Abstract
A fundamental design principle of microbial rhodopsins is that they share the same basic light-induced conversion between two conformers. Alternate access of the Schiff base to the outside and to the cytoplasm in the outwardly open “E” conformer and cytoplasmically open “C” conformer, respectively, combined with appropriate timing of pKa changes controlling Schiff base proton release and uptake make the proton path through the pumps vectorial. Phototaxis receptors in prokaryotes, sensory rhodopsins I and II, have evolved new chemical processes not found in their proton pump ancestors, to alter the consequences of the conformational change or modify the change itself. Like proton pumps, sensory rhodopsin II undergoes a photoinduced E → C transition, with the C conformer a transient intermediate in the photocycle. In contrast, one light-sensor (sensory rhodopsin I bound to its transducer HtrI) exists in the dark as the C conformer and undergoes a light-induced C → E transition, with the E conformer a transient photocycle intermediate. Current results indicate that algal phototaxis receptors channelrhodopsins undergo redirected Schiff base proton transfers and a modified E → C transition which, contrary to the proton pumps and other sensory rhodopsins, is not accompanied by the closure of the external half-channel. The article will review our current understanding of how the shared basic structure and chemistry of microbial rhodopsins have been modified during evolution to create diverse molecular functions: light-driven ion transport and photosensory signaling by protein-protein interaction and light-gated ion channel activity.
Keywords: microbial rhodopsins, Schiff base connectivity, proton transfer, photosensory transduction, phototaxis, optogenetics
1. Introduction
The large family of microbial rhodopsins provides a vivid example of evolution modifying a single protein scaffold to produce diverse new functions (for reviews, see [1–6]). Family members share a membrane-embedded seven-helix architecture forming an internal pocket for the chromophore retinal bound in a protonated Schiff base linkage to a lysyl residue in the middle of the seventh helix. Similar photochemical reactions energized by photoisomerization of retinal have been engineered by nature to drive distinctly different processes in different microbial rhodopsins: light-driven outward proton transport, inward chloride transport, and as reported very recently outward sodium ion transport [7], photosensory signaling by protein-protein interaction, and light-gated ion channel conduction.
As microbial rhodopsins with new functions have been discovered it has been natural to analyze their physical and chemical properties in terms of their similarities and differences to those of the light-driven proton pump bacteriorhodopsin (BR), the first found and best characterized member of the family (for review, see [2, 8]). For the prokaryotic sensory rhodopsins, SRI and SRII, subunits of phototaxis signaling complexes, such comparative analysis has been particularly informative. Their use of steps in the proton transport mechanism for signal relay and their latent proton transport activity when separated from other signaling complex subunits provide compelling evidence for their evolution from a light-driven proton pump [3, 9]. The generalization of this evolutionary progression, i.e. proton pumps as the earliest microbial rhodopsins, is consistent with phylogenetic analysis [10], and a possible scenario is that proton-pumping rhodopsins appeared first in evolution, underwent extensive lateral gene transfer, and in multiple cells independently evolved interactions with their signal transduction machinery to acquire sensory functions. This notion may be reinforced or negated as our knowledge of rhodopsin photosensor mechanisms increases. In either case it is instructive to consider to what extent microbial rhodopsins with newfound functions share mechanistic processes with light-driven proton transporters, for which these processes have been worked out in considerable, in several aspects atomic, detail.
In this minireview we address aspects of the light-driven pumping mechanism of BR that are shared and new aspects that have emerged in the two types of light-sensors whose physiological functions have been identified: the prokaryotic phototaxis receptors sensory rhodopsins I and II (SRI and SRII) and the algal phototaxis receptors channelrhodopsins (ChRs). We consider the roles of key processes in the proton pump mechanism in these rhodopsins whose functions are other than proton pumping. The emerging information regarding conserved features and new molecular processes in these members of the microbial rhodopsin family provides intriguing insights into how the proteins work as well as how they have evolved.
2. The ion pumping mechanism
2.1. Proton transfers and the Schiff base connectivity switch
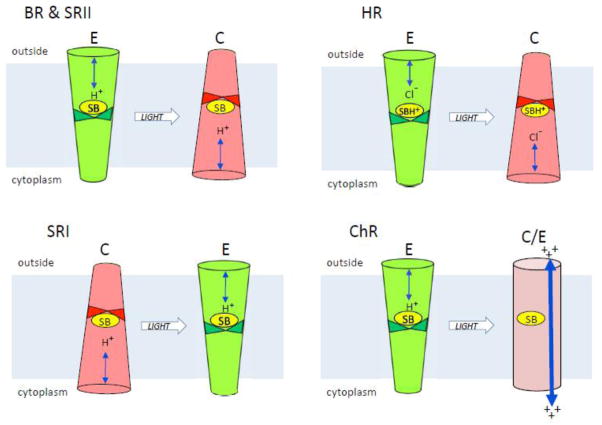
In proton pumps, as first shown for BR from Halobacterium salinarum, the dark conformation exhibits an outwardly-connected protonated Schiff base poised for proton release to an exterior half-channel. This conformation is denoted in this minireview as the E conformer (Figure 1). Light induces release of the proton to a counterion of the Schiff base, an anionic aspartyl residue (Asp85) in the exterior channel, forming the blue-shifted photocycle intermediate M, named after the mammalian visual pigment’s deprotonated Schiff base photoproduct “metarhodopsin”. In HsBR M formation is accompanied by an almost simultaneous release of the proton to the outside medium from a proton release group. The electrogenic Schiff base proton transfer to Asp85 is the first step in the pumping process. The protein then undergoes a conformational change during the lifetime of M (the M1 to M2 conversion) in which (i) a half-channel forms from the retinal chromophore’s deprotonated Schiff base to the cytoplasm and (ii) the Schiff base switches its connection (i.e. accessibility) to the cytoplasmic side (the C conformer). A second aspartyl residue (Asp96) in the cytoplasmic channel serves as a proton donor to the Schiff base. The alternate access of the Schiff base in the E and C conformers combined with appropriate timing of pKa changes controlling Schiff base proton release and uptake make the proton path through the protein vectorial [2, 8].
Figure 1. Microbial rhodopsin conformers.
The figure depicts light-induced conformer transitions in the indicated microbial rhodopsins in their native functional state. BR, bacteriorhodopsin; HR, halorhodopsin; SRI, sensory rhodopsin I; SRII, sensory rhodopsin II; ChR, channelrhodopsin. E (green), conformer with externally-connected Schiff base and exterior half-channelopen; C (red), conformer with cytoplasmically-connected Schiff base and cytoplasmic half-channel open; C/E (purple), conformer with an open channel from the extracellular to cytoplasmic surfaces of the protein.
The inward pumping of chloride ions by halorhodopsin (HR) can be explained by the same Schiff base connectivity switch mechanism that results in outward proton pumping by BR [11]. HR contains a threonine residue at the corresponding position of Asp85 in BR. As in the D85T mutant of BR, the absence of an anionic proton acceptor at the 85 position inhibits deprotonation from the Schiff base. HR contains a chloride ion bound as a counterion to the protonated Schiff base near the threonine in the external half channel, and when the protonated Schiff base undergoes the photoinduced switch in connectivity from the external to the cytoplasmic half channel the chloride ion follows the positive charge, thereby being actively transported inward across the membrane. A striking confirmation that the same alternating access switch that accomplishes outward proton pumping in BR is capable of driving inward chloride pumping is that BR with the single mutation D85T exhibits light-driven inward chloride transport activity [11].
Schiff base connectivity can be defined empirically by electrophysiological measurement of the direction of current produced by the light-induced release of the proton from the Schiff base and its reprotonation. In BR and other light-driven proton pumps both currents are outwardly directed indicating that reprotonation occurs from the opposite side of the membrane than the side to which the proton was released (i.e. a Schiff base connectivity switch occurred). Equivalently, in HR the same direction of currents as in BR (positive outward movement) is observed due to the inward displacements of chloride ion. Such measurements performed in other rhodopsins have been informative as described below in elucidating the significance of connectivity switching in sensory signaling as well as transport mechanisms.
2.2. Helix movement in the conformational change
The largest structural change in the E → C conversion is a laterally outward movement of the cytoplasmic half of helix F [12–13]. Cryoelectron crystallography of natural functional 2-D crystals of BR frozen within 1 ms after illumination to trap the C conformer was used to construct a projection difference Fourier map at near-atomic resolution [14]. This projection structure revealed a significant lateral displacement of helix F density by 3.5 Å. Based on the projection difference maps and a low resolution 3-D difference map, Subramaniam and Henderson proposed that the main features of the structural change in the E → C photoconversion were likely to be an ordering of helix G at the cytoplasmic end and an outward ~6-degree tilt of helix F, with Pro186, buried in the membrane-embedded portion of the helix, likely to serve as a hinge residue [15].
The lateral displacement of helix F toward the periphery of the protein would be expected to expand the structure on the cytoplasmic side thereby opening a proton-conducting channel. The tilting of helix F has been further defined by EPR using dipolar coupling distance measurements [16–18] and by direct and dynamic visualization using high-speed AFM [19]. Elegant time-resolved molecular spectroscopic studies have identified also residue changes and water molecule movements in the E → C transition in BR [20–22], but to test the generality of the conformational change in the microbial rhodopsin family, the two well-established properties of the C conformer considered here are (i) the connection of the Schiff base to the cytoplasmic side of the protein and (ii) an open channel from the Schiff base to the cytoplasm, detectable structurally as a tilting of the cytoplasmic portion of helix F away from neighboring helices.
3. Sensory rhodopsin II: something old and something new
The isolated SRII protein in the dark is in the E conformation, as shown by (i) its near superimposable helix positions to the BR E conformer [23], (ii) its light-induced Schiff base proton release outward to the aspartate residue corresponding to Asp85 in BR [24–25], (iii) its light-induced E → C transition according to helix F motion assessed by EPR [26–27], (iv) the similarity of late photocycle backbone changes of BR and SRII measured by FTIR [28], and (v) its ability to pump protons when free of its transducer HtrII, as first found for transducer-free SRI [29–30] showing that these sensory rhodopsins must switch Schiff base connectivity during the conformational change [6, 9]. In both SRI and SRII, the binding of their cognate Htr transducers block their proton pumping activity [31–32]. In HtrII-free SRII, unlike in HtrI-free SRI, strong pumping occurs only in the presence of azide, or after the mutation F86D, in the position corresponding to Asp96 in BR [33]. Like SRI, pumping by SRII/F86D is suppressed by complexation with its cognate Htr transducer [34]. The structure of SRII bound to HtrII is indistinguishable at 2Å resolution from that of the free form, except for one SRII surface residue that makes a crystal contact in the latter [23, 35]. The similarities of SRII to BR raised the question whether the E → C transition is sufficient for phototaxis signaling. If so, the light-induced E → C transition of BR, mutated at 2 positions on its lipid-facing surface to mimic SRII’s bonded contacts with HtrII, might activate the transducer. Such a double mutant of BR was found to bind to HtrII, but no phototaxis was observed [36].
In parallel work a steric interaction between the isomerizing retinal and residues in the retinal binding pocket, detected by Hideki Kandori’s laboratory by cryo-FTIR [37], was found to be essential for SRII signaling, since mutations that eliminated the steric conflict (e.g. T204A or Y174F), evident in FTIR spectra of the first SRII photointermediate K, eliminated phototaxis without major effects on SRII expression nor on the SRII photocycle [38]. An analogous steric interaction does not occur in BR, which contains Ala215 at the corresponding position of Thr204, the interacting residue in SRII [39]. Remarkably, simply substituting Thr for Ala (mutation A215T [40]) into the HtrII-bound double mutant of BR produced the triple mutant “BR-T” that exhibits a steric conflict during retinal photoisomerization chemically very similar to that in SRII [41] and exhibits robust phototaxis signaling through HtrII [36]. This result demonstrated a causative role of the steric conflict, a “steric trigger” for signaling. The results indicate a model in which the canonical conformational change combines with the structural consequence of the steric trigger to transfer the photosignal to HtrII (Figure 2).
Figure 2. Photoisomerization-induced steric-trigger in the SRII-HtrII complex.
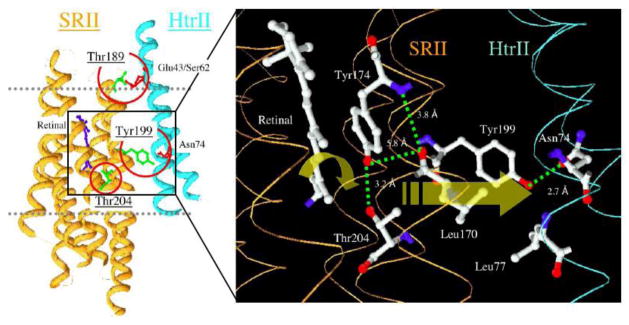
SRII, sensory rhodopsin II; HtrII, haloarchaeal transducer for SRII. The three residues in SRII circled in red are those which, when engineered into bacteriorhodopsin, enable bacteriorhodopsin-mediated phototaxis signaling through HtrII [36]. Redrawn from reference 30.
4. Sensory rhodopsin I: opposite signaling by running the conformational change in reverse
Sensory rhodopsin I (SRI) also exhibits a steric trigger as a new feature not found in BR. A steric interaction in SRI occurs between the 13-methyl group of the retinal and a protein residue [42], very likely Leu84 based on modeling the SRI structure using BR as a template [43]. Without this interaction SRI does not form a primary photoproduct and returns from the excited state to the all-trans retinal ground state without conformational changes or signaling function. Results from low temperature flash photolysis suggest a model in which the retinylidene 13-methyl group steric contact with Leu84 functions as a fulcrum to permit movement of one or both ends of retinal to overcome an energy barrier against isomerization [44]. Note that the steric trigger in SRI is very different from that in SRII in that in the latter the steric conflict occurs between residue Thr204 and C14H in the retinylidene polyene chain [39], and its absence does not prevent retinal isomerization nor a photochemical reaction cycle including deprotonation of the retinylidene Schiff base, but does prevent signal relay to HtrII [36, 38].
Sensory rhodopsin I when free of its normally tightly bound transducer HtrI functions as a light-driven proton pump undergoing, like BR, a light-induced E → C conformer transition, and binding of HtrI inhibits this activity [30, 45]. Over the past few years, it has become clear that SRI when bound to HtrI in the attractant phototaxis complex exhibits the two defining properties of the C conformer: (i) transducer-bound SRI undergoes photorelease of the Schiff base proton to the cytoplasmic side of the protein [45–46], unlike BR, transducer-free SRI, and SRII (with or without HtrII) which all release the proton towards the exterior diagnostic of the E conformer; (ii) SRI exhibits photoinduced inward tilting of the cytoplasmic portion of helix F toward the protein center [27] as shown by the same type of EPR dipolar coupling distance measurements that revealed an outward tilting movement of helix F in BR [16–18] and SRII [26–27]. Furthermore, Asp76, the exteriorly located residue corresponding to the counterion to the protonated Schiff base and proton acceptor in BR and in SRII, is protonated in the dark attractant receptor state at physiological pH in the SRI-HtrI complex as it is in the C conformer photointermediates of BR and SRII [46–47]. Finally, SRI bound to the mutant transducer HtrI_E56Q exhibits the opposite properties (extracellular connectivity of the Schiff base, untilted helix F, low Asp76 pKa) compared to the native attractant complex, and also exhibits inverted (repellent) signaling [27, 45–46]. Evidently in the SRI-Htr_E56Q complex the SRI dark form is the E conformer and the photoinduced E → C conversion generates a repellent (CheA kinase activating) signal, whereas in the wildtype SRI-HtrI complex the photoinduced C → E conversion mediates an attractant (CheA kinase inhibiting) signal.
In summary, SRI and SRII undergo closely similar photoreactions as BR exhibiting light-induced transitions between E and C conformers, switching of Schiff base connectivity, and similar structural changes (although in SRI the changes are in the opposite direction) in spite of the absence of vectorial proton translocation by these photosensors when bound as subunits in their natural complexes. Also both sensors have developed steric interactions with the retinal during photoisomerization not present in BR and essential for their signaling functions.
5. Channelrhodopsins
5.1. Background
Besides the prokaryotic SRs, the only other microbial rhodopsins with a firmly established sensory function in their native cells are the phototaxis receptors in green flagellate algae [48–50]. When expressed in animal cells, these algal sensory rhodopsins act as light-gated cation channels, and were therefore named “channelrhodopsins” (ChRs) to emphasize this unique property, unknown in other microbial rhodopsins or in fact in any other proteins [51–52]. This discovery provided a boost to the field of optogenetics, i.e., using genetically encoded tools to control activity of specific cell types by light with high temporal and spatial resolution (reviewed by [53–56]). Heterologous expression also opened the possibility to study ChRs in experimental systems under voltage clamp and defined ionic conditions and made possible purification of ChRs for spectroscopic analysis [57–58] and crystallization [59–60], difficult to achieve directly from algae, which contain only ~105 ChR molecules per cell [49].
5.2. Light-induced proton transfers
The mean amplitude of whole-cell channel currents generated by different ChRs in heterologous systems differ by as much as 10-fold, and this difference cannot be explained only by a difference in their expression levels [61]. In ChRs with relatively low channel efficiency (such as CaChR1 from Chlamydomonas augustae, VcChR1 from Volvox carteri and DsChR1 from Dunaliella salina) laser flash excitation elicits fast current components that precede channel opening [61]. These components are similar to those well-characterized in BR and other rhodopsin pumps (reviewed in [62–63]), beginning with an initial unresolved inward current that in BR corresponds to the early stages of the photocycle associated with the formation of K and L intermediates, and is attributed to the isomerization of the chromophore and a coupled motion of the Arg82 residue [64].
In three low efficiency ChRs tested, the initial inward current is followed by a fast outwardly-directed weakly voltage-dependent signal in the time window of M intermediate formation attributable to a transfer of the Schiff base proton to an outwardly located acceptor [61]. Hence, at least in those ChRs an E-conformation of the dark state in cell membranes is confirmed experimentally.
The complex Schiff base counterion in ChRs includes two conserved carboxylate residues, homologous to Asp85 and Asp212 in BR, although the position of the side chain of the Arg82 homolog is closer to that in NpSRII [23, 60]. Neutralization of either Asp85 and Asp212 leads to a block or severe inhibition of formation of the M intermediate in BR [65–66]. In contrast, in CaChR1 [67], M formation was observed in both corresponding mutants with even greater yields than in the wild type [61]. Correspondingly, the outward transfer of the Schiff base proton was absent in both BR mutants [68], whereas in both CaChR1 mutants this transfer was observed. Electrophysiological analysis of the respective mutants of VcChR1 and DsChR1, in which the Asp85 position is naturally occupied by Ala but could be reintroduced by mutation, showed similar results. Therefore, in contrast to BR, two alternative acceptors of the Schiff base proton exist at least in low-efficiency ChRs.
This conclusion is further corroborated by a clear correlation between changes in the kinetics of the outwardly directed fast current and M formation induced by the counterion mutations in CaChR1. Neutralization of the Asp85 homolog resulted in retardation of both processes, whereas neutralization of the Asp212 homolog brought about their acceleration [61]. The presence of a second proton acceptor in addition to the Asp85 homolog in ChRs makes them similar to blue-absorbing proteorhodopsin (BPR), in which the same conclusion was deduced from pH titration of its absorption spectrum [69] and analysis of photoelectric signals generated by this pigment and its mutants in E. coli cells [25].
The existence of the initial step of the outward electrogenic proton transport in low-efficiency ChRs [61] fits the notion that they are “leaky proton pumps”. Small photoinduced currents measured at zero voltage from CrChR2 expressed in electrofused giant HEK293 cells or incorporated in liposomes attached to planar lipid bilayers have been interpreted as proton pumping activity [70]. However, in CrChR2 and other high-efficiency ChRs (such as MvChR1 from Mesostigma viride and PsChR from Platymonas subcordiformis) no outwardly directed proton transfer currents were detected [61]. A possible explanation for their apparent absence is that the direction of the Schiff base proton transfer in high-efficiency ChRs strongly depends on the electrochemical gradient and therefore cannot be easily resolved from the channel current; in other words, unlike in BR, SRI, and SRII, a Schiff base connectivity switch may not be required for their molecular function, in this case channel opening. Taking into account these observations, the earlier reported currents attributed to pumping by CrChR2 [70] may reflect passive ion transport driven by residual transmembrane ion gradients, because their kinetics were very similar to that of channel currents. On the other hand, we cannot exclude that in high-efficiency ChRs the outward proton transfer current occurs but is screened by a high mobility of other charges in the Schiff base environment. An inverse relationship between outward proton transfer and channel currents revealed by comparative analysis of different ChRs suggests that the former is not necessary for the latter and may reflect the evolutionary transition from active to passive ion transport in microbial rhodopsins.
A time-resolved FTIR study identified the Asp212 homolog as the primary proton acceptor in CrChR2, whereas no protonation changes could be attributed to the Asp85 homolog [71]. However, neutralization of either the Asp85 or Asp212 homolog in CrChR2 produces very similar changes in photoelectric currents: both mutants exhibit a large unresolved negative signal and accelerated and reduced channel currents (authors, manuscript in preparation). Also, both mutations induce a red shift of the action spectrum ([72] and authors’ unpublished observations). Finally, formation of the M intermediate is almost unperturbed by neutralization of the Asp212 homolog [71], which is inconsistent with its role as a single proton acceptor. Taken together, these results suggest the existence of alternative acceptors of the Schiff base proton also in highly efficient ChRs, such as CrChR2.
5.3. The conductive state and light-induced conformational change
The P520 intermediate is generally accepted to be a conducting state in CrChR2, because its decay (~10 ms measured in detergent-purified pigment) roughly correlates to channel closing (measured in HEK cells and oocytes) after switching off the light, and because additional illumination with green light closes the channel that is opened in response to blue light stimulation [57–58, 73]. However, opening of the channel during the previous P390 state has also been suggested, although the rise of this intermediate is much faster than the rise of the channel current [74]. Channel opening initiated in M is supported by the observation of the extremely long-lived M state in CaChR1, which decays roughly in parallel with channel closing [61]. Therefore, an interesting possibility is that the channel opens during a spectrally silent transition between two different substates of P390, similar to the M1 → M2 transition (equivalently E → C conformational change) in BR. The presence of such substates, with the transition between them linked to the onset of protein backbone alterations, was inferred from time-resolved FTIR data [71]. Passive ion conductance of ChRs requires opening of a cytoplasmic half-channel (e.g. formation of the C conformer) without closing of the extracellular half-channel.
As mentioned above, a major conformational change that occurs during the M1 → M2 transition in BR is the outward movement of helix F, which is accompanied by more subtle rearrangements of the cytoplasmic moieties of helices C, E, and G. It is noteworthy that an outward radial movement of helix F is the principal large-scale change also associated with activation of vertebrate visual rhodopsin (e.g., [75–76]), even in the absence of sequence homology between microbial and animal (type 1 and type 2) rhodopsins [1]. An interesting hypothesis is that helix F movement may also contribute to channel opening in ChRs. Pro186, which is implicated in the movement of helix F in BR, is conserved in all so far known ChR sequences. However, experimental data have not been reported testing this hypothesis. A high-resolution crystal structure of chimeric ChR in the dark (E conformer) state is available [60], but no structures of intermediates have so far been resolved. A putative cation-conducting pathway appears to be formed by helices A, B, C and G. It is open towards the extracellular side, but its cytoplasmic side is occluded by two constrictions. Movement of the C-terminal end of helix A (possibly transmitted from the photoactive site via movements of helices B, C and/or G) was suggested to open the pore exit upon photoexcitation [60].
5.4. The second function of ChRs observed in vivo
There is no doubt that ChRs act in their native algal cells to depolarize the plasma membrane upon illumination thereby initiating photomotility responses [77]. This depolarization can be measured either in individual cells by the suction pipette technique [78], or in cell populations by a suspension assay [79]. The direct light-gated channel activity of these pigments in animal cells has been interpreted as eliminating the need for any chemical signal amplification in algal phototaxis [50], in contrast to, for example, animal vision. However, the notion that the channel activity observed in ChRs expressed in animal cells is sufficient for algal phototaxis is inconsistent with studies in algal cells.
It was shown more than two decades ago that the photoreceptor current in algal cells is comprised of two components [80]. The fast (early) current has no measurable lag period and saturates at intensities corresponding to excitation of all ChR molecules, which indicates that it is generated by the photoreceptor molecules themselves. The magnitude of this current in native algal cells corresponds to the value calculated from the unitary conductance of heterologously expressed CrChR2 estimated by noise analysis ([70] and our unpublished observations) and the number of ChR molecules in the C. reinhardtii cell [49]. Therefore this early saturating current, observed at high light intensities, matches the activity expected from heterologous expression of ChRs in animal cells. However, the second (late) current has a light-dependent delay, saturates at ~1,000-fold lower light intensities, and is carried specifically by Ca2+ ions, permeability for which in ChRs is very low [81]. This amplified Ca2+current plays a major role in the membrane depolarization that causes photomotility responses in flagellate algae extending the photosensitivity of the algae by 3 orders of magnitude [77, 82–83].
RNAi knock-down experiments demonstrated that out of two ChRs in C. reinhardtii, short wavelength-absorbing ChR2 predominantly contributes to the delayed high-sensitivity photocurrent [48]. However, the longer wavelength-absorbing CrChR1 is also involved in control of Ca2+channels, because the phototaxis action spectrum comprises a band corresponding to CrChR1 absorption even at low light intensities, when the contribution of direct channel activity to the membrane depolarization is negligible. The mechanisms by which photoexcitation of ChRs causes activation of these unidentified Ca2+ channels are not yet clear. Voltage and/or Ca2+gating seem unlikely because such gating would lead to an all-or-none electrical response, whereas the late photoreceptor current is gradual. The Ca2+ channels may be activated directly by photoactivated ChRs or via intermediate enzymatic steps, either of which is consistent with the short duration (<0.5 ms) of the delay between the laser flash and the appearance of the late receptor current (see model in Figure 3). The mechanism of the 1000-fold amplification of depolarizing current in the algae remains to be elucidated, and is potentially of great utility in optogenetics if it can be reproduced in animal cells.
Figure 3. Channelrhodopsin functions in vivo.
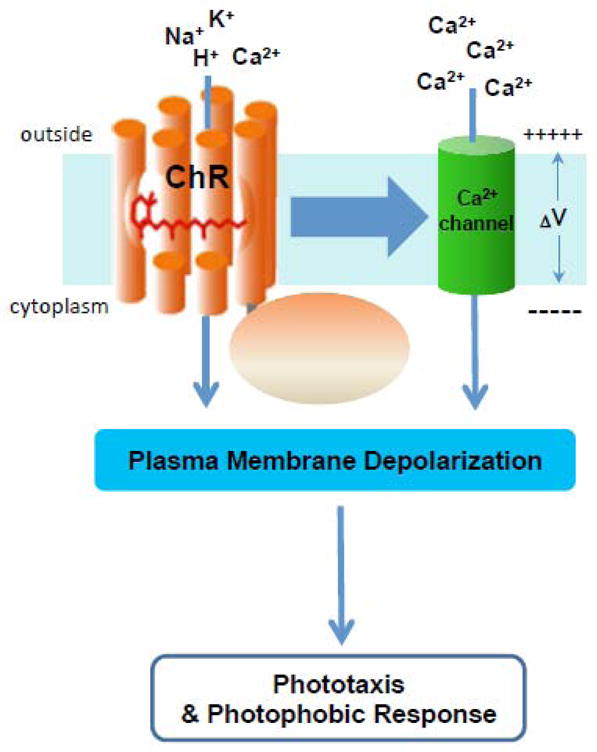
The figure depicts conclusions from studies of ChRfunction in Chlamydomonas reinhardtii and related algae (reviewed in [77, 82–83]) that ChRs depolarize algal plasma membranes with two distinct mechanisms, a direct light-gated channel activity as depicted in Figure 1 attributable to the 7-helix domain, and an amplified current dependent on an unidentified Ca2+ channel activated by ChRs either by direct protein-protein interaction or through intermediare components.
Besides green flagellate algae, similar photoreceptor currents have also been recorded from suspensions of the phylogenetically distant freshwater cryptophyte alga Cryptomonas sp. [84]. The genome of the related marine cryptomonad, Gulliardia theta, has been completely sequenced. It contains at least seven type 1 opsin genes, but none of them belong to the channelopsin subfamily. This raises the interesting possibility that structurally unrelated rhodopsins may activate similar amplification cascades in phototactic flagellates of different evolutionary origin.
Highlights.
Rhodopsin phototaxis sensors and light-driven ion pumps share aspects of mechanism
Proton pumps (e.g. bacteriorhodopsin) exhibit a light-induced E C transition
Prokaryotic photosensor signaling entails light-induced E C and C E transitions
Prokaryotic signals also entail steric triggers during retinal photoisomerization
Algal phototaxis receptors use two distinct mechanisms for membrane depolarization
Acknowledgments
Work by the authors was supported by Grant R01GM027750 from the National Institute of General Medical Sciences, Grant R21MH098288 from the National Institute of Mental Health, the Hermann Eye Fund, and Endowed Chair AU-0009 from the Robert A. Welch Foundation.
Abbreviations
- AFM
atomic force microscopy
- BR
bacteriorhodopsin
- BPR
blue-absorbing proteorhodopsin
- EPR
electron paramagnetic resonance
- FTIR
Fourier-transform infrared
- HR
halorhodopsin
- HtrI
haloarchaeal transducer for SRI
- HtrII
haloarchaeal transducer for SRII
- RNAi
RNA interference
- SRI
sensory rhodopsin I
- SRII
sensory rhodopsin II
- CaChR1
Chlamydomonas augustae channelrhodopsin 1
- CrChR2
Chlamydomonas reinhardtii channelrhodopsin 2
- DsChR1
Dunaliella salina channelrhodopsin 1
- MvChR1
Mesostigma viride channelrhodopsin 1
- PsChR
Platymonas subcordiformis channelrhodopsin
- VcChR1
Volvox carteri channelrhodopsin 1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Spudich JL, Yang CS, Jung KH, Spudich EN. Retinylidene proteins: structures and functions from archaea to humans. Annu Rev Cell Dev Biol. 2000;16:365–392. doi: 10.1146/annurev.cellbio.16.1.365. [DOI] [PubMed] [Google Scholar]
- 2.Lanyi JK, Luecke H. Bacteriorhodopsin. Curr Opin Struct Biol. 2001;11:415–419. doi: 10.1016/s0959-440x(00)00226-8. [DOI] [PubMed] [Google Scholar]
- 3.Spudich JL. The multitalented microbial sensory rhodopsins. Trends Microbiol. 2006;14:480–487. doi: 10.1016/j.tim.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Brown LS, Jung KH. Bacteriorhodopsin-like proteins of eubacteria and fungi: the extent of conservation of the haloarchaeal proton-pumping mechanism. Photochem Photobiol Sci. 2006;5:538–546. doi: 10.1039/b514537f. [DOI] [PubMed] [Google Scholar]
- 5.Jung KH. The distinct signaling mechanisms of microbial sensory rhodopsins in Archaea, Eubacteria and Eukarya. Photochem Photobiol. 2007;83:63–69. doi: 10.1562/2006-03-20-IR-853. [DOI] [PubMed] [Google Scholar]
- 6.Klare JP, Chizhov I, Engelhard M. Microbial rhodopsins: scaffolds for ion pumps, channels, and sensors. Results Probl Cell Differ. 2008;45:73–122. doi: 10.1007/400_2007_041. [DOI] [PubMed] [Google Scholar]
- 7.Inoue K, Ono H, Abe-Yoshizumi R, Yoshizawa S, Ito H, Kogure K, Kandori H. A light-driven sodium ion pump in marine bacteria. Nat Commun. 2013;4:1678. doi: 10.1038/ncomms2689. [DOI] [PubMed] [Google Scholar]
- 8.Lanyi JK. Proton transfers in the bacteriorhodopsin photocycle. Biochim Biophys Acta. 2006;1757:1012–1018. doi: 10.1016/j.bbabio.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 9.Sasaki J, Spudich JL. Signal transfer in haloarchaeal sensory rhodopsin-transducer complexes. Photochem Photobiol. 2008;84:863–868. doi: 10.1111/j.1751-1097.2008.00314.x. [DOI] [PubMed] [Google Scholar]
- 10.Sharma AK, Spudich JL, Doolittle WF. Microbial rhodopsins: functional versatility and genetic mobility. Trends Microbiol. 2006;14:463–469. doi: 10.1016/j.tim.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 11.Sasaki J, Brown LS, Chon YS, Kandori H, Maeda A, Needleman R, Lanyi JK. Conversion of bacteriorhodopsin into a chloride ion pump. Science. 1995;269:73–75. doi: 10.1126/science.7604281. [DOI] [PubMed] [Google Scholar]
- 12.Subramaniam S, Gerstein M, Oesterhelt D, Henderson R. Electron diffraction analysis of structural changes in the photocycle of bacteriorhodopsin. EMBO J. 1993;12:1–8. doi: 10.1002/j.1460-2075.1993.tb05625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vonck J. A three-dimensional difference map of the N intermediate in the bacteriorhodopsin photocycle: part of the F helix tilts in the M to N transition. Biochemistry. 1996;35:5870–5878. doi: 10.1021/bi952663c. [DOI] [PubMed] [Google Scholar]
- 14.Subramaniam S, Henderson R. Electron crystallography of bacteriorhodopsin with millisecond time resolution. J Struct Biol. 1999;128:19–25. doi: 10.1006/jsbi.1999.4178. [DOI] [PubMed] [Google Scholar]
- 15.Subramaniam S, Henderson R. Molecular mechanism of vectorial proton translocation by bacteriorhodopsin. Nature. 2000;406:653–657. doi: 10.1038/35020614. [DOI] [PubMed] [Google Scholar]
- 16.Thorgeirsson TE, Xiao W, Brown LS, Needleman R, Lanyi JK, Shin YK. Transient channel-opening in bacteriorhodopsin: an EPR study. J Mol Biol. 1997;273:951–957. doi: 10.1006/jmbi.1997.1362. [DOI] [PubMed] [Google Scholar]
- 17.Xiao W, Brown LS, Needleman R, Lanyi JK, Shin YK. Light-induced rotation of a transmembrane alpha-helix in bacteriorhodopsin. J Mol Biol. 2000;304:715–721. doi: 10.1006/jmbi.2000.4255. [DOI] [PubMed] [Google Scholar]
- 18.Radzwill N, Gerwert K, Steinhoff HJ. Time-resolved detection of transient movement of helices F and G in doubly spin-labeled bacteriorhodopsin. Biophys J. 2001;80:2856–2866. doi: 10.1016/S0006-3495(01)76252-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shibata M, Yamashita H, Uchihashi T, Kandori H, Ando T. High-speed atomic force microscopy shows dynamic molecular processes in photoactivated bacteriorhodopsin. Nat Nanotechnol. 2010;5:208–212. doi: 10.1038/nnano.2010.7. [DOI] [PubMed] [Google Scholar]
- 20.Kandori H. Hydration switch model for the proton transfer in the Schiff base region of bacteriorhodopsin. Biochim Biophys Acta. 2004;1658:72–79. doi: 10.1016/j.bbabio.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 21.Kötting C, Gerwert K. Proteins in action monitored by time-resolved FTIR spectroscopy. Chemphyschem. 2005;6:881–888. doi: 10.1002/cphc.200400504. [DOI] [PubMed] [Google Scholar]
- 22.Clair EC, Ogren JI, Mamaev S, Kralj JM, Rothschild KJ. Conformational changes in the archaerhodopsin-3 proton pump: detection of conserved strongly hydrogen bonded water networks. J Biol Phys. 2012;38:153–168. doi: 10.1007/s10867-011-9246-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luecke H, Schobert B, Lanyi JK, Spudich EN, Spudich JL. Crystal structure of sensory rhodopsin II at 2.4 angstroms: insights into color tuning and transducer interaction. Science. 2001;293:1499–1503. doi: 10.1126/science.1062977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bergo V, Spudich EN, Scott KL, Spudich JL, Rothschild KJ. FTIR analysis of the SII540 intermediate of sensory rhodopsin II: Asp73 is the Schiff base proton acceptor. Biochemistry. 2000;39:2823–2830. doi: 10.1021/bi991676d. [DOI] [PubMed] [Google Scholar]
- 25.Sineshchekov OA, Spudich JL. Light-induced intramolecular charge movements in microbial rhodopsins in intact E. coli cells. Photochem Photobiol Sci. 2004;3:548–554. doi: 10.1039/b316207a. [DOI] [PubMed] [Google Scholar]
- 26.Wegener AA, Chizhov I, Engelhard M, Steinhoff HJ. Time-resolved detection of transient movement of helix F in spin-labelled pharaonis sensory rhodopsin II. J Mol Biol. 2000;301:881–891. doi: 10.1006/jmbi.2000.4008. [DOI] [PubMed] [Google Scholar]
- 27.Sasaki J, Tsai AL, Spudich JL. Opposite displacement of helix F in attractant and repellent signaling by sensory rhodopsin-Htr complexes. J Biol Chem. 2011;286:18868–18877. doi: 10.1074/jbc.M110.200345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Braiman MS, Bousché O, Rothschild KJ. Protein dynamics in the bacteriorhodopsin photocycle: submillisecond Fourier transform infrared spectra of the L, M, and N photointermediates. Proc Natl Acad Sci USA. 1991;88:2388–2392. doi: 10.1073/pnas.88.6.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bogomolni RA, Stoeckenius W, Szundi I, Perozo E, Olson KD, Spudich JL. Removal of transducer HtrI allows electrogenic proton translocation by sensory rhodopsin I. Proc Natl Acad Sci USA. 1994;91:10188–10192. doi: 10.1073/pnas.91.21.10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spudich JL. Protein-protein interaction converts a proton pump into a sensory receptor. Cell. 1994;79:747–750. doi: 10.1016/0092-8674(94)90064-7. [DOI] [PubMed] [Google Scholar]
- 31.Olson KD, Spudich JL. Removal of the transducer protein from sensory rhodopsin-I exposes sites of proton release and uptake during the receptor photocycle. Biophys J. 1993;65:2578–2585. doi: 10.1016/S0006-3495(93)81295-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sudo Y, Iwamoto M, Shimono K, Sumi M, Kamo N. Photo-induced proton transport of pharaonis phoborhodopsin (sensory rhodopsin II) is ceased by association with the transducer. Biophys J. 2001;80:916–922. doi: 10.1016/S0006-3495(01)76070-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmies G, Luttenberg B, Chizhov I, Engelhard M, Becker A, Bamberg E. Sensory rhodopsin II from the haloalkaliphilic Natronobacterium pharaonis: Light-activated proton transfer reactions. Biophys J. 2000;78:967–976. doi: 10.1016/S0006-3495(00)76654-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmies G, Engelhard M, Wood PG, Nagel G, Bamberg E. Electrophysiological characterization of specific interactions between bacterial sensory rhodopsins and their transducers. Proc Natl Acad Sci USA. 2001;98:1555–1559. doi: 10.1073/pnas.031562298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gordeliy VI, Labahn J, Moukhametzianov R, Efremov R, Granzin J, Schlesinger R, Buldt G, Savopol T, Scheidig AJ, Klare JP, Engelhard M. Molecular basis of transmembrane signalling by sensory rhodopsin II-transducer complex. Nature. 2002;419:484–487. doi: 10.1038/nature01109. [DOI] [PubMed] [Google Scholar]
- 36.Sudo Y, Spudich JL. Three strategically placed hydrogen-bonding residues convert a proton pump into a sensory receptor. Proc Natl Acad Sci USA. 2006;103:16129–16134. doi: 10.1073/pnas.0607467103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sudo Y, Furutani Y, Shimono K, Kamo N, Kandori H. Hydrogen bonding alteration of Thr-204 in the complex between pharaonis phoborhodopsin and its transducer protein. Biochemistry. 2003;42:14166–14172. doi: 10.1021/bi035678g. [DOI] [PubMed] [Google Scholar]
- 38.Sudo Y, Furutani Y, Kandori H, Spudich JL. Functional importance of the interhelical hydrogen bond between Thr204 and Tyr174 of sensory rhodopsin II and its alteration during the signaling process. J Biol Chem. 2006;281:34239–34245. doi: 10.1074/jbc.M605907200. [DOI] [PubMed] [Google Scholar]
- 39.Ito M, Sudo Y, Furutani Y, Okitsu T, Wada A, Homma M, Spudich JL, Kandori H. Steric constraint in the primary photoproduct of sensory rhodopsin II is a prerequisite for light-signal transfer to HtrII. Biochemistry. 2008;47:6208–6215. doi: 10.1021/bi8003507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spudich EN, Ozorowski G, Schow EV, Tobias DJ, Spudich JL, Luecke H. A transporter converted into a sensor, a phototaxis signaling mutant of bacteriorhodopsin at 3.0 A. J Mol Biol. 2012;415:455–463. doi: 10.1016/j.jmb.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sudo Y, Furutani Y, Spudich JL, Kandori H. Early photocycle structural changes in a bacteriorhodopsin mutant engineered to transmit photosensory signals. J Biol Chem. 2007;282:15550–15558. doi: 10.1074/jbc.M701271200. [DOI] [PubMed] [Google Scholar]
- 42.Yan B, Nakanishi K, Spudich JL. Mechanism of activation of sensory rhodopsin I: evidence for a steric trigger. Proc Natl Acad Sci USA. 1991;88:9412–9416. doi: 10.1073/pnas.88.21.9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoff WD, Jung KH, Spudich JL. Molecular mechanism of photosignaling by archaeal sensory rhodopsins. Annu Rev Biophys Biomol Struct. 1997;26:223–258. doi: 10.1146/annurev.biophys.26.1.223. [DOI] [PubMed] [Google Scholar]
- 44.Yan B, Xie A, Nienhaus GU, Katsuta Y, Spudich JL. Steric constraints in the retinal binding pocket of sensory rhopdopsin I. Biochemistry. 1993;32:10224–10232. doi: 10.1021/bi00089a044. [DOI] [PubMed] [Google Scholar]
- 45.Sineshchekov OA, Sasaki J, Phillips BJ, Spudich JL. A Schiff base connectivity switch in sensory rhodopsin signaling. Proc Natl Acad Sci USA. 2008;105:16159–16164. doi: 10.1073/pnas.0807486105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sineshchekov OA, Sasaki J, Wang J, Spudich JL. Attractant and repellent signaling conformers of sensory rhodopsin-transducer complexes. Biochemistry. 2010;49:6696–6704. doi: 10.1021/bi100798w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sasaki J, Takahashi H, Furutani Y, Kandori H, Spudich JL. Sensory rhodopsin-I as a bidirectional switch: opposite conformational changes from the same photoisomerization. Biophys J. 2011;100:2178–2183. doi: 10.1016/j.bpj.2011.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sineshchekov OA, Jung KH, Spudich JL. Two rhodopsins mediate phototaxis to low- and high-intensity light in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 2002;99:8689–8694. doi: 10.1073/pnas.122243399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Govorunova EG, Jung KW, Sineshchekov OA, Spudich JL. Chlamydomonas sensory rhodopsins A and B: Cellular content and role in photophobic responses. Biophys J. 2004;86:2342–2349. doi: 10.1016/S0006-3495(04)74291-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hegemann P. Algal sensory photoreceptors. Annu Rev Plant Biol. 2008;59:167–189. doi: 10.1146/annurev.arplant.59.032607.092847. [DOI] [PubMed] [Google Scholar]
- 51.Nagel G, Ollig D, Fuhrmann M, Kateriya S, Musti AM, Bamberg E, Hegemann P. Channelrhodopsin-1: a light-gated proton channel in green algae. Science. 2002;296:2395–2398. doi: 10.1126/science.1072068. [DOI] [PubMed] [Google Scholar]
- 52.Nagel G, Szellas T, Huhn W, Kateriya S, Adeishvili N, Berthold P, Ollig D, Hegemann P, Bamberg E. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc Natl Acad Sci USA. 2003;100:13940–13945. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deisseroth K. Optogenetics. Nat Methods. 2011;8:26–29. doi: 10.1038/nmeth.f.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mei Y, Zhang F. Molecular tools and approaches for optogenetics. Biol Psychiatry. 2012 doi: 10.1016/j.biopsych.2012.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hegemann P, Nagel G. From channelrhodopsins to optogenetics. EMBO Mol Med. 2013;5:1–4. doi: 10.1002/emmm.201202387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yawo H, Asano T, Sakai S, Ishizuka T. Optogenetic manipulation of neural and non-neural functions. Dev Growth Differ. 2013 doi: 10.1111/dgd.12053. in press. [DOI] [PubMed] [Google Scholar]
- 57.Bamann C, Kirsch T, Nagel G, Bamberg E. Spectral characteristics of the photocycle of channelrhodopsin-2 and its implication for channel function. J Mol Biol. 2008;375:686–694. doi: 10.1016/j.jmb.2007.10.072. [DOI] [PubMed] [Google Scholar]
- 58.Ritter E, Stehfest K, Berndt A, Hegemann P, Bartl FJ. Monitoring light-induced structural changes of Channelrhodopsin-2 by UV-visible and Fourier transform infrared spectroscopy. J Biol Chem. 2008;283:35033–35041. doi: 10.1074/jbc.M806353200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Muller M, Bamann C, Bamberg E, Kuhlbrandt W. Projection structure of channelrhodopsin-2 at 6 Å resolution by electron crystallography. J Mol Biol. 2011;414:86–95. doi: 10.1016/j.jmb.2011.09.049. [DOI] [PubMed] [Google Scholar]
- 60.Kato HE, Zhang F, Yizhar O, Ramakrishnan C, Nishizawa T, Hirata K, Ito J, Aita Y, Tsukazaki T, Hayashi S, Hegemann P, Maturana AD, Ishitani R, Deisseroth K, Nureki O. Crystal structure of the channelrhodopsin light-gated cation channel. Nature. 2012;482:369–374. doi: 10.1038/nature10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sineshchekov OA, Govorunova EG, Wang J, Li H, Spudich JL. Intramolecular proton transfer in channelrhodopsins. Biophys J. 2013;104:807–817. doi: 10.1016/j.bpj.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kaulen AD. Electrogenic processes and protein conformational changes accompanying the bacteriorhodopsin photocycle. Biochim Biophys Acta - Bioenergetics. 2000;1460:204–219. doi: 10.1016/s0005-2728(00)00140-7. [DOI] [PubMed] [Google Scholar]
- 63.Der A, Keszthelyi L. Charge motion during the photocycle of bacteriorhodopsin. Biochemistry (Mosc) 2001;66:1234–1248. doi: 10.1023/a:1013179101782. [DOI] [PubMed] [Google Scholar]
- 64.Xu J, Stickrath AB, Bhattacharya P, Nees J, Varo G, Hillebrecht JR, Ren L, Birge RR. Direct measurement of the photoelectric response time of bacteriorhodopsin via electro-optic sampling. Biophys J. 2003;85:1128–1134. doi: 10.1016/S0006-3495(03)74549-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stern LJ, Ahl PL, Marti T, Mogi T, Dunach M, Berkowitz S, Rothschild KJ, Khorana HG. Substitution of membrane-embedded aspartic acids in bacteriorhodopsin causes specific changes in different steps of the photochemical cycle. Biochemistry. 1989;28:10035–10042. doi: 10.1021/bi00452a023. [DOI] [PubMed] [Google Scholar]
- 66.Otto H, Marti T, Holz M, Mogi T, Stern LJ, Engel F, Khorana HG, Heyn MP. Substitution of amino acids Asp-85, Asp-212, and Arg-82 in bacteriorhodopsin affects the proton release phase of the pump and the pK of the Schiff base. Proc Natl Acad Sci USA. 1990;87:1018–1022. doi: 10.1073/pnas.87.3.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hou SY, Govorunova EG, Ntefidou M, Lane CE, Spudich EN, Sineshchekov OA, Spudich JL. Diversity of Chlamydomonas channelrhodopsins. Photochem Photobiol. 2012;88:119–128. doi: 10.1111/j.1751-1097.2011.01027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gergely C, Ganea C, Száraz S, Váró G. Charge motions studied in the bacteriorhodopsin mutants D85N and D212N. J Photochem Photobiol B: Biol. 1995;27:27–32. [Google Scholar]
- 69.Wang WW, Sineshchekov OA, Spudich EN, Spudich JL. Spectroscopic and photochemical characterization of a deep ocean proteorhodopsin. J Biol Chem. 2003;278:33985–33991. doi: 10.1074/jbc.M305716200. [DOI] [PubMed] [Google Scholar]
- 70.Feldbauer K, Zimmermann D, Pintschovius V, Spitz J, Bamann C, Bamberg E. Channelrhodopsin-2 is a leaky proton pump. Proc Natl Acad Sci USA. 2009;106:12317–12322. doi: 10.1073/pnas.0905852106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lorenz-Fonfria VA, Resler T, Krause N, Nack M, Gossing M, Fischer von Mollard G, Bamann C, Bamberg E, Schlesinger R, Heberle J. Transient protonation changes in channelrhodopsin-2 and their relevance to channel gating. Proc Natl Acad Sci USA. 2013;110:E1273–1281. doi: 10.1073/pnas.1219502110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gunaydin LA, Yizhar O, Berndt A, Sohal VS, Deisseroth K, Hegemann P. Ultrafast optogenetic control. Nat Neurosci. 2010;13:387–392. doi: 10.1038/nn.2495. [DOI] [PubMed] [Google Scholar]
- 73.Stehfest K, Hegemann P. Evolution of the channelrhodopsin photocycle model. Chemphyschem. 2010;11:1120–1126. doi: 10.1002/cphc.200900980. [DOI] [PubMed] [Google Scholar]
- 74.Bamann C, Gueta R, Kleinlogel S, Nagel G, Bamberg E. Structural guidance of the photocycle of channelrhodopsin-2 by an interhelical hydrogen bond. Biochemistry. 2010;49:267–278. doi: 10.1021/bi901634p. [DOI] [PubMed] [Google Scholar]
- 75.Farrens DL, Altenbach C, Yang K, Hubbell WL, Khorana HG. Requirement of rigid-body motion of transmembrane helices for light activation of rhodopsin. Science. 1996;274:768–770. doi: 10.1126/science.274.5288.768. [DOI] [PubMed] [Google Scholar]
- 76.Altenbach C, Kusnetzow AK, Ernst OP, Hofmann KP, Hubbell WL. High-resolution distance mapping in rhodopsin reveals the pattern of helix movement due to activation. Proc Natl Acad Sci USA. 2008;105:7439–7444. doi: 10.1073/pnas.0802515105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sineshchekov OA, Spudich JL. Handbook of Photosensory Receptors. Wiley-VCH; Weinheim: 2005. Sensory rhodopsin signaling in green flagellate algae; pp. 25–42. [Google Scholar]
- 78.Litvin FF, Sineshchekov OA, Sineshchekov VA. Photoreceptor electric potential in the phototaxis of the alga Haematococcus pluvialis. Nature. 1978;271:476–478. doi: 10.1038/271476a0. [DOI] [PubMed] [Google Scholar]
- 79.Sineshchekov OA, Govorunova EG, Der A, Keszthelyi L, Nultsch W. Photoelectric responses in phototactic flagellated algae measured in cell suspension. J Photochem Photobiol B: Biol. 1992;13:119–134. [Google Scholar]
- 80.Sineshchekov OA, Litvin FF, Keszthelyi L. Two components of photoreceptor potential of the flagellated green alga Haematococcus pluvialis. Biophys J. 1990;57:33–39. doi: 10.1016/S0006-3495(90)82504-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tsunoda SP, Hegemann P. Glu 87 of channelrhodopsin-1 causes pH-dependent color tuning and fast photocurrent inactivation. Photochem Photobiol. 2009;85:564–569. doi: 10.1111/j.1751-1097.2008.00519.x. [DOI] [PubMed] [Google Scholar]
- 82.Sineshchekov OA, Govorunova EG. Rhodopsin-mediated photosensing in green flagellated algae. Trends Plant Sci. 1999;4:58–63. doi: 10.1016/s1360-1385(98)01370-3. [DOI] [PubMed] [Google Scholar]
- 83.Sineshchekov OA, Govorunova EG, Spudich JL. Photosensory functions of channelrhodopsins in native algal cells. Photochem Photobiol. 2009;85:556–563. doi: 10.1111/j.1751-1097.2008.00524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sineshchekov OA, Govorunova EG, Jung KH, Zauner S, Maier UG, Spudich JL. Rhodopsin-mediated photoreception in cryptophyte flagellates. Biophys J. 2005;89:4310–4319. doi: 10.1529/biophysj.105.070920. [DOI] [PMC free article] [PubMed] [Google Scholar]