Abstract
Wnt proteins have been implicated in regulating a variety of developmental processes in the central nervous system (CNS). Secreted Frizzled-related protein 3 (sFRP3) is a member of the sFRP family that can inhibit the Wnt signaling by binding directly to Wnts via their regions of homology to the Wnt-binding domain of Frizzleds. Recent studies suggested that sFRP3 plays an important role in cell proliferation and differentiation in various tissues. To understand the role of sFRP3 in neural development, we carried out detailed studies on the expression of sFRP3 in the developing nervous system. Our results revealed that sFRP3 is initially expressed in the ventricular zone of spinal cord and dorsal root ganglia (DRG), and later in the dorsal horn of spinal cord and subpopulation of DRG neurons. The spatiotemporally dynamic expression of sFRP3 strongly suggests that sFRP3 has potential functions in the sensory neuron genesis and sensory circuitry formation.
Keywords: secreted frizzled-related protein 3, spinal cord, dorsal root ganglia, sensory circuit
INTRODUCTION
Wnts are a family of secreted proteins involved in many aspects of central nervous system (CNS) development, including neural induction, patterning, cell fate specification, cell proliferation, migration, axon guidance and synaptogenesis (Cadigan and Nusse, 1997; Stern, 2001; Ciani and Salinas, 2005; Bovolenta et al., 2006). Wnts transduce their signals through the Frizzled family of Wnt receptors by at least three different intracellular signaling pathways, known as the canonical Wnt/β-catenin pathway, the Wnt/Ca2+ pathway and the planar cell polarity pathway (Bhanot et al., 1996; Cadigan and Liu, 2006; Gordon and Nusse, 2006).
Secreted Wnt antagonists, classified as secreted Frizzled-related protein (sFRP) family, are potential negative modulators of Wnt signaling (Kawano et al., 2003). It has been shown that sFRP proteins can inhibit Wnt signaling by binding directly to Wnts via their region of homology to the Wnt-binding domain of Frizzleds (Rattner et al., 1997; Dennis et al., 1999; Ladher et al., 2000). sFRP3, also known as frizzled-related protein 1, or Frzb1 or FrzB, has been independently identified from the bovine cartilage extracts and the Spemann's organizer cells from Xenopus laevis (Hoang et al., 1996; Leyns et al., 1997; Wang et al., 1997). The sFRP3 protein shares the N-terminal cysteine-rich domain (CRD) with other members in the Frizzled family of Wnt receptors, but lacks the putative seven-transmembrane domain (Bhanot et al., 1996; Wang et al., 1996; Yang-Snyder et al.,1996).
The inhibitory activity of sFRP3 appears to be specific, since sFRP3 blocks WNT-1 and WNT-8 signaling in both X laevis embryos and a mammalian cell line (Lin et al., 1997; Wang et al., 1997; Leyns et al., 1997), but not Wnt-3A or -5A signaling (Wang et al., 1997). Murine sFRP3 gene is expressed in the primitive streak, presomitic mesoderm, somites, and brain at early embryonic stages (Hoang et al., 1998). At later stages, it exhibits sharp boundaries of expression in the limb bud, branchial arches, facial mesenchyme, and in cartilaginous elements of the appendicular skeleton (Hu et al., 1998; Hoang et al., 1998). sFRP3 has been implicated in osteoblast proliferation, intimal vascular disease, fibroblast proliferation and foci formation (Chung et al., 2004; Schumann et al., 2000; Mao et al., 2000; Lories et al., 2007; Scardigli et al., 2008). Although Wnts signaling pathways are known to be important for the development of various tissues, the functions of sFRP3 in neural development are not well understood.
In the present study, we describe the spatiotemporal pattern of sFRP3 expression in the developing mouse spinal cord and dorsal root ganglia. The dynamic expression of sFRP3 in spinal cord and dorsal root ganglia suggests that sFRP3 has potential functions in sensory neuron genesis and sensory circuit formation.
EXPERIMENTAL PROCEDURES
Animal
C57BL/6N mice were obtained from The Jackson Laboratory. All experimental procedures conformed to National Institutes of Health guidelines and were approved by the Institutional Animal Care and Use Committee at the University of Louisville.
In situ hybridization
In situ hybridization (ISH) was performed according to Scheer et al. (2001). Animals were deeply anesthetized and perfused with 4% paraformaldehyde (PFA), and tissues were isolated and postfixed in 4% PFA at 4°C overnight. Fixed spinal cord tissues were embedded in OCT medium and sectioned on a cryostat. Frozen sections (16 μm thick) were subject to in situ hybridization with digoxigenin-labeled riboprobes according to Schaeren-Wiemers and Gerfin-Moser (1993) with minor modifications. The information of all riboprobes used for ISH is listed in Table 1.
Table 1.
The information of riboprobes used for ISH.
| Gene | Primer (5'–3') | Length of probe (bp) |
|---|---|---|
| Sfrp1 | CTTTAAGTGATACTGGGGCGGACT | 764 bp |
| AAGCAGACCCTTGCCTGGCATCC | ||
| Sfrp2 | TCGAGTACCAGAACATGCGGCTGC | 1097 bp |
| CAGACTGACCAAAGAGAGAAAATGCC | ||
| Sfrp3 | CGCCGTTGTGGAAGTGAAGG | 1137 bp |
| GAGCATCCAACAATGGGTTT | ||
| Sfrp4 | TCTATGACCGTGGAGTTTGTATCT | 1109 bp |
| AGTATCATTTCCCCTGCCTTTATC | ||
| Sfrp5 | CCACCTACTTGGACGACCCTAGTA | 589 bp |
| GGTCGTTGTCCAGGGGGAACTTGT | ||
| Wnt1 | CTTCGAGAAATCGCCCAACTTCTGC | 827 bp |
| AGGCATGGTTCACAGCTGTTCAATGG | ||
| Wnt3a | GTTCCTACTTGGAGGGGTCTCTTAC | 913 bp |
| CTATCATACGAGGCTGTCATGCC | ||
| Wnt4 | GAGAAGTTTGACGGTGCCACGGAG | 1029 bp |
| TCACGGTTGCTTGCCTGGAATCTA | ||
| Wnt5a | AGTCCCACTCCCAGGACCCACT | 857 bp |
| \GGCTAACACAAGATTATGG | ||
| Wnt7a | GGACAGTCTCCAGTGCCTAGC | 810 bp |
| AGTGTGGTCCAGCACGTCTTAGTG | ||
| Wnt7b | ACGCAATGGTGGTCTGGTACC | 978 bp |
| TGAGGAAATGGGACATTAAGCTTC | ||
| Wnt8b | CATCTCTCAATGTTTGAGTCGCT | 825 bp |
| GTCTTGTTCTCCAGGCAGTAGTCTG | ||
| Fzd3 | CAGATTCCGTTACCCTGAAAGAC | 894 bp |
| GAGCACCTGCCGGCTCTCATTCAC | ||
| Fzd7 | CGCTTCTACCACAGACTCAGCC | 745 bp |
| ACCAACTTCACGCTAGGACTCT | ||
| Fzd8 | CTCTACAACCGCGTCAAGACC | 776 bp |
| ACGAAGCCAGCTAACAGAAACATAGTC | ||
| Fzd10 | CAAGGACATCGGCTACAACACC | 790 bp |
| CTCCAGTCCTTCCTGGATAACATAC | ||
| Dbx1 | GATTCTGATGAGGATGAGGAGGG | 554 bp |
| GCTTGCTTGATAGTGCTTTCC | ||
| Dbx2 | CACAGCTTTTTCAAGAGAAGAACACGG | 529 bp |
| TCAAATGGTGCTCTGGAGTCCATG | ||
| Mash1 | CTAACAGGCAGGGCTGGAAGCGCG | 737 bp |
| AAGGGGTGGGTGTGAGGGGGAAGGC | ||
| Drg11 | CCGTCGGCGATGTTTTATTTCCACT | 1311 bp |
| CCAAGAAGTTCAGTAAGCCGTAAGT | ||
| Grpr | TCTACCTGTACCGTTCCTACCACT | 1219 bp |
| CAGTTAACCCTAAGCAAATACAC |
For double in situ hybridization, tissue sections were first subjected to sFRP3 ISH, followed by anti-Pax3, anti-Pax6, anti-Olig2, anti-Nkx6.1, anti-TrkA or IB4 immunohistochemical staining with ABC kit individually, as described previously (Zhao et al., 2007). Rabbit anti-Olig2 (a gift from Dr. Charles Stiles) was used at a final concentration of 2 μg/ml (Lu et al., 2001); Mouse anti-Pax3, Mouse anti-Pax6 and Mouse anti-Nkx6.1 (DSHB Inc.) at a final concentration of 20 μg/ml; Rabbit anti-TrkA (Epitomics) and Rabbit IB4-Biotin (Sigma) at a final concentration of 2 μg/ml. For double in situ hybridization experiments, tissues were hybridized simultaneously with digoxigenin-labeled GrprDrg11, sFRP1, sFRP2, Wnt7b, FZD7, FZD8 or FZD10 and fluorescin-labeled sFRP3 riboprobes.
It is noted that DRG tissues were not included in some sections of early embryos due to slight positional differences in tissues. Therefore, expression of some genes in DRGs may be missed in some sections.
RESULTS
Selective expression of sFRP3 in the ventricular zone of spinal cord at early embryonic stages
To examine sFRP3 expression in the developing spinal cord, we performed RNA in situ hybridization in wild-type mouse spinal cord tissues at the early stages of neurogenesis. After the time of neural tube closure (E9.5), sFRP3 was initially expressed in the ventricular zone (VZ) of the middle region of spinal cord (Fig. 1B). At E11.5, the expression of sFRP3 was strongly detected in the ventricular neural progenitor cells of the middle spinal cord, but weak expression was also observed in more dorsal VZ of the spinal cord including the roof plate (Fig. 1C). At the same stage, two related members, sFRP1 and sFRP2 were also expressed in the VZ of spinal cord (Fig. 2A B) and overlapped with sFRP3 (Fig. 2I J). In contrast, sFRP4 and sFRP5 did not have detectable expression in the developing spinal cord (Fig. 2C–D G–H). As development proceeded, sFRP3 signal remained strong in the middle VZ, but its expression in the dorsal VZ was progressively reduced (Fig. 1D–E) and eventually disappeared after E14.5 (Fig. 4A B).
Fig. 1.
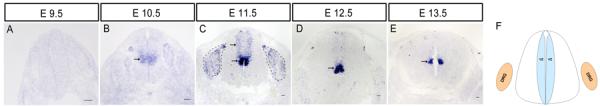
Dynamic expression of sFRP3 in spinal cord and dorsal root ganglia at early embryo stages. A–E: Spinal cord sections from E9.5, E10.5, E11.5, E12.5, E13.5 were subjected to ISH with sFRP3 riboprobe. F: Scheme of VZ and DRG in spinal cord. sFRP3 expression was primarily detected in middle region of the spinal cord at E10.5. At E11.5, sFRP3 expression in ventricular zone (VZ) was expanded to the entire dorsal half of the spinal cord and observed in most of dorsal root ganglia (DRG). By E12.5, sFRP3 expression was down-regulated in dorsal VZ and DRG, but persisted in the middle VZ. Expression of sFRP3 in VZ is indicated by arrows, and its expression in DRG is outlined by black dashed lines. Scale bar = 50 μm.
Fig. 2.
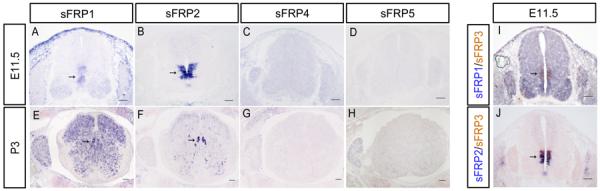
Expression of other sFRPs in spinal cord and dorsal root ganglia. A–D: E11.5 spinal cord sections were subjected to ISH with sFRP1, sFRP2, sFRP4 and sFRP5 riboprobes. E–H: P3 spinal cord sections were subjected to ISH with sFRP1, sFRP2, sFRP4 and sFRP5 riboprobes. I–J: Spinal cord sections from E11.5 embryos were subjected to ISH double labeling with digoxigenin-labeled sFRP1 or sFRP2 riboprobe (in blue) and fluorescein-labeled sFRP3 riboprobe (in red). Expression of sFRPs and co-localization of sFRP1/2 with sFRP3 in the ventricular cells are indicated by arrows. DRG is outlined by black dashed lines. Scale bar = 100 μm.
Fig. 4.
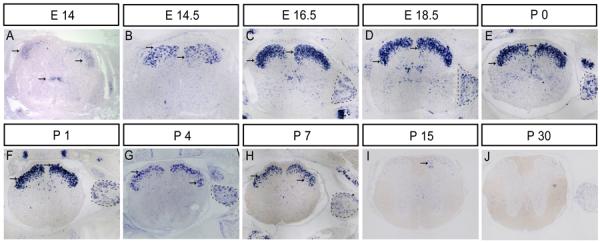
sFRP3 expression in spinal cords and dorsal root ganglia at later developmental stages. A–J: Spinal cord sections from E14, E14.5, E16.5, E18.5, P0, P1, P4, P7, P15 and P30 were subjected to ISH with sFRP3 riboprobe. sFRP3 expression was detected in spinal cord dorsal horn after E14 (indicated by arrows), and persisted till P30. sFRP3 expression in the dorsal root ganglia (DRG) was up-regulated again at E16.5 (C). Arrows denote areas of sFRP3 localization in spinal cord. DRG is outlined by black dashed lines. Scale bar = 100 μm.
sFRP3 is expressed in the dp6 – p1 domains of neural progenitors at neurogenesis stage
During early neural development, the ventricular zone of the spinal cord can be divided into six distinct dorsal progenitor domains and five ventral domains. To determine the precise domains of sFRP3 expression, we chose the E12.5 spinal cord to compare its expression with that of several neuroprogenitor genes (Pax3, Pax6, Mash1, Dbx1, Dbx2, Nkx6.1 and Olig2) that define discrete progenitor domains along the dorsoventral axis of the neural tube (Goulding et al. 1991; Ericson et al. 1997; Pierani et al. 1999). Comparative expression analysis on consecutive sections revealed that the domain of sFRP3 expression is nearly identical to that of Dbx2, but broader than that of Dbx1 (Fig. 3A B), suggesting that sFRP3 is expressed in dp6, p0 and p1 domains of neural tube. Consistently, the dorsal boundary of sFRP3 expression is positioned immediately ventral to the expression of Mash1 (Fig. 3C), which has been shown to be expressed in dp3–p5 domains of dorsal neural tube. In further support, the ventral boundary of sFRP3 expression is immediately dorsal to the Nkx6.1 expression domain, but separated from the Olig2 expression domain (Fig. 3D E). In addition, Pax3 expression extends ventrally into the sFRP3 expression domain (Fig. 3G), and Pax6 expression encloses the domain of sFRP3 expression (Fig. 3F). Together, these results demonstrate that sFRP3 is expressed weakly in dorsal domains dp1–dp5 and strongly in dp6, p0 and p1 domains of VZ; however, its dorsal expression is down-regulated after E12.5.
Fig. 3.
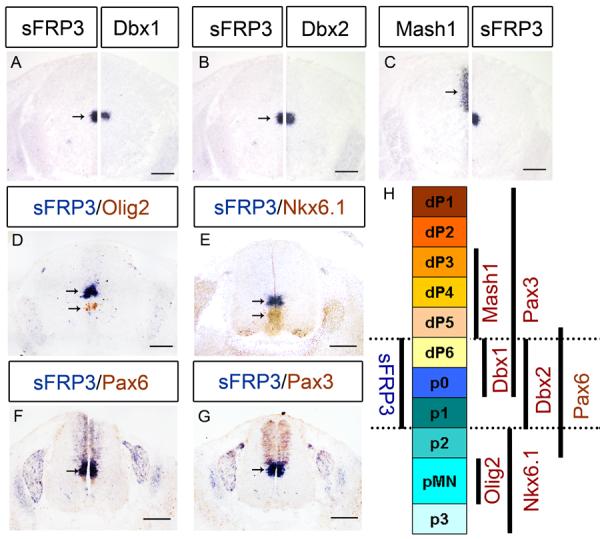
sFRP3 was expressed dp6 to p1 domains of ventricular zone at E12.5. A–C: Immediately adjacent sections from E12.5 spinal cord were subjected to in situ hybridization with sFRP3, Dbx1, Dbx2 and Mash1 riboprobes, and half sections were aligned at the midline. D–G: Spinal cords from E12.5 embryos were double-labeled with sFRP3 (blue) and anti-Olig2 (brown in D), anti-Nkx6.1 (brown in E), anti-Pax6 (brown in F) or anti-Pax3 (brown in G). H: Scheme of sFRP3 expression in progenitor domains labeled by various progenitor identity genes. Arrows indicate the ISH positive cells. Scale bar = 100 μm.
sFRP3 is expressed in lamina I to II of spinal cord dorsal horn at later embryonic stages
We next examined sFRP3 expression in the developing spinal cord at later embryonic stages. At E14, sFRP3 expression started to be detected in dorsal horn of spinal cord (Fig. 4A), while its expression in the middle VZ rapidly diminished and completely vanished at E14.5 (Fig. 4B). At later stages, sFRP3 expression continued to accumulate in the dorsal horn of spinal cord, and this expression pattern was maintained until P7 (Fig. 4C–H), prior to its rapid down-regulation at P15 and P30 (Fig. 4I–J), sFRP3 was specifically expressed in the lamina of spinal cord dorsal horn as compared to the other members the sFRP family (sFRP1, sFRP2, sFRP4 and sFRP5) (Fig. 2E–H).
The persistent expression of sFRP3 in the dorsal horn of spinal cord has raised the possibility that sFRP3 is expressed by subpopulations of sensory neurons. To examine this possibility, we performed the double ISH in E16.5 spinal cords with sFRP3, Grpr and Drg11 riboprobes. Grpr is expressed in primary sensory neurons and in their projection area in the lamina I of superficial dorsal horn (Saito et al., 1995; Sun and Chen, 2007). Drg11 is a transcription factor that is necessary for the assembly of the nociceptive circuitry in the spinal cord dorsal horn and is expressed in the lamina I–III of spinal dorsal horn. As shown in Fig. 5, double labeling experiments revealed that sFRP3+ cells occupied lamina I and II of superficial dorsal horn of the spinal cord (Fig. 5). It is also evident that a subpopulation of sFRP3+ cells in superficial dorsal horn co-expressed Grpr and Drg11 (Fig. 5).
Fig. 5.
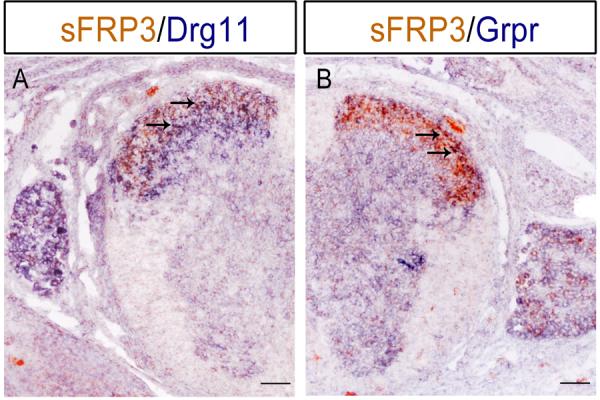
Co-expression of sFRP3 with sensory neuronal markers in the dorsal spinal cord. Spinal cord sections from E16.5 embryos were subjected to ISH double labeling with digoxigenin-labeled Drg11 or Grpr riboprobe (in blue) and fluorescein-labeled sFRP3 riboprobe. Representative double positive cells are indicated by arrows. Scale bar = 100 μm.
Dynamic expression of sFRP3 in dorsal root ganglia
In the primary sensory neurons of DRG, the expression of sFRP3 was first detected at E10.5 (Fig. 1B). At E11.5, the sFRP3 signals were mostly distributed to the periphery of DRG neurons (Fig. 1C). Expression of other members of the sFRP family was not detected at this stage (Fig 2). As development proceeded, similar to that in the dorsal VZ of spinal cord, the number of sFRP3+ cells in DRG was gradually decreased, and no sFRP3+ cells were detected at E13.5 (Fig. 1D–E). Notably, the expression of sFRP3 was up-regulated again at E16.5 (Fig. 4C) in a subpopulation of DRG neurons, including those expressing TrkA, a receptor for nerve growth factor (Fig. 6A), which labels peptidergic neurons in adult DRG. The expression sFRP3 in TrkA + neurons was maintained at postnatal stages (Fig. 6B). At P8, many sFRP3+ DRG neurons co-expressed IB4 (Fig. 6C), a molecular marker for non-peptidergic neurons. Thus, in postnatal DRG, sFRP3 is expressed in both peptidergic and non-peptidergic subpopulations of neurons.
Fig. 6.
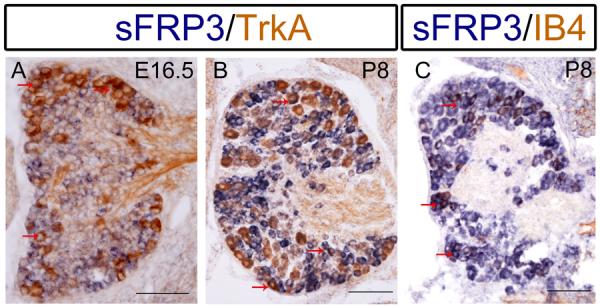
Co-expression of sFRP3 with TrkA and IB4 in DRG neurons. DRG sections from E16.5 and P8 mice were subjected to sFRP3 in situ hybridization (blue) followed by anti-TrkA (brown in A, B) or anti-IB4 (brown in C) immunostaining. Representative double positive cells are indicated by arrows. Scale bar = 100 μm.
Expression of Wnt molecules and receptors in relation to sFPR3 expression in the spinal cord
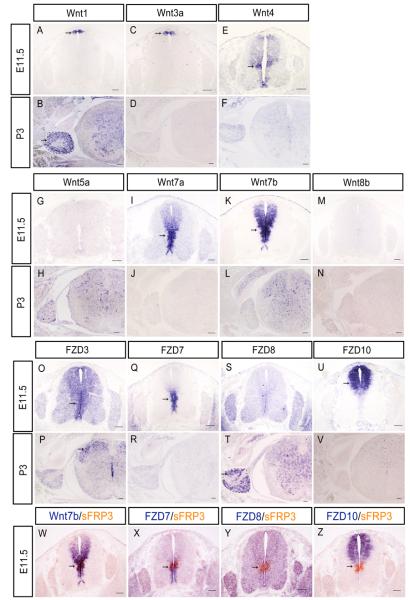
Given that sFRP3 functions as an antagonist of Wnt signaling, it would be important to determine the potential downstream target Wnt pathway of sFRP3. Therefore we next examined the expression of various Wnt molecules and receptors in the developing spinal cord. At E11.5, of all Wnt molecules examined, Wnt4, Wnt7a and Wnt7b displayed expression in the VZ of spinal cord (Fig7), including the sFRP3 expression domain in the middle VZ (Fig. 7I K W). This result suggested that sFPR3 may function to antagonize the Wnt4- and Wnt7-mediated signaling pathway. Consistently, among all FZD molecules examined (Fig 7 O–U), only Wnt7a receptor FZD7 is highly expressed in the VZ of the middle region (Fig. 7Q X). FZD3, the receptor for Wnt3a and Wnt4 had a weak expression in the VZ along the entire D-V axis (Fig 7O). FZD10, the receptor of Wnt7b, is highly expressed in the dorsal VZ but did not have overlapping with sFRP3 expression domain (Fig. 7U Z). At P3, of all Wnt molecules and receptors examined, only FZD3 showed weak and specific expression in the dorsal horn interneurons overlapping the expression of sFRP3 (Fig 7P). Examination of their expression in DRG at this stage revealed the specific expression of Wnt1, Wnt4 and FDZ8 in the peripheral DRG neurons (Fig 7B, E, T) where sFRP3 is expressed.
Fig. 7.
RNA in situ hybridization analysis of Wnts and their receptor gene expression in mouse embryonic (E11.5) and postanal (P3) spinal cord and DRG. A–N: E11.5 and P3 spinal cord sections were subjected to ISH with Wnt1, Wnt3a, Wint4, Wnt5a, Wnt7a, Wnt7b and Wnt8b riboprobes. O–V: E11.5 and P3 spinal cord sections were subjected to ISH with FZD3, FZD7, FZD8 and FZD10 riboprobes. W–Z: Spinal cord sections from E11.5 embryos were subjected to ISH double labeling with digoxigenin-labeled Wnt7b, FZD7, FZD8 or FZD10 riboprobe (in blue) and fluorescein-labeled sFRP3 riboprobe (in red). The ISH positive cells are indicated by arrows, DRG is outlined by black dashed lines. Scale bar = 100 μm.
DISCUSSION
Previous study has demonstrated that sFRP3 has restricted expression in the developing forebrain and midbrain, and suggested that it plays an important role in the early development of these CNS regions (Hoang et al., 1998). However, its expression and function in the spinal cord or DRG have not been determined. Here, we report the detailed spatiotemporal pattern of sFRP3 expression in the spinal cord and DRG, and suggest that sFRP3 may be involved in sensory neuron genesis and the formation of sensory circuitry.
Differential expression in VZ implies distinct functions of sFRP3
In mouse spinal cord, six distinct dorsal interneuron (dI) populations, termed dI1–dI6 are specified by E10, on the basis of the repertoire of transcription factors that they express. The dI1–dI3 neurons migrate ventrally, whereas the dI4 and a subset of the dI5 neurons migrate laterally to populate the deep dorsal horn. Beginning at E12, three additional populations of neurons are born later (Mattise 2002). These late-born populations are derived from dp4 and dp5 neural progenitor cells, migrate to the superficial laminae of the dorsal horn, where they mediate the sensing of pain and temperature (Caspary and Anderson, 2003). The transient and weak expression of sFRP3 in dp1–dp5 domains of VZ from E11.5 to E12.5 (Fig. 1) suggests that sFRP3 as an antagonist of the Wnt signaling pathway may be involved in dorsal interneuron generation.
The strong and persistent expression of sFRP3 in the Dbx1/2 expressing dp6–p1 domains of VZ from E11.5 to E14 (Fig. 3H) raises the possibility that sFRP3 participates in V0 and V1 interneuron fate specification or consolidation (Fig 1, 3). Two related members, sFRP1 and 2, are similarly expressed in these domains (Fig 2I J), and the other two members sFRP4 and 5 are not expressed in spinal cord and DRG (Fig 2C D G H). Interestingly, at these stages, Wnt7a and its receptor FZD7 are highly expressed in the VZ of the middle and ventral spinal cord (Fig 7I Q), including the dp6–p1 domains of sFRP3 expression. Wnt7a is known to play a critical role in control of neuronal progenitor maturation (Grand et al., 2009; Horn Z et al., 2007). Wnt7b is strongly expressed in these regions as well, but its receptor FDZ10 has restricted expression in the dorsal VZ (Fig. 7). Based on the strong and overlapping expression of sFRP1–3 and Wnt7a/Wnt7b/FZD7 in dp6–p1, it is possible that sFRP1–3 may have an important role in the fate specification of V0 and V1 interneurons by modulating the Wnt7 signaling in neural progenitor cells during early spinal cord development. The involvement of sFRPs and Wnt7/FDZ7 genes in V0 and V1 interneuron development can be tested with future genetic and experimental studies. However, since sFRP1–3 proteins are secreted molecules and their range of biological activity might well be broader, it is plausible that non-overlapping, neighboring expression domains of Wnts/Wnt-receptors may be influenced by sFRP1–3 proteins.
sFRP3 is required for spinal cutaneous sensation circuitry
sFRP3+ cells occupied lamina I and II (Fig 4) where pain and temperature are conveyed through thinner, unmyelinated axon bundles that project to this area (Altman et al., 1984; Caspary and Anderson, 2003). Double staining experiments revealed that sFRP3 is co-expressed with Grpr and Drg11 in this area (Fig 5, 6). Drg11 is required for the projection of cutaneous sensory afferent fibers to the dorsal horn (Chen et al., 2001), and Grpr is essential for mediating the itch sensation at the spinal cord level (Sun et al., 2007). The persistent expression of sFRP3 in the laminae I and II from E14 to postnatal stage P7 suggests that sFRP3-mediated inhibition of Wnt signaling may participate in the circuitry formation of cutaneous sensory neurons or itch neurons in the dorsal horn of spinal cord. In support of this concept, a recent study showed that Wnt signaling is indeed involved in the formation of spinal cutaneous sensation circuitry, as sFRP3 mutants displayed a reduced sensory innervation in the superficial dorsal horn from TrkA+ or aquaporin1+ DRG neurons (John et al., 2012)
It is noticed that in perinatal DRG neurons, there is a significant overlapping between the expression of sFRP3 and TrkA (Fig. 6A). Since TrkA expression in DRG is not affected in sFRP3 mutants (John et al., 2012), it is tempting to speculate at this stage that the expression of sFRP3 in sensory neurons of dorsal horn and DRG may serve as a molecular identity for specific connections of these TrkA+ DRG neurons. It should be noted, however, that most of the sFRP3+ neurons in P8 DRG are excluded from TrkA expression (Fig. 6B), and instead labeled by IB4 (Fig. 6C), a molecular marker for non-peptidergic neurons as compared to those TrkA-expressing peptidergic neurons in adult DRG. This could presumably suggest a distinct role for sFRP3 and Wnt signaling in the DRG neuron development during postnatal and adult stage. Consistently, Wnt1, Wnt4 and FZD8 signals are found in postnatal DRG neurons where sFRP3 is expressed (Fig. 7B F T). Future studies with conditional knockouts and transgenic approaches are necessary to elucidate the functions of sFRP3 and related Wnt/FZD signaling pathways in sensory neuron development and circuitry formation.
Highlights
We studied the expression pattern of sFRP3 in the spinal cord and DRG in detail.
sFRP3 is transiently expressed in dp1-dp5 domains of VZ at early embryonic stages.
sFRP3 is strongly expressed in the laminae I/II of the dorsal horn of spinal cord.
sFRP3 may participate in the circuitry formation of sensory neurons.
There is a distinct role of sFRP3 in the postnatal development of DRG neurons.
ACKNOWLEDGMENT
We thank Dr. Charles Stiles for generously providing the anti-Olig2 antibody.
Grant sponsors: The National Key Basic Research Program of China (2013CB531300); National Natural Sciences Foundation of China (Grant No: 31000488; 31071879), Zhejiang Provincial Natural Science Foundation of China (Z2100730) and Major Project of Science and Technology Department of Zhejiang Province (2011C13030). NIH (R01NS37717).
Abbreviations
- CRD
cysteine-rich domain
- CNS
central nervous system
- dI
dorsal interneuron
- DRG
dorsal root ganglia
- ISH
In situ hybridization
- PFA
paraformaldehyde
- FZD
Frizzled family of Wnt receptor
- sFRP
Secreted Frizzled-related protein
- VZ
ventricular zone
- dp1–6
dorsal progenitor domains 1–6
- P0–30
postnatal day 0–30
- E9.5–18.5
Embryo day 9.5–18.5.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Altman J, Bayer SA. The development of the rat spinal cord. Adv Anat Embryol Cell Biol. 1984;85:1–164. doi: 10.1007/978-3-642-69537-7. [DOI] [PubMed] [Google Scholar]
- Bhanot P, Brink M, Samos CH, Hsieh JC, Wang Y, Macke JP, Andrew D, Nathans J, Nusse R. A new member of the frizzled family from Drosophila functions as a Wingless receptor. Nature. 1996;382(6588):225–230. doi: 10.1038/382225a0. [DOI] [PubMed] [Google Scholar]
- Bovolenta P, Rodriguez J, Esteve P. Frizzled/RYK mediated signalling in axon guidance. Development. 2006;133:4399–4408. doi: 10.1242/dev.02592. [DOI] [PubMed] [Google Scholar]
- Briscoe J, Pierani A, Jessell TM, Ericson J. A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell. 2000;101:435–445. doi: 10.1016/s0092-8674(00)80853-3. [DOI] [PubMed] [Google Scholar]
- Burstyn-Cohen T, Stanleigh J, Sela-Donenfeld D, Kalcheim C. Canonical Wnt activity regulates trunk neural crest delamination linking BMP/noggin signaling with G1/S transition. Development. 2004;131:5327–5339. doi: 10.1242/dev.01424. [DOI] [PubMed] [Google Scholar]
- Cadigan KM, Liu YI. Wnt signaling: complexity at the surface. J Cell Sci. 2006;119:395–402. doi: 10.1242/jcs.02826. [DOI] [PubMed] [Google Scholar]
- Cadigan KM, Nusse R. Wnt signaling: a common theme in animal development. Genes Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- Caspary T, Anderson KV. Patterning cell types in the dorsal spinal cord: what the mouse mutants say. Nat Rev Neurosci. 2003;4:289–297. doi: 10.1038/nrn1073. [DOI] [PubMed] [Google Scholar]
- Chapman SC, Brown R, Lees L, Schoenwolf GC, Lumsden A. Expression analysis of chick Wnt and frizzled genes and selected inhibitors in early chick patterning. Dev Dyn. 2004;229:668–676. doi: 10.1002/dvdy.10491. [DOI] [PubMed] [Google Scholar]
- Chen ZF, Rebelo S, White F, Malmberg AB, Baba H, Lima D, Woolf CJ, Basbaum AI, Anderson DJ. The paired homeodomain protein DRG11 is required for the projection of cutaneous sensory afferent fibers to the dorsal spinal cord. Neuron. 2001;31(1):4–6. doi: 10.1016/s0896-6273(01)00341-5. [DOI] [PubMed] [Google Scholar]
- Ciani L, Salinas PC. WNTs in the vertebrate nervous system: from patterning to neuronal connectivity. Nat Rev Neurosci. 2005;6:351–362. doi: 10.1038/nrn1665. [DOI] [PubMed] [Google Scholar]
- Dennis S, Aikawa M, Szeto W, d'Amore PA, Papkoff J. A secreted frizzled related protein, FrzA, selectively associates with Wnt-1 protein and regulates wnt-1 signaling. J Cell Sci. 1999;112:3815–3820. doi: 10.1242/jcs.112.21.3815. [DOI] [PubMed] [Google Scholar]
- Ericson J, Morton S, Kawakami A, Roelink H, Jessell TM. Two critical periods of Sonic Hedgehog signaling required for the specification of motor neuron identity. Cell. 1996;87:661–673. doi: 10.1016/s0092-8674(00)81386-0. [DOI] [PubMed] [Google Scholar]
- Ericson J, Rashbass P, Schedl A, Brenner-Morton S, Kawakami A, van Heyningen V, Jessell TM, Briscoe J. Pax6 controls progenitor cell identity and neuronal fate in response to graded Shh signaling. Cell. 1997;90:169–180. doi: 10.1016/s0092-8674(00)80323-2. [DOI] [PubMed] [Google Scholar]
- Esteve P, Morcillo J, Bovolenta P. Early and dynamic expression of cSfrp1 during chick embryo development. Mech Dev. 2000;97:217–221. doi: 10.1016/s0925-4773(00)00421-4. [DOI] [PubMed] [Google Scholar]
- Gordon MD, Nusse R. Wnt signaling: multiple pathways, multiple receptors, and multiple transcription factors. J Biol Chem. 2006;281:22429–22433. doi: 10.1074/jbc.R600015200. [DOI] [PubMed] [Google Scholar]
- Goulding MD, Chalepakis G, Deutsch U, Erselius JR, Gruss P. Pax-3, a novel murine DNA binding protein expressed during early neurogenesis. EMBO J. 1991;10:1135–1147. doi: 10.1002/j.1460-2075.1991.tb08054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang B, Moos M, Jr, Vukicevic S, Luyten FP. Primary structure and tissue distribution of FRZB/SFRP3, a novel protein related to Drosophila frizzled, suggest a role in skeletal morphogenesis. J Biol Chem. 1996;271:26131–26137. doi: 10.1074/jbc.271.42.26131. [DOI] [PubMed] [Google Scholar]
- Hoang BH, Thomas JT, Abdul-Karim FW, Correia KM, Conlon RA, Luyten FP, Ballock RT. Expression pattern of two Frizzled-related genes, Frzb-1 and Sfrp-1, during mouse embryogenesis suggests a role for modulating action of Wnt family members. Dev Dyn. 1998;212:364–372. doi: 10.1002/(SICI)1097-0177(199807)212:3<364::AID-AJA4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Horn Z, Papachristou P, Shariatmadari M, Peyronnet J, Eriksson B, Ringstedt T. Wnt7a overexpression delays beta-tubulin III expression in transgenic mouse embryos. Brain Res. 2007;1130(1):67–72. doi: 10.1016/j.brainres.2006.10.090. [DOI] [PubMed] [Google Scholar]
- Ikeya M, Lee SM, Johnson JE, McMahon AP, Takada S. Wnt signalling required for expansion of neural crest and CNS progenitors. Nature. 1997;389:966–970. doi: 10.1038/40146. [DOI] [PubMed] [Google Scholar]
- Jessell TM. Neuronal specification in the spinal cord; inductive signals and transcriptional codes. Nature Rev Genet. 2001;1:20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- John A, Brylka H, Wiegreffe C, Simon R, Liu P, Jüttner R, Crenshaw EB, 3rd, Luyten FP, Jenkins NA, Copeland NG, Birchmeier C, Britsch S. Bcl11a is required for neuronal morphogenesis and sensory circuit formation in dorsal spinal cord development. Development. 2012;139:1831–1841. doi: 10.1242/dev.072850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano Y, Kypta R. Secreted antagonists of the Wnt signaling pathway. J Cell Sci. 2003;116:2627–2634. doi: 10.1242/jcs.00623. [DOI] [PubMed] [Google Scholar]
- Ladher RK, Church VL, Allen S, Robson L, Abdelfattah A, Brown NA, Hattersley G, Rosen V, Luyten FP, Dale L, Francis-West PH. Cloning and expression of the Wnt antagonists Sfrp-2 and Frzb during chick development. Dev Biol. 2000;218:183–198. doi: 10.1006/dbio.1999.9586. [DOI] [PubMed] [Google Scholar]
- Le Grand F, Jones AE, Seale V, Scimè A, Rudnicki MA. Wnt7a activates the planar cell polarity pathway to drive the symmetric expansion of satellite stem cells. Cell Stem Cell. 2009;4:535. doi: 10.1016/j.stem.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leimeister C, Bach A, Gessler M. Developmental expression patterns of mouse sFRP genes encoding members of the secreted frizzled related protein family. Mech Dev. 1998;75:29–42. doi: 10.1016/s0925-4773(98)00072-0. [DOI] [PubMed] [Google Scholar]
- Leyns L, Bouwmeester T, Kim SH, Piccolo S, De Robertis EM. Frzb-1 is a secreted antagonist of Wnt signaling expressed in the Spemann organizer. Cell. 1997;88:747–756. doi: 10.1016/s0092-8674(00)81921-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K, Wang S, Julius MA, Kitajewski J, Moos M, Jr, Luyten FP. The cysteine-rich frizzled domain of Frzb-1 is required and sufficient for modulation of Wnt signaling. Proc Natl Acad Sci USA. 1997;94:11196–11200. doi: 10.1073/pnas.94.21.11196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu QR, Cai L, Rowitch D, Cepko CL, Stiles CD. Ectopic expression of Olig1 promote oligodendrocyte formation an reduces neuronal survival in developing mouse cortex. Nat Neurosci. 2001;4:973–974. doi: 10.1038/nn718. [DOI] [PubMed] [Google Scholar]
- Lyuksyutova AI, Lu CC, Milanesio N, King LA, Guo N, Wang Y, Nathans J, Tessier-Lavigne M, Zou Y. Anterior-posterior guidance of commissural axons by Wnt-frizzled signaling. Science. 2003;302:1984–1988. doi: 10.1126/science.1089610. [DOI] [PubMed] [Google Scholar]
- Mattise M. A Dorsal Elaboration in the Spinal Cord. Neuron. 2002;34:491–497. doi: 10.1016/s0896-6273(02)00710-9. [DOI] [PubMed] [Google Scholar]
- Pierani A, Brenner-Morton S, Chiang C, Jessell TM. A sonic hedgehog-independent, retinoid-activated pathway of neurogenesis in the ventral spinal cord. Cell. 1999;97:903–915. doi: 10.1016/s0092-8674(00)80802-8. [DOI] [PubMed] [Google Scholar]
- Rattner A, Hsieh JC, Smallwood PM, Gilbert DJ, Copeland NG, Jenkins NA, Nathans J. A family of secreted proteins contains homology to the cysteine-rich ligand-binding domain of frizzled receptors. Proc Natl Acad Sci USA. 1997;94:2859–2863. doi: 10.1073/pnas.94.7.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Greenwood A, Sun Q, Anderson DJ. Identification by differential RT-PCR of a novel paired homeodomain protein specifically expressed in sensory neurons and a subset of their CNS targets. Mol Cell Neurosci. 1995;6:280–292. doi: 10.1006/mcne.1995.1022. [DOI] [PubMed] [Google Scholar]
- Sander M, Paydar S, Ericson J, Briscoe J, Berber E, German M, Jessell TM, Rubenstein JLR. Ventral neural patterning by Nkx homeobox genes Nkx6.1 controls somatic motor neuron and ventral interneuron fates. Genes Dev. 2000;14:2134–2139. doi: 10.1101/gad.820400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeren-Wiemers N, Gerfin-Moser A. A single protocol to detect transcripts of various types and expression levels in neural tissue and cultured cells: in situ hybridization using digoxigenin-labeled cRNA probes. Histochemistry. 1993;100:431–440. doi: 10.1007/BF00267823. [DOI] [PubMed] [Google Scholar]
- Scheer N, Groth A, Hans S, Campos-Ortega JA. An instructive function for Notch in promoting gliogenesis in the zebrafish retina. Development. 2001;128:1099–1107. doi: 10.1242/dev.128.7.1099. [DOI] [PubMed] [Google Scholar]
- Shimizu T, Kagawa T, Wada T, Muroyama Y, Takada S, Ikenaka K. Wnt signaling controls the timing of oligodendrocyte development in the spinal cord. Dev Biol. 2005;282:397–410. doi: 10.1016/j.ydbio.2005.03.020. [DOI] [PubMed] [Google Scholar]
- Stern CD. Initial patterning of the central nervous system: how many organizers? Nat Rev Neurosci. 2001;2:92–98. doi: 10.1038/35053563. [DOI] [PubMed] [Google Scholar]
- Sun YG, Chen ZF. A gastrin-releasing peptide receptor mediates the itch sensation in the spinal cord. Nature. 2007;448:700–703. doi: 10.1038/nature06029. [DOI] [PubMed] [Google Scholar]
- Terry K, Magan H, Baranski M, Burrus LW. Sfrp-1 and sfrp-2 are expressed in overlapping and distinct domains during chick development. Mech Dev. 2000;97:177–182. doi: 10.1016/s0925-4773(00)00407-x. [DOI] [PubMed] [Google Scholar]
- Vallstedt A, Muhr J, Pattyn A, Pierani A, Mendelsohn M, Sander M, Jessell TM, Ericson J. Different levels of repressor activity assign redundant and specific roles to Nkx6 genes in motor neuron and interneuron specification. Neuron. 2001;31(5):743–755. doi: 10.1016/s0896-6273(01)00412-3. [DOI] [PubMed] [Google Scholar]
- Wang S, Krinks M, Lin K, Luyten FP, Moos M., Jr Frzb, a secreted protein expressed in the Spemann organizer, binds and inhibits Wnt-8. Cell. 1997;88:757–766. doi: 10.1016/s0092-8674(00)81922-4. [DOI] [PubMed] [Google Scholar]
- Wang Y, Macke JP, Abella BS, Andreasson K, Worley P, Gilbert DJ, Copeland NG, Jenkins NA, Nathans J. A large family of putative transmembrane receptors homologous to the product of the Drosophila tissue polarity gene frizzled. J Biol Chem. 1996;271:4468–4476. doi: 10.1074/jbc.271.8.4468. [DOI] [PubMed] [Google Scholar]
- Wang YK, Samos CH, Peoples R, Pérez-Jurado LA, Nusse R, Francke U. A novel human homologue of the Drosophila frizzled wnt receptor gene binds wingless protein and is in the Williams syndrome deletion at 7q11.23. Hum Mol Genet. 1997;6:465–472. doi: 10.1093/hmg/6.3.465. [DOI] [PubMed] [Google Scholar]
- Wang Z, Shu W, Lu MM, Morrisey EE. Wnt7b activates canonical signaling in epithelial and vascular smooth muscle cells through interactions with Fzd1, Fzd10, and LRP5. Mol. Cell. Biol. 2005;25(12):5022–5030. doi: 10.1128/MCB.25.12.5022-5030.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang-Snyder J, Miller JR, Brown JD, Lai CJ, Moon RT. A frizzled homolog functions in a vertebrate Wnt signaling pathway. Curr Biol. 1996;6:1302–1306. doi: 10.1016/s0960-9822(02)70716-1. [DOI] [PubMed] [Google Scholar]
- Yuko Muroyama, Motoyuki Fujihara, Makoto Ikeya, Hisato Kondoh, Shinji Takada. Wnt signaling plays an essential role in neuronal specification of the dorsal spinal cord. Genes Dev. 2002;16(5):548–553. doi: 10.1101/gad.937102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Hu X, Park J, Zhu Y, Zhu Q, Li H, Luo C, Han R, Cooper N, Qiu M. Selective expression of LDLR and VLDLR in myelinating oligodendrocytes. Dev Dyn. 2007;236:2708–2712. doi: 10.1002/dvdy.21283. [DOI] [PubMed] [Google Scholar]