; after incubation with EncM). The color code refers to panel b. 7/7’ were only observed in presence of glycerol (here 20%, v/v). No intermediates could be detected. b, Proposed EncM-catalytic mechanism involving substrate oxygenation via a flavin-N5-oxoammonium species. The resultant electrophilic C4-ketone of 12 triggers the Favorskii-type rearrangement and lactone formation (see Fig. 1 for the detailed analogous reactions during natural enterocin biosynthesis), while the formed Flred reacts with O2 and restores the N5-oxide. The stepwise dual oxidation is supported by anaerobic single turnover experiments (Supplementary Fig. 16). The C7-hydroxyl is shown in green, and oxygen atoms derived from O2 and H2O are color coded red and blue, respectively. Roman numerals indicate reaction steps as discussed in the main text. c, UV-Vis spectra of EncM's oxidized flavin as isolated (Flox[O], catalytically active, purple curve) and after multiple substrate turnovers (Flox, catalytically inactive, blue curve). Extinction coefficients are shown. d, Compounds used for structure-activity relationship analyses.
The monooxygenase activity of EncM was evaluated by following the incorporation of oxygen atoms from 18O2 into 5/5’ and 7/7’ at C4. In contrast, isotope labeling from H218O was only associated with the non-enzymatic retro-Claisen cleavage of 6 to 5/5’ (Supplementary Figs 8 and 9). These measurements suggest that lactone formation during enterocin biosynthesis is controlled by the C7-hydroxyl via direct intramolecular attack (Fig. 1). Further support for this biosynthetic model came from the structure analysis of the EncM ligand-binding tunnel that can only accommodate the (R)-enantiomer of 3 (Supplementary Fig. 10), which is consistent with the observed retention of the C4-hydroxyl configuration in the final product enterocin (Fig. 1).
Surprisingly, EncM became inactivated after several turnovers (Supplementary Fig. 11). Moreover, the oxidized flavin cofactor of inactivate EncM (EncM-Flox) exhibited distinct, stable changes in the UV-Vis spectrum (Fig. 3c). We speculated that these spectral perturbations are caused by the loss of an oxygenating species maintained in the enzyme's active state. This species, “EncM-Flox[O]”, is largely restored at the end of each catalytic cycle (Fig. 3b), thereby providing an explanation for the innate monooxygenase activity of EncM in the absence of exogenous reductants. We excluded the participation of active site residues in harboring this oxidant through site-directed mutagenesis and by showing that denatured EncM retained the Flox[O] spectrum (Supplementary Fig. 12). We therefore focused on the flavin cofactor as the carrier of the oxidizing species. Based on the spectral features of EncM-Flox[O], we ruled out a conventional C4a-peroxide17,18. Moreover, Flox[O] is extraordinarily stable (no detectable decay for >7 d at 4°C) and thus is vastly longer lived than even the most stable flavin-C4a-peroxides described to date (t1/2 of ≤30 min at 4°C19,20).
To further test the possible intermediacy and catalytic role of EncM-Flox[O], we anaerobically reduced the flavin cofactor and showed that only flavin reoxidation with molecular oxygen restored the EncM-Flox[O] species. In contrast, anoxic chemical reoxidation generated catalytically inactive EncM-Flox (Supplementary Fig. 13a). Significantly, EncM reoxidized with 18O2 formed EncM-Flox[18O], which converted 4 to [18O]- 5/5’ with 1:1 stoichiometry of Flox[18O] to [18O]- 5/5’ (Supplementary Fig. 13b). The collective structure-function analyses reported here currently support the catalytic use of a unique flavin oxygenating species that is consistent with a flavin-N5-oxide. This chemical species was introduced over 30 years ago as a possible intermediate in flavin monooxygenases21,22 before the conventional C4a-peroxide model was experimentally accepted. Crucially, spectrophotometric comparison of chemically synthesized flavin-N5-oxide and EncM-Flox[O] revealed many of the same spectral features23 and both can be chemically converted to oxidized flavin (Supplementary Fig. 12). Moreover, consistent with an N-oxide, EncM-Flox[O] required four electrons per flavin cofactor to complete reduction in dithionite titrations, whereas EncM-Flox only required two (Supplementary Fig. 14). Noteworthy, we could not observe this flavin modification crystallographically (see Fig. 2b), presumably due to X-radiation induced reduction24 of the flavin-N5-oxide, which is highly prone to undergo reduction23.
We propose that during EncM catalysis, the N5-oxide is first protonated by the hydroxyl proton of the C5-enol of substrate 4 (Fig. 3b, step I). Despite the generally low basicity of N-oxides, the proton transfer is likely enabled by the high acidity of the C5 enol and its appropriate positioning 3.4 Å from the N5 atom of the flavin (Fig. 2c). After protonation, tautomerization of the N5-hydroxylamine would lead to the electrophilic oxoammonium (step II). Subsequent oxygenation of substrate enolate 11 by the oxoammonium species may then occur via one of several possible routes (Supplementary Fig. 15), yielding Flox and a C4-hydroxylated intermediate (steps III and IV). Flox-mediated dehydrogenation of the introduced alcohol group then produces the C4-ketone 12 and Flred (step V). Anaerobic single turnover experiments with 4 support this reaction sequence (Supplementary Fig. 16). Finally, 12 would undergo the Favorskii-type rearrangement (step VI) and retro-Claisen transformation (step VII) to yield the observed products 5/5’ or 7/7’, while the reduced cofactor Flred reacts with O2 to regenerate EncM-Flox[O] and thus prime the enzyme for the next catalytic cycle (steps VIII). Alternative mechanisms, however, are also plausible (Supplementary Fig. 17). This extraordinary flavin cofactor-mediated dual oxidation vaguely resembles the role of flavins in the scarce “internal monooxygenases” (EC 1.13.12) that also use their substrate as an electron donor25.
In summary, we provide the first in-depth investigation of an enzymatic oxidation-induced Favorskii-type rearrangement. The exceptionally reactive poly(β-carbonyl) substrate requires EncM to direct the reaction along a defined mechanistic trajectory by sequestration of reactants from bulk solvent, spatial separation of reactive functional groups, rapid “one-step” generation of a new electrophilic center, and expulsion of solvent from the active site to prevent retro-Claisen ring cleavage. The discovery that EncM utilizes a stable flavin-N5-oxide for oxygenation rather than the universally accepted flavin peroxide suggests that this species may have been overlooked in the flavin biochemical literature. Further studies are underway to explore the factors that govern enzymatic formation of the flavin-N5-oxide. In short, the archetypal dual oxidase EncM employs unanticipated oxidative flavin biochemistry for NAD(P)H-independent processing of extremely reactive polyketides.
Methods
Gene cloning, heterologous protein expression, and purification procedures
Escherichia coli strain BL21 (DE3) (New England Biolabs, Ipswich, MA, USA) and Streptomyces lividans TK24 were used for heterologous protein expression. The enterocin enzymes holo-EncC26, EncA-EncB26, EncD6, and EncN27 from Streptomyces maritimus, and FabD28 from Streptomyces glaucescens were prepared as His-tagged recombinant proteins as previously described6, 26-28. The plasmid encoding FabD was provided by Professor K. A. Reynolds. The EncM gene was amplified from pXY200-EncM6 with the following primer: 5’-AAAACCATGGGCAGTTCCCACAGCTCGAC-3’ and 5’-TTTTGAATTCTCAGGGGCTGCTCGGG-3’ (NcoI and EcoRI restriction sites are underlined) and then inserted between the NcoI and EcoRI sites of the expression vector pHIS829. E. coli BL21 (DE3) harboring pHIS8-EncM plasmid was grown at 28 °C in 4 L of lysogeny broth containing 50 μg/ml kanamycin until the D600nm reached approximately 0.5. Isopropyl-β-D-thiogalactoside (IPTG, μM) was then added to induce recombinant protein expression under control of T7 RNA polymerase induced using a modified lac promoter. Cells were grown for an additional 24 h at 28 °C and harvested by centrifugation. Cell pellets were resuspended in lysis buffer (50 mM sodium phosphate (pH 7.7), 300 mM sodium chloride and 10% (v/v) glycerol supplemented with 10 mM imidazole, and lysed by sonication. After centrifugation, the supernatant was passed over a Ni2+-NTA column connected to a FPLC system. Unbound protein was removed by washing and the N-terminal octahistidine-tagged EncM was then eluted with lysis buffer supplemented with 500 mM imidazole. The protein was desalted and concentrated using PD-10 and Vivaspin 6 (30 kDa exclusion size) columns (both GE Healthcare, Uppsala, Sweden), respectively. For crystallization, EncM was further treated with thrombin to remove the His-tag, subjected to another round of His-trap purification, followed by ResourceQ™ (GE Healthcare) anion exchange chromatography using a linear gradient from 0-1 M NaCl over 30 min in 10 mM TES-Na+ buffer (pH 7.7), 10% (v/v) glycerol.
Hydrodynamic analysis of EncM by size exclusion chromatography
0.5 mg of EncM protein was loaded onto a HiLoad 26/60 Superdex 200 column equilibrated with buffer containing 20 mM TES-Na+ (pH 7.5), 0.15 M NaCl and 10% (v/v) glycerol. Eluting protein was observed by monitoring the absorbance at 280 nm. The column was calibrated with Bio-Rad (Hercules, CA, USA) standard proteins (thyroglobulin, 670 kDa; γ-globulin, 158 kDa; ovalbumin, 44 kDa; myoglobin, 17 kDa).
Molar extinction coefficients of EncM-Flox[O] and EncM-Flox
A solution of anaerobic dithionite in a gas-tight syringe was calibrated by titrating a known concentration of flavin mononucleotide to full reduction. The dithionite syringe was transferred to an anaerobic cuvette containing EncM-Flox and then titrated with the calibrated dithionite to complete reduction. The amount of dithionite needed to fully reduce EncM-Flox was used to determine the molar extinction coefficient (ε) of 11,900 M−1cm−1 at 450 nm based on the original absorbance spectrum. Subsequent exposure to O2 led to oxidation of reduced EncM to EncM-Flox[O], from which ε of 9,600 M−1cm−1 at 460 nm was calculated.
Site-directed mutagenesis
The expression plasmid pHIS8-EncM was used for site-directed mutagenesis with the QuikChange site-directed mutagenesis kit according to protocol (Stratagene, La Jolla, CA). The following oligonucleotides (and respective complementary primers) were used to obtain the EncM mutants R210E, Y249F, Q353A, E355A, E355Q, and N383A, respectively: 5’-GAGTTCGACCTCCACGAGGTCGGGCCCGTC-3’, 5’-CTGACCTGGGCGTTGTTTCTGCGCCTGGCAC-3’, 5’-GCCTCCCCCTTCACTGCGCTCGAACTGCTCTACC-3’, 5’-CCCTTCACTCAGCTCGCACTGCTCTACCTGGG-3’, 5’-CCCTTCACTCAGCTCCAACTGCTCTACCTGGG-3’, and 5’-CGCCGTTCGTGACCGCCCTGGCCGCCGC-3’. The mutations were confirmed by sequence analysis.
Crystallization, structure determination, and refinement
Crystals of EncM were grown from a 1:1 mixture of protein solution (5 mg mL−1 in 10 mM TES-Na+ (pH 7.7), 10% (v/v) glycerol), and a reservoir solution (2 mM DTT, 0.1 M HEPES-Na+ (pH 7.5), 0.2 M calcium acetate, and 20% (w/v) PEG3350) using hanging-drop vapor diffusion at 4 °C. For co-crystallization, EncM was incubated with 2 mM of the respective substrate analogs prior to mixing with the reservoir solution. The crystals were transferred into the reservoir solution containing 25% (v/v) glycerol as a cryoprotectant and flash-frozen in liquid nitrogen until X-ray data collection on beamlines 8.2.1 and 8.2.2 at the Advanced Light Source (ALS, Berkeley, CA, USA). All diffraction data were indexed, integrated and scaled using the program HKL200030 or iMosfilm31. The initial phases were determined by molecular replacement using the program Molrep32. The crystal structure of 6-hydroxy-D-nicotine oxidase (6HDNO) (PDB code 2BVG) was used as a search model and the programs ARP/wARP33, Coot34 and Refmac35 were used for automatic model building, visual inspection and manual rebuilding of the model, and for several rounds of energy minimization and individual B-factor refinement, respectively. Ramachandran statistics: EncM apo: favored region 98.0%, allowed region 1.5%, outlier region 0.4%; EncM with 26: favored region 98.8%, allowed region 1.1%, outlier region 0.1%; EncM with 4: favored region 98.8%, allowed region 1.0%, outlier region 0.2%. The figures were prepared using Pymol36. Occupancies and B-factors for EncM-bound substrate analogs were determined with Phenix37.
Enzyme assays (Fig. 3a, Supplementary Figure 11)
The kinetics for product formation were determined at 22 °C using two replicate assays containing 20 mM HEPES-Na+ (pH 7.5), 300 mM NaCl, ≥10% (v/v) glycerol, 0.7 mM 4, and 10 μM EncM. EncM concentrations were adjusted based on the molar extinction coefficient of EncM-Flox[O] (9,600 M−1 cm−1) at 460 nm. Samples were sequentially withdrawn and quenched after 1, 3, 6, 12, 20, 30, and 40 min. To determine native and mutant EncM activities, a final concentration of 3.4 μM of each EncM mutant was incubated with 0.6 mM 4 in 50 mM HEPES-Na+ (pH 7.5), 200 mM NaCl, 1 mM NADPH, and 10% (v/v) glycerol using three replicate assays. The reactions were quenched after 10 min (when <50% of the substrate had been converted) and the products quantified. All samples described in this section were analyzed by HPLC (see below).
EncM flavin oxidation with molecular oxygen (18O2 or 16O2) and 2,6-dichlorophenolindophenol (Supplementary Fig. 13)
20 μM EncM-Flox[O] active sites were completely reduced in an anaerobic cuvette with sodium dithionite prior to reoxidation by injection of either ~97% 18O2 gas (Sigma-Aldrich, Saint Louis, MO, USA), ~50% 18O2 gas (1:1 mixture of 18O2: 16O2), or air. Unreacted O2 was then thoroughly removed by repeated cycles of vacuum and argon treatment. 100 μM 4 was then added at room temperature. After complete consumption of 4, protein was removed through filtration and the samples acidified with 1 M HCl prior to LC-MS analysis. Alternatively, EncM was reoxidized anaerobically with the chemical oxidant 2,6-dichlorophenolindophenol instead of O2, producing catalytically inactive EncM-Flox (no products were detected after incubation with 4).
Model docking (Supplementary Figure 3c)
The homology model of EncC was generated by Swiss Model38 based on the solved structure of the ACP of actinorhodin biosynthesis from Streptomyces coelicolor (PDB code: 1AF8). The docking simulation was carried out with the GRAMM-X Protein-Protein Docking Web Server39, using the EncM structure and the EncC homology model. The resulting structure was then energy-minimized with Swiss-model viewer40.
In vitro reconstitution assay with the enterocin PKS (Supplementary Figure 4)
The activities of EncM and EncM-R210E were assayed using the fully reconstituted enc PKS enzyme set as previously reported6. The standard mixture contained 1 μM EncA-EncB, 8 μM EncC, 1.5 μM EncD, 2 μM EncM, 0.15 μM EncN, 0.015 μM FabD, 5 mM ATP, 5 mM MgCl2, 5 mM NADPH, 1 mM malonyl-CoA and 0.25 mM benzoic acid in a volume of 100 μl. After incubation at 30 °C for 2 h, the reactions were quenched by the addition of 10 μl of 2 M HCl. The products were then extracted with 2 × 200 μl EtOAc. The organic extracts were combined and evaporated to dryness. The residual material was resuspended in 30 ml MeCN and analyzed by HPLC and LC-ESI mass spectrometry. A Phenomenex 250 mm × 4.6 mm C18 column was used at a flow rate of 1.0 mL min−1 with a linear gradient of 5% to 80% (v/v) MeCN in water containing 0.01% (v/v) TFA over a period of 40 min.
UV-Vis spectrophotometry (Fig. 3c, Supplementary Figs 12-14)
The flavin absorption spectra of purified EncM were analyzed using an Agilent Cary 50 UV-Vis spectrophotometer or a Shimadzu UV-2501 PC. Untreated EncM (as isolated from E. coli) showed the EncM-Flox[O] spectrum. After incubation with substrate (and subsequent product removal using a PD-10 column), the spectrum of EncM-Flox was observed.
Analytic (Fig. 3a), semipreparative, and chiral HPLC
Samples from enzymatic assays were quenched in acidic MeOH and centrifuged. The supernatants were analyzed by reverse-phase HPLC (Agilent, 1200 series) using a Sync Polar RP column 4μ (150 mm × 4.6 mm, ES industries, West-Berlin, NJ, USA) with 10% (v/v) MeCN as liquid phase buffered in 90% (v/v) of 20 mM ammonium acetate (pH 5.0). The buffer was gradually exchanged for MeCN using a linear gradient from 10 to 95% (v/v) MeCN over 15 min at a flow rate of 1 mL min−1. Products were quantified based on D254nm using a standard curve. Semi-preparative reverse-phase HPLC was performed using a Waters 600 controller coupled to a Waters 990 photodiode array detector. Chiral HPLC was performed using a SPD-10A VP Shimadzu system.
Mass spectrometry
Samples were purified by HPLC as described above and then analyzed with HR-ESI-MS (positive mode) using a 6230 Accurate-Mass TOF MS system (Agilent). Alternatively, a 1290 Infinity LC system coupled to a 6530 Accurate-Mass Q-TOF MS system (both Agilent) was employed. HPLC was conducted using a Phenomenex (Torrence, CA, USA) Luna 5μ C18E (2) column (150 × 4.6 mm) using a MeCN gradient of 10-90% (v/v) over 25 min in 0.1% (v/v) formic acid. For synthesized 5 and 5` and intermediates, high-resolution mass spectra (HRMS) were recorded on an Agilent LC/MSD TOF mass spectrometer by electrospray ionization time-of-flight (ESI-TOF) reflectron experiments.
NMR spectroscopy
NMR spectra were recorded on Bruker DRX-600 and AMX-400 instruments and were calibrated using residual undeuterated solvent as an internal reference (CHCl3 @ 7.26 ppm 1H-NMR, 77.16 ppm 13C-NMR). The following abbreviations were used to explain NMR peak multiplicities: s = singlet, d = doublet, t = triplet, q = quartet, m = multiplet, br = broad.
Optical rotations and circular dichroism spectroscopy
Optical rotations were obtained on a Perkin-Elmer 341 polarimeter. Circular dichroism spectroscopy (CD) measurements were obtained on an Aviv circular dichroism spectrometer model 62DS.
Chemical syntheses
See Supplementary Information for full experimental details and procedures of all performed reactions of the syntheses of substrate analogs, as well as their full characterization (1H and 13C nuclear magnetic resonance, high-resolution mass spectrometry, infrared, optical rotation, melting point, and Rf value). All reactions were carried out under an inert nitrogen atmosphere with dry solvents under anhydrous conditions unless otherwise stated. Dry acetonitrile (MeCN), dichloromethane (DCM), diethyl ether (Et2O), tetrahydrofuran (THF), toluene (PhMe) and triethylamine (Et3N) were obtained by passing the previously degassed solvents through activated alumina columns. Reagents were purchased at the highest commercial quality and used without further purification, unless otherwise stated. Yields refer to chromatographically and spectroscopically (1H NMR) homogeneous material, unless otherwise stated. Reactions were monitored by thin layer chromatography (TLC) carried out on 0.25 mm E. Merck silica plates (60F-254), using UV light as the visualizing agent and an acidic solution of p-anisaldehyde and heat, ceric ammonium molybdate and heat, or KMnO4 and heat as developing agents.
Flash silica gel chromatography was performed using E. Merck silica gel (60, particle size 0.043–0.063 mm).
IR experiments were recorded on a Perkin-Elmer Spectrum 100 FT-IR spectrometer.
Melting points were recorded on a Fisher-Johns 12-144 melting point apparatus and are uncorrected.
Supplementary Material
Acknowledgements
This research was supported by US National Institutes of Health (NIH) grant R01AI47818 to B.S.M., NSF award nos. EEC-0813570 and MCB-0645794 and the Howard Hughes Medical Institute to J.P.N., NSF grant CHE-1213620 to B.P., and by fellowships to R.T. from the Deutsche Forschungsgemeinschaft (TE 931/1-1) and to A.M. from JSPS (21-644). We thank Marianne Bowman (Salk Institute) for technical assistance, Dr. Yongxuan Su (UCSD) for MS measurements, D-H. Huang and L. Pasternack (TSRI) for NMR spectroscopic assistance, A. Rheingold (UCSD) for X-ray crystallographic analysis, and Christian Hertweck for establishing the synthesis of 26.
Footnotes
Author Contributions. R.T., A.M., Q.M., F.S., G.L, and J.P.N. performed research; all authors designed research and analyzed data; and R.T. and B.M. wrote the paper. R.T., A.M., Q.M., F.S contributed equally to the work.
Author Information. The GenBank accession number of EncM is AAF81732.1. PDB data bank numbers of submitted structures are 3W8W (apo-EncM), 3W8X (EncM with bound 26); 3W8Z (EncM with bound 4). The Cambridge Crystallographic Data Centre numbers of crystallized substrate analogs are CCDC 922822 (4) and CCDC 922821 (10), and CCDC 949270 (26).
The authors declare no competing financial interests.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Walsh CT, Wencewicz TA. Flavoenzymes: Versatile catalysts in biosynthetic pathways. Nat. Prod. Rep. 2012;30:175–200. doi: 10.1039/c2np20069d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chaiyen P, Fraaije MW, Mattevi A. The enigmatic reaction of flavins with oxygen. Trends Biochem. Sci. 2012;37:373–380. doi: 10.1016/j.tibs.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 3.Massey V. Activation of molecular oxygen by flavins and flavoproteins. J. Biol. Chem. 1994;269:22459–22462. [PubMed] [Google Scholar]
- 4.Palfey BA, McDonald CA. Control of catalysis in flavin-dependent monooxygenases. Arch. Biochem. Biophys. 2010;493:26–36. doi: 10.1016/j.abb.2009.11.028. [DOI] [PubMed] [Google Scholar]
- 5.Xiang L, Kalaitzis JA, Moore BS. EncM, a versatile enterocin biosynthetic enzyme involved in Favorskii oxidative rearrangement, aldol condensation, and heterocycle-forming reactions. Proc. Natl. Acad. Sci. USA. 2004;101:15609–15614. doi: 10.1073/pnas.0405508101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng Q, Xiang L, Izumikawa M, Meluzzi D, Moore BS. Enzymatic total synthesis of enterocin polyketides. Nat. Chem. Biol. 2007;3:557–558. doi: 10.1038/nchembio.2007.22. [DOI] [PubMed] [Google Scholar]
- 7.Piel J, et al. Cloning, sequencing and analysis of the enterocin biosynthesis gene cluster from the marine isolate ‘Streptomyces maritimus’: evidence for the derailment of an aromatic polyketide synthase. Chem. Biol. 2000;7:943–955. doi: 10.1016/s1074-5521(00)00044-2. [DOI] [PubMed] [Google Scholar]
- 8.Seto H, Sato T, Urano S, Uzawa J, Yonehara H. Utilization of 13C-13C coupling in structural and biosynthetic studies. VII. The structure and biosynthesis of vulgamycin. Tetrahedron Lett. 1976;48:4367–4370. [Google Scholar]
- 9.Hertweck C, et al. Context-dependent behavior of the enterocin iterative polyketide synthase; a new model for ketoreduction. Chem. Biol. 2004;11:461–468. doi: 10.1016/j.chembiol.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 10.Hertweck C, Luzhetskyy A, Rebets Y, Bechthold A. Type II polyketide synthases: gaining a deeper insight into enzymatic teamwork. Nat. Prod. Rep. 2007;24:162–190. doi: 10.1039/b507395m. [DOI] [PubMed] [Google Scholar]
- 11.Koetter JW, Schulz GE. Crystal structure of 6-hydroxy-D-nicotine oxidase from Arthrobacter nicotinovorans. J. Mol. Biol. 2005;352:418–428. doi: 10.1016/j.jmb.2005.07.041. [DOI] [PubMed] [Google Scholar]
- 12.Huang CH, et al. Crystal structure of glucooligosaccharide oxidase from Acremonium strictum: a novel flavinylation of 6-S-cysteinyl, 8alpha-N1-histidyl FAD. J. Biol. Chem. 2005;280:38831–38838. doi: 10.1074/jbc.M506078200. [DOI] [PubMed] [Google Scholar]
- 13.Alexeev I, Sultana A, Mantsala P, Niemi J, Schneider G. Aclacinomycin oxidoreductase (AknOx) from the biosynthetic pathway of the antibiotic aclacinomycin is an unusual flavoenzyme with a dual active site. Proc. Natl. Acad. Sci. USA. 2007;104:6170–6175. doi: 10.1073/pnas.0700579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crosby J, Crump MP. The structural role of the carrier protein--active controller or passive carrier. Nat. Prod. Rep. 2012;29:1111–1137. doi: 10.1039/c2np20062g. [DOI] [PubMed] [Google Scholar]
- 15.Baumann K, Bacher M, Damont A, Steck A. Selective transformation of ascomycin into 11-epi-ascomycin. Tetrahedron Lett. 2004;45:549–551. [Google Scholar]
- 16.Takikawa H, Takada A, Hikita K, Suzuki K. Formation of alpha-hydroxy-beta-diketones through hydroxylation of isoxazolium salts: stereoselective approach to angular cis-diols in polycyclic systems. Angew. Chem. Int. Ed. Engl. 2008;47:7446–7449. doi: 10.1002/anie.200801586. [DOI] [PubMed] [Google Scholar]
- 17.Entsch B, Ballou DP, Massey V. Flavin-oxygen derivatives involved in hydroxylation by p-hydroxybenzoate hydroxylase. J. Biol. Chem. 1976;251:2550–2563. [PubMed] [Google Scholar]
- 18.Entsch B, Ballou DP. Purification, properties, and oxygen reactivity of phydroxybenzoate hydroxylase from Pseudomonas aeruginosa. Biochim. Biophys. Acta. 1989;999:313–322. doi: 10.1016/0167-4838(89)90014-9. [DOI] [PubMed] [Google Scholar]
- 19.Thotsaporn K, Chenprakhon P, Sucharitakul J, Mattevi A, Chaiyen P. Stabilization of C4a-hydroperoxyflavin in a two-component flavin-dependent monooxygenase is achieved through interactions at flavin N5 and C4a atoms. J. Biol. Chem. 2011;286:28170–28180. doi: 10.1074/jbc.M111.241836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valton J, Mathevon C, Fontecave M, Niviere V, Ballou DP. Mechanism and regulation of the Two-component FMN-dependent monooxygenase ActVA-ActVB from Streptomyces coelicolor. J. Biol. Chem. 2008;283:10287–10296. doi: 10.1074/jbc.M709730200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rastetter WH, Gadek TR, Tane JP, Frost JW. Oxidations and oxygen transfers effected by a flavin N(5)-oxide. A model for flavin-dependent monooxygenases. J. Am. Chem. Soc. 1979;101:2228–2231. [Google Scholar]
- 22.Orf HW, Dolphin D. Oxaziridines as possible intermediates in flavin monooxygenases. Proc. Natl. Acad. Sci. USA. 1974;71:2646–2650. doi: 10.1073/pnas.71.7.2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walsh C, et al. Chemical and enzymatic properties of riboflavin analogues. Biochemistry. 1978;17:1942–1951. doi: 10.1021/bi00603a022. [DOI] [PubMed] [Google Scholar]
- 24.Garman EF, Owen RL. Cryocooling and radiation damage in macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2006;62:32–47. doi: 10.1107/S0907444905034207. [DOI] [PubMed] [Google Scholar]
- 25.van Berkel WJ, Kamerbeek NM, Fraaije MW. Flavoprotein monooxygenases, a diverse class of oxidative biocatalysts. J. Biotechnol. 2006;124:670–689. doi: 10.1016/j.jbiotec.2006.03.044. [DOI] [PubMed] [Google Scholar]
- 26.Izumikawa M, Cheng Q, Moore BS. Priming type II polyketide synthases via a type II nonribosomal peptide synthetase mechanism. J. Am. Chem. Soc. 2006;128:1428–1429. doi: 10.1021/ja0559707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kalaitzis JA, Izumikawa M, Xiang L, Hertweck C, Moore BS. Mutasynthesis of enterocin and wailupemycin analogues. J. Am. Chem. Soc. 2003;125:9290–9291. doi: 10.1021/ja035973o. [DOI] [PubMed] [Google Scholar]
- 28.Han L, Lobo S, Reynolds KA. Characterization of beta-ketoacyl-acyl carrier protein synthase III from Streptomyces glaucescens and its role in initiation of fatty acid biosynthesis. J. Bacteriol. 1998;180:4481–4486. doi: 10.1128/jb.180.17.4481-4486.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jez JM, Ferrer JL, Bowman ME, Dixon RA, Noel JP. Dissection of malonylcoenzyme A decarboxylation from polyketide formation in the reaction mechanism of a plant polyketide synthase. Biochemistry. 2000;39:890–902. doi: 10.1021/bi991489f. [DOI] [PubMed] [Google Scholar]
- 30.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 31.Leslie AGW. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB News-letter on Protein Crystallography. 1992;26 [Google Scholar]
- 32.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 33.Morris RJ, Perrakis A, Lamzin VS. ARP/wARP's model-building algorithms. I. The main chain. Acta Crystallogr. D Biol. Crystallogr. 2002;58:968–975. doi: 10.1107/s0907444902005462. [DOI] [PubMed] [Google Scholar]
- 34.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 35.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 36.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific LLC; San Carlos, CA, USA: 2002. [Google Scholar]
- 37.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 39.Tovchigrechko A, Vakser IA. GRAMM-X public web server for protein-protein docking. Nucleic Acids Res. 2006;34:W310–W314. doi: 10.1093/nar/gkl206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.