Abstract
Genetically engineered mice are a valuable resource for studies of the behavioral effects of ethanol. However, for some behavioral tests of ethanol action, the rat is a superior model organism. Production of genetically engineered rats has been severely hampered due to technical limitations. Here we utilized a promising new technique for efficient site-specific gene modification to create a novel gene knockout rat line. This approach is based on Transcriptional Activator-Like Effector Nucleases (TALENs). TALENs function in pairs and bind DNA in a sequence-specific manner. Upon binding to the target sequence, a functional nuclease is reconstituted that creates double-stranded breaks in the DNA that are efficiently repaired by non-homologous end joining. This error-prone process often results in deletions of varying lengths at the targeted locus. The toll-like receptor 4 (Tlr4) gene was selected for TALEN-mediated gene inactivation. Tlr4 has been implicated in ethanol-induced neuroinflammation and neurodegeneration, as well as multiple ethanol-induced behavioral effects. To generate Tlr4 knockout rats, a pair of TALEN constructs was created that specifically target Exon 1 immediately downstream of the start of translation. TALEN mRNAs were microinjected into the cytoplasm of one-cell Wistar rat embryos. Of 13 live-born pups that resulted, one harbored a mutation in Exon 1 of Tlr4. The mutated allele consisted of a 13 base-pair deletion that was predicted to create a frameshift mutation after amino acid 25. This founder rat successfully transmitted the mutation to F1 offspring. Heterozygous F1 offspring were interbred to produce homozygous F2 animals. Homozygous mutants expressed the 13-bp deletion in Tlr4 mRNA. In contrast to control rats that produced a robust increase in plasma tumor necrosis factor alpha in response to a lipopolysaccharide challenge, homozygous rats had a markedly attenuated response. Thus, the mutant Tlr4 allele generated by TALEN-mediated gene inactivation represents a null allele. This knockout rat line will be valuable for studies of ethanol action as well as more general inflammatory conditions including septic shock. In conclusion, TALEN-mediated gene targeting in rat zygotes represents an inexpensive, efficient, and rapid method for creating genetically engineered rats.
Keywords: gene targeted rats, toll-like receptor 4 (Tlr4), transcriptional activator-like effector nuclease (TALEN), knockout rats, genome editing
Introduction
The laboratory mouse has quickly become the principal mammalian model organism for alcohol research. The widespread use of mice has been fueled in large part because of the wide availability of genetically engineered (i.e., knockout, knockin) mouse lines. Such animals are an invaluable resource for studies of the cellular and behavioral effects of alcohol (for review, see Crabbe, Phillips, Harris, Arends, & Koob, 2006).
However, because of the rat’s more sophisticated behavioral repertoire, the rat may be a superior model organism, at least for some studies of alcohol action. For example, some aspects of alcohol dependence, such as relapse liability, habit formation, and cue-induced drug seeking require operant behavioral approaches which are readily achieved with rats, but not with mice (Corbit, Nie, & Janak, 2012; Pickens et al., 2011; von der Goltz et al., 2009).
The production of genetically engineered rats has been hampered by technical limitations. Whereas embryonic stem cell lines that are widely used to produce gene targeted mice have been available since 1981 (Evans & Kaufman, 1981; Martin, 1981), only recently have germline-competent rat embryonic stem cell lines become available (Li et al., 2008). Although these new cell lines have been used to create several knockout rat strains (e.g., Tong, Li, Wu, Yan, & Ying, 2010), their use has not been widespread because of the high cost and technically demanding nature of their use.
Very recently, several novel methods of efficient, targeted genome editing have emerged that circumvent the requirement for embryonic stem cells and open up the application of gene targeting to many species besides the mouse. These methods include zinc finger nucleases (Urnov, Rebar, Holmes, Zhang, & Gregory, 2010), clustered regularly interspaced short palindromic repeat (CRISPR) (Shen et al., 2013), and transcription activator-like effector nucleases (TALENs) (Joung & Sander, 2013). These nucleases can be injected directly into embryos to efficiently modify the germline genome in a DNA sequence-specific manner.
TALEN proteins are customizable, engineered nucleases that can be targeted to virtually any DNA sequence in the genome (for reviews, see Bogdanove & Voytas, 2011; Joung & Sander, 2013). Each TALEN is composed of 2 domains, a DNA binding domain and a nuclease domain. TALENs work as dimers with each monomer binding 15–20 bp of DNA that flank a 15–25 bp spacer region. When the monomers bind to their target sites 5′ and 3′ of the spacer region, a functional nuclease is reconstituted and a double-strand DNA break is created immediately within the spacer region. Double-strand DNA breaks are efficiently repaired by the cellular DNA repair process of non-homologous end joining. This error-prone process typically results in deletions of varying lengths but can also lead to insertions. When these deletions and insertions occur in a critical part of a gene such as an exon, the gene is often rendered nonfunctional (i.e., a knockout), especially if the mutation causes a frameshift.
To create a TALEN-mediated rat knockout line that would be of wide interest to the alcohol research community (and to many other areas of biomedical research), we focused on the toll-like receptor 4 (Tlr4). Tlr4 is widely known as the lipopolysaccharide (LPS) receptor and it has well-known functions in the immune system (for review, see Lu, Yeh, & Ohashi, 2008). Recently, neuroimmune signaling and Tlr4 in particular has garnered intense interest from the alcohol research community. For example, alcohol increases Tlr expression in the brain and increases its sensitivity to LPS (Crews, 2012; Crews, Qin, Sheedy, Vetreno, & Zou, 2013; Vetreno & Crews, 2012). In addition, Blednov et al. (2011) injected mice with LPS and found that a single injection produces long-lasting increases in alcohol consumption consistent with neuroimmune signaling mediating the reinforcing properties of alcohol. The single injection of LPS also increased the firing rate of dopamine neurons in the VTA, providing an example of how neuroimmune activation following peripheral LPS administration modulates brain reward circuitry (Blednov et al., 2011). Behavioral responses (loss of righting reflex, motor incoordination) to acute alcohol were reduced in mice lacking Tlr4 (Wu et al., 2012) and knockdown of Tlr4 in the rat amygdala using siRNA decreased alcohol self-administration (Liu et al., 2011). These results suggest that some effects of alcohol are due to activation of Tlr4 receptors. Null mutant rats will be important for defining these actions.
To test the feasibility of TALEN-mediated gene disruption to create gene knockout rats for studies of alcohol action, we attempted to create a mutant rat line that lacks Tlr4. We report that embryo microinjection of a TALEN pair targeted to Tlr4 efficiently led to the creation of a Tlr4 rat knockout line.
Materials and methods
TALEN Construction
A pair of TALENs targeting Exon 1 of the Tlr4 gene was created in-house. TALENs were designed using the first version of TALEN Targeter (Cermak et al., 2011) using the preset architecture of Cermak et al. (2011) with all default options checked except percent composition. Each TALEN binds to 20 bp of DNA and the binding sites are separated by a 14-bp spacer region as illustrated in Fig. 1A. TALENs were assembled using the TALE Toolkit (Addgene, catalog # 1000000019) according to published protocols (Sanjana et al., 2012). Final constructs were produced in the pTALEN_v2 (NG) backbone plasmid.
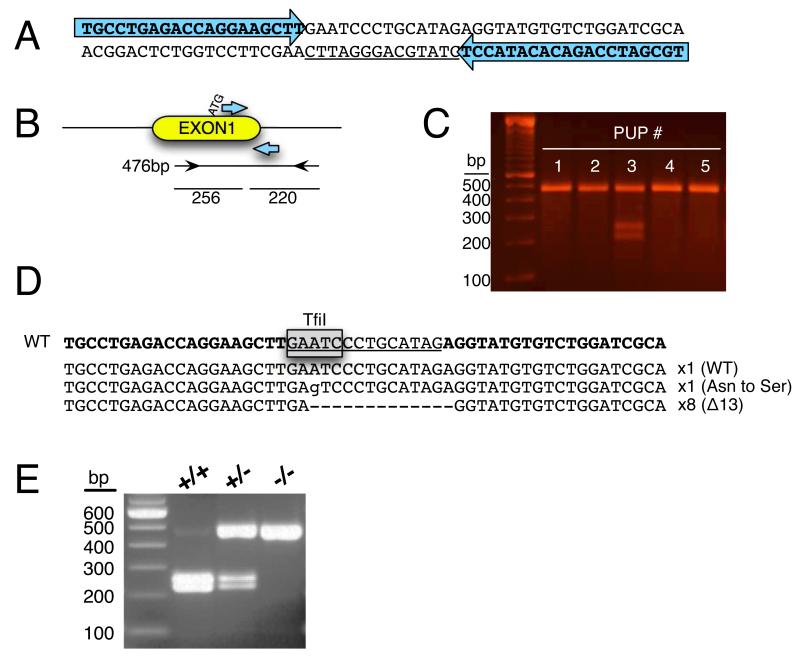
Figure 1.
Production of Tlr4 knockout rats by TALEN-mediated gene targeting A. Double-stranded DNA sequence of the Tlr4 locus that was targeted with TALENs. The TALEN binding sites are represented by the large arrows and the spacer region is underlined. B. Diagram of Tlr4 Exon 1 (yellow oval) showing the start of translation (ATG) and the TALEN binding sites (arrows). Immediately under the figure is the PCR amplicon used for the Surveyor Assay and for genotyping. Surveyor nuclease activity in the spacer region is predicted to cleave the PCR amplicon into ~220- and 256-bp fragments. C. Agarose gel photograph of Surveyor Nuclease Assay demonstrating digestion products of the predicted size from pup #3. D. DNA sequence of the wild type (WT) sequence of Tlr4 with TALEN binding sites in bold, the spacer region underlined, and the TfiI restriction site used for genotyping boxed. Immediately beneath the wild type sequence is the sequence derived from cloned PCR products from founder rat pup #3 demonstrating that 1 clone was wild type, 1 clone harbored an A to G substitution that is predicted to change an asparagine (Asn) to a serine (Ser), and that 8 clones harbored a 13-bp deletion (Δ13). E. Agarose gel photograph of genotyping results after PCR products from wild type (+/+), heterozygous (+/−), and homozygous (−/−) rats were digested with TfiI. The wild type allele produces TfiI products of 226 and 250 bp. The 463-bp Δ13 allele lacks the TfiI site and is undigested.
TALEN plasmids were linearized with SmaI and used as templates for in vitro transcription using a mMessage mMachine T7 Ultra Kit (Ambion), according to the manufacturer’s instructions. Capped, polyA-tailed mRNAs were cleaned up with a MEGAclear Kit (Ambion). mRNAs were precipitated, washed, and resuspended at 1 μg/μL in DEPC-treated H2O. TALEN mRNAs were subsequently diluted in 0.1× TE buffer at a final concentration of 10 ng/μL, dispensed into aliquots, and stored at −80 °C until used for embryo injection.
Embryo Manipulation
All animal-based experiments were reviewed and approved in advance by the Institutional Animal Care and Use Committee at the University of Pittsburgh and were conducted in accordance with the NIH Guide for the Care and Use of Laboratory Animals.
Wistar rats (Charles River) were housed under specific pathogen-free conditions with a 12-h light/dark cycle (lights on at 7:00 AM). Female embryo donors were superovulated with 25 IU of pregnant mare serum gonadotropin (Sigma #G4877) between 11:00 AM and 12:00 noon, followed by 25 IU of human chorionic gonadotropin (Sigma #CG5-1VL) 24 hours later and subsequently individually caged with a male stud rat. The following morning, donors were sacrificed and embryos collected from the oviducts. Embryos were cultured in M16 (Millipore) at 37 °C in 5% CO2/95% air. Fertilized one-cell embryos were transferred to M2 medium (Millipore) for microinjection. TALEN mRNAs were injected into the cytoplasm using glass injection pipettes. Embryos that survived the injection procedure were surgically transferred to the oviduct of day 0.5 postcoitum psuedopregnant recipient Wistar females that had successfully mated with vasectomized males.
Mutation Analysis
Offspring from injected embryos were screened for mutations in the Tlr4 locus using the Surveyor Mutation Detection Kit for Standard Gel Electrophoresis (Transgenomic Inc., Omaha, NE). Briefly, DNA was prepared from tail snips (~0.5 cm) using QuickExtract DNA Extraction Solution (Epicentre Biotechnologies, Madison, WI). Tails were lysed in 150 μL QuickExtract Solution by heating to 68 °C for 20 min and 95 °C for 8 min. A portion of the Tlr4 locus that overlaps the TALEN spacer region was amplified by PCR. Amplification primers were F1 5′-AAGGTTGGCACTCTCACTTCCTCTTGCT-3′ and R1 5′-AACAAGACACCACTGACTGCCTGATCCT-3′. PCR reactions used Platinum Taq DNA Polymerase High Fidelity (Life Technologies, Grand Island, NY), and amplification consisting of: 95 °C 2 min; 40 cycles of 95 °C 30 sec, 60 °C 30 sec, and 72 °C 90 sec; and a final extension at 72 °C for 5 min. PCR products (10–16 μL) were processed according to the Surveyor Kit protocol using 1 μL Enhancer and 1 μL Nuclease in 1× PCR buffer with 0.5 mM MgSO4. Digested products were analyzed on ethidium bromide-stained 2% agarose gels in Tris-Borate-EDTA buffer.
In addition, Tlr4 PCR products from the founder rat were subcloned into pCR2.1 vector (Life Technologies) and sequenced using the PCR primers described above.
F1 and F2 offspring were genotyped by FastDigest TfiI restriction analysis (Thermo Fisher Scientific, Waltham, MA) of PCR products.
mRNA Analysis
Total RNA was prepared from liver samples using Trizol (Life Technologies) according to the manufacturer’s instructions. mRNA was reverse transcribed using an iScript cDNA Synthesis Kit (Bio-Rad Laboratories, Hercules, CA). Tlr4 cDNA was amplified using two different forward primers (F1 [described above] or F3 5′-GCTTGAATCCCTGCATAGAGG-3′) separately with the same reverse primer (R4 5′-TGTCTCCACAGCCACCAGATTCTC-3′). PCR reactions were cycled as described above. PCR products were analyzed by agarose gel electrophoresis or by DNA sequence analysis using primer F1.
Lipopolysaccharide Challenge
Blood samples were collected from the tail vein immediately prior to intraperitoneal injection with LPS (500 μg/kg in saline; catalog # L3129, Sigma-Aldrich Corp., St. Louis, MO) and from the heart 2 h post-injection. Plasma samples were analyzed for TNFα levels using a Rat TNFα Quantikine ELISA Kit (R&D Systems, Inc., Minneapolis, MN) according to the manufacturer’s instructions. The stated sensitivity of the assay was 5 pg/mL. All tested samples that fell below this minimum detection level were assigned a value of 5 pg/mL. Results were analyzed by repeated measures two-way ANOVA with main effects of treatment (LPS) and genotype. A Bonferroni post hoc test was used to identify group differences.
Results
TALEN constructs were created that target the DNA sequence of the rat Tlr4 gene (see Fig. 1A) as illustrated graphically in Fig. 1B. TALEN mRNAs transcribed in vitro were injected into the cytoplasm of 1-cell Wistar rat embryos. Following transfer of injected embryos to pseudopregnant recipients, 13 live-born offspring were produced. Offspring were screened for targeted disruption of Tlr4 Exon 1 as illustrated in Fig. 1B. A 476-bp PCR product was amplified from each animal and subjected to Surveyor Nuclease digestion. Samples that are not mutated are resistant to digestion. In contrast, mutations in the amplicon result in heteroduplex formation that is sensitive to nuclease digestion and is predicted to result in ~220- and 256-bp cleavage products. Of the 13 live-born pups produced, 1 yielded Surveyor Nuclease digestion results indicative of mutation in the Tlr4 gene (Fig. 1C).
The nature of the mutation in this founder animal was characterized by DNA sequence analysis. Following subcloning of the PCR products from this animal, 10 subclones were sequenced. One subclone was wild type, one subclone had an A to G substitution, and 8 clones had a 13-bp deletion (Δ13) in the spacer region (Fig. 1D).
This male founder rat was mated to wild type Wistar females. This founder rat transmitted the Δ13 allele to 41 of 148 (28%) F1 offspring. Heterozygous F1 offspring were interbred and genotypes of the F2 generation were consistent with that predicted by Mendelian genetics (n = 12:25:16, wild type: heterozygote: homozygote, respectively). Homozygous rats were morphologically normal and overtly indistinguishable from wild type and heterozygous littermates.
All F1 and F2 offspring were genotyped by TfiI restriction digestion of the PCR product generated with the same primers used for the Surveyor assay. As illustrated in Fig. 1D, there is a single TfiI site in the wild type allele that is deleted in the Δ13 allele. This assay results in DNA fragments of 226 and 250 bp from the wild type allele and 463 bp from the Δ13 allele (Fig. 1E).
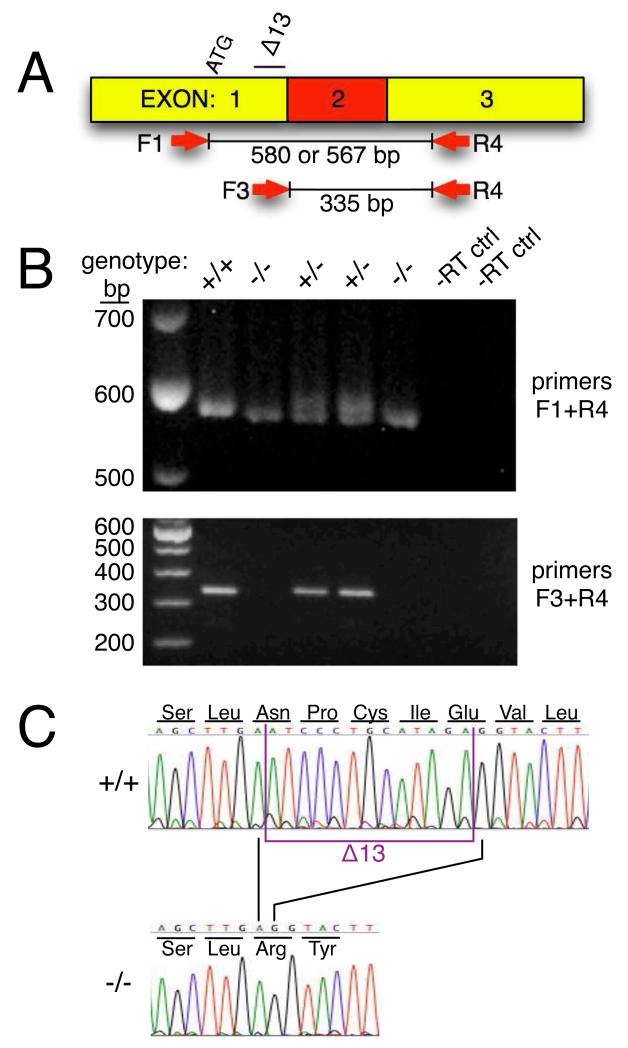
Tlr4 mRNA was analyzed by RT-PCR. Primers F1 and R4 (which span the Δ13 deletion) generated a 580-bp product from wild type samples and a 567-bp product from homozygous knockout samples (Fig. 2B). Heterozygous samples have both the 567- and 580-bp products. In contrast, amplification with primers F3 (which overlaps the Δ13 deletion) and R4 only resulted in PCR products from wild type and heterozygous samples; homozygous samples did not amplify with this primer set (Fig. 2B).
Figure 2.
Analysis of Tlr4 mRNA A. Diagram of Tlr4 cDNA illustrating position of start codon (ATG), 13-bp deletion (Δ13), and PCR primers used for RT-PCR analyses. B. Top panel is an agarose gel photograph of RT-PCR products using primers F1 + R4. Note that samples of all genotypes amplified with this primer set. The wild type allele produces a 580-bp product whereas the mutant allele produces a 567-bp product. Samples processed without the addition of reverse transcriptase to the reactions (-RT ctrl) did not amplify. Bottom panel is an agarose gel photograph of RT-PCR products using primers F3 + R4. Note that wild type (+/+) and heterozygous (+/−) samples produced the 335-bp product but the 2 homozygous mutant (−/−) samples failed to amplify. C. DNA sequence chromatograms of F1 + R4 RT-PCR products demonstrating the 13-bp deletion in the −/− sample.
RT-PCR products were further analyzed by DNA sequence analysis. As shown in Fig. 2C, the wild type sequence was identical to that in the rat genome database (transcript ID: ENSRNOT00000014020). In contrast, sequences from the homozygous knockout samples revealed the Δ13 deletion. These RT-PCR analyses establish that the Δ13 deletion is present in the Tlr4 mRNA expressed in the knockout rats. As illustrated in Fig. 2C, the Δ13 deletion is predicted to result in a frameshift mutation that results in a protein product containing the first 25 amino acids of Tlr4 followed by 28 nonsense amino acids and an inframe stop codon. For comparison, full-length wild type Tlr4 protein is 835 amino acids in length.
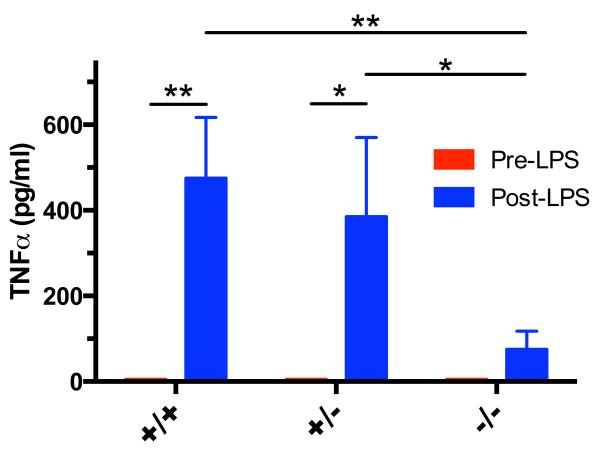
To functionally validate inactivation of Tlr4, we challenged rats with an intraperitoneal injection of LPS. Tlr4 is the primary LPS receptor, and LPS signaling through Tlr4 induces a robust inflammatory reaction (Lu, Yeh, & Ohashi, 2008). Mice with a defective Tlr4 gene fail to respond to LPS (Hoshino et al., 1999). We used plasma TNFα as a marker for the inflammatory reaction. As shown in Fig. 3, plasma levels of TNFα were not detectable in any genotype prior to LPS injection. One heterozygous rat was an outlier and excluded from the analysis because TNFα levels (2684 pg/mL) following LPS were greater than 2 standard deviations from the mean. Repeated-measures two-way ANOVA revealed an effect of LPS treatment (p ≤ 0.001) and a trend for genotype (p = 0.068) and interaction of treatment with genotype (p = 0.068). Post hoc analysis revealed that LPS induced a significant increase in TNFα production in wild type (p ≤ 0.01) and heterozygous rats (p < 0.05), but not in homozygous knockouts. TNFα levels in response to LPS were reduced in knockouts compared to heterozygotes (p < 0.05) and wild type (p ≤ 0.01) rats. The near failure of homozygous knockouts to respond to the LPS challenge indicates that Tlr4 is nonfunctional in these animals.
Figure 3.
TNFα production in response to LPS challenge A. TNFα levels collected from plasma immediately prior to (pre-LPS) or 2 h following (post-LPS) intraperitoneal injection with LPS (500 μg/kg). TNFα levels were undetectable in all samples prior to LPS injection. LPS robustly induced TNFα levels in wild type (p ≤ 0.01; n = 4) and heterozygous (p < 0.05; n = 4) rats but not in the homozygous rats (n = 6). LPS-induced TNFα levels in homozygous rats were reduced compared to heterozygous (p < 0.05) and wild type (p ≤ 0.01) rats. *p < 0.05; **p ≤ 0.01.
Discussion
This study was designed to explore the use of an emerging genome editing technology (TALEN) to create gene knockout models in rats that would be useful for studies of alcohol action. In recent years, in the alcohol field (and many areas of biomedical research), the mouse has been the predominant model species because techniques have long been available for producing genetically engineered mouse models such as knockouts and knockins. The techniques for creating these engineered models have been restricted to the mouse because of difficulties developing germline competent embryonic stem cell lines in species other than mouse. However, new technologies have emerged recently that obviate the need for embryonic stem cell lines. These new genome editing approaches are so efficient that targeted genome modifications can be created directly in 1-cell embryos.
We employed TALEN technology to create a Tlr4 gene knockout rat line. To accomplish this, a pair of TALENs targeting Exon 1 of Tlr4 was injected into recently fertilized, 1-cell rat zygotes. Of 13 live-born animals derived from injected embryos, 1 harbored a mutation in the Tlr4 gene. This founder rat was able to transmit the mutation to the F1 generation. Subsequent breeding of heterozygous F1 animals led to the production of F2 generation animals that were homozygous for the mutant allele.
DNA sequence analysis revealed the mutation to be a 13-bp deletion in Exon 1. This deletion was predicted to result in a shift of the reading frame such that the mutant locus would only produce the first 25 amino acids of Tlr4 followed by 28 nonsense amino acids and a premature translation termination codon. This severely truncated protein (for comparison, wild type Tlr4 is 835 amino acids) is predicted to be a nonfunctional, null allele.
To establish that the Δ13 allele results in a null allele, we first confirmed that the Δ13 mutation is present in the mRNA that is expressed in the animals (Fig. 2). We then tested for Tlr4 function by challenging the animals with an injection of LPS. LPS is known to signal through Tlr4 and induce a robust inflammatory response that can be measured by induction of various cytokines in the circulation. As shown in Fig. 3, in contrast to the robust LPS stimulated increase of TNFα in serum of wild type rats, animals with the Δ13 mutation in Tlr4 had a markedly reduced response to LPS. Thus, at the DNA, mRNA, and functional levels, these animals are Tlr4 null.
The genome editing tool we have used here, TALENs, shows great promise for efficiently creating genetically engineered models in many different species. To date, TALENs have been used to create mutants in yeast, plants, mouse, rat, cow, pig, human, xenopus, zebrafish, drosophila, cricket, roundworm, and silkworm (Joung & Sander, 2013 and references therein). A single report has been published about the rat demonstrating TALEN-mediated inactivation of the IgM locus (Tesson et al., 2011).
One concern with new genome editing tools is the potential for cleavage at unwanted, off-target sites as recently highlighted for CRISPR technology (Fu et al., 2013). Although we did not survey for off-target mutations following TALEN-mediated inactivation of Tlr4, previous studies with TALENs have revealed very few off-target events in rats (Tesson et al., 2011), mice (Sung et al., 2013), zebrafish (Dahlem et al., 2012), or human cells (Hockemeyer et al., 2011).
The model gene we targeted in this study was Tlr4. Although Tlr4 has long been studied for its involvement in immune system function, great interest in Tlr4 has recently developed in relation to its effects on nervous system function and alcohol action (Mayfield, Ferguson, & Harris, 2013). However, as discussed above, the relevance of Tlr4 for some key aspects of alcohol dependence, such as relapse and habit formation, requires the use of rat models. The availability of rats lacking Tlr4 will allow operant behavioral studies to explore aspects of dependence not accessible in mice. In addition, Tlr4 may be important for actions of other drugs of addiction, including opioids (Hutchinson et al., 2012), and our new rat model will facilitate understanding of these roles of Tlr4.
Acknowledgments
A grant from the National Institute on Alcohol Abuse and Alcoholism (AA020889) provided funding for this project as part of the INIA-West consortium. The authors would like to acknowledge the support and encouragement of all INIA-West investigators. This Tlr4 KO rat line is now available to the scientific community through the Rat Resource & Research Center (Strain #694).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Blednov YA, Benavidez JM, Geil C, Perra S, Morikawa H, Harris RA. Activation of inflammatory signaling by lipopolysaccharide produces a prolonged increase of voluntary alcohol intake in mice. Brain, Behavior, and Immunity. 2011;25(Suppl 1):S92–S105. doi: 10.1016/j.bbi.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanove AJ, Voytas DF. TAL effectors: customizable proteins for DNA targeting. Science. 2011;333:1843–1846. doi: 10.1126/science.1204094. [DOI] [PubMed] [Google Scholar]
- Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Research. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbit LH, Nie H, Janak PH. Habitual alcohol seeking: time course and the contribution of subregions of the dorsal striatum. Biological Psychiatry. 2012;72:389–395. doi: 10.1016/j.biopsych.2012.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe JC, Phillips TJ, Harris RA, Arends MA, Koob GF. Alcohol-related genes: contributions from studies with genetically engineered mice. Addiction Biology. 2006;11:195–269. doi: 10.1111/j.1369-1600.2006.00038.x. [DOI] [PubMed] [Google Scholar]
- Crews FT. Immune function genes, genetics, and the neurobiology of addiction. Alcohol research: current reviews. 2012;34:355–361. [PMC free article] [PubMed] [Google Scholar]
- Crews FT, Qin L, Sheedy D, Vetreno RP, Zou J. High mobility group box 1/Toll-like receptor danger signaling increases brain neuroimmune activation in alcohol dependence. Biological Psychiatry. 2013;73:602–612. doi: 10.1016/j.biopsych.2012.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlem TJ, Hoshijima K, Jurynec MJ, Gunther D, Starker CG, Locke AS, et al. Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS Genetics. 2012;8:e1002861. doi: 10.1371/journal.pgen.1002861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nature Biotechnology. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nature. Biotechnology. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino K, Takeuchi O, Kawai T, Sanjo H, Ogawa T, Takeda Y, et al. Cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the Lps gene product. Journal of Immunology. 1999;162:3749–3752. [PubMed] [Google Scholar]
- Hutchinson MR, Northcutt AL, Hiranita T, Wang X, Lewis SS, Thomas J, et al. Opioid activation of toll-like receptor 4 contributes to drug reinforcement. The Journal of Neuroscience. 2012;32:11187–11200. doi: 10.1523/JNEUROSCI.0684-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung JK, Sander JD. TALENs: a widely applicable technology for targeted genome editing. Nature Reviews. Molecular Cell Biology. 2013;14:49–55. doi: 10.1038/nrm3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Tong C, Mehrian-Shai R, Jia L, Wu N, Yan Y, et al. Germline competent embryonic stem cells derived from rat blastocysts. Cell. 2008;135:1299–1310. doi: 10.1016/j.cell.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yang AR, Kelly T, Puche A, Esoga C, June HL, Jr., et al. Binge alcohol drinking is associated with GABAA alpha2-regulated Toll-like receptor 4 (TLR4) expression in the central amygdala. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:4465–4470. doi: 10.1073/pnas.1019020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y-C, Yeh W-C, Ohashi PS. LPS/TLR4 signal transduction pathway. Cytokine. 2008;42:145–151. doi: 10.1016/j.cyto.2008.01.006. [DOI] [PubMed] [Google Scholar]
- Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proceedings of the National Academy of Sciences of the United States of America. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield J, Ferguson L, Harris RA. Neuroimmune signaling: a key component of alcohol abuse. Current Opinion in Neurobiology. 2013;23:513–520. doi: 10.1016/j.conb.2013.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickens CL, Airavaara M, Theberge F, Fanous S, Hope BT, Shaham Y. Neurobiology of the incubation of drug craving. Trends in Neurosciences. 2011;34:411–420. doi: 10.1016/j.tins.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Cong L, Zhou Y, Cunniff MM, Feng G, Zhang F. A transcription activator-like effector toolbox for genome engineering. Nature Protocols. 2012;7:171–192. doi: 10.1038/nprot.2011.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Zhang J, Wu H, Wang J, Ma K, Li Z, et al. Generation of genemodified mice via Cas9/RNA-mediated gene targeting. Cell Research. 2013;23:720–723. doi: 10.1038/cr.2013.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung YH, Baek IJ, Kim DH, Jeon J, Lee J, Lee K, et al. Knockout mice created by TALEN-mediated gene targeting. Nature Biotechnology. 2013;31:23–24. doi: 10.1038/nbt.2477. [DOI] [PubMed] [Google Scholar]
- Tesson L, Usal C, Ménoret S, Leung E, Niles BJ, Remy S, et al. Knockout rats generated by embryo microinjection of TALENs. Nature Biotechnology. 2011;29:695–696. doi: 10.1038/nbt.1940. [DOI] [PubMed] [Google Scholar]
- Tong C, Li P, Wu NL, Yan Y, Ying QL. Production of p53 gene knockout rats by homologous recombination in embryonic stem cells. Nature. 2010;467:211–213. doi: 10.1038/nature09368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nature Reviews. Genetics. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- Vetreno RP, Crews FT. Adolescent binge drinking increases expression of the danger signal receptor agonist HMGB1 and Toll-like receptors in the adult prefrontal cortex. Neuroscience. 2012;226:475–488. doi: 10.1016/j.neuroscience.2012.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Goltz C, Vengeliene V, Bilbao A, Perreau-Lenz S, Pawlak CR, Kiefer F, et al. Cue-induced alcohol-seeking behaviour is reduced by disrupting the reconsolidation of alcohol-related memories. Psychopharmacology (Berl) 2009;205:389–397. doi: 10.1007/s00213-009-1544-1. [DOI] [PubMed] [Google Scholar]
- Wu Y, Lousberg EL, Moldenhauer LM, Hayball JD, Coller JK, Rice KC, et al. Inhibiting the TLR4-MyD88 signalling cascade by genetic or pharmacological strategies reduces acute alcohol-induced sedation and motor impairment in mice. British Journal of Pharmacology. 2012;165:1319–1329. doi: 10.1111/j.1476-5381.2011.01572.x. [DOI] [PMC free article] [PubMed] [Google Scholar]