Abstract
Using the miniprotein Trp-cage as a model, we show that D-amino acids can be used to facilitate the delineation of protein folding mechanism. Specifically, we study the folding-unfolding kinetics of three Trp-cage mutants where the native glycine residue near the C-terminus of the α-helix is replaced by a D-amino acid. A previous study showed that these mutations increase the Trp-cage stability, due to a terminal capping effect. Our results show that the stabilizing effect of D-asparagine and D-glutamine originates almost exclusively from a decrease in the unfolding rate, while the D-alanine mutation results in a similar decrease in the unfolding rate, but it also increases the folding rate. Together, these results support a folding mechanism wherein the α-helix formation in the transition state is nucleated at the N-terminus, whereas those long-range native interactions stabilizing this helix are developed at the downhill side of the folding free energy barrier.
1. Introduction
Loops and turns in proteins not only ensure connectivity of the polypeptide chain, but they also, in many cases, play an important role in directing the process of folding. A survey of the available protein structures in the Protein Data Bank (PDB) indicated that the most frequently found amino acids in loops are glycine (Gly), serine (Ser), aspartate (Asp), asparagine (Asn), and proline (Pro).1 In particular, Gly is frequently found to be at the C-terminus of α-helices in proteins, serving as a secondary structure breaker and directing the main chain in a new direction. This is due to the fact that Gly can extensively populate backbone conformations that are normally forbidden for other L-amino acids, as seen in the Ramachandran plot. For the same reason, however, there is a relatively large conformational entropy penalty associated with restricting a Gly residue in a small region of phi and psi conformations. Therefore, Gly has recently become a popular target in protein design to improve the stability of proteins of interest via mutations that could alleviate this entropy penalty.2 For example, it has been shown that Pro, which has low backbone conformational entropy, can in some cases replace Gly and, as a result, further stabilize the native fold.3 A different, but perhaps more elegant and more effective approach, is to replace Gly with various D-amino acids,2,4–6 the enantiomeric relative of L-amino acids, which can occupy conformations on the Ramachandran plot that otherwise only glycine could. For example, D-alanine (D-Ala), which has no sidechain rotamer preferences and hence a smaller entropy penalty upon folding, has been used, in a number of cases, to increase the stability of proteins,4,6 to reconstitute an active and specific ion channel,5 and to create novel folds.7
In a more recent study,8 Rodriguez-Granillo et al. have shown that D-Ala, D-Asn, and D-Gln all show a stabilizing effect, although to a different extent, on the folding free energy of the mini-protein Trp-cage (Figure 1), when replacing the Gly residue at position 10 (Specifically, the TC5b variant, sequence: NLYIQWLKDGGPSSGRPPPS, was used). Interestingly, both D-Ala and D-Gln were found to significantly stabilize the native fold, raising the thermal melting temperature (Tm) of Trp-cage by about 20 °C, whereas D-Asn only increased the Tm by about 10 °C. This observation was interpreted as due to a combination of effects, including the difference in sidechain-backbone hydrogen-bonding interaction ability and conformational entropy of these D-amino acids. The hydrogen bonding ability of the D-Gln sidechain was found to be greater than that of D-Asn, while the sidechain conformational entropy penalties for both D-Gln and D-Asn are larger than for D-Ala. While this study demonstrated the effectiveness, especially when placed at the C-terminus of α-helices, of several D-amino acids to alleviate the intrinsic conformational limitations of naturally occurring L-amino acids, and offered a thermodynamic rationalization of the stabilizing effect of these D-amino acids on the Trp-cage fold, it was unable to yield a mechanistic and dynamic view of how this is achieved, i.e., through an increase in the folding rate, a decrease in the unfolding rate, or even a change in the folding mechanism. Herein, we use a laser-induced temperature-jump (T-jump) infrared (IR) technique to study the conformational dynamics of several previously characterized D-amino acid mutants of TC5b, including 5b-G10DA, 5b-G10DN, and 5b-G10DQ, aiming to elucidate the kinetic role of these D-amino acids. In addition, Culik et al. have recently put forward a folding mechanism for Trp-cage, based on extensive spectroscopic studies and amino acid mutations,9 in which the transition state of the main folding event, i.e., the formation of the cage structure, involves the consolidation of the α-helix, from amino acids 2–9. Since the abovementioned D-amino acids are expected to provide further stabilization to the α-helix,4,6 studying the folding kinetics of these -D-amino acid mutants will help improve our understanding of the folding mechanism of this extremely popular protein folding model.10–48
Fig. 1.
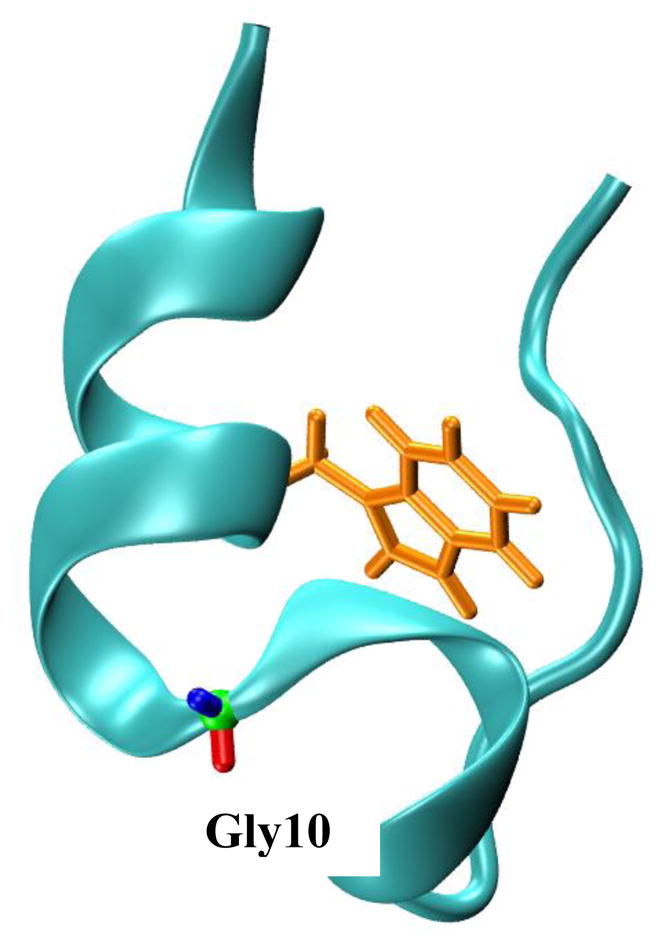
The NMR structure of Trp-cage (PDB 1L2Y). The tryptophan sidechain and the glycine residue that was mutated to various D-amino acids in the current study are highlighted.
2. Materials and methods
Peptides were synthesized using standard solid-phase Fmoc methods at the Tufts University Core Facility (http://www.tucf.org). All peptides were acetylated at the N-terminus and amidated at the C-terminus. Peptides were purified using reversed-phase high-performance liquid chromatography (HPLC), and peptide identities were verified using mass spectrometry. Triflouroacetic acid (TFA) removal and H-D exchange were achieved by multiple rounds of lyophilization. Fourier transform infrared (FTIR) spectra were collected on a Magna-IR 860 spectrometer (Nicolet, WI) using a home made, two-compartment CaF2 sample cell of 56 μm. The details of the T-jump IR setup have been described elsewhere.49 For both static and time-resolved IR measurements, the peptide sample was prepared by directly dissolving lyophilized solids in 20 mM phosphate D2O buffer (pH* 7) and the final peptide concentration was between 1–2 mM.
3. Results and discussion
A number of studies have shown that the short 310-helix in the Trp-cage fold is much less stable than the global cage structure9,22,48 and its folding can proceed independently and with a faster rate.9 Therefore, in the following discussion, we focus only on the effect of the corresponding D-amino acid mutations on the two-state folding and unfolding kinetics of the cage, which consists of several key structural elements: an α-helix, a polyproline stretch, and a hydrophobic core that encloses a tryptophan residue (Figure 1). In addition, all the thermodynamic parameters used to analyze the T-jump-induced relaxation kinetics are taken from a previous study8 and are summarized in Table 1.
Table 1.
Unfolding thermodynamic parameters of TC5b and its D-amino acid mutants, adapted from ref 8. Also listed are the folding and unfolding times of these peptides at 30 °C.
| Peptide | ΔHm (kcal mol−1) | ΔSm (cal mol−1 K−1) | ΔCp (cal mol−1 K−1) | Tm (°C) | τf (μs) | τu (μs) |
|---|---|---|---|---|---|---|
| TC5b | 12.9 | 40.4 | 72.1 | 45.9 | 3.0 | 7.6 |
| 5b-10GDA | 17.1 | 50.4 | 53.6 | 66.6 | 1.7 | 33.4 |
| 5b-10GDQ | 16.5 | 48.2 | 63.9 | 69.3 | 2.5 | 44.0 |
| 5b-10GDN | 15.8 | 47.8 | 91.6 | 56.4 | 3.7 | 25.5 |
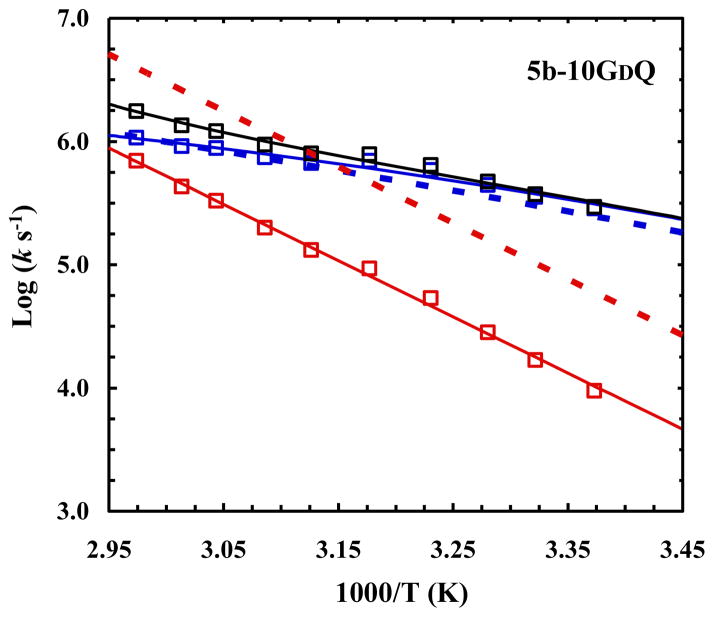
5b-10GDQ
As indicated (Table 1), substitution of Gly10 in TC5b with D-Gln significantly improves Trp-cage stability, increasing its Tm from 45.9 to 69.3 °C. It has been hypothesized and subsequently computationally verified that this stabilizing effect of D-Gln arises from, among other factors, its ability to form hydrogen bonds between the backbone amine of D-Gln and the backbone carbonyl of Leu7, and another between the backbone carbonyl of D-Gln and the backbone amine of Ser13. From a kinetic perspective, however, the higher stability of 5b-10GDQ, compared to that of the wild type, could result from either a faster folding rate or a slower unfolding rate, or both. Since the CD spectrum of 5b-10GDQ indicates that it has a higher helical content than the wild type,8 it is reasonable, based on the notion that the α-helix is populated in the folding transition state of the cage,9 to assume that the α-helix stabilizing effect of D-Gln would lower the free energy of the transition state and, as a result, speed up the rate of folding. As shown (Figure 2), however, the folding rate of 5b-10GDQ is, within our experimental uncertainties, identical to that of the wild type, indicating that the stabilizing interactions stemming from the D-Gln mutation only start to develop at the downhill side of the major folding free energy barrier. In other words, these interactions act primarily to decrease the free energy of the folded state, making the protein unfold slower as a result of an increased unfolding free energy barrier, as observed (Figure 2). This picture is entirely consistent with earlier studies9,13,17,20,24,25,30,48 that indicated that the developments of several key structural elements that involve residues near this part of the sequence, e.g., the formation of the D9-R16 salt-bridge and the burial of the Trp6 sidechain, occurs after the folding transition state. Furthermore, this result is in accord with the observation that stabilizing sidechain-sidechain and/or sidechain-backbone interactions can decrease the unfolding rate of α-helices in isolation.50
Fig. 2.
Arrhenius plots of the relaxation (black), folding (blue), and unfolding (red) rate constants of 5b-10GDQ. The dotted lines correspond to the folding (blue) and unfolding (red) rate constants of the wild type, adapted from ref 9.
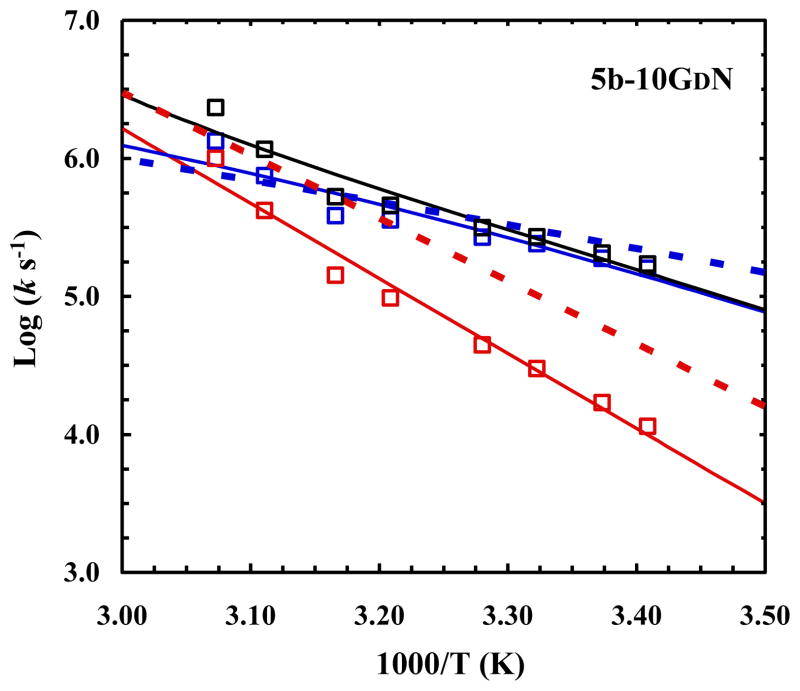
5b-10GDN
Similar to D-Gln, D-Asn was also found to form backbone-backbone hydrogen bonds with Leu7 and Ser13. However, its stabilizing effect is less pronounced compared to D-Gln (Table 1). For example, the Tm of 5b-10GDN was determined to be 56 °C, only 10 °C higher than the wild type. This improvement in stability has been attributed to the difference in backbone hydrogen bonding ability of D-Asn, in comparison to that of D-Gln,8 due to the difference in sidechain-backbone sterics.3 Despite this thermodynamic difference between D-Gln and D-Asn however, we expect that these amino acid substitutions would exert similar impacts on the folding/unfolding kinetics of the cage because of their structural similarity. Indeed, as shown (Figure 3), substitution of Gly10 with D-Asn does not, within our experimental uncertainty, change the folding rate of TC5b; instead it decreases the unfolding rate. Previous studies have shown that the α-helix of TC5b in isolation is unfolded.37 Thus, folding of this α-helix in the absence of other native stabilizing interactions would place the system at a higher free energy position, as concluded previously.9,17,24,30,48 However, such a free energy ‘penalty’ is warranted and necessary, as it prepares and ensures the polypeptide chain to rapidly evolve toward the native state. Since D-Gln and D-Asn are able to act, in the current case, as a C-terminal helix cap, one could have assumed that both would be able to lower the folding free energy barrier, making the protein fold faster. The fact that this is not the case suggests that these mutations involve interactions that stabilize not only the α-helix, through capping, but also other parts of the cage structure. This is consistent with previous MD simulations of these mutants, which found that backbone fluctuations in the D-amino acid mutants are reduced, especially from residues 9 to 15.8 However, these ‘nonlocal’ interactions are expected to be intimately coupled to the structural evolution of the cage, not the α-helix. Thus, the native rotameric state of these D-amino acids in the corresponding mutants would be expected to be significantly populated only after the major folding free energy barrier.
Fig. 3.
Arrhenius plots of the relaxation (black), folding (blue), and unfolding (red) rate constants of 5b-10GDN. The dotted lines correspond to the folding (blue) and unfolding (red) rate constants of the wild type, adapted from ref 9.
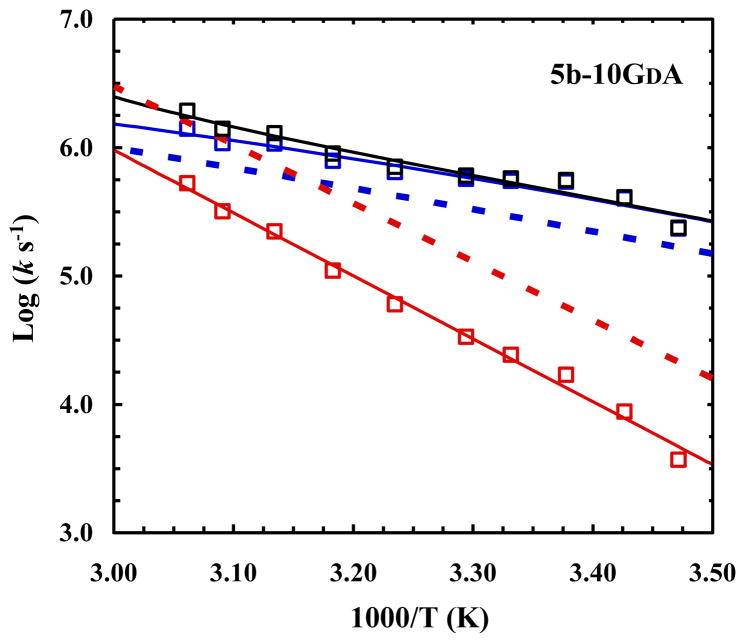
5b-10GDA
As shown (Table 1), D-Ala is almost as effective as D-Gln in stabilizing the Trp-cage fold at position 10. One of the factors this has been attributed to is that D-Ala has less sidechain conformational entropy than both D-Gln and D-Asn, which highlights the well-known importance of entropy-enthalpy compensation in protein folding and design. In light of the above discussion and the fact that D-Ala does not have a sidechain rotamer preference, one might expect that this mutation would lead to an increase the folding rate, in addition to a decrease in the unfolding rate. As shown (Figure 4), indeed, while the unfolding rate of 5b-10GDA is significantly slower than TC5b, similar to that observed for 5b-10GDQ and 5b-10GDN, its folding rate also shows a marginal, but detectable, increase. These results thus confirm the above hypothesis that, unlike D-Gln and D-Asn, D-Ala helps to stabilize, although only to a small extent, the folding transition state or the nascent α-helix. A simple calculation indicates that at 30 °C the D-Ala mutation stabilizes the folding transition state by approximately 0.33 kcal/mol, suggesting that D-Ala already becomes partially native in the transition state, again owing to the relatively small number of configurations the residue can sample. Thus, taken together, the results obtained from these D-amino acids demonstrate their utility in helping reveal certain fine details of the folding mechanism of proteins of interest.
Fig. 4.
Arrhenius plots of the relaxation (black), folding (blue), and unfolding (red) rate constants of 5b-10GDA. The dotted lines correspond to the folding (blue) and unfolding (red) rate constants of the wild type, adapted from ref 9.
4. Conclusions
Many previous studies have elegantly demonstrated the versatility of D-amino acids in protein design, in particular to increase the conformational stability of the targeted folds. However, what is lacking in our current understanding of the effectiveness of specific D-amino acids in this regard is their kinetic mechanism of action. Herein, we use the miniprotein Trp-cage as a model to examine the role of three D-amino acids, i.e., D-Gln, D-Asn, and D-Ala in altering the folding/unfolding kinetics of this protein, based on an earlier study that showed that these D-amino acids can significantly increase the stability of Trp-cage, when replacing the Gly residue in the loop region at position 10. Our results show that these mutations almost exclusively reduce the unfolding rate of the Trp-cage, although the D-Ala mutation also shows a marginal increase in the folding rate. In light of the ability of these D-amino acids to stabilize the α-helix in this miniprotein via a C-capping effect, these results indicate that this α-helix, while being populated in the folding transition state as suggested by a previous study, is nucleated at its N-terminus when passing through the folding free energy barrier.
The relaxation kinetics of three D-amino acid mutants of Trp-cage were measured.
These mutations stabilize the fold.
These mutations primarily increase the folding rate.
The results suggest that the folding of the α-helix in this miniprotein is nucleated at the N-terminus.
Acknowledgments
We thank the National Institutes of Health (GM-065978, RR01348, GM-008275, and GM-089949) for funding. R.M.C. is an NIH Ruth Kirschstein Predoctoral Fellow.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Costantini S, Colonna G, Facchiano AM. Biochem Biophys Res Commun. 2006;342:441. doi: 10.1016/j.bbrc.2006.01.159. [DOI] [PubMed] [Google Scholar]
- 2.Bang D, Gribenko AV, Tereshko V, Kossiakoff AA, Kent SB, Makhatadze GI. Nature Chem Biol. 2006;2:139. doi: 10.1038/nchembio766. [DOI] [PubMed] [Google Scholar]
- 3.Stites WE, Pranata J. Proteins. 1995;22:132. doi: 10.1002/prot.340220206. [DOI] [PubMed] [Google Scholar]
- 4.Anil B, Song BB, Tang YF, Raleigh DP. J Am Chem Soc. 2004;126:13194. doi: 10.1021/ja047119i. [DOI] [PubMed] [Google Scholar]
- 5.Valiyaveetil FI, Sekedat M, MacKinnon R, Muir TW. Proc Natl Acad Sci USA. 2004;101:17045. doi: 10.1073/pnas.0407820101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williams DV, Barua B, Andersen NH. Org Biomol Chem. 2008;6:4287. doi: 10.1039/b814314e. [DOI] [PubMed] [Google Scholar]
- 7.Struthers MD, Cheng RP, Imperiali B. Science. 1996;271:342. doi: 10.1126/science.271.5247.342. [DOI] [PubMed] [Google Scholar]
- 8.Rodriguez-Granillo A, Annavarapu S, Zhang L, Koder RL, Nanda V. J Am Chem Soc. 2011;133:18750. doi: 10.1021/ja205609c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Culik RM, Serrano AL, Bunagan MR, Gai F. Angew Chem Int Ed. 2011;50:10884. doi: 10.1002/anie.201104085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neidigh JW, Fesinmeyer RM, Andersen NH. Nat Struct Biol. 2002;9:425. doi: 10.1038/nsb798. [DOI] [PubMed] [Google Scholar]
- 11.Qiu LL, Pabit SA, Roitberg AE, Hagen SJ. J Am Chem Soc. 2002;124:12952. doi: 10.1021/ja0279141. [DOI] [PubMed] [Google Scholar]
- 12.Snow CD, Zagrovic B, Pande VS. J Am Chem Soc. 2002;124:14548. doi: 10.1021/ja028604l. [DOI] [PubMed] [Google Scholar]
- 13.Simmerling C, Strockbine B, Roitberg AE. J Am Chem Soc. 2002;124:11258. doi: 10.1021/ja0273851. [DOI] [PubMed] [Google Scholar]
- 14.Zhou RH. Proc Nat Acad Sci USA. 2003;100:13280. [Google Scholar]
- 15.Chowdhury S, Lee MC, Xiong GM, Duan Y. J Mol Biol. 2003;327:711. doi: 10.1016/s0022-2836(03)00177-3. [DOI] [PubMed] [Google Scholar]
- 16.Pitera JW, Swope W. Proc Natl Acad Sci USA. 2003;100:7587. doi: 10.1073/pnas.1330954100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nikiforovich GV, Andersen NH, Fesinmeyer RM, Frieden C. Proteins. 2003;52:292. doi: 10.1002/prot.10409. [DOI] [PubMed] [Google Scholar]
- 18.Schug A, Herges T, Wenzel W. Phys Rev Lett. 2003;91:158102. doi: 10.1103/PhysRevLett.91.158102. [DOI] [PubMed] [Google Scholar]
- 19.Carnevali P, Toth G, Toubassi G, Meshkat SN. J Am Chem Soc. 2003;125:14244. doi: 10.1021/ja036647b. [DOI] [PubMed] [Google Scholar]
- 20.Ota M, Ikeguchi M, Kidera A. Proc Natl Acad Sci USA. 2004;101:17658. doi: 10.1073/pnas.0407015102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steinbach PJ. Proteins. 2004;57:665. doi: 10.1002/prot.20247. [DOI] [PubMed] [Google Scholar]
- 22.Ahmed Z, Beta IA, Mikhonin AV, Asher SA. J Am Chem Soc. 2005;127:10943. doi: 10.1021/ja050664e. [DOI] [PubMed] [Google Scholar]
- 23.Neuweiler H, Doose S, Sauer M. Proc Natl Acad Sci USA. 2005;102:16650. doi: 10.1073/pnas.0507351102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding F, Buldyrev SV, Dokholyan NV. Biophys J. 2005;88:147. doi: 10.1529/biophysj.104.046375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Linhananta A, Boer J, MacKay I. J Chem Phys. 2005;122:114901. doi: 10.1063/1.1874812. [DOI] [PubMed] [Google Scholar]
- 26.Irback A, Mohanty S. Biophys J. 2005;88:1560. doi: 10.1529/biophysj.104.050427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alonso JL, Echenique P. Biophys Chem. 2005;115:159. doi: 10.1016/j.bpc.2004.12.021. [DOI] [PubMed] [Google Scholar]
- 28.Chen J, Im W, Brooks CL. J Am Chem Soc. 2006;128:3728. doi: 10.1021/ja057216r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Juraszek J, Bolhuis PG. Proc Natl Acad Sci USA. 2006;103:15859. doi: 10.1073/pnas.0606692103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang L, Grubb MP, Gao YQ. J Chem Phys. 2007;126:125102. doi: 10.1063/1.2709639. [DOI] [PubMed] [Google Scholar]
- 31.Beck DAC, White GWN, Daggett V. J Struct Biol. 2007;157:514. doi: 10.1016/j.jsb.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhan LX, Chen JZY, Liu WK. Proteins. 2007;66:436. doi: 10.1002/prot.21157. [DOI] [PubMed] [Google Scholar]
- 33.Piana S, Laio A. J Phys Chem B. 2007;111:4553. doi: 10.1021/jp067873l. [DOI] [PubMed] [Google Scholar]
- 34.Copps J, Murphy RF, Lovas S. Biopolymers. 2007;88:427. doi: 10.1002/bip.20709. [DOI] [PubMed] [Google Scholar]
- 35.Kentsis A, Gindin T, Mezei M, Osman R. PLoS ONE. 2007;2:e446. doi: 10.1371/journal.pone.0000446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barua B, Lin JC, Williams VD, Kummler P, Neidigh JW, Andersen NH. Protein Eng, Des Sel. 2008;21:171. doi: 10.1093/protein/gzm082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stewart JM, Lin JC, Andersen NH. Chem Commun. 2008:4765. doi: 10.1039/b807101b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu ZH, Tang YH, Wang HF, Zhang X, Lei M. Arch Biochem Biophys. 2008;475:140. doi: 10.1016/j.abb.2008.04.024. [DOI] [PubMed] [Google Scholar]
- 39.Wu S, Zhuravlev PI, Papoian GA. Biophys J. 2008;95:5524. doi: 10.1529/biophysj.108.136697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hudaky P, Straner P, Farkas V, Varadi G, Toth G, Perczel A. Biochemistry. 2008;47:1007. doi: 10.1021/bi701371x. [DOI] [PubMed] [Google Scholar]
- 41.Xu WX, Mu YG. Biophys Chem. 2008;137:116. doi: 10.1016/j.bpc.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 42.Yao XQ, She ZS. Biochem Biophys Res Commun. 2008;373:64. doi: 10.1016/j.bbrc.2008.05.179. [DOI] [PubMed] [Google Scholar]
- 43.Kannan S, Zacharias M. Proteins. 2009;76:448. doi: 10.1002/prot.22359. [DOI] [PubMed] [Google Scholar]
- 44.Cerny J, Vondrasek J, Hobza P. J Phys Chem B. 2009;113:5657. doi: 10.1021/jp9004746. [DOI] [PubMed] [Google Scholar]
- 45.Chebaro Y, Dong X, Laghaei R, Derreumaux P, Mousseau N. J Phys Chem B. 2009;113:267. doi: 10.1021/jp805309e. [DOI] [PubMed] [Google Scholar]
- 46.Matthes D, de Groot BL. Biophys J. 2009;97:599. doi: 10.1016/j.bpj.2009.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marinelli F, Pietrucci F, Laio A, Piana S. PLoS Comput Biol. 2009;5:e1000452. doi: 10.1371/journal.pcbi.1000452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Day R, Paschek D, Garcia AE. Proteins. 2010;78:1889. doi: 10.1002/prot.22702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Serrano AL, Waegele MM, Gai F. Prot Sci. 2012;21:157. doi: 10.1002/pro.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mukherjee S, Chowdhury P, Bunagan MR, Gai F. J Phys Chem B. 2008;112:9146. doi: 10.1021/jp801721p. [DOI] [PubMed] [Google Scholar]