Abstract
Closely related peptide epitopes can be recognized by the same T cells and contribute to the immune response against pathogens encoding those epitopes, but sometimes cross-reactive epitopes share little homology. The degree of structural homology required for such disparate ligands to be recognized by cross-reactive T cell receptors remains unclear. Here, we examined the mechanistic basis for cross-reactive T cell responses between epitopes from unrelated and pathogenic viruses, lymphocytic choriomeningitis virus (LCMV) and vaccinia virus (VV). Our results show that the LCMV-cross-reactive T cell response towards VV is dominated by a shared asparagine residue, together with other shared structural elements conserved in the crystal structures of Kb-VV-A11R and Kb-LCMV-GP34. Based on analysis of the crystal structures and the specificity determinants for the cross-reactive T cell response, we were able to manipulate the degree of cross-reactivity of the T cell response, and to predict and generate a LCMV-cross-reactive response towards a variant of a null ovalbumin-derived peptide. These results indicate that protective heterologous immune responses can occur for disparate epitopes from unrelated viruses.
Introduction
Memory T cell populations generated against a previously encountered pathogen can alter the outcome of a subsequent exposure to an unrelated pathogen (1-3). This phenomenon, known as heterologous immunity, has been well-documented in humans and mice for both related and unrelated pathogens (4-9). In humans, T cell cross-reactivity has been found to mediate heterologous immunity between influenza A virus and either hepatitis C virus (4) or Epstein-Barr virus (7). T cell cross-reactivity has also been found associated with immunopathology following sequential infections with different dengue virus serotypes (10). In mice, functional cross-reactive T cell responses between the closely related arenaviruses Pichinde virus and lymphocytic choriomeningitis virus (LCMV) (6, 11), or between two completely unrelated viruses, LCMV and vaccinia virus (VV), have been well characterized (8, 12). For LCMV and VV, previous exposure to LCMV results in either protective immunity or altered immunopathology in mice that are challenged with VV (13, 14). The demonstrated impact on the overall immune response for T cell cross-reactivity highlights the importance of understanding the underlying mechanisms.
VV challenge of LCMV-immune mice results in proliferative T cell responses towards an immunodominant LCMV-GP34 epitope (See Table 1) (15). A prior study showed that adoptive transfer of T cell lines derived from LCMV-immune mice and cross-reactive towards LCMV-GP34 and VV-A11R protects against VV challenge in vivo (8). Our previous results showed that cross-reactivity between VV-A11R and LCMV-GP34 is mediated by T cell receptors (TCR) that could recognize both epitopes (16). The sequence disparity between LCMV-GP34 (AVYNFATM) and VV-A11R (AIVNYANL), which share only three of eight residues (underlined), made it seemingly unlikely that structural mimicry could be the underlying mechanism.
Table 1.
Relevant epitopes used in this study.
| Source | Protein | Abbreviation | Sequence |
|---|---|---|---|
| VV | Nonstructural protein A11R198-206 | A11R | AIVNYANL |
| VV | E7R130-137 | E7R | STNLFNNL |
| VV | B8R20-27 | B8R | TSYKFESV |
| LCMV | Glycoprotein34-41 | GP34 | AVYNFATMa |
| LCMV | Nucleoprotein205-212 | NP205 | YTVKYPNL |
| LCMV | Nucleoprotein396-404 | NP396 | FQPQNGQFI |
| - | Designed sequence | SIY | SIYRYYGL |
| - | Ovalbumin257-264 | OVA | SIINFEKL |
| - | Designed sequence | OVA-AA | SIINFAAL |
| - | Designed sequence | OVA-AT | SIINFATL |
In principle, there are two ways by which T cells can recognize cross-reactive peptide-MHC complexes. T cells can express T cell receptors that are individually cross-reactive towards two or more peptide-MHC antigens (17). Alternatively, cross-reactive T cell responses might be mediated by a subset of T cells carrying two different TCRs on their surface, thereby allowing for the independent recognition of two cross-reactive peptide-MHC complexes. Dual TCR expression on a single T cell can occur in the absence of allelic exclusion, where the non-selected TCR has been found to mount functional responses in the periphery (18-20). Alternatively, dual TCR expression has been suggested to occur through TCR sharing, where two clonotypically different T cells transfer cell surface TCRs amongst each other (21). In this study we are examining T cells expressing receptors that are individually cross-reactive towards LCMV-GP34 and VV-A11R.
The ligand requirements for cross-reactive TCR recognition may depend on structural similarities between the different peptide-MHC ligands, or structural reconfiguration of the peptide and/or the MHC after binding TCR (22). However, the degree of structural homology required prior to TCR engagement remains unclear, as many studies have been directed towards peptide epitopes with at least 50% sequence homology (22-24). Furthermore, many studies of molecular mimicry have characterized aberrant auto-reactive immune responses, which have different affinities and functional characteristics (22, 24-26).
The recognition of cross-reactive peptide-MHC complexes may also occur through structural rearrangements of TCR (26, 27). One example is the cross-reactive TCR BM3.3, which was found to modify its CDR loops to accommodate three different peptides, all presented by H-2Kb, using the same overall docking strategy (26). Another example is the alloreactive 2C TCR, which through globally repositioning of its TCRα and TCRβ chains, is able to recognize a self and foreign peptide presented by two different MHC molecules (27).
To study protective heterologous immune responses towards two unrelated pathogens, we examined the mechanistic basis for the LCMV-VV cross-reactive T cell response by comparing the recognition determinants for LCMV-GP34 and VV-A11R using site-specific mutations at the predicted non-MHC binding residues in both epitopes. We studied the impact of these mutations by using a combination of intracellular cytokine staining (ICS), MHC tetramer staining, and in vivo cytotoxicity assays. The studies were conducted on either acute LCMV-infected mice or GP34-A11R cross-reactive T cell lines, which were derived from LCMV-immune mice and expanded in vitro with peptide-pulsed targets. In addition, we determined X-ray crystal structures of the Kb-LCMV-GP34 and Kb-VV-A11R peptide-MHC complexes recognized by cross-reactive T cells, and identified peptide sequence requirements for cross-reactivity. This analysis revealed that shared aspects of the Kb-LCMV-GP34 and Kb-VV-A11R molecular surfaces underlie the protective heterologous immune response between LCMV and VV. Using the functional and structural requirements for the cross-reactive LCMV-VV T cell response, we were able to predict and generate a cross-reactive response towards a variant of an unrelated chicken ovalbumin peptide. Overall our results highlight how dissimilar peptides may be cross-reactive and how one can use structural information to predict or engineer cross-reactivity.
Materials and Methods
Production of Class I H-2Kb complexes
Extracellular domains of the murine MHC class I H-2Kb heavy chain and full length human light chain β2 – microglobulin, were expressed separately as inclusion bodies in Escherichia coli and were folded in vitro by dilution in the presence of excess peptide, as previously described for human class I MHCs (28). Synthetic peptides purified by reverse phase-HPLC were purchased from 21st Century Biochemicals. Folded H-2Kb monomers were purified by anion exchange chromatography on Poros HQ columns (Roche) using a gradient of NaCl from 0-0.5M in 20 mM Tris buffer (pH 8.0). The concentration of each H-2Kb monomer was calculated by absorbance spectroscopy after anion exchange chromatography using ε280 = 74955 cm−1 M−1 for H-2Kb heavy chain, ε280 = 20003 cm−1 M−1 for β2 – microglobulin light chain and varied ε280 for peptides depending on sequence. Purified H-2Kb monomers were adjusted to a concentration of 10-20 mg/mL using regenerated cellulose filters (Amicon) and stored at −80°C.
RMA-S Stabilization Assay
Peptide-MHC binding assays were performed using transporter associated with antigen (TAP)-deficient RMA-S cells as previously described (29). Briefly, 5e5 RMA-S cells were pulsed with different concentrations of peptide in RPMI medium at 27°C overnight in a 5% CO2 incubator. Cells were subsequently washed to get rid of excess peptide and incubated in RPMI medium at 37°C for 1 hour. Cells were washed twice with PBS and surface expression of H-2Kb was detected by using anti-H-2Kb (clone AF6-88.5, BD Pharmingen). Thereafter, cells were washed twice with staining buffer and fixed in Cytofix (BD Pharmingen). Samples were analyzed using a Becton Dickinson LSRII flow cytometer (BD Biosciences) and FlowJo software (Tree Star).
Analysis of Peptide-MHC Complex Stability using Differential Scanning Fluorimetry
Peptide-MHC complex stability was assessed using differential scanning fluorimetry (thermofluor) which utilizes SYPRO Orange (Invitrogen) as previously described (30). Briefly, purified peptide-MHC complexes and SYPRO Orange dye were added to a 96-well PCR plate and sealed using optical sealing tape (Bio-Rad). A real-time PCR detection system (Bio-Rad) was utilized to adjust the temperature from 20°C to 95°C and the fluorescence of the sample was recorded every 0.5°C. The melting temperature for each peptide-MHC complex was determined based on the maximum of the first derivative of the melting curve.
Isolation of antigen-specific CTL
LCMV (Armstrong strain), an RNA virus in the Old World arenavirus family, was propagated in BHK-21 baby hamster kidney cells as previously described (31). VV (Western Reserve) was propagated in mouse L929 cells as previously described (32). C57BL/6 (B6) mice were infected intraperitoneally (i.p.) with a non-lethal dose of 5 × 104 PFU of LCMV or 106 PFU of VV as previously described (31, 32). Mice were considered immune 6 weeks or more postinfection. Control naïve mice were age matched and were not inoculated. Mice were considered immune at greater than 6 weeks after infection (12). Splenocytes from LCMV-immune mice were co-cultured with mouse RMA cells that were pulsed with 1 μM VV-A11R peptide, washed and then γ-irradiated (3000 rads) as previously described (12). RMA is a Kb-positive, Rauscher virus-induced, T-lymphoma cell line of B6 origin. Briefly, the co-culture of splenocytes and VV-A11R-pulsed RMA cells were grown in RPMI medium supplemented with 100 U/ml penicillin G, 100 μg/ml streptomycin sulfate, 2 mM L-glutamine, 10 mM HEPES, 1 mM sodium pyruvate, 0.1 mM MEM nonessential amino acids, 0.05 mM β-mercaptoethanol, and 10% FBS for 5 days at 37°C at 5% CO2. Following this initial culture period, cells were harvested and stimulated with VV-A11R peptide-pulsed RMA cells in the presence of 10% BD T-Stim (BD Biosciences), an IL-2 culture supplement. VV-A11R peptide-pulsed RMA stimulation was repeated every 4 to 5 days. After 30 to 35 days of stimulation (six or seven stimulations), T-cell lines were characterized by intracellular cytokine staining or MHC tetramer staining.
Cell surface and MHC staining by flow cytometry
Cell suspensions were passed through lympholyte M gradient (Cedarlane) to exclude dead cells and subsequently washed and incubated in staining buffer (phosphate-buffered saline containing 1% FBS and 0.2% sodium azide). Cell suspensions were incubated in staining buffer containing anti-mouse CD16/CD32 (Fc-block, clone 2.4G2). Cells were washed once with staining buffer and then stained with H-2Kb tetramers for 40 min followed by cell-surface antibody staining with anti-CD8β (clone 53-6.8), anti-CD44 (clone IM7) and Live/Dead aqua exclusion dye (Invitrogen) for 20 min. Thereafter, cells were washed twice with staining buffer and fixed in Cytofix (BD Pharmingen). Samples were analyzed using a Becton Dickinson LSRII flow cytometer (BD Biosciences) and FlowJo software (Tree Star).
Intracellular cytokine staining (ICS)
A suspension of 106 cells was stimulated with 1 μM synthetic peptide or a medium only control Stimulations were performed for 5 h at 37°C in a total volume of 200 μl RPMI medium supplemented with 10% FBS, 10 U/ml of human recombinant interleukin-2 (IL-2), and 0.2 μM of brefeldin A (GolgiPlug; BD Pharmingen). After incubation, cell-surface antibody staining with anti-CD8β (clone 53-6.8), anti-CD44 (clone IM7), and Live/Dead aqua (Invitrogen) was performed. Thereafter, cells were washed twice with staining buffer, and then fixed and permeabilized (Cytofix/Cytoperm; BD Pharmingen). Intracellular-cytokine-producing cells were detected with PE-labeled anti-mouse interferon-gamma (IFN-γ, clone XMG1.2) and APC-labeled anti-mouse tumor necrosis factor alpha (TNF-α, clone MP6-XT22) monoclonal antibodies. Antibodies were purchased from BD Pharmingen. The samples were analyzed as described above for cell surface staining.
In vivo CTL assay
RBC-lysed, single-cell spleen suspensions from naive B6 mice were pulsed at 107 cells/ml with 1 μM peptide RPMI containing 10% FBS for 45 min at 37°C. For a four peak CTL assay (Figure 1B), each peptide-pulsed spleen cell population was labeled with a dilution of carboxyfluorescein succinimidyl ester (CFSE, Invitrogen) at 5μM, 2.5μM, 1.25nM and 0.63nM along with 0.5 μM of 7-hydroxy-9H-(1,3-dichloro-9,9-dimethylacridin-2-one) (DDAO, Invitrogen) at a cell density of 2 × 107 cells/ml in HBSS. Labeling was stopped by addition of excess HBSS. For an eight peak CTL assay (Figure 3A), 1μM of Violet (Invitrogen) was used in addition to the CFSE and DDAO. One million cells of each peptide-pulsed population were mixed together, and 500μL was injected intravenously (i.v.) into LCMV infected and uninfected syngeneic mice.
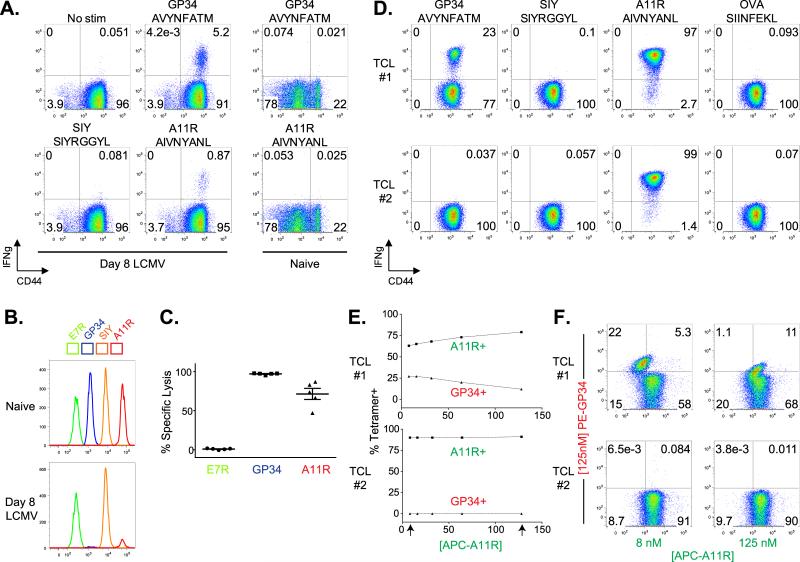
Figure 1. VV-A11R and LCMV-GP34 are Cross-reactive CD8+ T cell Epitopes Elicited by LCMV Infection.
(A). CD8+ T cells from acute day 8 LCMV-infected mice (n=15) and uninfected mice (n=3) were isolated and analyzed for IFNγ production using an ICS assay, with results from a representative mouse presented as dot plots. (B and C). A four peak in vivo cytotoxicity assay was set up using two different fluorophores as described in the materials and methods. Briefly, naïve B6 splenocytes were peptide-pulsed and then labeled with a dilution of CFSE and DDAO. Peptide-pulsed, and CFSE and DDAO double-labeled cells were injected (i.v.) into uninfected (naïve, n=3) or LCMV-infected (D8 LCMV, n=5) mice. Three hours after transfer, spleens were harvested and analyzed by flow cytometry. Plots from a representative infected and naïve mouse are shown in B. Percent specific lysis was calculated based on the “SIY” peptide-pulsed population and plotted in C. (D). Splenocytes from an LCMV-immune mouse were isolated and expanded in vitro using VV-A11R peptide-pulsed targets to generate a GP34-A11R cross-reactive T cell line (TCL #1) and a control T cell line (TCL #2). T cell lines were analyzed for IFNγ production in response to the indicated peptides, with the results presented as dot plots. The results are representative of three different T cell lines. (E and F). The T cell lines from panel d were stained with R-PE or APC-labeled streptavidin-based Kb tetramers folded with VV-A11R or LCMV-GP34 peptides. Double MHC tetramer staining experiments were set up using a PE-labeled LCMV-GP34-Kb tetramer and an APC-labeled VV-A11R-Kb tetramer. The VV-A11R tetramers were used at concentrations starting at 125 nM titrated down two-fold to 8 nM while the LCMV-GP34 tetramer concentration was maintained constant at 125 nM. The percent tetramer positive for every concentration is plotted in f.
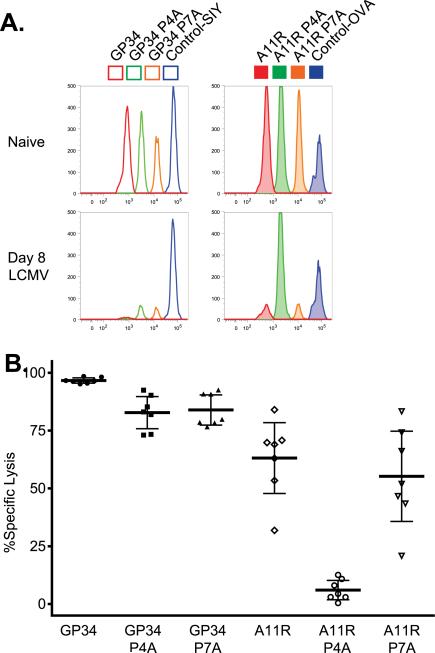
Figure 3. Cross-reactive T cells Require the P4N for Specific Lysis of VV-A11R Peptide-Pulsed Targets.
(A and B). To test the necessary contact residues at P4 and P7 in vivo, an eight peak cytotoxic T lymphocyte assay was set up like previously described in Figure 1. Briefly, the 8 peak assay was split into two simultaneous four peak assays using dilutions of either CFSE vs. DDAO or CFSE vs. violet as described in the Materials and Methods. Target cell populations were injected (i.v.) into acute LCMV-infected mice (n=7) or uninfected mice (n=3). Percent specific lysis in panel F was calculated based on either the “SIY” peptide-pulsed population or the “OVA” peptide-pulsed population.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism software. Comparisons between two groups (mean values) were performed using a tukey's test. P values less than 0.05 were considered statistically significant.
Crystallization and data collection
H-2Kb complexes were crystallized in 14-16% PEG 8K, 0.1 M Na cacodylate, 0.2 M magnesium acetate at 4°C. Typically, 1 μl of a 10 mg/ml protein solution in 10 mM Tris Cl (pH 7.0) was mixed with equal volumes of the crystallization reservoir. Crystals were transferred to a reservoir solution containing 30% glycerol and flash frozen using liquid nitrogen. X-ray diffraction data were collected as 1° oscillations at 100 K with 1.08 Å radiation at beamline X29A in the National Synchrotron Light Source. Data were processed and scaled using HKL2000 (33). Data collection and refinement statistics are shown in Table 1.
Structure determination and refinement
Phases were estimated by molecular replacement using PHASER (34) with PDB ID# 1S7R, peptide GNYSFYAL removed) as a search model. To validate the molecular replacement solution, composite omit maps (CNS) (35) were generated and inspected using COOT (36). During refinement, the peptide was omitted to reduce model bias. The peptide was later built into the observed electron density, which was unambiguous within the peptide binding groove. Crystallographic rigid body, positional, B-factor, and TLS refinement was performed in PHENIX (37). Ramachandran statistics showed all residues in the allowed regions. Diffraction data and coordinates were deposited into the Protein Data Bank (PDB ID# 3TID and 3TIE) (38).
Results
CD8+ T cells Elicited by LCMV Infection Recognize Cross-reactive VV-A11R and LCMV-GP34 Epitopes
Previous work indicated that VV challenge of LCMV-immune mice stimulates expansion of T cell populations recognizing LCMV-GP34 (15). To show that these cross-reactive T cell responses were elicited in mice infected with only LCMV, we infected B6 mice with LCMV for 8 days, and at the peak of the immune response, performed an ex vivo intracellular cytokine staining (ICS) assay for cross-reactive VV-A11R T cell responses in immunized as compared to control naïve B6 mice. The results from a representative LCMV-infected mouse show an IFNγ response to the cross-reactive VV-A11R peptide at ~1% of total CD8+ T cells, with only background levels of VV-A11R cross-reactivity observed in a representative naïve mouse (Figure 1A). A T cell response to the control-SIY peptide was not observed, and the response to cognate LCMV-GP34 was observed at ~5% of total CD8+ T cells (Figure 1A). Both LCMV-GP34 and VV-A11R form stable complexes with H-2Kb, as judged by a thermal denaturation assay (30) (Supplemental Figure 2). Tm values for Kb-LCMV-GP34 and Kb-VV-A11R complexes (54°C and 53°C, respectively) were at least as high as for a control Kb-OVA complex (51.5 °C), and substantially higher than for a known weak-binding antigenic peptide complex Db-SMCY (42.5 °C) (39).
To determine if VV-A11R is recognized in vivo in acute LCMV-infected mice, we set up an in vivo cytotoxicity assay using uninfected mice as controls. Briefly, splenocytes from syngeneic B6 mice were isolated, peptide-pulsed and then fluorescently-labeled with different dilutions of carboxyfluorescein succinimidyl ester (CFSE) and a constant concentration of 7-hydroxy-9H-(1,3-dichloro-9,9-dimethylacridin-2-one) (DDAO) to monitor the target cell populations. The peptide-pulsed, DDAO- and CFSE-double labeled target populations were then injected intravenously into either acute LCMV-infected mice or uninfected controls. The cytolytic response was monitored by flow cytometry 3 hours after injection. Flow cytometry data from a representative mouse are shown in Figure 1B, with percent specific lysis calculated for 5 mice plotted in Figure 1C. The results show that both VV-A11R and LCMV-GP34 peptide-pulsed targets are specifically lysed in LCMV-infected mice (Figure 1B). Killing was not observed in naïve mice, and no killing was observed with the non-cross-reactive peptide VV-E7R (Figure 1B). These data clearly show that LCMV infection generates a specific T cell population which is cross-reactive in vivo against VV-A11R.
To study the cross-reactive T cell response specifically between VV-A11R and LCMV-GP34, we generated cross-reactive T cell lines, which were tested for TCR specificity using an ICS assay. From an LCMV-immune mouse, we generated a VV-cross-reactive T cell line (TCL), which was cross-reactive between LCMV-GP34 and VV-A11R (TCL #1). We also generated a non-cross-reactive TCL, also from an LCMV-immune mouse, responding to VV-A11R but not to LCMV-GP34 (TCL #2) for use as a control. No VV-A11R reactive TCL could be generated from LCMV-immune mice. We performed ICS assays to show specific cross-reactivity between VV-A11R and LCMV-GP34. For the ICS assay, the peptides utilized were VV-A11R, LCMV-GP34 and the control peptides SIY and OVA. The results indicate that nearly the entire (97%) memory phenotype (CD44+) CD8+ T cell population in the cross-reactive TCL #1 produced IFNγ in response to the VV-A11R peptide (Figure 1D). In response to the LCMV-GP34 peptide, 23% of these T cells produced IFNγ, and this was not observed in the control T cell line (TCL #2). Because essentially the entire T cell population in TCL #1 reacted against VV-A11R, any T cells that were reactive against LCMV-GP34 must therefore have been cross-reactive against VV-A11R.
To test for the presence of cross-reactive TCRs, we designed a MHC tetramer competition experiment. For the MHC tetramer competition assay, we utilized a fixed concentration of 125 nM LCMV-GP34 tetramer and 2-fold dilutions of VV-A11R tetramer ranging from 125 nM down to 8 nM and stained the GP34-A11R cross-reactive TCL (TCL#1) and control TCL (TCL#2) (Figure 1E,F). We reasoned that if cross-reactive TCRs are present in the cross-reactive TCL, the binding of the VV-A11R tetramer would alter the binding of the LCMV-GP34 tetramer, due to tetramer competition for the same pool of cross-reactive TCRs. However, if two non-cross-reactive TCRs are present, the binding of LCMV-GP34 tetramer and the VV-A11R tetramer would be mutually exclusive, due to two different populations of cell surface TCRs. In this experiment, the LCMV-GP34 tetramer was prepared using streptavidin coupled to R-phycoerythrin (PE) with the VV-A11R tetramer prepared using streptavidin coupled to allophycocyanin (APC) so that binding of both tetramers could be monitored simultaneously. The overall MHC tetramer binding MFI for APC-A11R (A11R+) versus PE-GP34 (GP34+) is plotted in Figure 1E. The arrows represent two representative FACs plots for 8 nM APC-A11R or 125 nM APC-A11R versus the fixed amount of 125 nM PE-GP34 (shown in Figure 1F). With increasing concentrations of the APC-A11R tetramer, specific GP34 tetramer staining of the cross-reactive TCL (#1) decreased (Figure 1E, top panel), whereas the specific GP34 tetramer staining was not observed at any concentration of APC-A11R tetramer (Figure 1E, bottom panel). This demonstrates the presence of cross-reactive TCRs on the cross-reactive TCL (#1) which dually recognizes VV-A11R and LCMV-GP34.
Recognition Determinants for the Cross-reactive T cells
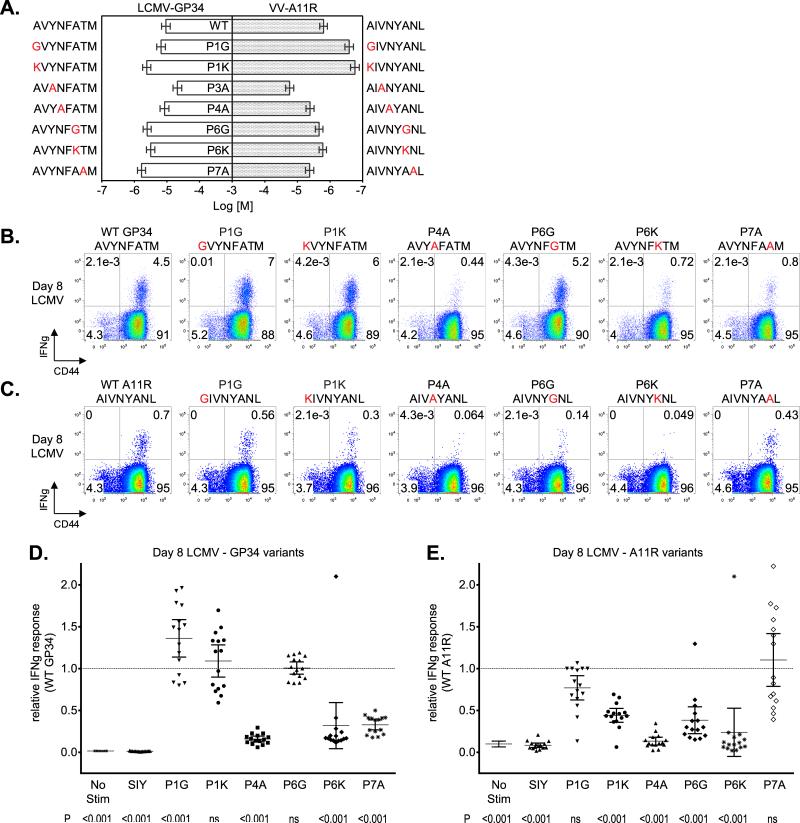
To further characterize the cross-reactive T cell responses between LCMV-GP34 and VV-A11R, we compared the recognition determinants for LCMV-GP34 and VV-A11R individually. We reasoned that a comparison of the important TCR contact residues for both epitopes would highlight similarities and differences in recognition determinants, and this might hint at the mode(s) of TCR engagement. Peptide binding to MHC class I H-2Kb depends on the peptide positions 5 (P5) and 8 (P8) as primary anchors, and peptide positions 2 (P2) and 3 (P3) as secondary anchors (40, 41). Alanine substitutions were generated at the predicted non-MHC binding residues (underlined) in LCMV-GP34 (AVYNFATM) and VV-A11R (AIVNYANL). We modified the other positions and included the P3 secondary anchor position, in order to test the potential TCR contact residues. Residues with alanines present already were mutated to either glycine for a conservative substitution or to lysine for a non-conservative substitution.
We tested the substituted LCMV-GP34 and VV-A11R peptides for binding to Kb using an RMA-S stabilization assay (29). EC50 plots are shown in Figure 2A. We observed that the substituted LCMV-GP34 and VV-A11R peptides do not bind significantly differently from the WT peptides (Figure 2A), with the exception of the P3A variants, which were not utilized further.
Figure 2. Recognition Determinants for Cross-reactive T cells.
Recognition determinants for CD8+ T cell responses against VV-A11R and LCMV-GP34 were mapped by mutational analysis. Non-H-2Kb binding residues from LCMV-GP34 and VV-A11R, which include P1, P4, P6 and P7, were mutated to alanine, with alanine residues mutated to either glycine or lysine. (A). An RMA-S stabilization assay was utilized to measure relative binding affinity of WT and variant peptides from LCMV-GP34 and VV-A11R. EC50 values were calculated for each peptide titration series (n=3) and represented as bar graphs. For each LCMV-GP34 and VV-A11R peptide variant, the full sequence and abbreviation is shown with the substituted residues colored in red. (B and C). CD8+ T cells from acute LCMV-infected mice were tested for IFNγ production in response to 1 μM of the indicated peptides with representative results from a single mouse shown as dot plots (n=15). (D and E). IFNγ production results from acute LCMV-infected mice (n=15) were normalized to WT LCMV-GP34 for the LCMV-GP34 variants or WT VV-A11R for the VV-A11R variants and shown in the graph. The p values for IFNγ production were calculated using the means and indicated below for each peptide
Next, we performed an ICS assay to test the substituted peptides from LCMV-GP34 and VV-A11R for reactivity with T cells from acutely LCMV-infected mice. Flow cytometry data from a representative mouse are shown for the LCMV-GP34 and VV-A11R peptide variants in Figures 2B and 2C, respectively. Relative T cell responses for 15 mice are shown in Figures 2D and 2E, for the LCMV-GP34 and VV-A11R peptide variants, respectively. For each mouse, the IFNγ production in response to a given peptide was normalized to the WT GP34 or WT A11R response (these responses varied from 5-10% and 0.5-2.5% for WT-GP34 and WT-A11R, respectively). Substitutions in LCMV-GP34 at the P4N, the P6A and the P7T residues resulted in loss of IFNγ production (Figures 2B and 2D). For LCMV-GP34 P6A, glycine (P6G) did not alter T cell recognition, suggesting that the P6A side chain is not a direct TCR contact (Figure 2B). However, P6A substitution by lysine (P6K) reduced T cell recognition, suggesting TCR might be in proximity of the P6 position so that a large residue could interfere with productive binding, or possibly that the lysine substitution induces a conformational change. As for the cross-reactive ligand VV-A11R, a different pattern was observed, where alanine substitution at the P4 and P6 but not P7 positions resulted in loss of IFNγ production, and P1A substitution resulted in loss of IFNγ production for lysine (P1K) but not glycine (P1G) substitutions (Figures 2C and 2E). Overall, the effects of substitution were similar at the P4 position, but different elsewhere. The shared recognition determinant at P4 suggests that cross-reactive TCRs utilize this residue for T cell responses. At P6, both VV-A11R P6G and P6K substitution resulted in decreased IFNγ production, differently from LCMV-GP34 where the P6G mutation had no effect. Finally, the VV-A11R P7 residue was not important for IFNγ production on average, but there was a broad range of reactivity amongst the different mice tested. In summary, cognate recognition of LCMV-GP34 is dependent on P4N and P7T whereas cross-reactive recognition of VV-A11R depends mainly on P4N.
To determine whether the P4 and the P7 residues are required for the cross-reactive recognition of VV-A11R in vivo, we performed a dual label in vivo cytotoxicity assay as described in the methods, utilizing dilutions of CFSE and DDAO for target cells pulsed with the GP34 variants or dilutions of CFSE and Celltrace violet for cells pulsed with the A11R variants (Figures 3A and 3B). Flow cytometry data from a representative naïve mouse and LCMV-infected mouse is shown in Figure 3A, with percent specific lysis calculated for 7 LCMV-infected mice plotted in Figure 3B. We asked if CD8+ T cells from acute LCMV-infected mice could specifically lyse WT, P4A and P7A variants of LCMV-GP34 and VV-A11R, using the non-cross-reactive peptides OVA and SIY as controls. The P4A substitution in VV-A11R resulted in a log reduction in percent specific lysis (Figure 3B), showing that T cells which cross-react with VV-A11R from LCMV-infected mice require the P4N in VV-A11R for specific lysis in vivo. These functional data demonstrate that P4N is a major recognition determinant that is required for the polyclonal LCMV-cross-reactive T cell response against VV-A11R. To confirm that cross-reactive T cell responses towards VV-A11R are not unique to one T cell clone, we evaluated the TCR repertoire from LCMV-infected mice that respond towards VV-A11R (Supplemental Figure 1). A polyclonal response including at least six different TCRβ genes was observed, indicating that the polyclonal, cross-reactive T responses towards VV-A11R require the P4N as a major recognition determinant
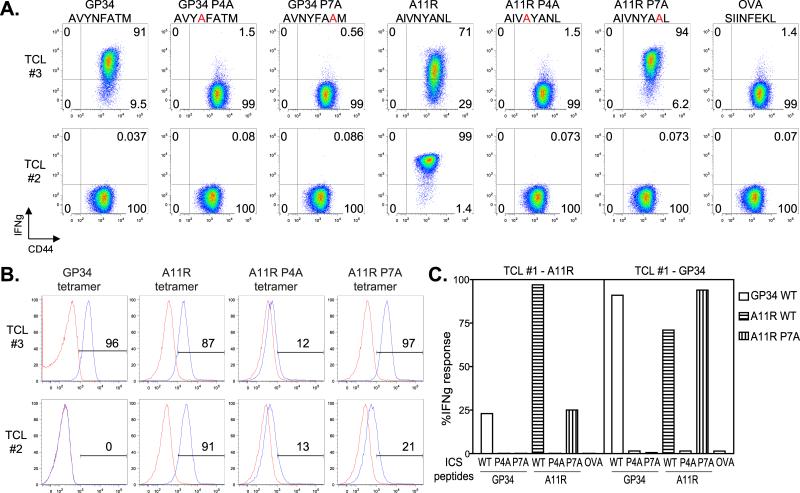
Although a clear role for the P4N in LCMV-cross-reactive T cell responses against VV-A11R was observed in vivo, the experiments reported in Figure 2 used highly polyclonal populations, making it difficult to evaluate the contribution of cross-reactive TCRs relative to recognition by different T cell populations. Therefore, we tested for the P4N dependence on cross-reactive T cell lines that have narrowed TCR repertoires due to repeated in vitro expansion (results from previous in vitro expanded TCLs, Figure 5). We performed ICS assays followed by tetramer staining assays using the P4A and P7A variants of both LCMV-GP34 and VV-A11R. For both assays, we tested the variant peptides on an LCMV-GP34 and VV-A11R cross-reactive T cell line (TCL #3) and a control T cell line (TCL #2). The results of the ICS assay show that for the cross-reactive TCL #3, IFNγ production in response to LCMV-GP34 was dependent on both P4N and P7T, while recognition of the VV-A11R was dependent only on P4N (Figure 4A), consistent with the in vivo results. In contrast, in the control TCL #2, recognition of VV-A11R was dependent on both P4N and P7N.
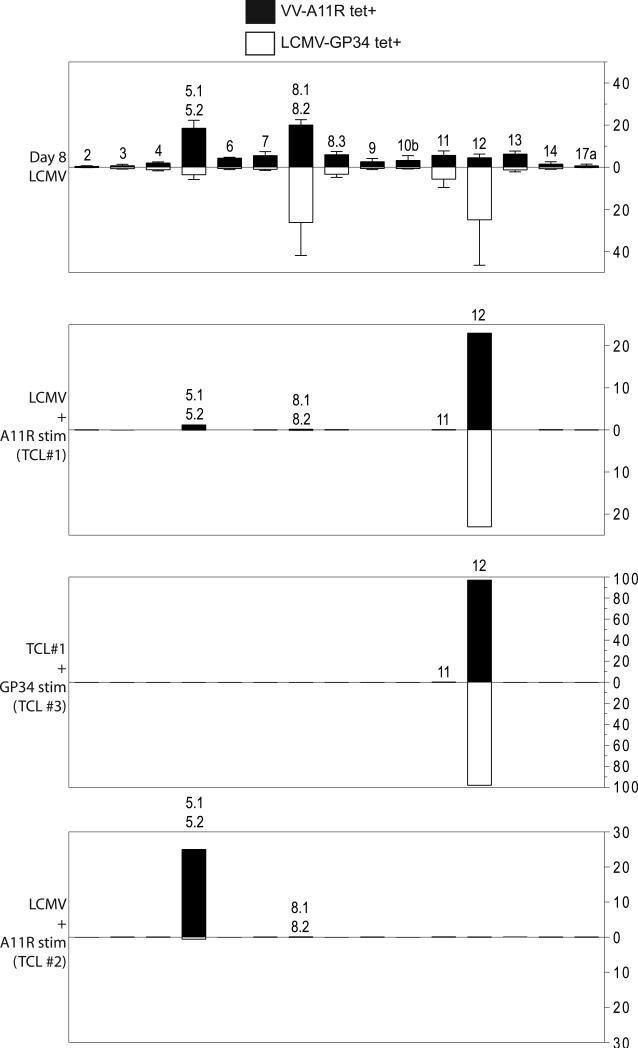
Figure 5. Analysis of the LCMV-GP34 specific versus the LCMV-GP34 and VV-A11R cross-reactive TCR repertoires.
CD8+ T cells were isolated from either acute day 8 LCMV-infected mice (n=4) or from antigen-specific T cell lines (TCLs #1, 2, 3) used in Figures 1 and 4. CD8+ T cells were stained using 125 nM APC-labeled strepavidin-based Kb tetramers folded with VV-A11R or LCMV-GP34 peptides in combination with FITC/PE-labeled TCR Vβ antibodies. The percentage of each TCR Vβ utilized in the recognition of either LCMV-GP34 tetramer (open bars) or VV-A11R tetramer (closed bars) is plotted.
Figure 4. Recognition Determinants for GP34-A11R Cross-reactive T cell receptors.
GP34-A11R cross-reactive (TCL #3) and control (TCL #2) T cell lines were generated as described for Figure 1. (A). Cross-reactive T cell lines were tested for IFNγ production against WT, P4A or P7A variant peptides from LCMV-GP34 and VV-A11R along with the control-peptide, OVA. The results are represented as dot plots and are representative of three independent experiments. (B). Single MHC tetramer staining experiments were performed on both the GP34-A11R cross-reactive (TCL #3) and control (TCL #2) T cell lines. Briefly, WT LCMV-GP34, VV-A11R (WT, P4A, and P7A) and control-OVA peptide-MHC tetramers were tested at 125 nM with the results represented as histograms. In each histogram, the indicated tetramer (blue trace) was overlaid with the control-OVA tetramer (red trace). (C). The GP34-A11R cross-reactive T cell line from Figure 1 (TCL #1) was re-stimulated in vitro using LCMV-GP34 peptide-pulsed targets, to selectively expand the GP34-A11R cross-reactive population (TCL #1 - GP34). For comparison, TCL #1 was also re-stimulated in vitro using VV-A11R peptide-pulsed targets (TCL #1 - A11R). After two rounds of stimulation, CD8+ T cells were analyzed for IFNγ production in response to WT, P4A and P7A variants of both LCMV-GP34 peptide and VV-A11R peptide along with the control-OVA peptide. The data is represented in the bar graph.
For the tetramer staining assay, we monitored binding of LCMV-GP34 WT, VV-A11R WT, VV-A11R P4A, and VV-A11R P7A or control OVA tetramers on the LCMV-GP34 and VV-A11R cross-reactive (TCL #3), and a control (TCL #2) T cell lines (Figure 4B). The tetramer staining patterns paralleled the results from the ICS assay. The GP34-A11R cross-reactive TCL #3 stains with WT GP34, WT A11R and A11R P7A tetramers, while the control T cell line (TCL #2) stains only with the WT A11R tetramer (Figure 4B). Taken together, these results show that cross-reactive TCRs that recognize LCMV-GP34 depend on the P4N for the cross-reactive recognition of VV-A11R.
Using the notion that the LCMV-cross-reactive T cell response directed towards VV-A11R is dominated by the P4N, we hypothesized that an increased LCMV-GP34, VV-A11R, cross-reactive T cell response would correspond with an increased dependence on the P4N. To address this, we used the GP34-A11R cross-reactive T cell line from Figure 1 (TCL #1), which was nearly 100% reactive towards VV-A11R, and we selectively expanded the population that also recognizes LCMV-GP34 (23%) by stimulating in vitro with the LCMV-GP34 peptide for two additional passages (TCL #1-GP34, Figure 4C, right panel). For comparison, we stimulated the same T cell line using the VV-A11R peptide for two additional passages (TCL #1-A11R, Figure 4C, left panel). We tested for functional responses by performing an ICS for IFNγ production in response to WT, P4A or P7A variants of the LCMV-GP34 or the VV-A11R peptides, with OVA used as a control (Figure 4C). Cross-reactive T cells stimulated with A11R only responded to LCMV-GP34 WT (~25%, plain bars), VV-A11R WT (~100%, horizontally striped bars) and VV-A11R P7A (~25%, vertically striped bars). After the same TCL was expanded in vitro with the LCMV-GP34 peptide to select for the LCMV-GP34 and VV-A11R cross-reactive T cells, we observed an increase in IFNγ production in response to LCMV-GP34 WT and VV-A11R P7A. By skewing the T cell response in favor of LCMV-GP34 reactivity in this entirely VV-A11R cross-reactive TCL, we observed a proportional decrease in the dependence on the P7 residue in the VV-A11R peptide. These results indicate that VV-A11R cross-reactive T cells that respond towards LCMV-GP34 do not require the P7 residue.
To address whether cross-reactive TCR recognition could also depend on contacts with the H-2Kb molecule, we tested previously identified LCMV- and VV-derived Kb epitopes which have the same important P4N or the P7 residue with either LCMV-GP34 or VV-A11R (Supplementary Table 1). These peptides have all been tested for T cell responses in ICS assays, which measured IFNγ production on T cells from either acutely LCMV-infected mice or GP34-A11R cross-reactive TCLs. Cross-reactive T cell responses have not been observed with any of these peptides (data not shown), indicating that the shared aspects leading to cross-reactive T cell responses depend predominantly on peptide contacts with TCR. This result highlights the importance of the P4N that is shared between LCMV-GP34 and VV-A11R.
TCR Repertoire Overlap between the LCMV-GP34-specific and the LCMV-GP34-cross-reactive T cell Populations
We evaluated the relationship between T cell populations specific for LCMV-GP34 and populations cross-reactive with both LCMV-GP34 and VV-A11R, using TCR Vβ repertoire analysis in combination with Kb-VV-A11R or Kb-LCMV-GP34 tetramer staining. We reasoned that the LCMV-GP34 and VV-A11R cross-reactive T cell population should be a subset of the total T cell population recognizing LCMV-GP34. To address this, we compared the TCR Vβ repertoire of LCMV-GP34 and VV-A11R tetramer-positive CD8+ T cells from acute LCMV-infected mice, two different GP34-A11R cross-reactive TCLs and a VV-A11R cross-reactive T cell line that does not react towards LCMV-GP34 (Figure 5). Acute LCMV infection generates a broad repertoire of TCR Vβs that are utilized in the recognition of LCMV-GP34 (Day 8 LCMV, open bars). The population recognizing VV-A11R also utilized a broad repertoire of TCR Vβs (Day 8 LCMV, closed bars). However, when we expanded the VV-A11R cross-reactive T cell population from an LCMV-immune mouse using VV-A11R peptide-pulsed targets, we observed narrowing of the TCR Vβ repertoire to mainly TCR Vβ 12 for the recognition of both LCMV-GP34 and VV-A11R (TCL #1, Figure 5). Interestingly, when we selectively expanded the GP34-A11R cross-reactive T cells from TCL #1 by re-stimulating T cells with LCMV-GP34 peptide-pulsed targets, we observed exclusive expansion of the TCR Vβ 12 population (TCL #3, Figure 5). This finding clearly indicates that GP34-A11R cross-reactive T cells from TCL #1 and TCL #3 are utilizing TCR Vβ 12 for the recognition of LCMV-GP34 and VV-A11R. To confirm that TCR Vβ 12 is utilized in LCMV-GP34 and VV-A11R cross-reactive T cell responses, we tested another cross-reactive TCL (TCL #2). In this LCMV-GP34 null and VV-A11R cross-reactive TCL, we observed utilization of TCR Vβ 5.1 5.2 for the recognition of VV-A11R tetramer (Figure 5, TCL #2, closed bars). LCMV-GP34 tetramer staining was absent, which is consistent with the absence of LCMV-GP34 reactivity (see TCL #2, see Figure 4A). Together, these results confirm that the subset of T cells that cross-react against LCMV-GP34 and VV-A11R are derived from a subset of the total population of LCMV-specific T cells elicited by LCMV infection. More importantly, the results highlight TCR Vβ 12 as a potential target for screening GP34-A11R cross-reactive T cell responses in vivo.
Analysis of the Kb-VV-A11R and Kb-LCMV-GP34 Molecular Surfaces
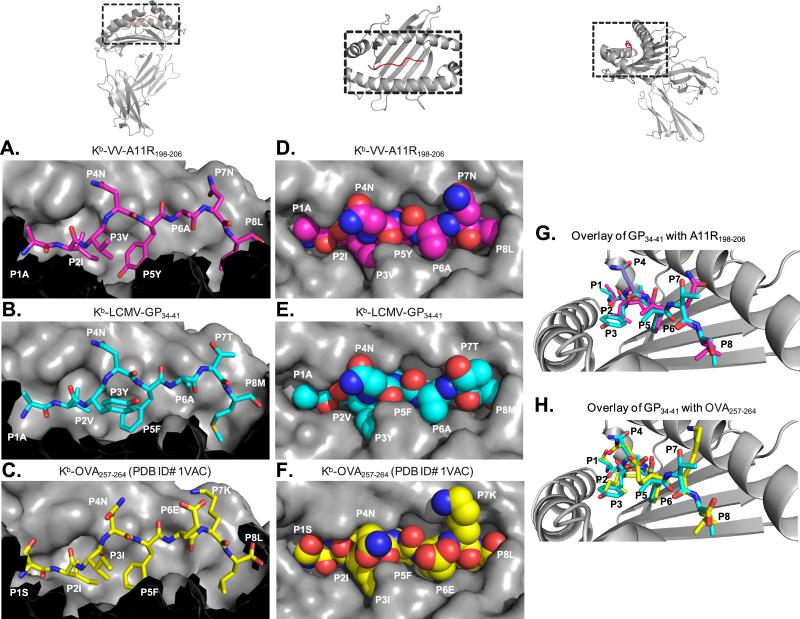
To understand the dependence of P4N for cross-reactive T cell recognition, we determined the crystal structures of Kb-VV-A11R (PDB ID# 3TIE) and Kb-LCMV-GP34 (PDB ID# 3TID) complexes, using data extending to 2.2Å and 1.7Å resolution, respectively (Table 2). The overall structures of Kb-VV-A11R and Kb-LCMV-GP34 differ very little, with an RMSD of 0.472 Å. A side profile comparison of the antigen-binding cleft from Kb-VV-A11R and Kb-LCMV-GP34 reveal that both VV-A11R and LCMV-GP34 are bound and presented similarly in Kb (Figures 6A and 6B). Both peptides at P4 and P7 have solvent exposed side chains that protrude away from the Kb heavy chain and are available for TCR contact (Figures 6A and 6B). The main chain at P6 also appears solvent accessible, while side chains at P2, P3, P5 and P8 are buried in the Kb molecule (Figures 6A and 6B). In the top views of Kb-VV-A11R and Kb-LCMV-GP34 (Figures 6D and 6E), it is clear that most of the surfaces of LCMV-GP34 and VV-A11R are virtually identical when presented by Kb, particularly at the asparagine at P4, which appears to be in the same conformation (Figure 6G). The P7 side chain is a notable exception, with differences clearly observed when the Kb-VV-A11R and the Kb-LCMV-GP34 structures are overlaid (Figure 6G).
Table 2.
Data Collection and Refinement Statistics.
| Data Collection | Kb-LCMV-GP34 (AVYNFATM) | Kb-VV-A11R (AIVNYANL) |
|---|---|---|
| Space group | C2 | P21 |
| Wavelength | 1.0809 | 1.0809 |
| Unit cell parameters | a= 172.261, b= 47.649, c= 70.539 Å, β= 106.39° | a= 69.264, b= 84.710, c= 87.895 Å, β= 98.12° |
| Resolution (Å) | 50-1.7 | 50-2.2 |
| Rsym (%) | 5(26) | 10(28) |
| I/sI | 22.1 (7.8) | 15.1 (5.7) |
| Completeness (%) | 98 (96.7) | 99.4 (99.3) |
| Redundancy | 3.8 (3.8) | 6.2 (6.3) |
| Refinement | ||
| Resolution (Å) | 40-1.7 | 40-2.2 |
| No. reflections | 59,404 | 50,967 |
| Rwork/Rfree (%) | 0.167/0.190 | 0.177/0.219 |
| No. atoms | ||
| Protein | 3081 | 6160 |
| Water | 665 | 464 |
| Average B-factor (A2) | 20.7 | 37.4 |
| R.M.S. Deviations | ||
| Bond length (Å) | 0.006 | 0.008 |
| Bond angles (°) | 1.036 | 1.054 |
| Ramachandran Plot | ||
| Most favored (%) | 98.9 | 98.5 |
| Allowed (%) | 1.1 | 1.4 |
| Disallowed (%) | 0 | 0.1 |
Figure 6. Analysis of the Kb-VV-A11R198-206, Kb-LCMV-GP34-41 molecular surfaces.
(A-C). The peptide-MHC structures of Kb-VV-A11R198-206, Kb-LCMV-GP34-41, and Kb-OVA257-264 (PDB ID# 1VAC) are shown from a side profile view. In each panel, the peptide is represented as a stick model, while the H-2Kb molecule is represented by its surface. Residues T143, K144, K146, W147, A150, G151, E152, R155, L156, A158, Y159, T163, and C164 from H-2Kb are excluded. (D-F). The peptide-MHC structures of Kb-VV-A11R198-206, Kb-LCMV-GP34-41, and Kb-OVA257-264 (PDB ID# 1VAC) are represented as surfaces and viewed from above. The peptides are colored magenta for VV-A11R198-206, cyan for LCMV-GP34-41 and yellow for OVA257-264 with the H-2Kb molecule shown in grey. The residues in the peptide are labeled as “P” followed by the peptide position. (G). The peptide-MHC structures of Kb-VV-A11R198-206 and Kb-LCMV-GP34-41 have been overlaid for comparison. The presented view is looking down towards the N terminus of the peptide. The peptide in each structure is represented as stick model and colored using the same coloring scheme used in A-C, while the H-2Kb molecule is shown as cartoon and colored grey. (H). The peptide-MHC structures of Kb-OVA257-264 and Kb-LCMV-GP34-41 have been overlaid for comparison using the same view from F. Pymol was used to generate all the images.
As a comparison, we overlaid the previously reported crystal structure of the highly defined and studied Kb-OVA, which was not recognized by T cells cross-reactive with LCMV-GP34 or VV-A11R. Similar to the Kb-LCMV-GP34 and the Kb-VV-A11R structures, the OVA peptide has solvent accessible side chains at P4, P6 and P7, whereas the P2, P3, P5 and P8 residues are all buried in Kb (Figure 6C). The Kb-OVA surface appears quite different than those of Kb-LCMV-GP34 and the Kb-VV-A11R, with bulky side chains placed at P6 and P7. These might sterically inhibit interactions with the TCR (Figures 6F and 6H). Furthermore, the P4N, which is shared by all the peptides in OVA, adopts a different rotamer as compared to VV-A11R and LCMV-GP34 (Figures 6F and 6H). The combination of the bulky side chains at P6 and P7 with the P4N in a different conformation may help to explain the absence of a cross-reactive T cell response towards OVA. In summary, the structures of Kb-VV-A11R and Kb-LCMV-GP34 have markedly similar surfaces for TCR recognition and contrast greatly with that of Kb-OVA.
Skewing Cross-reactive T cell Responses against VV-A11R and LCMV-GP34
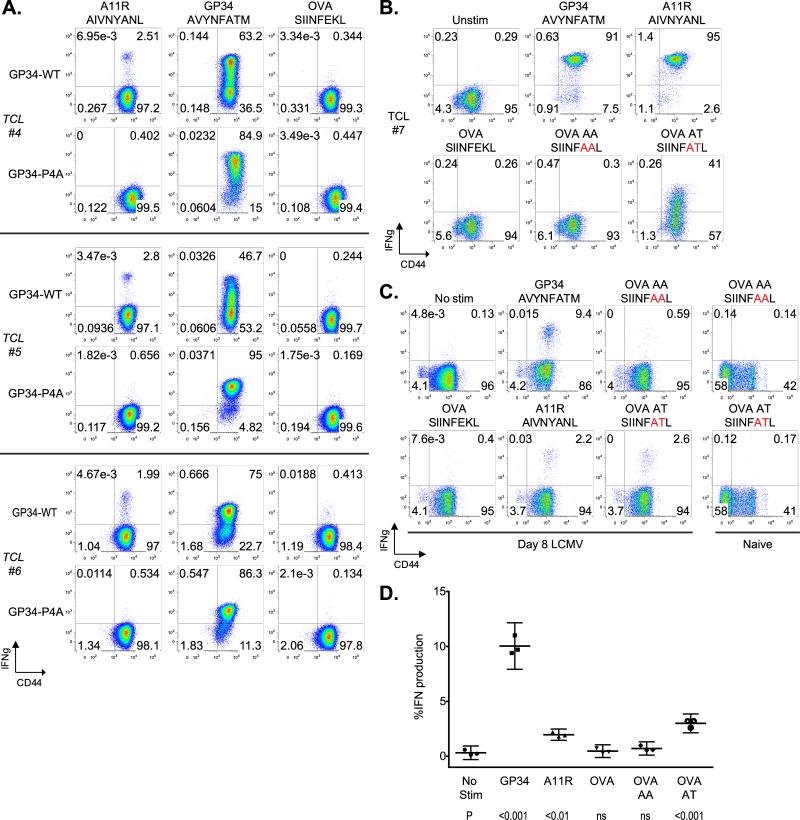
The functional and structural data presented thus far support the idea that the cross-reactive T cell response against VV-A11R is dominated by the P4 asparagine. We hypothesized that VV-A11R cross-reactive T cell responses would be selected against by in vitro stimulation with a P4A variant lacking the important asparagine contact. To test for this, we generated T cell lines from three LCMV-immune mice (Figure 7A) and expanded them in vitro using either the LCMV-GP34 WT peptide or the LCMV-GP34 P4A mutant peptide (TCLs #4,5,6). In each case, expansion with the LCMV-GP34 P4A peptide selects against a cross-reactive VV-A11R T cell response. Interestingly, the cognate response towards LCMV-GP34 was maintained, perhaps through selecting for T cells which recognize the P7T residue.
Figure 7. Skewing of Cross-reactive Responses against both A11R and GP34.
(A). T cell lines (TCL #4,5,6) were generated using the splenocytes from three LCMV-immune mice. T cells were expanded in vitro for four passages by stimulation with either WT-GP34 or GP34 P4A peptide-pulsed targets. T cell lines were analyzed for IFNγ production in response to VV-A11R peptide, LCMV-GP34 peptide and OVA peptide with the results presented as dot plots. (B). Cross-reactive T cell lines were tested for IFNγ production against LCMV-GP34, VV-A11R, OVA or the OVA hybrid peptides, OVA-AA and OVA-AT. The results are represented as dot plots and are representative of three independent experiments. (C and D). CD8+ T cells from acute LCMV-infected mice (n=3) or uninfected mice (n=2) were tested for IFNγ production in response to 1 μM of the indicated peptides with the results from a representative results from a single mouse shown in dot plots. (D). Total CD8+ T cell responses from acute LCMV-infected mice are plotted. The p values for IFNγ production were calculated using the means and indicated below for each peptide.
Predicting and Creating A New Cross-Reactive Epitope Using Structural Information
Next, we wanted to utilize the requirement for the P4N in our system to generate a novel cross-reactive T cell response. To address this, we utilized the OVA peptide, which contains a P4N, but has never been found to be cross-reactive. Our structural analysis indicates that the cross-reactive T cell response against OVA might be absent because of the underlined bulky residues at P6 and P7 in OVA (SIINFEKL), which might sterically inhibit TCR docking. To address this, we generated OVA variants with the P6 and P7 residues from LCMV-GP34 (OVA-AT, SIINFATL), or with both P6 and P7 as alanines (OVA-AA, SIINFAAL). We performed an ICS assay on a GP34-A11R cross-reactive T cell line (TCL #7) to test for the recognition of VV-A11R, LCMV-GP34, OVA, OVA-AA and OVA-AT (Figure 7B). The results show that while the native OVA peptide does not cross-react, 41% of the CD8+ T cells respond towards the modified OVA-AT peptide (Figure 7B). Because >90% of T cells respond to both VV-A11R and LCMV-GP34, most cells that respond against VV-A11R or LCMV-GP34 must also cross-react against OVA-AT. Interestingly, we found that OVA-AA is not cross-reactive, indicating that the cross-reactive recognition of OVA-AT requires the P7T. Because the hybrid OVA peptides have not been reported previously, we needed to exclude the possibility that these cross-reactive responses were unique to our in vitro T cell line. To test if the hybrid OVA peptides could be recognized ex vivo, we performed an ICS assay on splenocytes from acute-LCMV infected mice or naïve mice as controls (Figures 7C and 7D). T cell responses were observed towards the OVA-AT peptide, with IFNγ production measured at ~3%. Similar results were obtained for TNFα production (data not shown). T cell responses were not observed towards the OVA-AA peptide, matching the results in vitro from the GP34-A11R cross-reactive TCL (Figure 7B). T cell responses towards VV-A11R (~2%) and LCMV-GP34 (~10%) were elevated, but within the range that has been previously observed (data not shown). Taken together, these results highlight the pervasive nature of cross-reactive T cell responses, as even the null OVA peptide can be made cross-reactive simply through substitution of the side chains that may inhibit TCR engagement.
Nonreciprocal Nature of LCMV-GP34 and VV-A11R Cross-reactive T cell Epitopes
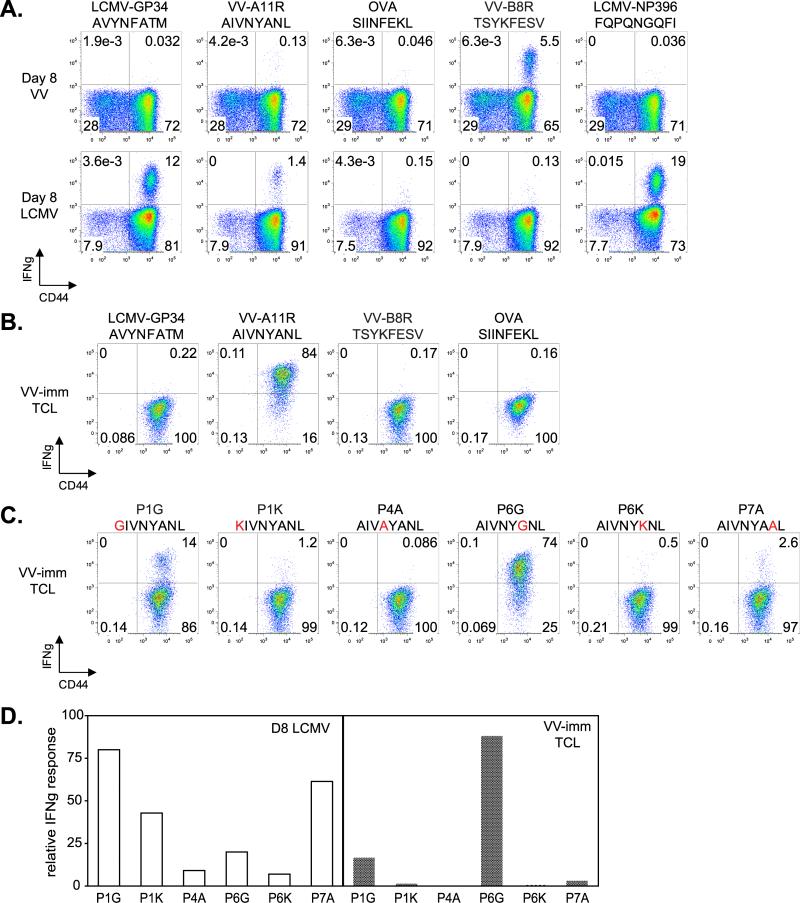
While prior exposure to LCMV protects against subsequent VV infection, prior exposure to VV does not protect against subsequent LCMV infection (42). The mechanism for the non-reciprocal protection is unclear, but might depend on the epitopes elicited during primary LCMV versus primary VV infection. To investigate if the nonreciprocal nature of cross-reactive T cell responses might be apparent at the epitope level, we infected mice with VV and LCMV and monitored T cell responses to various VV, LCMV, or control epitopes in an ICS assay (Figure 8A). VV infection did not elicit cross-reactive T cell responses towards LCMV-GP34, and cognate VV-A11R responses were barely above background, although a robust response to another VV-derived peptide, B8R, was observed (Figure 8A). As before, LCMV infection elicited strong responses to both cognate (LCMV-GP34, -NP396) and cross-reactive (VV-A11R) peptides. Thus, the lack of VV-induced protection against LCMV could be due in part to the relatively weak response to the VV-A11R peptide elicited by VV infection. To study the cross-reactivity of the VV-A11R-specific T cell population that was elicited, we expanded these cells using repeated stimulation in vitro with VV-A11R peptide-pulsed targets, and tested for T cell responses to VV, LCMV, and control peptides (Figure 8B). The expanded T cell line was highly specific for VV-A11R, but no cross-reactivity against LCMV-GP34 was detected. We reasoned that the lack of a cross-reactive T cell response towards LCMV-GP34 might be due to the recognition of non-conserved residues, and so we tested specificity determinants as before using a set of substituted A11R peptides (Figure 8C). To facilitate comparison of the fine specificity of VV-A11R specific T cell response elicited by LCMV and VV infection (from Figure 2B and 8C, respectively) we replotted T cell responses to the substituted peptides as a percentage of the response to the wild-type A11R peptide (Figure 8D). For cells elicited by VV infection, all substitutions except P6G resulted in substantial loss of IFNγ production (Figure 8C), a different pattern than we had observed for A11R-specific T cells elicited by LCMV infection, for which P1 and P7 substitutions were tolerated (Figure 8D). As noted above, distinct differences in the Kb-VV-A11R and Kb-LCMV-GP34 surfaces are restricted to the region around P7. Thus, another aspect of the non-reciprocal nature of LCMV and VV infection is a difference in the fine specificity of the primary response, which for LCMV elicits T cells that recognize VV-A11R through interactions with conserved residues, but for VV elicits T cells that focus on nonconserved VV-A11R residues and so do not recognize LCMV-GP34.
Figure 8. The T cell response directed towards VV-A11R has different recognition determinants depending on whether it was elicited by LCMV or VV infection.
(A). CD8+ T cells from acute day 8 LCMV-infected mice (n=3) and day 8 VV-infected mice (n=3) were isolated and analyzed for IFNγ production using an ICS assay, with results from a representative mouse presented as dot plots. (B,C).VV-A11R specific T cell lines (VV-imm TCL, n=3) were generated using splenocytes from a VV-immune mice and expanded in vitro with VV-A11R peptide-pulsed targets. VV-A11R specific T cell lines were tested for IFNγ production in response to the VV, LCMV, and control peptides shown, in an ICS assay with the results represented as dot plots. (D). ICS data from panel C (VV-imm TCL) and figure 2B (D8 LCMV) were replotted to facilitate comparison of the recognition determinants of VV-A11R specific T cells elicited from LCMV versus VV infection. Responses of each cell line to the A11R variants were normalized to the wild-type A11R response.
Discussion
Protective heterologous immunity plays an important role in the immune response, but ligand requirements for T cell recognition of disparate epitopes from unrelated pathogens are unclear. In this study, we observed that LCMV-GP34 and VV-A11R tetramers compete with each other for TCR. This clearly indicates the presence of individually cross-reactive TCRs that can dually recognize both LCMV-GP34 and VV-A11R (Figure 1E). We compared the recognition determinants for LCMV-GP34 and VV-A11R by generating site-specific mutations in both epitopes at potential TCR contact residues. We tested the mutated peptides for IFNγ production on either acute LCMV-infected mice or cross-reactive TCLs and found different recognition patterns for LCMV-GP34 and VV-A11R. The cognate response towards LCMV-GP34 was dependent on the P4N and P7T, whereas the cross-reactive T cell response towards VV-A11R depended mainly on P4N. Interestingly, we found that the P4A variant of VV-A11R protected against cytotoxic T cell responses in acutely LCMV-infected mice, in vivo. Analysis of the Kb-LCMV-GP34 and Kb-VV-A11R crystal structures revealed that these cross-reactive peptide-MHC complexes have nearly identical surface structures, with the P4N conserved in both structures. Structural mimicry was somewhat unexpected due to the sequence disparity between LCMV-GP34 (AVYNFATM) and VV-A11R (AIVNYANL), which share only three of eight residues (underlined). Combined, our results clearly indicate that the P4N, which is shared between LCMV-GP34 and VV-A11R, mediates the LCMV-cross-reactive T cell response towards VV-A11R.
Primary LCMV infection elicits cognate T cell responses towards LCMV-NP205 and LCMV-GP118 in addition to LCMV-GP34 (8). While the focus of this study is the cross-reactive T cell response between the immunodominant LCMV epitope GP34 with VV-A11R, cross-reactive T cell responses with LCMV-NP205 or LCMV-GP118 and VV-A11R have also been reported (8). In our studies using VV-A11R peptide-pulsed T cell lines, T cell responses towards LCMV-NP205 were observed during the first 2-3 passages during in vitro culture, but specific responses towards LCMV-NP205 disappeared as T cell population specificity increased for VV-A11R. In contrast, cross-reactive T cell responses between VV-A11R and LCMV-GP118 commonly were observed in antigen-specific T cell lines. In fact, the control TCL#2 that recognizes VV-A11R but not LCMV-GP34 does react with LCMV-GP118 (Supplemental Figure 3), explaining why it arose following LCMV infection. It has been reported that extended in vitro culture of previously unprimed CTLs with HIV envelope peptides can lead to de novo CTL responses (43). However, with respect to LCMV and VV-A11R cross-reactive T cell responses, priming was required. When splenocytes from naïve mice were stimulated with VV-A11R peptide-pulsed targets in vitro, we were not able to maintain a culture beyond 5 passages (data not shown). This finding is consistent with previous work in this system which also showed that VV-A11R cross-reactivity was dependent on LCMV priming (12).
Using information about the requirement for P4N in the VV-A11R cross-reactive T cell response, we were able to selectively eliminate the VV-A11R cross-reactive T cell response while maintaining a non-cross-reactive cognate LCMV-GP34 T cell response by expanding T cells in vitro with the LCMV-GP34 P4A peptide. Additionally, we were able to predict and generate a novel cross-reactive T cell response by simply substituting the P6 and P7 residues of the null OVA peptide. These modifications probably facilitated recognition of the important P4N. Collectively, our results support the concept that shared aspects of the molecular surface formed by peptide-MHC complexes can be utilized for cross-reactive T cell recognition. These findings highlight the pervasive nature of cross-reactive T cell responses, but more importantly, demonstrate that cross-reactive T cell responses can be controlled and even manipulated. The ability to skew cross-reactive T cell responses in a specific manner may improve the potential for vaccine design.
VV-A11R-specific T cells are not efficiently generated during primary VV infection and even after expansion in vitro do not cross-react appreciably against LCMV-GP34. By contrast LCMV infection efficiently generates VV-A11R cross-reactive T cells. During a primary LCMV infection, abundant LCMV-GP34 specific T cell populations requiring either the exposed P4N or P7T residues expand, but only the P4N-dependent population cross-reacts with VV-A11R. In contrast, during VV-infection essentially all of the VV-A11R specific T cells elicited require each of the P1A, P4N and P7N residues, and so do not cross-react with LCMV-GP34, which has a different P7 residue. Thus different recognition determinants of the primary response could help explain the observed non-reciprocity in addition to differences in the magnitude of the T cell response and the breadth of the respective T cell repertoires elicited by infection.
The CD8 memory population is not stable in vivo, as infections with cross-reactive viruses can elicit expansions of subsets of the antigen specific memory pool and result in memory pools with altered affinities to and protective capacities against the first-encountered pathogen (6, 11). This is an important issue to evaluate when designing vaccines against viruses known to have highly mutable epitopes or epitopes cross-reactive with other pathogens. Of note, we have shown that altering the epitope used in an ICS assay by a peptide substitution can sometimes lead to a much higher T cell response to that epitope, indicating that a cross-reactive response may sometimes be more detectable and effective than the response to a cognate peptide. Thus, by understanding the parameters of cross-reactivity one might be able to construct a vaccine with an altered peptide that would stimulate an improved response against the desired targeted epitope.
In conclusion, the results presented in this study highlight the pervasive nature of cross-reactive T cell responses. A detailed understanding of the LCMV-GP34 and VV-A11R cross-reactive T cell response revealed that the shared P4N residue is utilized for cross-reactive TCR recognition. Using our understanding of this system, we were able to selectively eliminate VV-A11R cross-reactive T cell responses or generate novel cross-reactive T cell responses to a variant of the OVA peptide. A better mechanistic understanding of protective heterologous immune responses is needed to improve vaccine design strategies.
Supplementary Material
Acknowledgements
We would like to thank Dr. Liisa Selin for helpful discussions, Dr. Michael Brehm for advice on maintenance of cross-reactive T cell lines, and Guoqi Li for kindly providing preparations of human β2-microglobulin.
Footnotes
This work was supported by NIH grants R01 AI38996 and U19 AI57319
References
- 1.Selin LK, Brehm MA, Naumov YN, Cornberg M, Kim SK, Clute SC, Welsh RM. Memory of mice and men: CD8+ T-cell cross-reactivity and heterologous immunity. Immunol Rev. 2006;211:164–181. doi: 10.1111/j.0105-2896.2006.00394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Selin LK, Cornberg M, Brehm MA, Kim SK, Calcagno C, Ghersi D, Puzone R, Celada F, Welsh RM. CD8 memory T cells: cross-reactivity and heterologous immunity. Semin Immunol. 2004;16:335–347. doi: 10.1016/j.smim.2004.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Welsh RM, Che JW, Brehm MA, Selin LK. Heterologous immunity between viruses. Immunol Rev. 2010;235:244–266. doi: 10.1111/j.0105-2896.2010.00897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wedemeyer H, Mizukoshi E, Davis AR, Bennink JR, Rehermann B. Cross-reactivity between hepatitis C virus and Influenza A virus determinant-specific cytotoxic T cells. J Virol. 2001;75:11392–11400. doi: 10.1128/JVI.75.23.11392-11400.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Urbani S, Amadei B, Fisicaro P, Pilli M, Missale G, Bertoletti A, Ferrari C. Heterologous T cell immunity in severe hepatitis C virus infection. J Exp Med. 2005;201:675–680. doi: 10.1084/jem.20041058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen AT, Cornberg M, Gras S, Guillonneau C, Rossjohn J, Trees A, Emonet S, de la Torre JC, Welsh RM, Selin LK. Loss of anti-viral immunity by infection with a virus encoding a cross-reactive pathogenic epitope. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clute SC, Watkin LB, Cornberg M, Naumov YN, Sullivan JL, Luzuriaga K, Welsh RM, Selin LK. Cross-reactive influenza virus-specific CD8+ T cells contribute to lymphoproliferation in Epstein-Barr virus-associated infectious mononucleosis. J Clin Invest. 2005;115:3602–3612. doi: 10.1172/JCI25078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cornberg M, Clute SC, Watkin LB, Saccoccio FM, Kim SK, Naumov YN, Brehm MA, Aslan N, Welsh RM, Selin LK. CD8 T cell cross-reactivity networks mediate heterologous immunity in human EBV and murine vaccinia virus infections. J Immunol. 2010;184:2825–2838. doi: 10.4049/jimmunol.0902168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mathew A, Kurane I, Green S, Stephens HA, Vaughn DW, Kalayanarooj S, Suntayakorn S, Chandanayingyong D, Ennis FA, Rothman AL. Predominance of HLA-restricted cytotoxic T-lymphocyte responses to serotype-cross-reactive epitopes on nonstructural proteins following natural secondary dengue virus infection. J Virol. 1998;72:3999–4004. doi: 10.1128/jvi.72.5.3999-4004.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rothman AL. T lymphocyte responses to heterologous secondary dengue virus infections. Ann N Y Acad Sci. 2009;1171(Suppl 1):E36–41. doi: 10.1111/j.1749-6632.2009.05055.x. [DOI] [PubMed] [Google Scholar]
- 11.Cornberg M, Chen AT, Wilkinson LA, Brehm MA, Kim SK, Calcagno C, Ghersi D, Puzone R, Celada F, Welsh RM, Selin LK. Narrowed TCR repertoire and viral escape as a consequence of heterologous immunity. J Clin Invest. 2006;116:1443–1456. doi: 10.1172/JCI27804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cornberg M, Sheridan BS, Saccoccio FM, Brehm MA, Selin LK. Protection against vaccinia virus challenge by CD8 memory T cells resolved by molecular mimicry. J Virol. 2007;81:934–944. doi: 10.1128/JVI.01280-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen HD, Fraire AE, Joris I, Brehm MA, Welsh RM, Selin LK. Memory CD8+ T cells in heterologous antiviral immunity and immunopathology in the lung. Nat Immunol. 2001;2:1067–1076. doi: 10.1038/ni727. [DOI] [PubMed] [Google Scholar]
- 14.Selin LK, Vergilis K, Welsh RM, Nahill SR. Reduction of otherwise remarkably stable virus-specific cytotoxic T lymphocyte memory by heterologous viral infections. J Exp Med. 1996;183:2489–2499. doi: 10.1084/jem.183.6.2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim SK, Cornberg M, Wang XZ, Chen HD, Selin LK, Welsh RM. Private specificities of CD8 T cell responses control patterns of heterologous immunity. J Exp Med. 2005;201:523–533. doi: 10.1084/jem.20041337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen ZT, Brehm MA, Daniels KA, Sigalov AB, Selin LK, Welsh RM, Stern LJ. Bi-specific MHC heterodimers for characterization of cross-reactive T cells. J Biol Chem. 2010;285:33144–33153. doi: 10.1074/jbc.M110.141051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yin Y, Mariuzza RA. The multiple mechanisms of T cell receptor cross-reactivity. Immunity. 2009;31:849–851. doi: 10.1016/j.immuni.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 18.Hardardottir F, Baron JL, Janeway CA., Jr. T cells with two functional antigen-specific receptors. Proc Natl Acad Sci U S A. 1995;92:354–358. doi: 10.1073/pnas.92.2.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He X, Janeway CA, Jr., Levine M, Robinson E, Preston-Hurlburt P, Viret C, Bottomly K. Dual receptor T cells extend the immune repertoire for foreign antigens. Nat Immunol. 2002;3:127–134. doi: 10.1038/ni751. [DOI] [PubMed] [Google Scholar]
- 20.Zal T, Weiss S, Mellor A, Stockinger B. Expression of a second receptor rescues self-specific T cells from thymic deletion and allows activation of autoreactive effector function. Proc Natl Acad Sci U S A. 1996;93:9102–9107. doi: 10.1073/pnas.93.17.9102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaudhri G, Quah BJ, Wang Y, Tan AH, Zhou J, Karupiah G, Parish CR. T cell receptor sharing by cytotoxic T lymphocytes facilitates efficient virus control. Proc Natl Acad Sci U S A. 2009;106:14984–14989. doi: 10.1073/pnas.0906554106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Macdonald WA, Chen Z, Gras S, Archbold JK, Tynan FE, Clements CS, Bharadwaj M, Kjer-Nielsen L, Saunders PM, Wilce MC, Crawford F, Stadinsky B, Jackson D, Brooks AG, Purcell AW, Kappler JW, Burrows SR, Rossjohn J, McCluskey J. T cell allorecognition via molecular mimicry. Immunity. 2009;31:897–908. doi: 10.1016/j.immuni.2009.09.025. [DOI] [PubMed] [Google Scholar]
- 23.Borbulevych OY, Piepenbrink KH, Gloor BE, Scott DR, Sommese RF, Cole DK, Sewell AK, Baker BM. T cell receptor cross-reactivity directed by antigen-dependent tuning of peptide-MHC molecular flexibility. Immunity. 2009;31:885–896. doi: 10.1016/j.immuni.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borbulevych OY, Santhanagopolan SM, Hossain M, Baker BM. TCRs used in cancer gene therapy cross-react with MART-1/Melan-A tumor antigens via distinct mechanisms. J Immunol. 2011;187:2453–2463. doi: 10.4049/jimmunol.1101268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Borbulevych OY, Piepenbrink KH, Baker BM. Conformational melding permits a conserved binding geometry in TCR recognition of foreign and self molecular mimics. J Immunol. 2011;186:2950–2958. doi: 10.4049/jimmunol.1003150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mazza C, Auphan-Anezin N, Gregoire C, Guimezanes A, Kellenberger C, Roussel A, Kearney A, van der Merwe PA, Schmitt-Verhulst AM, Malissen B. How much can a T-cell antigen receptor adapt to structurally distinct antigenic peptides? EMBO J. 2007;26:1972–1983. doi: 10.1038/sj.emboj.7601605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Colf LA, Bankovich AJ, Hanick NA, Bowerman NA, Jones LL, Kranz DM, Garcia KC. How a single T cell receptor recognizes both self and foreign MHC. Cell. 2007;129:135–146. doi: 10.1016/j.cell.2007.01.048. [DOI] [PubMed] [Google Scholar]
- 28.Garboczi DN, Hung DT, Wiley DC. HLA-A2-peptide complexes: refolding and crystallization of molecules expressed in Escherichia coli and complexed with single antigenic peptides. Proc Natl Acad Sci U S A. 1992;89:3429–3433. doi: 10.1073/pnas.89.8.3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schumacher TN, Heemels MT, Neefjes JJ, Kast WM, Melief CJ, Ploegh HL. Direct binding of peptide to empty MHC class I molecules on intact cells and in vitro. Cell. 1990;62:563–567. doi: 10.1016/0092-8674(90)90020-f. [DOI] [PubMed] [Google Scholar]
- 30.Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc. 2007;2:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 31.Selin LK, Nahill SR, Welsh RM. Cross-reactivities in memory cytotoxic T lymphocyte recognition of heterologous viruses. J Exp Med. 1994;179:1933–1943. doi: 10.1084/jem.179.6.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang HY, Dundon PL, Nahill SR, Welsh RM. Virus-induced polyclonal cytotoxic T lymphocyte stimulation. J Immunol. 1989;142:1710–1718. [PubMed] [Google Scholar]
- 33.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 34.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 36.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 37.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moskophidis D, Lechner F, Pircher H, Zinkernagel RM. Virus persistence in acutely infected immunocompetent mice by exhaustion of antiviral cytotoxic effector T cells. Nature. 1993;362:758–761. doi: 10.1038/362758a0. [DOI] [PubMed] [Google Scholar]
- 39.Santori FR, Arsov I, Vukmanovic S. Modulation of CD8+ T cell response to antigen by the levels of self MHC class I. J Immunol. 2001;166:5416–5421. doi: 10.4049/jimmunol.166.9.5416. [DOI] [PubMed] [Google Scholar]
- 40.Fremont DH, Matsumura M, Stura EA, Peterson PA, Wilson IA. Crystal structures of two viral peptides in complex with murine MHC class I H-2Kb. Science. 1992;257:919–927. doi: 10.1126/science.1323877. [DOI] [PubMed] [Google Scholar]
- 41.Matsumura M, Fremont DH, Peterson PA, Wilson IA. Emerging principles for the recognition of peptide antigens by MHC class I molecules. Science. 1992;257:927–934. doi: 10.1126/science.1323878. [DOI] [PubMed] [Google Scholar]
- 42.Kim SK, Brehm MA, Welsh RM, Selin LK. Dynamics of memory T cell proliferation under conditions of heterologous immunity and bystander stimulation. J Immunol. 2002;169:90–98. doi: 10.4049/jimmunol.169.1.90. [DOI] [PubMed] [Google Scholar]
- 43.Riberdy JM, Zirkel A, Surman S, Hurwitz JL, Doherty PC. Cutting edge: culture with high doses of viral peptide induces previously unprimed CD8(+) T cells to produce cytokine. J Immunol. 2001;167:2437–2440. doi: 10.4049/jimmunol.167.5.2437. [DOI] [PubMed] [Google Scholar]
- 44.Achour A, Michaelsson J, Harris RA, Odeberg J, Grufman P, Sandberg JK, Levitsky V, Karre K, Sandalova T, Schneider G. A structural basis for LCMV immune evasion: subversion of H-2D(b) and H-2K(b) presentation of gp33 revealed by comparative crystal structure.Analyses. Immunity. 2002;17:757–768. doi: 10.1016/s1074-7613(02)00478-8. [DOI] [PubMed] [Google Scholar]
- 45.Khanolkar A, Fuller MJ, Zajac AJ. CD4 T cell-dependent CD8 T cell maturation. J Immunol. 2004;172:2834–2844. doi: 10.4049/jimmunol.172.5.2834. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.