Abstract
With the development of new technologies in genome sequencing, gene expression profiling, genotyping, and high-throughput screening of chemical compound libraries, small molecules are playing increasingly important roles in studying gene expression regulation, gene-gene interaction, and gene function. Here we briefly review and discuss some recent advancements in drug target identification and phenotype characterization using combinations of high-throughput screening of small-molecule libraries and various genome-wide methods such as whole genome sequencing, genome-wide association studies, and genome-wide expressional analysis. These approaches can be used to search for new drugs against parasitic infections, to identify drug targets or drug-resistance genes, and to infer gene function.
Keywords: High-throughput screening, genetics, genomics, gene function
What is meant by chemical genomics?
Chemical genomics is a means to study biology by recapitulating the effect of changes in gene integrity (e.g., mutations) and chromatin structure (e.g., epigenetic gene regulation) through the pharmacologic effect of a small molecule (SM) [1, 2]. Effects elicited by SMs can occur either through direct interaction with a target protein or indirectly by modulating expression or post transcriptional/translational modification [2]. Experimentally, a cell line(s) is treated with various SMs, and then changes in mRNA or protein expression are studied using genome-wide methods such as microarray hybridization or mass spectrometry. As more genomic data and tools to characterize genome-wide genetic variations become available, the concept of chemical genomics is also evolving. Genomics is the study of the genomes of organisms, focusing on efforts to determine the entire DNA sequences of organisms and to develop systematic approaches such as genome-wide association studies (GWAS) to analyze genome variations. Consequently, chemical genomics can be extended to include emerging disciplines that combine screening a large number of cell lines for phenotypic changes after treatment with chemical compounds and detection of genome-wide expressional and/or genetic variations using microarrays, proteomics, parallel DNA sequencing, or GWAS to study gene function and interaction.
SMs as tools for phenotypic screens
Changes in a genome can have different effects on a cell. Phenotypic differences between individual parts of an organism are largely controlled by differences in their genomic DNA. Genetic variations such as nucleotide substitution, insertion/deletion (indel), copy number variation (CNV), or epigenetic modifications likely affect cell growth or survival to some degree. At the molecular level, if a change occurs in the coding region of a gene, the change may alter protein structure or enzyme activity; if a change is in a non-protein coding region or caused by an epigenetic modification, the change may affect gene expression. At the cell level, some mutations are deleterious, leading to death or reduced fitness of a cell, whereas other changes can be neutral or even beneficial. Moreover, the effects of these substitutions are probably conditional. That is, a neutral mutation can become beneficial, deleterious, or even lethal to a cell under different physiologic conditions. For example, a mutation may not have any effect on a microbe grown in rich medium but can be lethal or lead to retarded growth in a restricted medium. Treatment of a cell with SMs can tip the balance of a physiologic state and cause a neutral mutation to manifest an observable phenotype that is not present under normal growth conditions. SMs can inhibit protein activity or block a biologic process, leading to observable phenotypic changes. One exciting example of changing the physiologic state by SMs is the recent induction of pluripotent stem cells from mouse somatic cells using a combination of seven SM compounds [3]. In principle, large numbers of phenotypic variations can be revealed through treatment of a cell line with SMs, and the molecular changes underlying the phenotypic variation can be studied using genetic and genomics approaches. A major hurtle in applying this concept is the ability to reveal or measure a phenotype.
Traditionally, the primary goal of screening SMs is to identify chemical leads to develop into drugs for treating infections or disorders. Indeed, various assays have been developed to screen for antiparasitic drugs [4–11]. SMs are now being recognized as an important tool for studying gene functions and disease mechanisms [12]. Unlike genetic approaches such as RNA interference (RNAi) or gene knockout (KO), SMs can be added and removed at will so that temporal modulation of a protein or pathway can be achieved [2, 13]. SMs usually act on their target rapidly, allowing the effects to be measured in real time, and the degree of the effects on cell viability can be controlled simply through changes of SM concentrations. Additionally, genetic manipulation of a target gene such as gene KO or RNAi may not be applicable to all genes or some organisms. Many genes are essential for the survival of a cell or an organism and therefore cannot be disrupted. RNAi can theoretically be used to study some essential genes by reducing the gene expression level; unfortunately, many organisms—including malaria parasites—do not have the required enzymes for RNAi [14]. SMs therefore can be an alternative for situations that prevent the use of traditional genetic methods. For example, the Mek1/2 inhibitor U0126 was used to study notochord differentiation in zebrafish and mice, which is difficult to investigate using gene disruption due to early lethality (Mek1−/−) and functional redundancy (Mek2−/−) in the KO animals [15]. Identification of the inhibitors illustrates the usefulness of SMs in dissecting complex biologic processes.
The strategy of using SMs to expose phenotypes with genetically encoded reporters is particularly useful for studying such microorganisms as bacteria, viruses, protozoan parasites, and fungi because of their small sizes and limited distinguishable morphologic phenotypes. As more functional assays are being developed for growing and testing microorganisms, SM screening will become an increasingly useful means for studying gene function and gene interactions in these organisms.
High-throughput screening (HTS) and genome-wide approaches to study targets of SMs
Most bioactive SMs carry out their pharmacologic action through binding to protein targets. Therefore, identification of protein targets is important in our understanding of the mode of action and the mechanisms underlying SM efficacy. Various biochemical approaches such as photo-crosslinking, radioisotope labeling, and affinity chromatography have been used to identify SM targets [16]; however, these methods are labor intensive, time consuming, and often generate inconclusive experimental results. Recent advances in high-throughput screens (HTS) and genome-wide techniques in monitoring genome modification and gene expression—including using microarray to detect variation in mRNA transcript variation [17, 18], mass spectrometry-based methods to monitor changes in protein expression [19–23] and metabolite production [24, 25], and chromatin immunoprecipitation-microarray (ChIP-chip) and chromatin immunoprecipitation-sequencing (ChiP-Seq) to detect chromatin modification [26, 27]—have significantly improved our ability to identify SM targets and study gene function (Figure 1). Ideally, more than one genomic approach such as expression profiling and genomic association/linkage mapping should be employed so that mutations in protein coding regions affecting protein activity and those in non-coding regions affecting expression level can be simultaneously characterized.
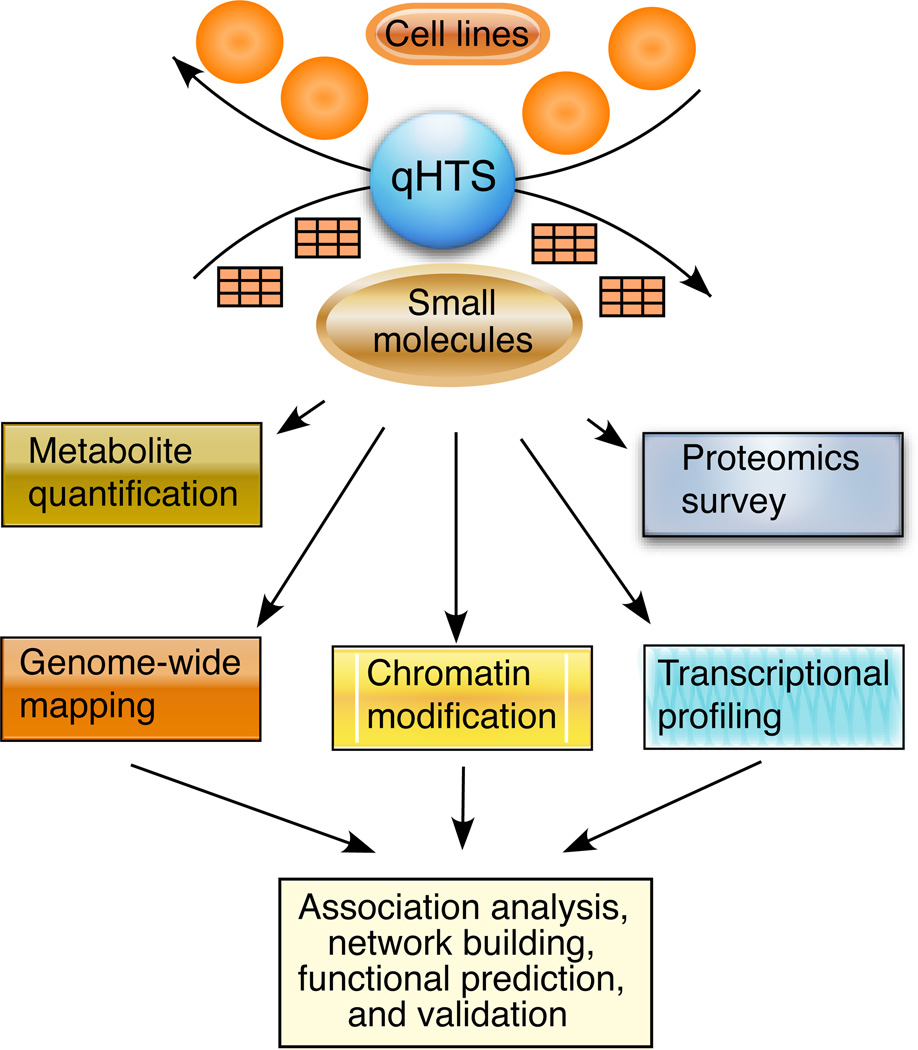
Figure 1.
Strategies for identifying chemical targets and developing gene interaction networks. Cell lines or individual isolates of microbes can be screened or treated with small molecules, and variations in their responses can be studied using genome-wide approaches. Association analysis and construction of gene-interaction networks will provide important information for predicting gene function and gene interaction. Abbreviation: qHTS, quantitative high-throughput screening.
Chemical transcriptomics
One particularly active field of SM research is the application of microarrays to detect transcriptional changes after treatment of a cell line with different SMs. Comparison of gene expression patterns between drug-resistant and sensitive cell lines may reveal genes involved in drug metabolism or transport. In a study of the budding yeast Saccharomyces cerevisiae, a reference database of expression profiles for 300 diverse mutations and chemical treatments was obtained, and cellular pathways affected by the chemical treatment were determined through pattern matching [17]. The expression profile from an uncharacterized perturbation, such as treatment of a new compound, was compared with a database of reference profiles, and the pathway(s) perturbed by the treatment could be determined if the profile is similar to a known profile of a cellular pathway. This approach has led to the identification of uncharacterized genes involved in sterol metabolism, cell wall function, mitochondrial respiration, or protein synthesis. In another study, microarray analysis was used to assess gene expression profiles in 60 human cancer cell lines, and the gene expression patterns were matched with patterns of the sensitivity of the cell lines to chemotherapeutic drugs [28–30]. Large databases on gene expression and molecular pharmacology were integrated to investigate the relationship of variations in the transcript levels of particular genes and mechanisms of drug susceptibility in the cell lines [18]. Although clustering of the cell lines based on 1376 gene expression profiles grouped cell lines into clusters of the same tissue origins, they were also clustered differently based on their sensitivity to 1400 drugs, suggesting common genes in responses to the drugs. Additionally, tumor cell lines expressing high levels of the multidrug-resistance gene ABCB1 (MDR1) had similar drug-response profiles [18]. Anticancer drugs with similar mechanisms of action may induce similar gene expression patterns, which can be explored for identification of candidate genes related to the sensitivity of cancer cells to some specific anticancer drugs [31].
In the malaria parasite Plasmodium falciparum, treatment of SMs may or may not change gene expression depending on which compounds are used and the molecular basis of the resistance mechanism. Treatment of P. falciparum asexual stages with chloroquine or antifolate WR99210 had only limited influence on gene expression [32, 33]. Resistance to chloroquine is largely caused by nucleotide substitutions in the gene encoding a putative chloroquine resistance transporter (pfcrt) [34], and the resistance to antifolates is mediated by mutations in the gene encoding dihydrofolate reductase [35–37]. It is not surprising to see little or no change in the expression levels of these genes after chloroquine or antifolate treatment. By contrast, changes in P. falciparum gene expression were induced using 20 compounds that inhibit growth of the schizont stage [38]. More than 3000 genes showed ≥threefold change in transcript level for at least one of the 23 time points after exposure to chemical stimulus. Based on gene expression patterns after chemical perturbation, sequence homology, domain conservation, and yeast two-hybrid data, networks of gene interaction were constructed, leading to functional prediction of some unknown genes. Additionally, 31 of 42 genes predicted to mediate parasite invasion of red blood cells were expressed in locations or parasite stages associated with invasion [38]. Treatment of P. falciparum asexual stages with sphingolipid analogue dl-threo-1-phenyl-2-palmitoyl-3-morpholino-1-propanol (PPMP) followed by microarray transcriptional analysis identified a protein that is necessary for tubovesicular network (TVN) assembly [39]. The combination of genome-wide expression profiling and large-scale chemical treatment/screening will provide useful information for constructing networks of gene interaction and for studying gene function.
Chemical genetics/genomics
Genetic mapping, either linkage analysis or association, has been used widely to identify genetic loci or mutations that contribute to biologic traits or phenotypic variations [40]. For genetic mapping, differences in phenotype are first identified from study populations or progeny of a genetic cross. DNA samples from individuals with different phenotypes (disease or normal) are genotyped using genetic markers such as microsatellites and single nucleotide polymorphisms (SNPs). Thus, genetic mapping involves genetic markers, phenotypic variations (traits), and methods to genotype DNA samples and to analyze the data obtained.
For malaria parasites, obtaining parasite isolates from patients or progeny from a genetic cross is often the rate-limiting step. Although several genetic crosses for the human malaria parasite P. falciparum have been performed [41], the process is time consuming, labor intensive, and expensive owing to involving nonhuman primates. GWAS is an alternative method, but it requires genotyping large numbers of parasite isolates and genetic markers. Collection and adaptation of parasites from patient blood samples for phenotype evaluation is laborious. Identification of phenotypic variations among limited numbers of culture-adapted parasites or from progeny of genetic crosses becomes an essential issue for genetic mapping in malaria parasites and will probably be a concern for many other pathogenic microbes, too.
SMs can be used to display (or magnify) phenotypic variations that are otherwise not amenable for characterization using traditional methods such as microscopic observation. The principle of using SMs to display differences between malaria parasites was demonstrated in a study involving seven P. falciparum malaria strains (Dd2, HB3, GB4, 7G8, W2, D10, and 3D7). In the study, the parasites were screened against the LOPAC 1280 library (Sigma) containing 1279 bioactive compounds using quantitative HTS (qHTS) (Box 1), and over 600 differential chemical phenotypes (DCPs), defined as pairwise IC50 differences of fivefold or more between parasite lines, were identified among the parasites.
Box 1. Quantitative high-throughput screening (qHTS).
Studying the effects of a large number of small molecules on a cell requires an efficient assay. Indeed, various high-throughput chemical screening strategies have been developed to identify leads for drug development [82–86], including assays for screening compounds against malaria parasites [80, 87–90]. To reduce the number of tests (and costs) to be performed, the majority of chemical screens examine only a single concentration for each compound (typically 10 µM) or even pool compounds in one test. Although the strategy of using a single fixed dose of chemical compound in the initial screening may save time and money, thousands of potential candidate compounds are usually obtained after initial screening, requiring subsequent rounds of screening and evaluation to eliminate false positives. A second screening of the initial hits provides an opportunity to reconfirm the hits independently; this approach, however, is prone to false negatives, thus limiting the comprehensive assessment of the chemical library. To address these issues, a quantitative high-throughput screening (qHTS) methodology was developed [91]. Using robotic platforms, each compound is tested at seven (or more) fivefold dilutions for chemical libraries now containing >350 000 compounds [92].
Concentration response data for each compound is automatically fit and categorized into four major classes using custom software (see http://ncgc.nih.gov/pub/openhts/curvefit/). For example, if a compound produces a complete response curve containing two asymptotes, it is classified into Class 1; compounds in Class 2 have incomplete response curves containing one asymptote, and so on (Figure I). The curve classifications and resulting EC50 and efficacy values enable the prioritization of follow-up chemical series for genetic and genomic analyses and the preliminary establishment of structure-activity relationships among compounds of related structure; therefore, compounds in Class 3 (weakly active) and 4 (inactive) can provide important structure-activity relationship information related to active chemotypes.
Linkage analysis of the IC50 values and hundreds of MS genotypes from the progeny of the Dd2×HB3 and GB4×7G8 crosses mapped several DCPs to loci on chromosome 4 and 5 containing the P. falciparum dihydrofolate reductase (pfdhfr) and multiple drug resistance gene 1 (pfmdr1) genes, respectively. Further fine-mapping using genotypes and phenotypes from field isolates and parasites with an allelic exchanged pfmdr1 confirmed that two mutations in the pfmdr1 could confer resistance to dihydroergotamine methanesulfonate. Similarly, mutations in the pfdhfr gene were shown to confer resistance to trimethoprim and triamterene. This study demonstrated a strategy of using qHTS of SMs and genetic mapping to study malaria traits and to identify potential drug targets. This approach of using SMs to detect genetic variations in response to compounds will be particularly useful for studying microorganisms.
The strategy of screening DCPs for genetic mapping was extended to parasite isolates collected from patients. Phenotypic differences obtained from progeny of a genetic cross will largely reflect the differences between the parents, although some new phenotypes may be generated after chromosome recombination and re-assortment. The genome of the P. falciparum parasite is quite diverse [42–46]; a collection of field parasite isolates will contain a relatively large number of polymorphisms in the genomes, some of which will be reflected in the parasite response to SMs. DCPs from a collection of parasite isolates can be determined, and loci or genes associated with responses to the DCPs can be identified using GWAS employing various high-density genotyping microarray or high-throughput parallel genome sequencing [47–49]. SMs of similar structures may target the same genes, and highly correlated responses (or patterns of response) from two or more SMs may point to the targets of similar functions or genes in related pathways. Genes in a pathway or interaction networks can be constructed if sufficient numbers of SMs targeting different metabolic pathways are tested (Figure 2). Using qHTS, Yuan, et al., tested 2816 compounds approved for human and animal use against 61 P. falciparum field isolates at eight dilutions each and generated an IC50 value for each compound (if active at concentrations used) against the parasites that were also genotyped with 3345 SNPs using a microarray [9]. Parasite responses to hundreds of compounds were associated with mutations in the pfmdr1, pfdhfr, and pfcrt genes after GWAS, and many of the associations were confirmed after linkage analysis using progeny from two genetic crosses. Compounds that were active against wild type or mutant pfcrt and pfmdr1 were also identified. Combinations of chloroquine with selected compounds that were active against chloroquine-resistant parasite Dd2 showed synergistic activity, suggesting potential use for treating chloroquine-resistant parasites. New genetic loci linked or associated with responses to many other compounds were also identified. Moreover, this approach can be applied to other microbes or protozoan parasites that have a sexual life-cycle or to collections of cell lines. Indeed, specific cancer-genomic alterations that can be targeted by SMs were detected after testing 354 SMs targeting different nodes in cell circuitry against 242 genomically characterized cancer cell lines [50].
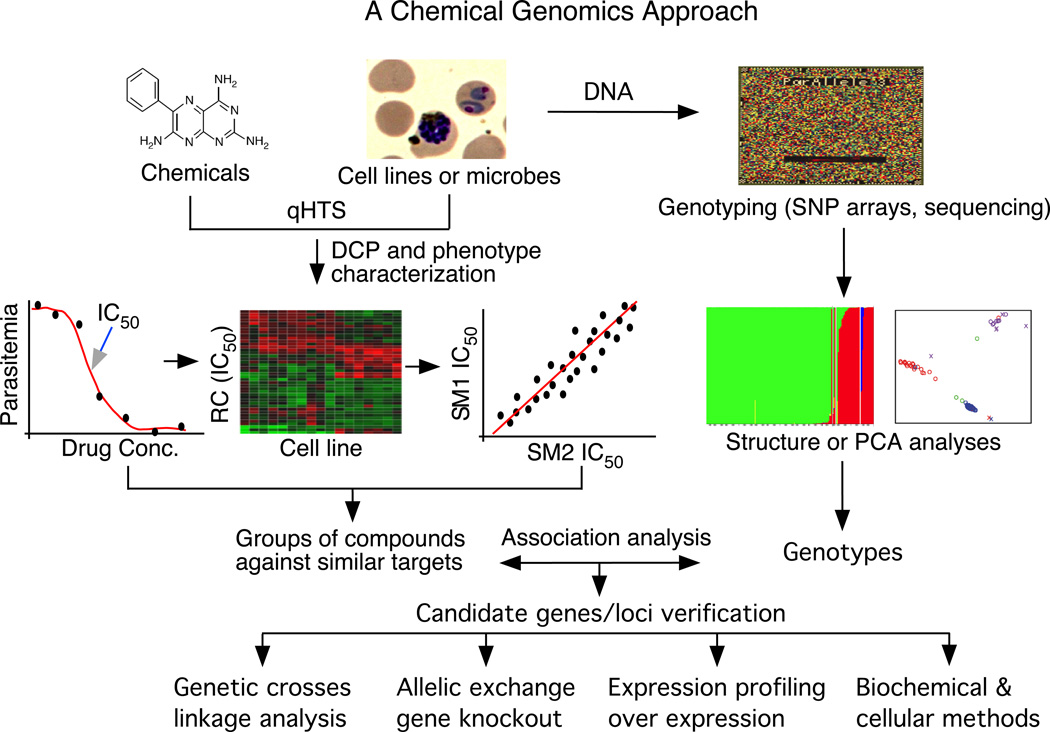
Figure 2.
A chemical genomics approach to identify targets of small molecules. Cell lines or microorganisms are screened against chemical libraries for differences in their responses to the chemicals (differential chemical phenotypes or DCPs). The compounds can be clustered based on the similarity of response patterns (or positive/negative correlation between pairs of compounds) from a panel of cell lines or isolates. Similar response patterns among compounds may suggest that they act on the same or similar targets. DNA samples from the cell lines or isolates are typed using high-throughput genotyping methods such as microarrays or parallel sequencing. Genotypes obtained are analyzed for potential population structures before association analysis. Candidate loci or genes can be further characterized using linkage analysis if progeny from genetic crosses are available. The candidate genes can also be genetically modified or overexpressed to evaluate phenotypic changes. Gene functions can be deduced after additional biochemical and cellular characterizations. Abbreviations: qHTS, quantitative high-throughput screening; RC, response to chemicals; SM1 and SM2, small molecules 1 and 2; SNP, single nucleotide polymorphism; PCA, principal component analysis.
Drug selection and genome sequencing
Another frequently used approach is to subject the parasite to prolonged drug or chemical pressure to select drug-resistant parasites. Changes in gene expression, CNV, or nucleotide substitutions in the genome are then evaluated and compared with the original sensitive parasite using high-density tiling microarrays and parallel sequencing. If the same mutations are detected from repeated independent selections using the same compounds, the mutations detected are likely to play a role in resistance to the compounds. This principle has been employed to identify Plasmodium targets of various SMs having antimalarial activities such as cladosporin and spiroindolones [51–53]. Genome-wide nucleotide substitutions and CNV were also detected using a tiling array, which showed that in vitro-derived fosmidomycin resistance was due to a CNV in the pfdxr gene (P. falciparum 1-deoxy-d-xylulose 5-phosphate reductoisomerase) [54]. Under pressure of artemisinin derivatives, P. falciparum parasites became resistant to the selection agents; gene expression and CNV analysis showed that the resistant parasites also had amplification and increased expression of pfmdr1 [55, 56]. Elevation of the antioxidant defense network and increased expression of many chaperones were also observed in dihydroartemisinin-resistant parasites [56]. In a similar study, the P. falciparum Dd2 parasite was subjected to continuous piperaquine pressure in vitro, and parasites with IC50 values over 100-fold greater than that of the parent were obtained. In addition to a nucleotide change in the pfcrt gene, deamplification of an 82-kb region of chromosome 5 (that includes pfmdr1) and amplification of an adjacent 63-kb region were also found [57]. Using a similar approach, a two-step mutation process (A/T track-based DNA duplication followed by a homology-based expansion) in response to in vitro selection by a triazolopyrimidine-based dihydroorotate dehydrogenase inhibitor (DSM1) was recently proposed [58].
Chemical proteomics
Similar to chemical transcriptomics, changes in protein abundance and activity can be monitored after treatment of cells with SMs. Chemical proteomics is a very active field and has been reviewed extensively [21, 22], but limited information is available for parasites. Here we will limit our discussion on this subject to a couple of examples. There are many different experimental designs combining chemical treatment and diverse methods of protein detection. One approach is to treat a cell line with SMs; the protein profiles of the cell before and after treatment can be compared. In one study, pancreas carcinoma cells were treated with increasing concentrations of the cytotoxic agent daunorubicin, and the changes in protein expression were followed using two-dimensional gel electrophoresis and a matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometer [59]. Another approach, dependent on structure-activity relationship (SAR) guided synthetic modification of the ligand, is to immobilize bioactive compounds to a matrix such as sepharose or agarose and allow proteins from the cell lysate to bind to the compounds. After washes, affinity-captured proteins are specifically eluted with free ligand and digested with enzymes. Peptides from the digested proteins are determined using various mass spectrometry methods including liquid chromatography and tandem mass spectrometry or MALDI-TOF [22]. This strategy was recently used to search for inhibitors against kinases of Trypanosoma brucei and showed that T. brucei kinases were sensitive to typical kinase inhibitors at the nanomolar level [23]. Treatment of cells with large numbers of SMs and profiling their protein expression patterns using high-throughput protein detection methods such as multidimensional protein identification technology (MudPIT) may lead to target proteins or pathways of the SMs [19, 60–63].
Chemical metabolomics (metabonomics)
Metabolomics is the study of small-molecule metabolite profiles or the collection of all metabolites produced from cellular processes using analytical techniques that can simultaneously analyze hundreds to thousands of SM metabolites in biologic samples [25, 64]. SMs may inhibit or interfere with some metabolic pathways, leading to changes in metabolite profiles in a cell. Variation in metabolic profile after chemical treatments can be detected using gas chromatography-mass spectrometry, 1H nuclear magnetic resonance spectroscopy, liquid chromatography-mass spectrometry, or direct infusion-mass spectrometry [24]. Using direct-injection mass spectrometry, Allen, et al., showed that metabolic footprinting was an effective method to classify yeast strains and that unknown mutants could be clustered according to their genetic defect [65]. Different phases of growth were also shown to have specific metabolic footprints. In another recent study, gut microbiota of female mice were treated with antibiotic vancomycin, and the metabolic phenotype of the host was characterized using 1H NMR spectroscopy of urine and fecal extract samples [66]. The excretion of uracil, amino acids, and short chain fatty acids was associated with vancomycin treatment. Changes in metabolites can, in turn, be correlated with gene expression profiling or results from other analyses. Additionally, genome scale enzyme-metabolite and drug-target interactions may be predicted using various protein and chemical databases and machine-learning techniques [67]. Mass spectrometry has been employed to characterize the metabolomic profiles of P. falciparum-infected red blood cells [68] or parasite development throughout its 48 h intraerythrocytic developmental cycle [69] and can be applied to studying parasite response to SMs.
Chemical epigenomics
In addition to changes in nucleotide sequences, modification of DNA or chromatin can also affect gene expression and phenotype display. “The term ‘epigenetic’ defines all heritable changes in gene expression and chromatin structure that are not coded in the DNA sequence itself” [70]. Therefore, an epigenome consists of chemical modifications of the genome such as DNA/RNA methylation and histone modification that include histone methylation and histone acetylation; in broader terms, it should include RNA-mediated silencing. Treatment of a cell with a SM represents a change in environment that may result in changes in patterns of chromatin modifications that can be detected using high-throughput methods. Various genome-wide techniques have been developed to detect DNA methylation and histone modification. ChIP-chip is a technique that combines chromatin immunoprecipitation (ChIP) with microarray technology (chip) to identify the cistrome of DNA-associated proteins, whereas ChIP-Seq is a technique combining ChIP with massively parallel DNA sequencing [26, 27, 71, 72]. Combining DNA microarray or high-throughput sequencing with DNA methylation detection methods such as bisulfite conversion, methylation-sensitive restriction enzymes, methyl-binding proteins, and anti-methylcytosine antibodies, it is also feasible to map DNA methylation on a genome-wide scale [73]. In a recent study, human promoter tiling arrays along with ChIP were used to identify changes in histone acetylation profiles after exposing cells to environmental pollutant polyaromatic hydrocarbon benzo(a)pyrene [74]. SMs can inhibit or interfere with enzymes or proteins involved in chromatin modifications, leading to changes in gene expression. Addition of the histone deacetylase activity inhibitor apicidin induced profound transcriptional changes in P. falciparum intraerythrocytic stages [75] that were characterized by rapid activation and repression of a large percentage of the genome. A major component of this response was the induction of genes that are otherwise suppressed during specific developmental stages. In the schizont stage, apicidin induced hyperacetylation of histone lysine residues H3K9, H4K8, the tetra-acetyl H4 (H4Ac4), and demethylation of H3K4me3. Finally, a combination of genome-scale RNAi target-sequencing screens and application of anti-trypanosomal drugs allowed identification of known drug importers and linked more than fifty genes to drug action in T. brucei [76].
Chemical profiling of signaling networks and deletion mutants
Display of a phenotype is generally controlled by multiple genes in a pathway. Identification of SMs targeting different proteins in a biologic pathway may allow dissection of gene interactions in biologic pathways. Traditional SM screenings focus on a single protein target or molecular pathway, but perturbation of a particular protein or pathway can also alter the behavior of other pathways. This may result from binding to unexpected targets (off target) or unknown interactions between the target and other signaling pathways (Box 2). Therefore, the overall phenotypic changes caused by SMs may reflect both the on-target and various off-target interactions [77]. Exploring multiple signaling pathways or phenotypes using SMs may lead to better understanding of the complexity of biologic pathways and the nature of polypharmacology. In one study, 49 protein-fragment complementation assays across ten cell-signaling pathways were screened against 107 different drugs from six therapeutic areas [78]. The pharmacologic profiles showed both expected and unexpected activities of drugs. By comparing the profiles of these drugs and individual assays, “hidden” phenotypes of tested drugs and assay-phenotype relationships were identified. Moreover, the profiles also reveal high degrees of crosstalk across the pathways.
Box 2. Potential pitfalls of chemical genomics.
Use of SMs to perturb the metabolism of a cell and genome-wide approaches to characterize responses will provide invaluable information on gene function and interaction. However, a SM may also hit multiple targets or unknown targets (off-targets) [78]. Many SMs require specific transporters to move across cellular membranes, and mutations in relevant transporters, despite not being the primary target, may affect the concentration and effectiveness of a SM in the cell. For example, pfmdr1 is probably not the site of action of dihydroergotamine methanesulfonate, although mutations in pfmdr1 were shown to affect parasite response to the compound [37]. Additionally, some SMs may require metabolic activation, and variation in enzyme activities of this process will also affect the SM potency. Functional effects resulting from the off-target activity of a SM may not be as effective as the on-target activity [93].
False positive, negative, and artifactual activity is often associated with HTS [94]. The process requires carefully designed controls and follow-up validation of resulting primary active compound [95, 96]. Likewise, genome-wide association is know to have high ‘background’ and confounding effects, and the results generally require independent verification using different samples or population [97]. It is well-known that many factors can influence results from microarray analysis [98, 99], and the presence of large numbers of antisense transcripts require preparation of directional libraries for RNA-seq analysis [100]. Proteins expressed at low levels are unlikely to be detected using current proteomics technologies [101]. Even with the limitations, combinations of SM screening and genome-wide approaches will provide powerful tools for studying gene function and gene interaction.
Chemical profiling can also be explored to detect impacts of nonessential gene deletion. In another study, 1144 chemical genomic assays were performed against budding yeast (S. cerevisiae) whole-genome heterozygous and homozygous deletion collections [79]. Quantitative relationships between the functions of yeast “nonessential” genes and the growth fitness in response to chemical or environmental stress conditions were measured, and approximately 97% of yeast genes were found to be required for wild-type growth under at least one experimental condition, suggesting that most yeast genes are actually “essential.” Further gene cluster analysis revealed that gene products existing in the same signaling pathways or protein complexes tended to have similar co-fitness profiles. The functions of novel genes or new functions of known genes may be predicted by profile comparison. Testing of a genome-wide collection of heterozygous knockout yeast strains against cladosporin allowed the identification of a lysyl-tRNA synthetase as the target [52].
Chemical profiling has been employed to systematically investigate the molecular targets of uncharacterized SMs and predict their mechanisms of action. The basic concept of this approach is that compounds exhibiting similar activities against a pathway or a target are likely to show similar profiles across all the assays. A screen of ~ 1.7 million compounds for inhibition of P. falciparum malaria growth in vitro identified ~ 17 000 compounds [80]. These selected compounds were then evaluated across 131 cell-based and biochemical assays from pre-existing databases (PubChem and World Drug Index). With in silico activity profiling, the mechanisms of action of uncharacterized compounds were predicted, including uncharacterized compounds in the folate cluster that are likely to target DHFR in the proof-of-principle study. In another study, Kokel, et al., screened 14 000 well and un-characterized SMs against zebrafish embryos and looked for compounds that affect the photomotor response (PMR) [81]. They were able to cluster SMs by different phenotypic profiles called “behavior barcodes.” Because functionally related SMs should have similar behavior barcodes, the mechanisms of action of novel behaviormodifying compounds may be predicted.
Concluding remarks and future perspectives
The ultimate goal of chemical genomics is to link variations in the genome to differences in cell survival, growth, and differentiation. Although it will be difficult to detect mutations that are essential for cell survival, many nonessential but important genetic variations can be identified through chemical genomics approaches. If a sufficiently diverse array of pharmacologically active SMs is screened against a large number of cell lines or microbes, it is possible to build connections or networks that belong to a biologic pathway or related pathways. SMs that produce similar response patterns in a panel of cell lines are likely to interact with similar molecular targets or targets within a pathway in the cells; however, a gene contributing to a particular phenotype may not necessarily be the target of the linked compound (Box 2). Further analysis, such as enzyme or affinity chromatography assays, is required for validating bona fide SM-protein interactions. Other methods to verify a gene function include gene KO, gene overexpression, RNAi, and specific biochemical, biophysical, and molecular approaches. Incorporation of these “connections” with chemical structures and chemical properties of SMs will lead to better understanding of the biologic functions of genes and their products, and invariably to the design and development of more effective and safe antimicrobial drugs.
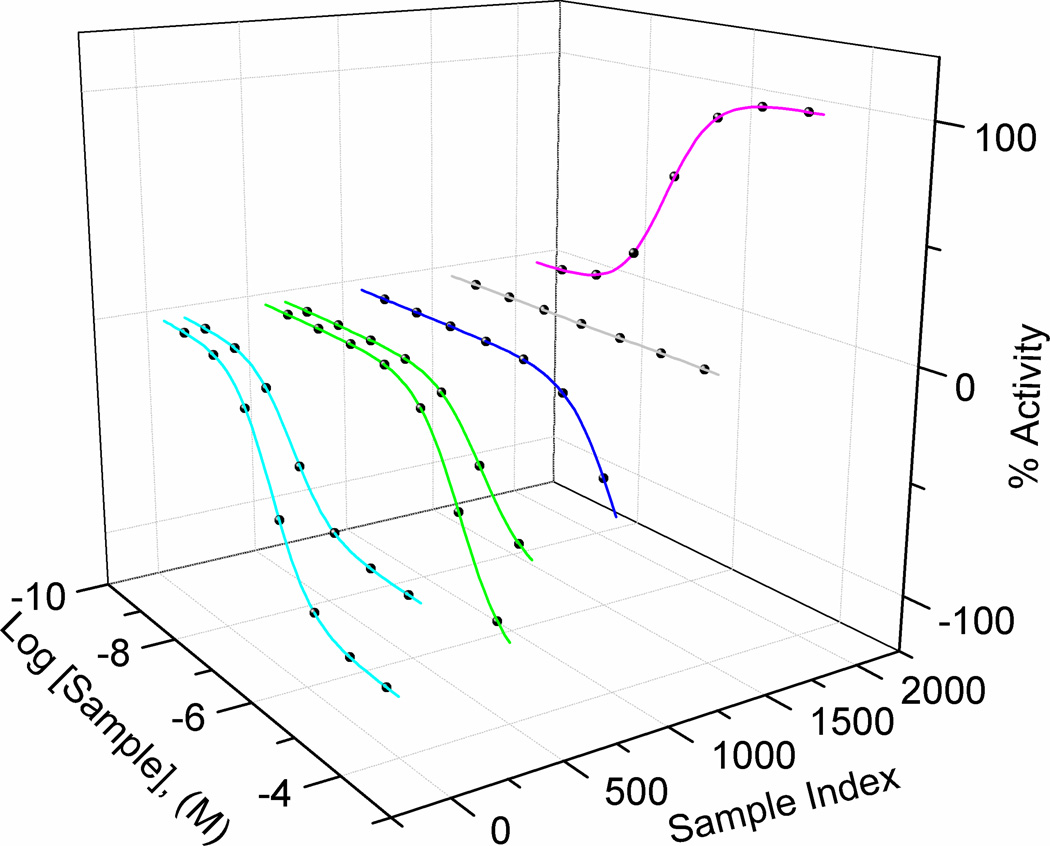
Figure I.
qHTS curve classification graphic. Concentration-response curves generated from 4-parameter fits to experimental data are categorized into curve class (CC) according to Inglese et al. [91], where magenta, CC=−1a and −1b (left to right); green, CC=−2a and −2b, dark blue, CC=−3; and grey, CC=4 (inactive). The sign (− or +) indicates inhibitory or stimulatory, for example, the purple curve is a stimulatory 1a curve. The ‘a’ and ‘b’ designate a fully efficacious (relative to control) vs. a partially efficacious effect, respectively.
Highlights.
Quantitative high throughput screen (qHTS) of small molecules (SMs)
SMs as a tool for studying phenotypes and gene function
Combination of SM screening and genome-wide approaches
Target identification and pathway/network building
Acknowledgments
This work was supported by the National Natural Science Foundation of China (#81220108019, #81271858, and #81201324), by Project 111 of the State Bureau of Foreign Experts and Ministry of Education of China (B06016), and by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Center for Advancing Translational Sciences, National Institutes of Health. We thank Dr. Ronald Johnson for comments, and intramural editor Brenda Rae Marshall, DPSS, NIAID, for assistance. We apologize to those whose work on screening for antiparasitic drugs cannot be cited here due to space constraints.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Roemer T, et al. Bugs, drugs and chemical genomics. Nat Chem Biol. 2012;8:46–56. doi: 10.1038/nchembio.744. [DOI] [PubMed] [Google Scholar]
- 2.Knight ZA, Shokat KM. Chemical genetics: where genetics and pharmacology meet. Cell. 2007;128:425–430. doi: 10.1016/j.cell.2007.01.021. [DOI] [PubMed] [Google Scholar]
- 3.Hou P, et al. Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science. 2013;341:651–654. doi: 10.1126/science.1239278. [DOI] [PubMed] [Google Scholar]
- 4.Sharlow ER, et al. A target-based high throughput screen yields Trypanosoma brucei hexokinase small molecule inhibitors with antiparasitic activity. PLoS Negl Trop Dis. 2010;4:e659. doi: 10.1371/journal.pntd.0000659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuhn I, et al. Identification by high-throughput screening of inhibitors of Schistosoma mansoni NAD(+) catabolizing enzyme. Bioorg Med Chem. 2010;18:7900–7910. doi: 10.1016/j.bmc.2010.09.041. [DOI] [PubMed] [Google Scholar]
- 6.Gut J, et al. An image-based assay for high throughput screening of Giardia lamblia. J Microbiol Methods. 2011;84:398–405. doi: 10.1016/j.mimet.2010.12.026. [DOI] [PubMed] [Google Scholar]
- 7.Siqueira-Neto JL, et al. Antileishmanial high-throughput drug screening reveals drug candidates with new scaffolds. PLoS Negl Trop Dis. 2010;4:e675. doi: 10.1371/journal.pntd.0000675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guiguemde WA, et al. Chemical genetics of Plasmodium falciparum. Nature. 2010;465:311–315. doi: 10.1038/nature09099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuan J, et al. Chemical genomic profiling for antimalarial therapies, response signatures, and molecular targets. Science. 2011;333:724–729. doi: 10.1126/science.1205216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meister S, et al. Imaging of Plasmodium liver stages to drive next-generation antimalarial drug discovery. Science. 2011;334:1372–1377. doi: 10.1126/science.1211936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frearson JA, et al. N-myristoyltransferase inhibitors as new leads to treat sleeping sickness. Nature. 2010;464:728–732. doi: 10.1038/nature08893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hasson SA, Inglese J. Innovation in academic chemical screening: filling the gaps in chemical biology. Curr Opin Chem Biol. 2013;17:329–338. doi: 10.1016/j.cbpa.2013.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Norambuena L, et al. Chemical genomics approaches in plant biology. Methods Mol Biol. 2009;553:345–354. doi: 10.1007/978-1-60327-563-7_18. [DOI] [PubMed] [Google Scholar]
- 14.Ullu E, et al. RNA interference in protozoan parasites. Cell Microbiol. 2004;6:509–519. doi: 10.1111/j.1462-5822.2004.00399.x. [DOI] [PubMed] [Google Scholar]
- 15.Hawkins TA, et al. The small molecule Mek1/2 inhibitor U0126 disrupts the chordamesoderm to notochord transition in zebrafish. BMC Dev Biol. 2008;8:42. doi: 10.1186/1471-213X-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Becker F, et al. A three-hybrid approach to scanning the proteome for targets of small molecule kinase inhibitors. Chem Biol. 2004;11:211–223. doi: 10.1016/j.chembiol.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 17.Hughes TR, et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 18.Scherf U, et al. A gene expression database for the molecular pharmacology of cancer. Nat Genet. 2000;24:236–244. doi: 10.1038/73439. [DOI] [PubMed] [Google Scholar]
- 19.Wolters DA, et al. An automated multidimensional protein identification technology for shotgun proteomics. Anal Chem. 2001;73:5683–5690. doi: 10.1021/ac010617e. [DOI] [PubMed] [Google Scholar]
- 20.Washburn MP, et al. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 21.Ovaa H, van Leeuwen F. Chemical biology approaches to probe the proteome. Chembiochem. 2008;9:2913–2919. doi: 10.1002/cbic.200800454. [DOI] [PubMed] [Google Scholar]
- 22.Rix U, Superti-Furga G. Target profiling of small molecules by chemical proteomics. Nat Chem Biol. 2009;5:616–624. doi: 10.1038/nchembio.216. [DOI] [PubMed] [Google Scholar]
- 23.Urbaniak MD, et al. Chemical proteomic analysis reveals the drugability of the kinome of Trypanosoma brucei. ACS Chem Biol. 2012;7:1858–1865. doi: 10.1021/cb300326z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roberts LD, et al. Metabolic phenotyping of a model of adipocyte differentiation. Physiol Genomics. 2009;39:109–119. doi: 10.1152/physiolgenomics.90365.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han J, et al. Metabolomics: towards understanding host-microbe interactions. Future Microbiol. 2010;5:153–161. doi: 10.2217/fmb.09.132. [DOI] [PubMed] [Google Scholar]
- 26.Huebert DJ, et al. Genome-wide analysis of histone modifications by ChIP-on-chip. Methods. 2006;40:365–369. doi: 10.1016/j.ymeth.2006.07.032. [DOI] [PubMed] [Google Scholar]
- 27.Wang Z, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ross DT, et al. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet. 2000;24:227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- 29.Nakatsu N, et al. Chemosensitivity profile of cancer cell lines and identification of genes determining chemosensitivity by an integrated bioinformatical approach using cDNA arrays. Mol Cancer Ther. 2005;4:399–412. doi: 10.1158/1535-7163.MCT-04-0234. [DOI] [PubMed] [Google Scholar]
- 30.Amundson SA, et al. Integrating global gene expression and radiation survival parameters across the 60 cell lines of the National Cancer Institute Anticancer Drug Screen. Cancer Res. 2008;68:415–424. doi: 10.1158/0008-5472.CAN-07-2120. [DOI] [PubMed] [Google Scholar]
- 31.Nakamura T, et al. Identification of candidate genes determining chemosensitivity to anti-cancer drugs of gastric cancer cell lines. Biol Pharm Bull. 2009;32:1936–1939. doi: 10.1248/bpb.32.1936. [DOI] [PubMed] [Google Scholar]
- 32.Jiang H, et al. Genome-wide compensatory changes accompany drug- selected mutations in the Plasmodium falciparum crt gene. PLoS ONE. 2008;3:e2484. doi: 10.1371/journal.pone.0002484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ganesan K, et al. A genetically hard-wired metabolic transcriptome in Plasmodium falciparum fails to mount protective responses to lethal antifolates. PLoS Pathog. 2008;4:e1000214. doi: 10.1371/journal.ppat.1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fidock DA, et al. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol Cell. 2000;6:861–871. doi: 10.1016/s1097-2765(05)00077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cowman AF, et al. Amino acid changes linked to pyrimethamine resistance in the dihydrofolate reductase-thymidylate synthase gene of Plasmodium falciparum. Proc Natl Acad Sci U S A. 1988;85:9109–9113. doi: 10.1073/pnas.85.23.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peterson DS, et al. Molecular basis of differential resistance to cycloguanil and pyrimethamine in Plasmodium falciparum malaria. Proc Natl Acad Sci U S A. 1990;87:3018–3022. doi: 10.1073/pnas.87.8.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuan J, et al. Genetic mapping of targets mediating differential chemical phenotypes in Plasmodium falciparum. Nat Chem Biol. 2009;5:765–771. doi: 10.1038/nchembio.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu G, et al. Transcriptional profiling of growth perturbations of the human malaria parasite Plasmodium falciparum. Nat Biotechnol. 2009;28:91–98. doi: 10.1038/nbt.1597. [DOI] [PubMed] [Google Scholar]
- 39.Tamez PA, et al. An erythrocyte vesicle protein exported by the malaria parasite promotes tubovesicular lipid import from the host cell surface. PLoS Pathog. 2008;4:e1000118. doi: 10.1371/journal.ppat.1000118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- 41.Su X-z, et al. Genetic linkage and association analyses for trait mapping in Plasmodium falciparum. Nat Rev Genet. 2007;8:497–506. doi: 10.1038/nrg2126. [DOI] [PubMed] [Google Scholar]
- 42.Volkman SK, et al. A genome-wide map of diversity in Plasmodium falciparum. Nat Genet. 2007;39:113–119. doi: 10.1038/ng1930. [DOI] [PubMed] [Google Scholar]
- 43.Jeffares DC, et al. Genome variation and evolution of the malaria parasite Plasmodium falciparum. Nat Genet. 2007;39:120–125. doi: 10.1038/ng1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mu J, et al. Genome-wide variation and identification of vaccine targets in the Plasmodium falciparum genome. Nat Genet. 2007;39:126–130. doi: 10.1038/ng1924. [DOI] [PubMed] [Google Scholar]
- 45.Manske M, et al. Analysis of Plasmodium falciparum diversity in natural infections by deep sequencing. Nature. 2012;487:375–379. doi: 10.1038/nature11174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miotto O, et al. Multiple populations of artemisinin-resistant Plasmodium falciparum in Cambodia. Nat Genet. 2013;45:648–655. doi: 10.1038/ng.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang H, et al. Detection of genome wide polymorphisms in the AT rich Plasmodium falciparum genome using a high density microarray. BMC Genomics. 2008;9:398. doi: 10.1186/1471-2164-9-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dharia NV, et al. Whole-genome sequencing and microarray analysis of ex vivo Plasmodium vivax reveal selective pressure on putative drug resistance genes. Proc Natl Acad Sci U S A. 2010;107:20045–20050. doi: 10.1073/pnas.1003776107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mu J, et al. Plasmodium falciparum genome-wide scans for positive selection, recombination hot spots and resistance to antimalarial drugs. Nat Genet. 2010;42:268–271. doi: 10.1038/ng.528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Basu A, et al. An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell. 2013;154:1151–1161. doi: 10.1016/j.cell.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rottmann M, et al. Spiroindolones, a potent compound class for the treatment of malaria. Science. 2010;329:1175–1180. doi: 10.1126/science.1193225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hoepfner D, et al. Selective and specific inhibition of the Plasmodium falciparum lysyl-tRNA synthetase by the fungal secondary metabolite cladosporin. Cell Host Microbe. 2012;11:654–663. doi: 10.1016/j.chom.2012.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Flannery EL, et al. Using genetic methods to define the targets of compounds with antimalarial activity. J Med Chem. 2013 doi: 10.1021/jm400325j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dharia NV, et al. Use of high-density tiling microarrays to identify mutations globally and elucidate mechanisms of drug resistance in Plasmodium falciparum. Genome Biol. 2009;10:R21. doi: 10.1186/gb-2009-10-2-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chavchich M, et al. Role of pfmdr1 amplification and expression in induction of resistance to artemisinin derivatives in Plasmodium falciparum. Antimicrob Agents Chemother. 2010;54:2455–2464. doi: 10.1128/AAC.00947-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cui L, et al. Mechanisms of in vitro resistance to dihydroartemisinin in Plasmodium falciparum. Mol Microbiol. 2012;86:111–128. doi: 10.1111/j.1365-2958.2012.08180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eastman RT, et al. Piperaquine resistance is associated with a copy number variation on chromosome 5 in drug-pressured Plasmodium falciparum parasites. Antimicrob Agents Chemother. 2011;55:3908–3916. doi: 10.1128/AAC.01793-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guler JL, et al. Asexual populations of the human malaria parasite, Plasmodium falciparum use a two-step genomic strategy to acquire accurate, beneficial DNA amplifications. PLoS Pathog. 2013;9:e1003375. doi: 10.1371/journal.ppat.1003375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moller A, et al. Two-dimensional gel electrophoresis: a powerful method to elucidate cellular responses to toxic compounds. Toxicology. 2001;160:129–138. doi: 10.1016/s0300-483x(00)00443-1. [DOI] [PubMed] [Google Scholar]
- 60.Petricoin EF, Liotta LA. SELDI-TOF-based serum proteomic pattern diagnostics for early detection of cancer. Curr Opin Biotechnol. 2004;15:24–30. doi: 10.1016/j.copbio.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 61.Delahunty CM, Yates JR., 3rd MudPIT: multidimensional protein identification technology. Biotechniques. 2007;43 563, 565, 567 passim. [PubMed] [Google Scholar]
- 62.Lu B, et al. Improving protein identification sensitivity by combining MS and MS/MS information for shotgun proteomics using LTQ-Orbitrap high mass accuracy data. Anal Chem. 2008;80:2018–2025. doi: 10.1021/ac701697w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jessani N, et al. A streamlined platform for high-content functional proteomics of primary human specimens. Nat Methods. 2005;2:691–697. doi: 10.1038/nmeth778. [DOI] [PubMed] [Google Scholar]
- 64.Griffin JL, Vidal-Puig A. Current challenges in metabolomics for diabetes research: a vital functional genomic tool or just a ploy for gaining funding? Physiol Genomics. 2008;34:1–5. doi: 10.1152/physiolgenomics.00009.2008. [DOI] [PubMed] [Google Scholar]
- 65.Allen J, et al. High-throughput classification of yeast mutants for functional genomics using metabolic footprinting. Nat Biotechnol. 2003;21:692–696. doi: 10.1038/nbt823. [DOI] [PubMed] [Google Scholar]
- 66.Yap IK, et al. Metabonomic and microbiological analysis of the dynamic effect of vancomycin-induced gut microbiota modification in the mouse. J Proteome Res. 2008;7:3718–3728. doi: 10.1021/pr700864x. [DOI] [PubMed] [Google Scholar]
- 67.Faulon JL, et al. Genome scale enzyme-metabolite and drug-target interaction predictions using the signature molecular descriptor. Bioinformatics. 2008;24:225–233. doi: 10.1093/bioinformatics/btm580. [DOI] [PubMed] [Google Scholar]
- 68.Sana TR, et al. Global mass spectrometry based metabolomics profiling of erythrocytes infected with Plasmodium falciparum. PLoS One. 2013;8:e60840. doi: 10.1371/journal.pone.0060840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Olszewski KL, et al. Host-parasite interactions revealed by Plasmodium falciparum metabolomics. Cell Host Microbe. 2009;5:191–199. doi: 10.1016/j.chom.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Herceg Z, Hainaut P. Genetic and epigenetic alterations as biomarkers for cancer detection, diagnosis and prognosis. Mol Oncol. 2007;1:26–41. doi: 10.1016/j.molonc.2007.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pokholok DK, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 72.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 73.Zilberman D, Henikoff S. Genome-wide analysis of DNA methylation patterns. Development. 2007;134:3959–3965. doi: 10.1242/dev.001131. [DOI] [PubMed] [Google Scholar]
- 74.Sadikovic B, et al. Genome-wide H3K9 histone acetylation profiles are altered in benzopyrene-treated MCF7 breast cancer cells. J Biol Chem. 2008;283:4051–4060. doi: 10.1074/jbc.M707506200. [DOI] [PubMed] [Google Scholar]
- 75.Chaal BK, et al. Histone deacetylases play a major role in the transcriptional regulation of the Plasmodium falciparum life cycle. PLoS Pathog. 2010;6:e1000737. doi: 10.1371/journal.ppat.1000737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Alsford S, et al. High-throughput decoding of antitrypanosomal drug efficacy and resistance. Nature. 2012;482:232–236. doi: 10.1038/nature10771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Abraham RT. Signalomic signatures enlighten drug profiling. Nat Chem Biol. 2006;2:295–296. doi: 10.1038/nchembio0606-295. [DOI] [PubMed] [Google Scholar]
- 78.MacDonald ML, et al. Identifying off-target effects and hidden phenotypes of drugs in human cells. Nat Chem Biol. 2006;2:329–337. doi: 10.1038/nchembio790. [DOI] [PubMed] [Google Scholar]
- 79.Hillenmeyer ME, et al. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science. 2008;320:362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Plouffe D, et al. In silico activity profiling reveals the mechanism of action of antimalarials discovered in a high-throughput screen. Proc Natl Acad Sci U S A. 2008;105:9059–9064. doi: 10.1073/pnas.0802982105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kokel D, et al. Rapid behavior-based identification of neuroactive small molecules in the zebrafish. Nat Chem Biol. 6:231–237. doi: 10.1038/nchembio.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hacksell U, et al. Chemical genomics: massively parallel technologies for rapid lead identification and target validation. Cytotechnology. 2002;38:3–10. doi: 10.1023/A:1021169023731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Burstein ES, et al. Integrative functional assays, chemical genomics and high throughput screening: harnessing signal transduction pathways to a common HTS readout. Curr Pharm Des. 2006;12:1717–1729. doi: 10.2174/138161206776873662. [DOI] [PubMed] [Google Scholar]
- 84.Snowden M, Green DV. The impact of diversity-based, high-throughput screening on drug discovery: "chance favours the prepared mind". Curr Opin Drug Discov Devel. 2008;11:553–558. [PubMed] [Google Scholar]
- 85.Mayr LM, Bojanic D. Novel trends in high-throughput screening. Curr Opin Pharmacol. 2009;9:580–588. doi: 10.1016/j.coph.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 86.Kainkaryam RM, Woolf PJ. Pooling in high-throughput drug screening. Curr Opin Drug Discov Devel. 2009;12:339–350. [PMC free article] [PubMed] [Google Scholar]
- 87.Weisman JL, et al. Searching for new antimalarial therapeutics amongst known drugs. Chem Biol Drug Des. 2006;67:409–416. doi: 10.1111/j.1747-0285.2006.00391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chong CR, et al. A clinical drug library screen identifies astemizole as an antimalarial agent. Nat Chem Biol. 2006;2:415–416. doi: 10.1038/nchembio806. [DOI] [PubMed] [Google Scholar]
- 89.Baniecki ML, et al. High-throughput Plasmodium falciparum growth assay for malaria drug discovery. Antimicrob Agents Chemother. 2007;51:716–723. doi: 10.1128/AAC.01144-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Buchholz K, et al. A high-throughput screen targeting malaria transmission stages opens new avenues for drug development. J Infect Dis. 2011;203:1445–1453. doi: 10.1093/infdis/jir037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Inglese J, et al. Quantitative high-throughput screening: a titration-based approach that efficiently identifies biological activities in large chemical libraries. Proc Natl Acad Sci U S A. 2006;103:11473–11478. doi: 10.1073/pnas.0604348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Michael S, et al. A robotic platform for quantitative high-throughput screening. Assay Drug Dev Technol. 2008;6:637–657. doi: 10.1089/adt.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Welsbie DS, et al. Functional genomic screening identifies dual leucine zipper kinase as a key mediator of retinal ganglion cell death. Proc Natl Acad Sci U S A. 2013;110:4045–4050. doi: 10.1073/pnas.1211284110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Thorne N, et al. Apparent activity in high-throughput screening: origins of compound-dependent assay interference. Curr Opin Chem Biol. 2010;14:315–324. doi: 10.1016/j.cbpa.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cheng KC, Inglese J. A coincidence reporter-gene system for high-throughput screening. Nat Methods. 2012;9:937. doi: 10.1038/nmeth.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Inglese J, et al. High-throughput screening assays for the identification of chemical probes. Nat Chem Biol. 2007;3:466–479. doi: 10.1038/nchembio.2007.17. [DOI] [PubMed] [Google Scholar]
- 97.Sillanpaa MJ. Overview of techniques to account for confounding due to population stratification and cryptic relatedness in genomic data association analyses. Heredity (Edinb) 2011;106:511–519. doi: 10.1038/hdy.2010.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schurmann C, et al. Analyzing illumina gene expression microarray data from different tissues: methodological aspects of data analysis in the metaxpress consortium. PLoS One. 2012;7:e50938. doi: 10.1371/journal.pone.0050938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Draghici S, et al. Reliability and reproducibility issues in DNA microarray measurements. Trends Genet. 2006;22:101–109. doi: 10.1016/j.tig.2005.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lopez-Barragan MJ, et al. Directional gene expression and antisense transcripts in sexual and asexual stages of Plasmodium falciparum. BMC Genomics. 2011;12:587. doi: 10.1186/1471-2164-12-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wilson R. Sensitivity and specificity: twin goals of proteomics assays. Can they be combined? Expert Rev Proteomics. 2013;10:135–149. doi: 10.1586/epr.13.7. [DOI] [PubMed] [Google Scholar]