Abstract
Multiple extracellular stimuli, such as growth factors and antigens, initiate signaling cascades through tyrosine phosphorylation and activation of phospholipase C (PLC)-γ isozymes. Like most other PLCs, PLC-γ1 is basally auto-inhibited by its X-Y linker, which separates the X-and Y-boxes of the catalytic core. The C-terminal SH2 (cSH2) domain within the X-Y linker is the critical determinant for auto-inhibition of phospholipase activity. Release of auto-inhibition requires an intramolecular interaction between the cSH2 domain and a phosphorylated tyrosine, Tyr783, also located within the X-Y linker. The molecular mechanisms that mediate auto-inhibition and phosphorylation-induced activation have not been defined. Here, we describe structures of the cSH2 domain both alone and bound to a PLC-γ1 peptide encompassing phosphorylated Tyr783. The cSH2 domain remains largely unaltered by peptide engagement. Point mutations in the cSH2 domain located at the interface with the peptide were sufficient to constitutively activate PLC-γ1 suggesting that peptide engagement directly interferes with the capacity of the cSH2 domain to block the lipase active site. This idea is supported by mutations in a complimentary surface of the catalytic core that also enhanced phospholipase activity.
Diverse extracellular stimuli including hormones, neurotransmitters, antigens, and growth factors, promote phospholipase C (PLC)-catalyzed hydrolysis of the minor membrane phospholipid phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2) to generate the intracellular second messengers inositol 1,4,5-trisphosphate (Ins(1,4,5)P3) and diacylglycerol1. Ins(1,4,5)P3 and diacylglycerol mobilize intracellular calcium and activate PKC isozymes, respectively, to regulate multiple cellular processes including fertilization, proliferation, differentiation, and chemotaxis2.
Six families of PLCs, PLC-β, -γ, -δ, -ε, -ζ, and –η, including thirteen distinct isozymes, exist in humans1. With the exception of the sperm-specific isozyme PLC-ζ, PLCs have a common core architecture consisting of a pleckstrin homology (PH) domain, a series of EF hands, a catalytic triose phosphate isomerase (TIM) barrel split into X- and Y-boxes by a variable length linker (X-Y linker), and a C2 domain. Most PLCs also contain additional domains, which engender isozyme-specific regulation. We have proposed a general model of regulation in which the X-Y linker basally auto-inhibits PLC isozymes. In many PLCs, the X-Y linker is disordered and negatively charged, and its deletion accelerates phospholipase activity in vitro and in cells3. We posit that auto-inhibition is due to electrostatic repulsion between the linker and membranes, as well as physical occlusion of the active site3–5. As a result of these observations, we proposed a model of interfacial activation in which PLCs are recruited to, and oriented at, membranes leading to a concomitant displacement of the X-Y linker from the active site and enhanced phospholipase activity.
PLC-γ isozymes (PLC-γ1 and -γ2) uniquely possess a highly elaborated X-Y linker, which contains two Src homology 2 (SH2) domains, an SH3 domain, and a split PH domain, suggesting that these isozymes exhibit a distinct mode of regulation. Indeed, this domain architecture engenders PLC-γ-specific activation by multiple tyrosine kinases, and tyrosine phosphorylation within the X-Y linker stimulates the activity of PLC-γ isozymes in vitro and in cells6–9. In particular, phosphorylation of Tyr783 in PLC-γ1 is critical for activation downstream of receptor tyrosine kinases (RTKs) and immune cell receptors7,9,10. We recently demonstrated that PLC-γ isozymes are basally auto-inhibited by the X-Y linker, and that deletion of the C-terminal SH2 (cSH2) domain within the X-Y linker was sufficient to constitutively activate PLC-γ1 similarly to deletion of the entire X-Y linker9. Therefore, the cSH2 domain represents the core element required for auto-inhibition of lipase activity. Further deletion-mapping defined 10 amino acids encompassing the BG loop and βG strand at the C-terminus of the cSH2 domain as critical for auto-inhibitory capacity. We also demonstrated that activation of PLC-γ1 requires that the cSH2 domain engage phosphorylated Tyr783, and that this engagement results in an allosteric rearrangement of the linker coupled to activation. This allosteric rearrangement and activation is recapitulated by deletion of the BG loop and βG strand. Therefore, we postulate that PLC-γ1 couples tyrosine phosphorylation to conformational rearrangements within the X-Y linker that drive phospholipase activity.
While this model explains many aspects of the phosphorylation-dependent activation of the PLC-γ isozymes, several questions remain unresolved. For example, the mechanism by which elements within the cSH2 domain, specifically the BG loop and βG strand, contribute to the auto-inhibition of PLC-γ1 activity is poorly understood. Here, we used a combination of structural biology and cell-based measures of phospholipase activity to propose that PLC-γ isozymes are regulated by direct competition of the cSH2 domain for interaction with the TIM barrel (auto-inhibited) and the phosphorylated X-Y linker (active).
MATERIALS AND METHODS
Cloning and purification of the cSH2 domain of PLC-γ1
The cSH2 domain (amino acids 664– 766) was amplified from full-length rat PLC-γ1 by PCR, then subcloned into a modified pET15b vector, which incorporates a His6 tag and a tobacco etch virus (TEV) protease site at the N-terminus of the expressed protein. The cSH2 domain was expressed in the BL21 strain of E. coli, and purified as described previously with the following modifications9. After TEV cleavage, the sample was applied to a 5 ml HisTrap HP column (GE Healthcare). Flow-through fractions were pooled and diluted 5-fold into 20 mM HEPES (7.5), 300 mM NaCl, and 5% (v/v) glycerol. The sample was further diluted 3-fold into buffer S1 (20 mM HEPES (7.5), 1 mM DTT, and 5% (v/v) glycerol) and applied to an 8 mL SourceS cation exchange column equilibrated in 10% buffer S2 (20 mM HEPES (7.5), 1 M NaCl, 1 mM DTT, and 5% (v/v) glycerol). Bound proteins were eluted with a linear gradient of 10%–100% buffer S2 over 20 column volumes.
Crystallization of the cSH2 domain of PLC-γ1
Diffraction quality crystals of the PLC-γ1 cSH2 domain were obtained by hanging drop vapor diffusion at 4 °C. Drops were formed by mixing 2 µL of the cSH2 domain at 7.5 mg/mL in 20 mM HEPES (7.5), 150 mM NaCl, 1 mM DTT, and 5% (v/v) glycerol with 1 µL of the reservoir solution (100 mM MES (5.5), 200 mM ammonium acetate, 25% (w/v) PEG 4,000). Single crystals appeared within two days, grew to maximum size (~200 µm on the longest edge) within one week, and had a Matthews coefficient (Vm) of 1.94 Å3 /Da. The space group and unit cell parameters are shown in Table 1. Crystals were cryo-protected by flash cooling in liquid nitrogen.
Table 1.
Data collection and refinement statistics for the PLC-γ1 cSH2 domain and the PLC-γ1 cSH2 domain bound to the pTyr783 peptide. Numbers in parenthesis refer to the highest resolution shell. Each dataset was collected from a single crystal.
| data collection | PLC-γ1 cSH2 | PLC-γ1 cSH2/ pTyr783 peptide |
|---|---|---|
| space group | C2221 | P212121 |
| unit cell dimensions | ||
| a, b, c (Å) | 48.1, 53.3, 150.3 | 29.0, 54.5, 59.9 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| resolution (Å) | 35.7–1.70 (1.73–1.70) | 40.3–1.50 (1.53–1.50) |
| unique reflections | 21,853 (1,066) | 15,900 (763) |
| Rmerge1 | 6.1 (46.3) | 6.7 (33.1) |
| <I/σ>2 | 33.3 (4.7) | 29.1 (3.8) |
| completeness (%) | 99.3 (91.1) | 99.8 (97.4) |
| redundancy | 8.0 (8.0) | 7.3 (4.7) |
| wavelength (Å) | 1.000 | 0.979 |
| no. of molecules in asymmetric unit | 2 | 1 |
| refinement | ||
| resolution (Å) | 35.7–1.70 | 40.3–1.50 |
| no. of reflections (test/working) | 1,120/20,696 | 790/15,060 |
| R/Rfree3 | 18.4/22.3 | 16.3/19.8 |
| no. of atoms | ||
| protein | 1,721 | 851 |
| peptide | - | 68 |
| water | 103 | 80 |
| R.M.S. deviations | ||
| bond lengths (Å) | 0.007 | 0.015 |
| bond angles (°) | 1.09 | 1.63 |
| average B-factors (Å2) | ||
| protein | 27.8 | 14.0 |
| peptide | - | 27.4 |
| water | 29.0 | 25.3 |
| Ramachandran plot (%) | ||
| favored | 96 | 97 |
| allowed | 4 | 3 |
| disallowed | 0 | 0 |
| PDB ID | 4K44 | 4K45 |
Rmerge=100×∑|I-<I>|/∑I, where I is the integrated intensity of a measured reflection.
<I/σ> is the mean signal to noise ratio, where I is the integrated intensity of a measured reflection, and σ is the estimated error in the measurement.
R=100×∑|Fo–Fc|/∑Fo, where Fo and Fc are the observed and calculated structure factor amplitudes, respectively. Rfree is calculated as R using 5% of the total reflections that have been randomly excluded from refinement.
Crystallization of the cSH2 domain of PLC-γ1 bound to the pTyr783 peptide
The PLC-γ1 cSH2 domain-pTyr783 peptide complex was formed by incubating the cSH2 domain (7.5 mg/mL) with a 1.1-fold molar excess of the pTyr783 peptide (1.4 mg/mL) in 20 mM HEPES (7.5), 150 mM NaCl, 1 mM DTT, and 5% (v/v) glycerol at 4 °C for one h prior to initiating crystallization trials. Crystals were obtained at 20 °C by microseeding hanging drops with crushed crystals of the apo-cSH2 domain. Crystals grew from drops formed by mixing 1 µL of protein solution with 2 µL of the reservoir solution (100 mM MES (6.0), 200 mM ammonium acetate, 35% (w/v) PEG 4,000), reached a maximum length of ~250 µm within two weeks, and had a Vm of 1.62 Å3/Da. The space group and unit cell parameters are shown in Table 1. Crystals were cryo-protected by flash cooling in liquid nitrogen.
Data collection and structure determination
Native sets of X-ray diffraction data were collected on single crystals of the apo-cSH2 domain and the cSH2 domain bound to the pTyr783 peptide on SER-CAT beamline 22-ID at the Advanced Photon Source at Argonne National Laboratory. Data were collected on a MAR300 CCD detector, at a crystal to detector distance of 150 mm. One scan about ω totaling 200° was collected for each crystal. Each frame consisted of a 0.5° rotation taken for one second. Data were indexed, integrated, and scaled using HKL200011. Phases were solved by molecular replacement using the program Phaser in the CCP4 program suite12,13. The coordinates of the cSH2 domain of PLC-γ1 (PDB ID: 3GQI) were used as a search model for the apo-cSH2 domain. After partial refinement of the apo-cSH2 domain structure, these phases were used to identify a solution for the cSH2 domain bound to the pTyr783 peptide. Both models were subjected to multiple rounds of restrained refinement and TLS refinement using Refmac5, as well as manual refinement in Coot14,15. A final round of restrained refinement and TLS refinement was carried out in Phenix16 . The stereochemical quality of each model was assessed with MolProbity17. Complete data collection and structure refinement statistics are shown in Table 1.
Cloning of PLC-γ1 point mutants
The R675E, K71 1E + K713E, R716E, N728E, Y747E + R748E, D342K, E347K, D370K, and D1019K point mutations were introduced into rat PLC-γ1 in a modified pcDNA3.1 vector, which incorporates an HA tag at the N-terminus of the expressed protein, by QuikChange site-directed mutagenesis (Stratagene). Incorporation of each mutation was confirmed by automated DNA sequencing of the entire PLC-γ1 open reading frame.
Measurement of PLC-γ1 activity
Cell-based assays to measure accumulation of [3H]inositol phosphates were performed as described previously3,5,9. Briefly, HEK293 cells were transfected with plasmids encoding HA-tagged PLC-γ1 constructs using Fugene-6 (Promega). Twenty-four h post-transfection, cells were metabolically labeled with 1 µCi of [3H]myo-inositol in serum-free, inositol-free DMEM for 16 h followed by a one h incubation in the presence of 10 mM LiCl. For experiments in which cells were treated with LiCl overnight, 10 mM LiCl was added to the radiolabeling medium. To measure PLC activity in response to EGF stimulation, cells were co-transfected with 200 ng of the indicated PLC-γ1 construct and 100 ng of plasmid encoding the human EGF receptor (a generous gift from Dr. H. Shelton Earp, The University of North Carolina at Chapel Hill). Cells were stimulated with recombinant human EGF (Invitrogen) for 30 min in the presence of 10 mM LiCl. Cells were lysed with formic acid, and following neutralization with ammonium hydroxide, the lysate was applied to Dowex columns. Accumulation of [3H]inositol phosphates was measured by liquid scintillation counting. Expression of PLC-γ1 constructs was determined by western blotting. Cell lysates were probed with a monoclonal antibody against the HA epitope (clone 3F10, Roche). As a loading control, lysates were also probed with an antibody against β-actin (clone AC-15, Sigma-Aldrich).
Homology modeling and electrostatic surface calculation
A homology model of PLC-γ1 (amino acids 1 to 1219) lacking the X-Y linker (amino acids 471 to 983) was generated using the Phyre server18. Electrostatic surfaces were calculated using APBS executed within PyMOL19,20.
Protein and Peptide Concentrations
Concentrations of the cSH2 domain were determined using the A280 and the extinction coefficient calculated using ProtParam (ε = 14,400 M−1cm−1). Concentrations of the pTyr783 peptide were calculated using the A267 and an extinction coefficient ε = 2,752 M−1cm−1 calculated by adding the extinction coefficient of tyrosine (ε = 1,050 M−1cm−1) and phosphotyrosine (ε = 652 M−1cm−1). The pTyr783 peptide was synthesized by Creative Peptides Inc., Shirley, NY and had the following sequence: acetyl-DYGALYEGRNPGFYVEAN-amide, with the phosphorylated tyrosine underlined.
RESULTS
Phosphorylation of PLC-γ1 at Tyr783 elevates lipase activity, and we previously suggested that this increase results from intramolecular interaction of the cSH2 domain with the phosphorylated form of Tyr783 found immediately C-terminal to the cSH2 domain. To better understand this regulation, we determined the crystal structure of the cSH2 domain of PLC-γ1 in isolation as well as in complex with a peptide encompassing the region surrounding pTyr783 (Fig. 1).
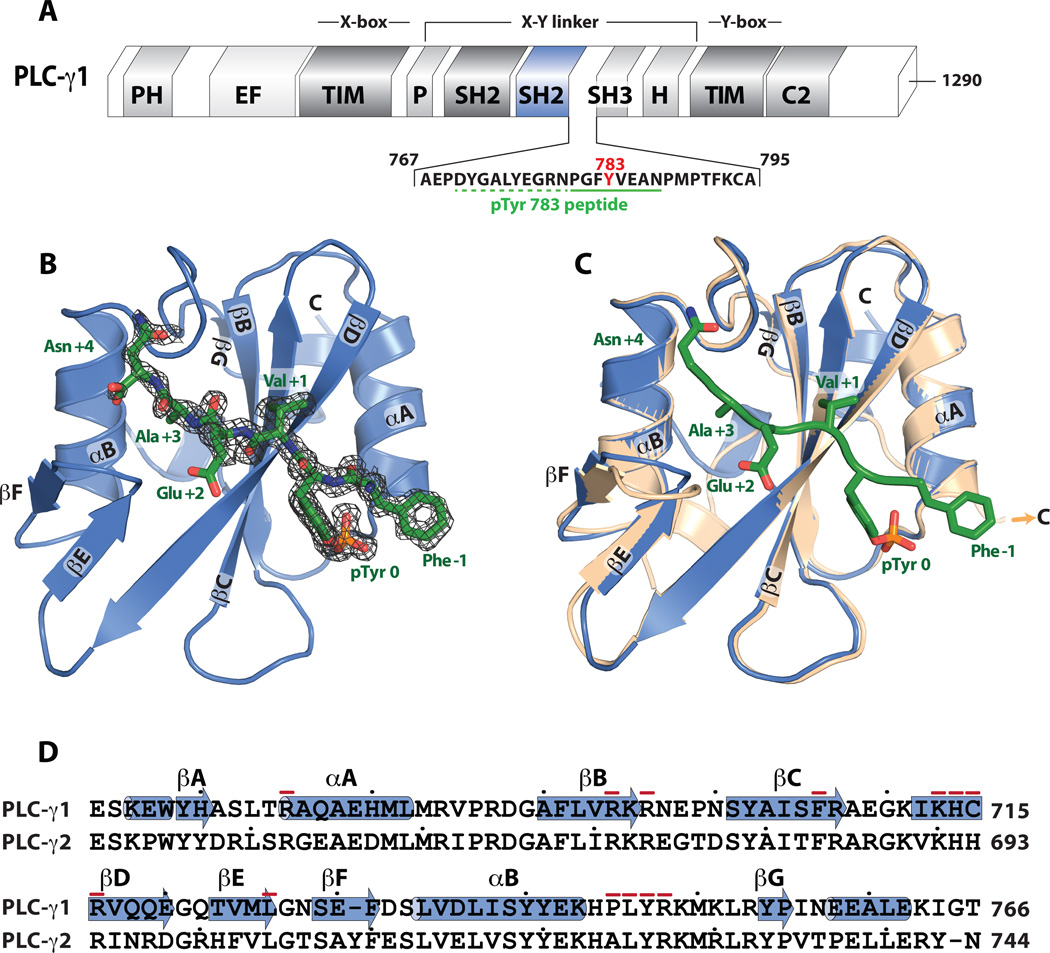
Fig. 1.
Crystal structures of the C-terminal SH2 domain of PLC-γ1. (A) Schematic representation of full-length rat PLC-γ1. Highlighted are the C-terminal SH2 domain (cSH2; blue) and primary sequence of the phosphorylated peptide (green underline, phosphorylated Tyr783 is red) used for structure determination. Dashed green line indicates portion of the peptide disordered in the crystal structure. (B) Structure of the cSH2 domain of PLC-γ1 bound to the pTyr783 peptide. Elements of secondary structure within the cSH2 domain are labeled according to established nomenclature. The bound peptide is green with associated electron density (2Fo-Fc map contoured at 1.0 σ) in black mesh. Positions of amino acids within the peptide are numbered relative to the phosphotyrosine. (C) Structural comparison of the cSH2 domain in the presence (blue) and absence (wheat) of the pTyr783 peptide. Structures superimposed (RMSD ~0.3 Å) for 77 equivalent Cα atoms. (D) Alignment of the primary sequences of the cSH2 domain from rat PLC-γ1 and PLC-γ2. α helices (cylinders) and β strands (arrows) are assigned using the structure of the peptide-bound form of the cSH2 domain. Dots demark every 10th residue and residues that contact the pTyr783 peptide are indicated with a red dash.
The structure of the apo-form of the cSH2 domain was solved by molecular replacement, using the structure of the cSH2 domain of PLC-γ1 (PDB ID: 3QGI) as a search model, and refined at a resolution of 1.7 Å (Table 1). Phases from this model were subsequently used to refine the 1.5-Å resolution structure of the peptide-bound form (Table 1).
The structure of the cSH2 domain is similar to other SH2 domains and consists of a central antiparallel β sheet flanked by two α helices (Fig. 1B). The phosphopeptide binds in an extended groove roughly orthogonal to the central β sheet and is anchored at one end by the phosphorylated tyrosine in a deep pocket on the cSH2 domain. Although the phosphopeptide consists of 18 residues, only the C-terminal eight have clear electron density in the structure and bury ~430 Å2 of solvent-accessible surface of the cSH2 domain upon complex formation. The remainder of the peptide is presumably disordered and not required for complex formation. The extended interface is recapitulated in other peptide:SH2 complexes21,22.
Conformational differences between the peptide-bound and unbound forms of the cSH2 domain (Fig. 1C) are unlikely to be functionally important. For instance, the largest differences between the two forms occur at the termini of the cSH2 domain on the opposite side of the domain relative to bound peptide. The N-terminal helix reorients and the C-terminal helix is missing in the unbound form. However, these conformational changes do not propagate to the site of the bound peptide, suggesting that the termini are inherently flexible. Additionally, the EF loop is translated ~2.5 Å toward the bound peptide relative to the unbound form, but this difference is ascribed to interactions of this loop with the C-terminus of a symmetry mate in the crystal of the unbound form.
The two structures superimpose with an RMSD ~0.3 Å for 77 equivalent Cα atoms. In particular, the conformations of the BG loop and βG strand are essentially identical between the two structures. We previously postulated that conformational changes centered within this region in response to engagement of phosphopeptide would be linked to the release of auto-inhibition of phospholipase activity9.
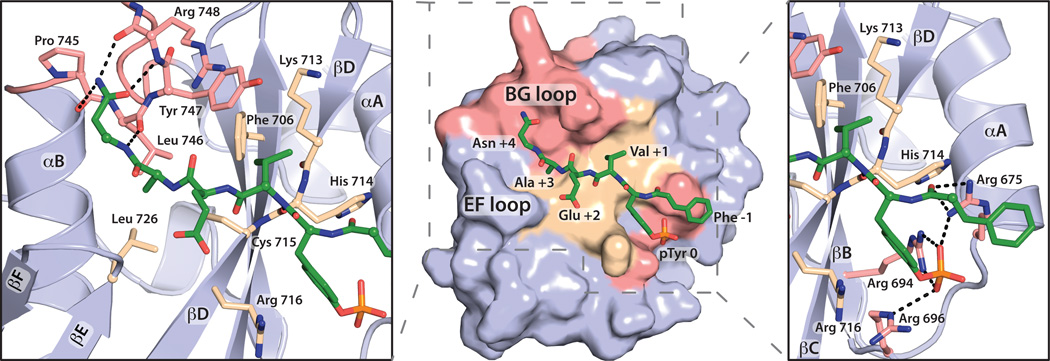
While the conformations of the BG loop and βG strand remain unaltered between the two structures, the BG loop substantially contributes to the interface with the phosphopeptide (Fig. 1D). For example, Asn +4 of the phosphopeptide, corresponding to residue 787 in full-length PLC-γ1, participates in four hydrogen bonds with the BG loop (Fig. 2). Leu746 within the BG loop also forms part of the hydrophobic groove that interacts with Ala +3 of the phosphopeptide. The remainder of the hydrophobic groove of the cSH2 domain is occupied by Val +1 of the phosphopeptide before terminating in the phosphotyrosine-binding pocket where pTyr783 interacts with four arginines (Arg675, Arg694, Arg696, Arg716).
Fig. 2.
The BG loop forms part of the interface with the pTyr738 peptide. The surface of the cSH2 domain (center panel) is colored to highlight the BG loop and phosphotyrosine-binding pocket (pink) as well as intervening residues (wheat) that interact with the peptide. Side panels detail interactions between the peptide and the cSH2 domain; dashed lines represent hydrogen bonds.
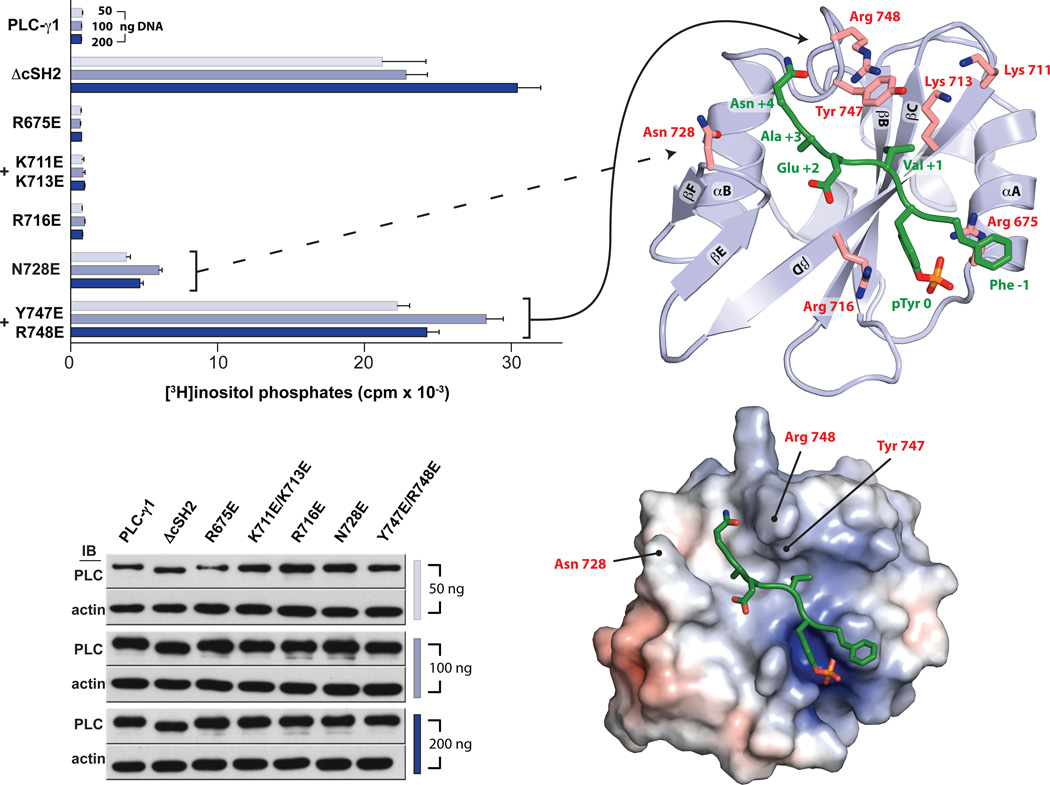
The structures of the bound and unbound forms of the cSH2 domain fail to highlight conformational changes that can be reasonably linked to enhancement of phospholipase activity. However, a likely alternative posits that the peptide-binding surface of the cSH2 domain directly occludes the active site of PLC-γ1. In this case, engagement of the cSH2 domain by a phosphopeptide would directly disrupt this occlusion to release auto-inhibition. This idea was tested by mutating the peptide-binding surface of the cSH2 domain within the context of full-length PLC-γ1 and measuring phospholipase activity in cells after metabolic labeling of phosphoinositide pools (Fig. 3). Several substitutions of positively charged residues to glutamic acid had no effect on basal phospholipase activity. This set includes the individual substitution of two arginines (Arg675 and Arg716) within the pocket that binds pTyr783 as well as double substitution of lysines (Lys711 and Lys713) adjacent to this pocket. In contrast, mutation of the EF loop (N728E) enhanced basal phospholipase activity approximately five-fold and a tandem substitution (Y747E + R748E) within the BG loop activated PLC-γ1 by greater than 30-fold. This elevated basal activity approximates the activity of PLC-γ1 lacking the entire cSH2 domain (ΔcSH2; Fig 3), which we have shown completely lacks auto-inhibition mediated by the X-Y linker9. These results further emphasize the importance of the BG loop and adjacent regions of the cSH2 domain in the auto-inhibition of PLC-γ isozymes and suggest that these regions may interact with the active site to auto-inhibit phospholipase activity. Conversely, the pocket required to bind pTyr783 likely does not interact with the active site to auto-inhibit PLC-γ1. However, this pocket is nevertheless essential for activation since the cSH2 domain must engage pTyr783 and surrounding peptide to relieve auto-inhibition as shown previously9.
Fig. 3.
The auto-inhibitory interface within the cSH2 domain maps to the electropositive EF and BG loops. HEK293 cells were transfected with the indicated amounts of plasmids encoding either wild-type or mutant forms of PLC-γ1 prior to quantification of accumulated [3H]inositol phosphates (top left). Mutated residues are highlighted on the structure of the cSH2 domain (top right). Data are presented as the means ± S.E.M. of triplicate samples from a single experiment and are representative of three independent experiments. Expression of PLC-γ1 constructs confirmed by western blots of cell lysates (bottom left). Mutated residues that increase basal phospholipase activity are indicated on the solvent accessible surface of the cSH2 domain (bottom right) colored according to electrostatic potential calculated without bound peptide (red: −5 kT/e, blue: +5kT/e).
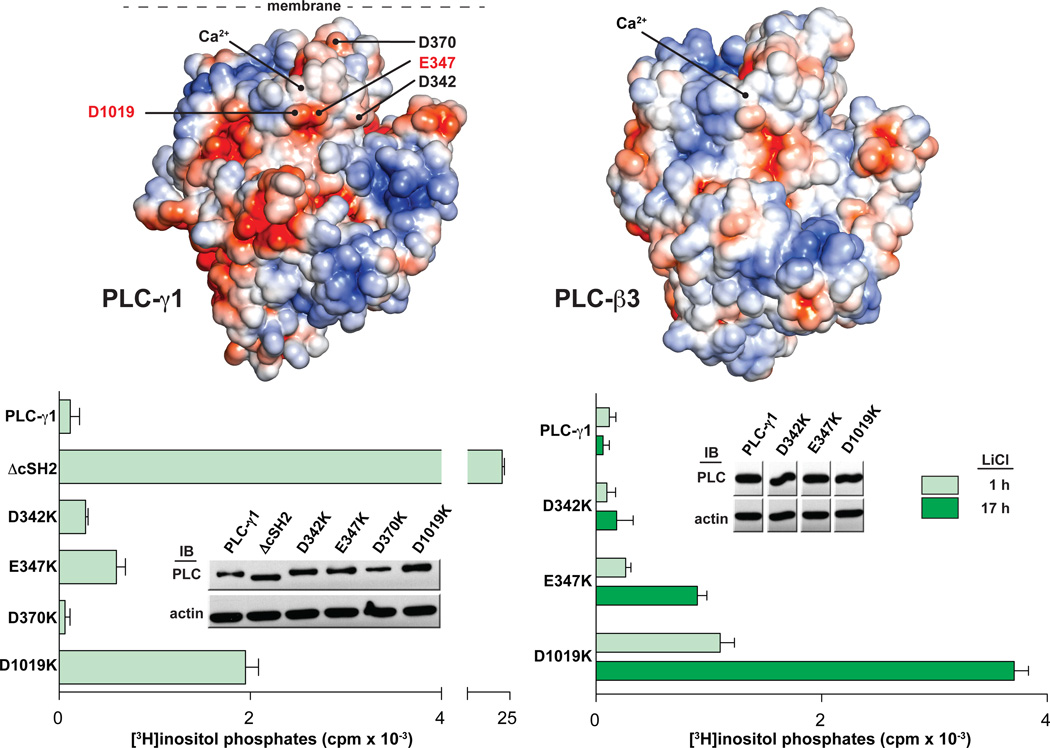
The surface of the cSH2 domain, including the BG loop, that engages the pTyr783 peptide is positively charged (Fig. 3). If this surface directly occludes the active site of PLC-γ1 to auto-inhibit phospholipase activity, it seems reasonable to expect the complementary surface of PLC-γ1 to be negatively charged. In an effort to identify potential areas near the active site of PLC-γ1 that interact with the cSH2 domain, a homology model of PLC-γ1 was generated based on the structure of PLC-β3 (Fig. 4). This model spans the PH to C2 domains but lacks the X-Y linker. Comparison of this model with the structure of PLC-β3 highlights several negative patches in the vicinity of the active site of PLC-γ1 that are not found in PLC-β3. Negatively charged residues within these areas of PLC-γ1 were mutated to lysine, and phospholipase activity of the resulting mutants was quantified in cells.
Fig. 4.
The catalytic core of PLC-γ1 contains a putative interface with the cSH2 domain. (A) The core of PLC-γ1 is electronegative. The solvent accessible surface of a homology model of PLC-γ1 lacking the X-Y linker (left) is colored according to electrostatic potential (red: −5 kT/e, blue: +5 kT/e). Residues selected for mutational analysis are labeled. The equivalent representation of PLC-β3 is shown at right. (B) Mutations near the active site of PLC-γ1 elevate basal activity. HEK293 cells were transfected with plasmids encoding either wild-type PLC-γ1 or PLC-γ1 harboring the indicated mutations in the TIM barrel. Accumulation of [3H]inositol phosphates was quantified after a one hour incubation with LiCl (left) or for the indicated times (right). The amount of radioactivity that accumulated in cells transfected with empty vector was subtracted from all measurements. Data are presented as the means ± S.E.M. of triplicate samples from a single experiment. Expression of PLC-γ1 constructs verified by western blots (insets).
Individual substitutions at two positions (D342K and E347K) produced modest enhancement (2–5x) of basal phospholipase activity relative to the wild-type isozyme (Fig. 4). More interestingly, substitution of Asp1019 to lysine enhanced basal phospholipase activity approximately 15-fold. In comparison, the phospholipase activity of PLC-γ1 (Δc SH2), which completely lacks auto-inhibition mediated by the X-Y linker, is elevated by approximately 130-fold. To assess further these relatively small increases in phospholipase activity, a second set of cells were treated identically except with a prolonged incubation (17 hours) with LiCl to enhance accumulation of inositol phosphates. Under these conditions, the phospholipase activity of PLC-γ1 (E347K) was elevated by approximately 15-fold relative to wild-type PLC-γ1; the activity of PLC-γ1(D1019K) was approximately 60-fold higher. These results are consistent with the proposal that Glu347 and Asp1019 interact with the cSH2 domain to auto-inhibit PLC-γ1. These two sites are separated by the X-Y linker in the primary sequence of PLC-γ1, but are adjacent to each other in the homology model.
The relative basal phospholipase activities of PLC-γ1 (Y747E + R748E) and PLC-γ1(D1019K) measured in intact cells were confirmed with purified proteins. These proteins, along with wild-type PLC-γ1, were purified to homogeneity after expression in insect cells and their specific activities compared using 30 µM [3H]PtdIns(4,5)P2. Consistent with the intact cell experiments, wild-type PLC-γ1 had low basal activity (0.72 ± 0.25 pmol/min/ng) while PLC-γ1(D1019K) possessed slightly elevated phospholipase activity (3.98 ± 0.23 pmol/min/ng) and PLC-γ1(Y747E + R748E) was highly active (42.5 ± 10.85 pmol/min/ng) (data not shown).
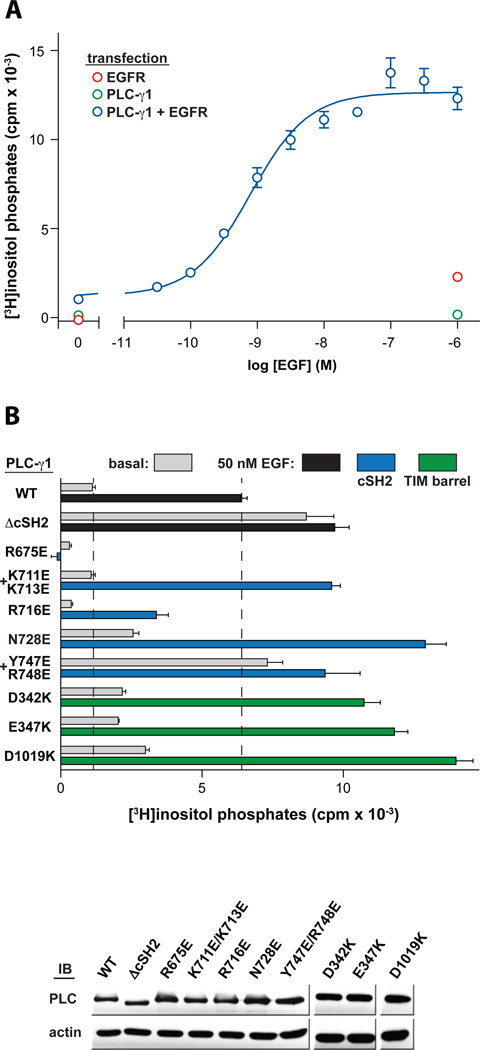
These mutational analyses suggest that the cSH2 domain directly interacts with the active site of PLC-γ1 to auto-inhibit phospholipase activity. However, it was also important to assess the effects of these mutations on phosphorylation-induced activation. Thus, we examined the capacity of the epidermal growth factor receptor (EGFR) to activate a panel of the PLC-γ1 mutants in HEK293 cells (Fig. 5).
Fig. 5.
PLC-γ1 is activated to a similar extent by either EGFR-dependent phosphorylation or mutation of the BG loop. (A) Dose-dependent accumulation of inositol phosphates by EGF in cells overexpressing EGFR and PLC-γ1. HEK293 cells were transfected with plasmids encoding PLC-γ1, EGFR, or both constructs as indicated prior to challenge with EGF for 30 minutes and measurement of [3H]inositol phosphates. In each case, the level of radioactivity that accrued in cells not treated with LiCl was subtracted from all values. (B) Stimulation of PLC-γ1 by EGF is not further enhanced by mutation of the BG loop. HEK293 cells were co-transfected with plasmids encoding EGFR and either wild-type PLC-γ1 or the indicated mutant forms. Accumulation of [3H]inositol phosphates was quantified after a 30 minute challenge with vehicle (gray bars) or 50 nM EGF (black, blue, and green bars). The amount of radioactivity that accumulated in cells overexpressing EGFR following treatment with either vehicle or EGF was subtracted from each basal or EGF-treated condition, respectively. Activities of PLC-γ1 forms harboring mutations in either the cSH2 domain or the TIM barrel were measured independently and the results combined into a single graph for clarity. Expression of PLC-γ1 constructs verified by western blots of cell lysates (bottom). Data are presented as the means ± S.E.M. of triplicate samples from single experiments and are representative of three independent experiments in panel B.
Concentration dependent activation of phospholipase C by epidermal growth factor (EGF) was observed in HEK293 cells co-transfected with wild-type PLC-γ1 and EGFR (Fig. 5A). Maximal activation was observed with approximately 50 nM EGF. In contrast, transient expression of either PLC-γ1 or EGFR alone resulted in minimal responses to EGF. As expected, PLC-γ1(ΔcSH2) was not further activated by 50 nM EGF when co-expressed with the EGFR (Fig. 5B). Of the panel of PLC-γ1 mutants harboring substitutions (Fig. 5B), only two (R675E and R716E) could not be activated to the same extent as wild-type PLC-γ1 by application of 50 nM EGF after co-transfection with EGFR. Both of these sites reside in the pocket of the cSH2 domain needed to engage phosphotyrosine and likely prevent activation by reducing the capacity of the cSH2 domain to bind pTyr783. Under identical conditions, the remaining substituted forms retained the capacity to be activated fully by addition of EGF. Indeed, these mutants, which include substitutions within both the cSH2 domain and TIM barrel, consistently exhibited phospholipase activity in response to EGF that surpassed the equivalent activity of wild-type PLC-γ1 and approached or exceeded the phospholipase activity of PLC-γ1(ΔcSH2). These data indicate a residual auto-inhibition of phosphorylated, wild-type PLC-γ1 that can be relieved by either deletion of the cSH2 domain or mutations that prevent its interaction with the active site.
DISCUSSION
The structure of the cSH2 domain is not substantially altered upon engagement of the pTyr783 peptide and in particular, the BG loop and βG strand adopt an essentially identical configuration in the presence and absence of the phosphopeptide (Fig. 1C). However, the BG loop makes a substantial contribution to the interface with the pTyr783 peptide (Fig. 2). Therefore, these data strongly suggest that phosphorylated sequences and the lipase active site directly compete for binding to overlapping surfaces of the cSH2 domain, and that in contrast to our previous supposition9, no conformational changes within the cSH2 domain are required to regulate phospholipase activity.
The model driven by the structures of the cSH2 domain with and without bound pTyr783 peptide shares many features with the auto-inhibition of class I PI 3-kinases23–25. Indeed, the enzymatic activity of the catalytic p110 subunit is auto-inhibited by tandem SH2 domains in the regulatory subunit, p85. Structures of p85-p110 complexes indicate that the nSH2 domain engages the helical and kinase domains of p110 through the same surface that binds phosphopeptides26,27. Biochemical studies suggest that positively charged residues that form the phosphopeptide-binding site on the nSH2 domain engage a cluster of acidic residues within the helical domain of p110 to auto-inhibit kinase activity28. These positively charged residues in the nSH2 domain also directly contact bound phosphopeptides, suggesting that binding of the nSH2 domain to p110 and phosphorylated target sequences is mutually exclusive29. Charge reversal mutations within the interface between the nSH2 domain and p110 are sufficient to elevate kinase activity in vitro28. Although the binding mode between the nSH2 domain and p110 is well-defined, the precise mechanism by which disruption of this auto-inhibitory interface is translated into enhanced enzymatic activity remains to be fully elucidated. Nevertheless, phosphorylated peptides derived from receptor tyrosine kinases and adaptor proteins activate PI 3-kinase in vitro, and oncogenic missense mutations in p110 were mapped to the auto-inhibitory interface with the nSH2 domain30–35.
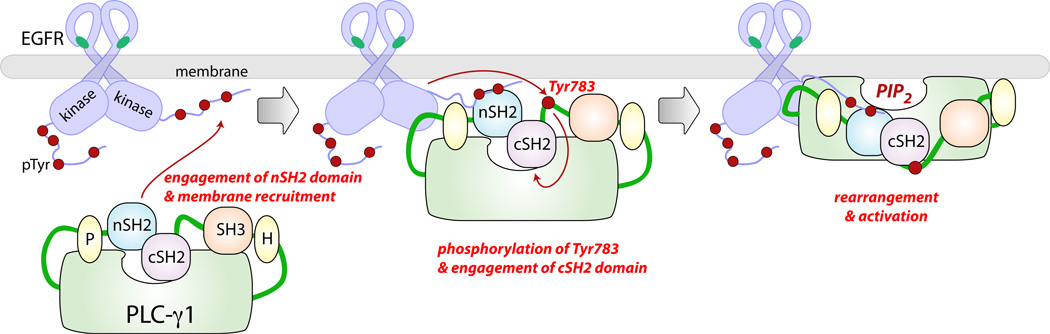
We have identified a positively charged surface of the cSH2 domain that is critically involved in auto-inhibition of PLC-γ1 activity (Fig. 3). The corresponding portion of the PLC-γ2 cSH2 domain is also highly electropositive, suggesting a conserved mode of regulation (data not shown). Mutational analysis of the TIM barrel suggests the presence of a complimentary, i.e. negatively charged, surface that is involved in the regulation of lipase activity (Fig. 4). The nSH2 domain of p85 auto-inhibits p110 activity in trans. In contrast, PLC-γ1 activity is auto-inhibited by an intramolecular interaction that requires the cSH2 domain. Nevertheless, we posit that the cSH2 domain of PLC-γ1 and the nSH2 domain of p85 auto-inhibit enzymatic activity through a similar mechanism. In the basal state, the surface of the cSH2 domain that binds phosphorylated sequences, specifically the BG and EF loops, directly engages the TIM barrel such that access of substrate to the lipase active site is precluded. Upon nSH2-dependent recruitment to activated RTKs, PLC-γ1 is phosphorylated on specific tyrosines, e.g. Tyr783. Phosphorylated Tyr783 then engages the cSH2 domain, disrupting the interaction with the TIM barrel, and allowing enhanced phospholipase activity (Fig. 6).
Fig. 6.
Model for the phosphorylation-dependent activation of PLC-γ1. In the basal state, the catalytic TIM barrel (light green) of PLC-γ1 is auto-inhibited by the cSH2 domain within the X-Y linker. The phosphorylated tails of activated RTKs (red circles), such as EGFR, provide docking sites for the nSH2 domain resulting in the recruitment of PLC-γ1 to the plasma membrane. PLC-γ1 is subsequently phosphorylated on Tyr783 within the X-Y linker, which results in an intramolecular association with the cSH2 domain. This interaction leads to conformational changes within the larger X-Y linker that are coupled to the release of auto-inhibition and enhanced hydrolysis of the substrate PtdIns(4,5)P2 (PIP2).
In-frame deletions in the cSH2 domain of PLC-γ2 were linked recently to immune system dysregulation in a cohort of patients with a dominantly inherited syndrome characterized by immunodeficiency and autoimmunity36. Cell-based overexpression studies as well as ex vivo analysis of primary immune cells from patients indicated that these deleted forms of PLC-γ2 are hyperactive. Genetic analysis of affected individuals identified two distinct deletion variants of PLC-γ2. One variant lacks a portion of the cSH2 domain encompassing the EF and BG loops. These loops play a central role in mediating the auto-inhibition of PLC-γ1 activity. Therefore, the absence of these elements provides a clear rational for the observed increases in PLC-γ2 activity. The other deleted form of PLC-γ2 lacks approximately 40 amino acids at the N-terminus of the cSH2 domain. In this case, elevated lipase activity is likely attributable to unfolding of the cSH2 domain and the concomitant loss of the surface required for auto-inhibition.
The auto-inhibitory interface within the cSH2 domain apparently is centered on the BG loop. However, a single point mutation in the EF loop, N728E, elevated basal PLC-γ1 activity approximately 5-fold (Fig. 3), suggesting that this loop also makes a contribution to the auto-inhibitory surface. Alanine-scanning mutagenesis of individual amino acids, including Asn728, across the EF loop, did not result in mutants of PLC-γ1 with altered basal phospholipase activity9. Apparently, the neutral N728A mutation is insufficient to disrupt the auto-inhibitory interface between the cSH2 domain and catalytic core.
Genetic evidence supports the notion that the surface of the cSH2 domain encompassing the EF loop is involved in the auto-inhibition of PLC-γ isozymes. A single missense mutation, S707Y, located in the βE strand of the PLC-γ2 cSH2 domain was linked to a distinct group of patients with dominantly inherited auto-inflammatory disease37. The equivalent residue in PLC-γ1, Ser729, is immediately adjacent to Asn728. Concordant with our observation that the N728E mutation elevates the basal activity of PLC-γ1, several lines of evidence suggest that S707Y is a gain-of-function substitution in PLC-γ2. Zhou and colleagues suggest that altered activity of PLC-γ2(S707Y) is due to insertion of a putative phosphorylation site within the cSH2 domain. However, we posit that point mutations in the EF loop (N728E in PLC-γ1) and βE strand (S707Y in PLC-γ2) elevate the activity of PLC-γ isozymes through a common mechanism: disruption of the auto-inhibitory interface between the cSH2 domain and the catalytic core.
Forward genetic screens in mice also implicate residues outside of the cSH2 domain in the regulation of PLC-γ2 activity. One such screen identified a missense mutation, D993G, in the TIM barrel of PLC-γ2 that is linked to spontaneous inflammation, autoimmunity, and arthritis38. B cells derived from mice heterozygous for this mutation are hyper-responsive to receptor activation ex vivo, suggesting that D993G is a gain-of-function mutation. Indeed, subsequent analysis of PLC-γ2(D993G) in an overexpression system demonstrated that this mutant isozyme exhibits higher basal and receptor-dependent activity than wild-type PLC-γ2 in cells39. Consistent with these findings, we observed that PLC-γ1 harboring a mutation at Asp1019, which corresponds to Asp993 in PLC-γ2, had elevated basal activity (Fig. 4). The D1019K mutation also potentiated the EGFR-dependent activation of PLC-γ1 in cells (Fig. 5). Given that these effects resulted from a charge reversal mutation, it is tempting to speculate that Asp1019 interfaces with the positively charged surface of the cSH2 domain to auto-inhibit PLC-γ1 activity.
The cSH2 domain of PLC-γ1 is the primary determinant mediating auto-inhibition of PLC-γ1, and we have focused on how this domain responds to the engagement of phosphorylated Tyr783 to regulate phospholipase activity. However, the cSH2 domain operates within the context of the larger X-Y linker, and structure determination of a full-length form of PLC-γ1 will be required to fully understand how tyrosine phosphorylation is coupled to the release of cSH2 domain-mediated auto-inhibition in holo-PLC-γ1.
ACKNOWLEDGEMENTS
We thank Dr. Mischa Machius and Dr. Michael Miley for assistance with the collection of X-ray diffraction data, and Dr. H. Shelton Earp for the gift of EGFR plasmid. We are also indebted to Matthew Barrett for assistance with phospholipase assays, and members of the Sondek and Harden laboratories for many helpful suggestions and discussions. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38.
This work was supported by National Institutes of Health Grants GM057391 (to J.S. and T.K.H.) and GM098894 (to J.S.).
ABBREVIATIONS
- cSH2 domain
C-terminal SH2 domain
- EGF
epidermal growth factor
- EGFR
epidermal growth factor receptor
- Ins(1,4,5)P3
inositol 1,4,5-trisphosphate
- nSH2 domain
N-terminal SH2 domain
- PtdIns(4,5)P2
phosphatidylinositol 4,5-bisphosphate
- PLC
phospholipase C
- PH
pleckstrin homology
- RTK
receptor tyrosine kinase
- SH
Src homology
- TIM
triose phosphate isomerase
REFERENCES
- 1.Harden TK, Sondek J. Regulation of phospholipase C isozymes by Ras superfamily GTPases. Annu. Rev. Pharmacol. Toxicol. 2006;46:355–379. doi: 10.1146/annurev.pharmtox.46.120604.141223. [DOI] [PubMed] [Google Scholar]
- 2.Rhee SG. Regulation of phosphoinositide-specific C. Annu. Rev. Biochem. 2001;70:281–312. doi: 10.1146/annurev.biochem.70.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hicks SN, Jezyk MR, Gershburg S, Seifert JP, Harden TK, Sondek J. General and versatile autoinhibition of PLC isozymes. Mol. Cell. 2008;31:383–394. doi: 10.1016/j.molcel.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jezyk MR, Snyder JT, Gershberg S, Worthylake DK, Harden TK, Sondek J. Crystal structure of Rac1 bound to its effector phospholipase C-β2. Nat. Struct. Mol. Biol. 2006;13:1135–1140. doi: 10.1038/nsmb1175. [DOI] [PubMed] [Google Scholar]
- 5.Waldo GL, Ricks TK, Hicks SN, Cheever ML, Kawano T, Tsuboi K, Wang X, Montell C, Kozasa T, Sondek J, Harden TK. Kinetic scaffolding mediated by a phospholipase C-β and Gq signaling complex. Science. 2010;330:974–980. doi: 10.1126/science.1193438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nishibe S, Wahl MI, Hernandez-Sotomayor SM, Tonks NK, Rhee SG, Carpenter G. Increase of the catalytic activity of phospholipase C-γ1 by tyrosine phosphorylation. Science. 1990;250:1253–1256. doi: 10.1126/science.1700866. [DOI] [PubMed] [Google Scholar]
- 7.Kim HK, Kim JW, Zilberstein A, Margolis B, Kim JG, Schlessinger J, Rhee SG. PDGF stimulation of inositol phospholipid hydrolysis requires PLC-γ1 phosphorylation on tyrosine residues 783 and 1254. Cell. 1991;65:435–441. doi: 10.1016/0092-8674(91)90461-7. [DOI] [PubMed] [Google Scholar]
- 8.Ozdener F, Dangelmaier C, Ashby B, Kunapuli SP, Daniel JL. Activation of phospholipase Cγ2 by tyrosine phosphorylation. Mol. Pharmacol. 2002;62:672–679. doi: 10.1124/mol.62.3.672. [DOI] [PubMed] [Google Scholar]
- 9.Gresset A, Hicks SN, Harden TK, Sondek J. Mechanism of phosphorylation-induced activation of phospholipase C-γ isozymes. J. Biol. Chem. 2010;285:35836–35847. doi: 10.1074/jbc.M110.166512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Serrano CJ, Graham L, DeBell K, Rawat R, Veri MC, Bonvini E, Rellahan BL, Reischl IG. A new tyrosine phosphorylation site in PLCγ1: the role of tyrosine 775 in immune receptor signaling. J. Immunol. 2005;174:6233–6237. doi: 10.4049/jimmunol.174.10.6233. [DOI] [PubMed] [Google Scholar]
- 11.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 12.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 16.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen VB, Arendall WB, Headd JJ, 3rd, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 19.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.The PyMOL Molecular Graphics System, Version 1.3. Schrödinger, LLC; [Google Scholar]
- 21.Lee CH, Kominos D, Jacques S, Margolis B, Schlessinger J, Shoelson SE, Kuriyan J. Crystal structures of peptide complexes of the amino-terminal SH2 domain of the Syp tyrosine phosphatase. Structure. 1994;2:423–438. doi: 10.1016/s0969-2126(00)00044-7. [DOI] [PubMed] [Google Scholar]
- 22.Pascal SM, Singer AU, Gish G, Yamazaki T, Shoelson SE, Pawson T, Kay LE, Forman-Kay JD. Nuclear magnetic resonance structure of an SH2 domain of phospholipase C-γ1 complexed with a high affinity binding peptide. Cell. 1994;77:461–472. doi: 10.1016/0092-8674(94)90160-0. [DOI] [PubMed] [Google Scholar]
- 23.Yu J, Zhang Y, McIlroy J, Rordorf-Nikolic T, Orr GA, Backer JM. Regulation of the p85/p110 phosphatidylinositol 3’-kinase: stabilization and inhibition of the p110α catalytic subunit by the p85 regulatory subunit. Mol. Cell Biol. 1998;18:1379–1387. doi: 10.1128/mcb.18.3.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu J, Wjasow C, Backer JM. Regulation of the p85/p110α phosphatidylinositol 3’-kinase. Distinct roles for the N-terminal and C-terminal SH2 domains. J. Biol. Chem. 1998;273:30199–30203. doi: 10.1074/jbc.273.46.30199. [DOI] [PubMed] [Google Scholar]
- 25.Vadas O, Burke JE, Zhang X, Berndt A, Williams RL. Structural basis for activation and inhibition of class I phosphoinositide 3-kinases. Sci. Signal. 2011;4:2. doi: 10.1126/scisignal.2002165. [DOI] [PubMed] [Google Scholar]
- 26.Huang CH, Mandelker D, Schmidt-Kittler O, Samuels Y, Velculescu VE, Kinzler KW, Vogelstein B, Gabelli SB, Amzel LM. The structure of a human p110α/p85α complex elucidates the effects of oncogenic PI3Kα mutations. Science. 2007;318:1744–1748. doi: 10.1126/science.1150799. [DOI] [PubMed] [Google Scholar]
- 27.Mandelker D, Gabelli SB, Schmidt-Kittler O, Zhu J, Cheong I, Huang CH, Kinzler KW, Vogelstein B, Amzel LM. A frequent kinase domain mutation that changes the interaction between PI3Kα and the membrane. Proc. Natl. Acad. Sci. U.S.A. 2009;106:16996–17001. doi: 10.1073/pnas.0908444106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miled N, Yan Y, Hon WC, Perisic O, Zvelebil M, Inbar Y, Schneidman-Duhovny D, Wolfson HJ, Backer JM, Williams RL. Mechanism of two classes of cancer mutations in the phosphoinositide 3-kinase catalytic subunit. Science. 2007;317:239–242. doi: 10.1126/science.1135394. [DOI] [PubMed] [Google Scholar]
- 29.Nolte RT, Eck MJ, Schlessinger J, Shoelson SE, Harrison SC. Crystal structure of the PI 3-kinase p85 amino-terminal SH2 domain and its phosphopeptide complexes. Nat. Struct. Biol. 1996;3:364–374. doi: 10.1038/nsb0496-364. [DOI] [PubMed] [Google Scholar]
- 30.Backer JM, Myers MG, Jr, Shoelson SE, Chin DJ, Sun XJ, Miralpeix M, Hu P, Margolis B, Skolnik EY, Schlessinger J. Phosphatidylinositol 3’-kinase is activated by association with IRS-1 during insulin stimulation. EMBO J. 1992;11:3469–3479. doi: 10.1002/j.1460-2075.1992.tb05426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carpenter CL, Auger KR, Chanudhuri M, Yoakim M, Schaffhausen B, Shoelson S, Cantley LC. Phosphoinositide 3-kinase is activated by phosphopeptides that bind to the SH2 domains of the 85-kDa subunit. J. Biol. Chem. 1993;268:9478–9483. [PubMed] [Google Scholar]
- 32.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, Willson JK, Markowitz S, Kinzler KW, Vogelstein B, Velculescu VE. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 33.Ikenoue T, Kanai F, Hikiba Y, Obata T, Tanaka Y, Imamura J, Ohta M, Jazag A, Guleng B, Tateishi K, Asaoka Y, Matsumura M, Kawabe T, Omata M. Functional analysis of PIK3CA gene mutations in human colorectal cancer. Cancer Res. 2005;65:4562–4567. doi: 10.1158/0008-5472.CAN-04-4114. [DOI] [PubMed] [Google Scholar]
- 34.Isakoff SJ, Engelman JA, Irie HY, Luo J, Brachmann SM, Pearline RV, Cantley LC, Brugge JS. Breast cancer-associated PIK3CA mutations are oncogenic in mammary epithelial cells. Cancer Res. 2005;65:10992–11000. doi: 10.1158/0008-5472.CAN-05-2612. [DOI] [PubMed] [Google Scholar]
- 35.Kang S, Bader AG, Vogt PK. Phosphatidylinositol 3-kinase mutations identified in human cancer are oncogenic. Proc. Natl. Acad. Sci. U.S.A. 2005;102:802–807. doi: 10.1073/pnas.0408864102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ombrello MJ, Remmers EF, Sun G, Freeman AF, Datta S, Torabi-Parizi P, Subramanian N, Bunney TD, Baxendale RW, Martins MS, Romberg N, Komarow H, Aksentijevich I, Kim HS, Ho J, Cruse G, Jung MY, Gilfillan AM, Metcalfe DD, Nelson C, O’Brien M, Wisch L, Stone K, Douek DC, Gandhi C, Wanderer AA, Lee H, Nelson SF, Shianna KV, Cirulli ET, Goldstein DB, Long EO, Moir S, Meffre E, Holland SM, Kastner DL, Katan M, Hoffman HM, Milner JD. Cold urticaria, immunodeficiency, and autoimmunity related to PLCG2 deletions. N. Engl. J. Med. 2012;366:330–338. doi: 10.1056/NEJMoa1102140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou Q, Lee GS, Brady J, Datta S, Katan M, Sheikh A, Martins MS, Bunney TD, Santich BH, Moir S, Kuhns DB, Long Priel DA, Ombrello A, Stone D, Ombrello MJ, Khan J, Milner JD, Kastner DL, Aksentijevich I. A hypermorphic missense mutation in PLCG2, encoding phospholipase Cγ2, causes a dominantly inherited autoinflammatory disease with immunodeficiency. Am. J. Hum. Genet. 2012;91:713–720. doi: 10.1016/j.ajhg.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu P, Constien R, Dear N, Katan M, Hanke P, Bunney TD, Kunder S, Quintanilla-Martinez L, Huffstadt U, Schroder A, Jones NP, Peters T, Fuchs H, de Angelis MH, Nehls M, Grosse J, Wabnitz P, Meyer TP, Yasuda K, Schiemann M, Schneider-Fresenius C, Jagla W, Russ A, Popp A, Josephs M, Marquardt A, Laufs J, Schmittwolf C, Wagner H, Pfeffer K, Mudde GC. Autoimmunity and inflammation due to a gain-of-function mutation in phospholipase Cγ2 that specifically increases external Ca2+ entry. Immunity. 2005;22:451–465. doi: 10.1016/j.immuni.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 39.Everett KL, Bunney TD, Yoon Y, Rodrigues-Lima F, Harris R, Driscoll PC, Abe K, Fuchs H, de Angelis MH, Yu P, Cho W, Katan M. Characterization of phospholipase Cγ enzymes with gain-of-function mutations. J. Biol. Chem. 2009;284:23083–23093. doi: 10.1074/jbc.M109.019265. [DOI] [PMC free article] [PubMed] [Google Scholar]