Summary
Holliday junctions (HJs), the DNA intermediates of homologous recombination need to be faithfully processed in order to preserve genome integrity. In human cells the BLM helicase complex promotes non-nucleolytic dissolution of double HJs. In vitro, HJs may be nucleolytically processed by MUS81-EME1, GEN1, and SLX4-SLX1. Here, we exploit human SLX4-null cells to examine the requirements for HJ resolution in vivo. Lack of BLM and SLX4 or GEN1 and SLX4 are synthetically lethal in the absence of exogenous DNA damage with lethality being a consequence of dysfunctional mitosis proceeding in the presence of unprocessed HJs. Thus, GEN1 activity cannot substitute for the SLX4-associated nucleases and one of the HJ resolvase activities, either those associated with SLX4 or GEN1 is required for cell viability even in the presence of BLM. In-vivo HJ resolution depends on both SLX4-associated MUS81-EME1 and SLX1, suggesting that they are acting in concert in the context of SLX4.
Introduction
Homologous recombination (HR) is a high-fidelity pathway for the repair of double-strand breaks, interstrand crosslinks (ICLs), and lesions arising during normal DNA replication (Li and Heyer, 2008). Deficiency in HR results in a greatly increased susceptibility to multiple cancers including those of the breast, ovary, pancreas, and blood. The key step in the completion of HR is processing of Holliday junctions (Holliday, 1964; Szostak et al., 1983). All organisms possess a specific set of DNA processing activities that can act on these HR intermediates. Distinct genetic interactions between the genes coding for these activities have been seen in different organisms and lack of or inappropriate repair of HJs results in diverse mutant phenotypes (Schwartz and Heyer, 2011). In mitotically-dividing human cells, at least four enzymatic activities are implicated in the processing of HJs. BLM-TOP3α-RMI1-RMI2 complex is well established as a Holliday junction dissolvase, able to branch migrate double HJs towards one another and decatenate the DNA strands without the use of structure specific endonucleases (Cejka et al., 2010; Wu and Hickson, 2003). Alternatively, the nucleases MUS81-EME1 (Chen et al., 2001; Constantinou et al., 2002; Taylor and McGowan, 2008), GEN1 (Ip et al., 2008), and SLX1 (Andersen et al., 2009; Fekairi et al., 2009; Munoz et al., 2009; Svendsen et al., 2009) have been shown to have nucleolytic activity on synthetic single HJs in vitro and there is evidence that they play a role in resolving HJs in vivo in human cells, although their respective contributions to in vivo HJ resolution are still undefined (Wechsler et al., 2011). Interestingly, both human MUS81 and SLX1 interact with SLX4, a scaffold protein that is implicated in enhancing the activity of these two nucleases as well as a third nuclease, XPF-ERCC1, which also binds to SLX4 (Andersen et al., 2009; Fekairi et al., 2009; Munoz et al., 2009; Svendsen et al., 2009).
We and others have reported mutations in SLX4 in patients with Fanconi anemia (Kim et al., 2011; Stoepker et al., 2011), a recessive disorder of bone marrow failure and cancer predisposition that arises due to an inability to repair DNA interstrand crosslinks (ICLs) (reviewed in Kottemann and Smogorzewska, 2013). Using complementation of the SLX4-null Fanconi anemia patient cell line with a number of SLX4 mutants deficient in binding to their nuclease partners, we have established that the in vivo activity of the XPF-ERCC1, MUS81- EME1, and SLX1 nucleases during DNA repair relies strictly on their association with SLX4 and that the nucleases are important for DNA repair of distinct DNA lesions (Kim et al., 2013). XPFSLX4 interaction is necessary for resistance to ICL agents and this nuclease acts in the incision step of ICL repair (Kuraoka et al., 2000). MUS81-SLX4 interaction is necessary for resistance to Topoisomerase I (TOP1) inhibitor Camptothecin, as well as to PARP inhibitor (KU0058948 or Olaparib) and it is most likely involved in processing of stalled replication forks prior to the HR step (Ray Chaudhuri et al., 2012). Finally lack of SLX1-SLX4 interaction results in intermediate sensitivity to ICL agents, CPT, and PARP inhibitor suggesting that SLX1-SLX4, although important, might be redundant with other activities in the HR pathway. The SLX4 complementation system we developed gives us a unique opportunity to assess the respective contributions of each of the SLX4-associated nucleases to in vivo HJ resolution and to study their genetic interactions with the other two HJ processing factors, GEN1 and BLM during unperturbed cell growth.
Results
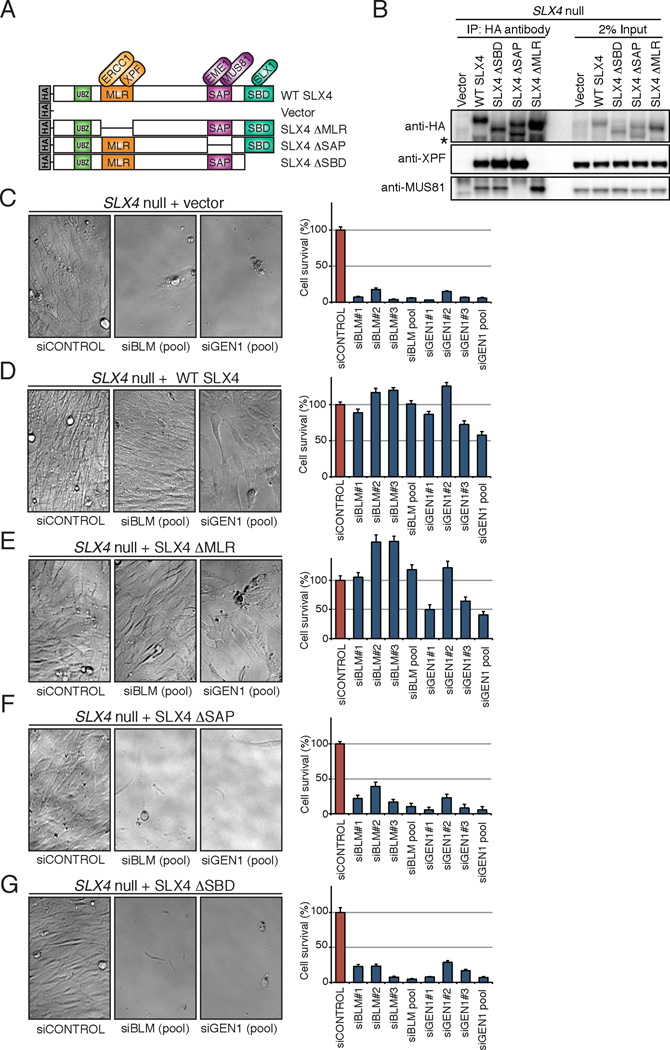
BLM or GEN1 were depleted in the SLX4 null human cell line (RA3331/E6E7/hTERT) (Kim et al., 2011) complemented with either an empty vector, wild type (WT) SLX4, SLX4 lacking interaction with XPF (SLX4ΔMLR), SLX4 lacking interaction with MUS81 (SLX4ΔSAP), or SLX4 lacking interaction with SLX1 (SLX4ΔSBD) (Kim et al., 2013), Figure 1A and B). We observed that the depletion of either BLM or GEN1 induced synthetic lethality in the absence of SLX4 and that the expression of WT SLX4 suppressed the lethality (Figure 1C and 1D). Moreover, both MUS81 and SLX1, but not XPF association with SLX4 were necessary for the suppression of synthetic lethality caused by BLM or GEN1 depletion (Figure 1E to G).
Figure 1. Depletion of BLM or GEN1 in the absence of SLX4 is synthetically lethal in human cells.
(A). Schematic of SLX4 illustrating select domains and interacting nucleases, along with the N-terminally HA-tagged SLX4 cDNAs used in all experiments. Although the interaction of SLX1 and SLX4 has been shown to be direct, SLX4-XPF-ERCC1 and SLX4-MUS81-EME1 might not be direct. (B). Western analysis of immunoprecipitated HA-tagged SLX4 and co-immunoprecipitated XPF or MUS81 from cell lines used in the experiments that follow. The lower band (*) indicates degradation products. (C-G) Survival of SLX4 null cells complemented with indicated cDNAs and treated with siRNA against Luciferase (siCONTROL), siBLM, and siGEN1. SLX4 null cells complemented with empty vector (C), WT SLX4 cDNA (D), SLX4ΔMLR lacking interaction with XPF (E), SLX4ΔSAP lacking interaction with MUS81 (F), and SLX4ΔSBD that lacks interaction with SLX1 (G). Three separate siRNAs or a pool of three siRNAs were used for these studies as indicated. Left panels show cell morphology and the decreased cell number as observed using a phase contrast microscope. Right panels show cell survival as measured with Cell Titer-Glo. Survival data show average of at least six replicates for each siRNA and the error bars indicate standard deviations between replicates. Efficiency of BLM and GEN1 depletion is shown in Figure S1.
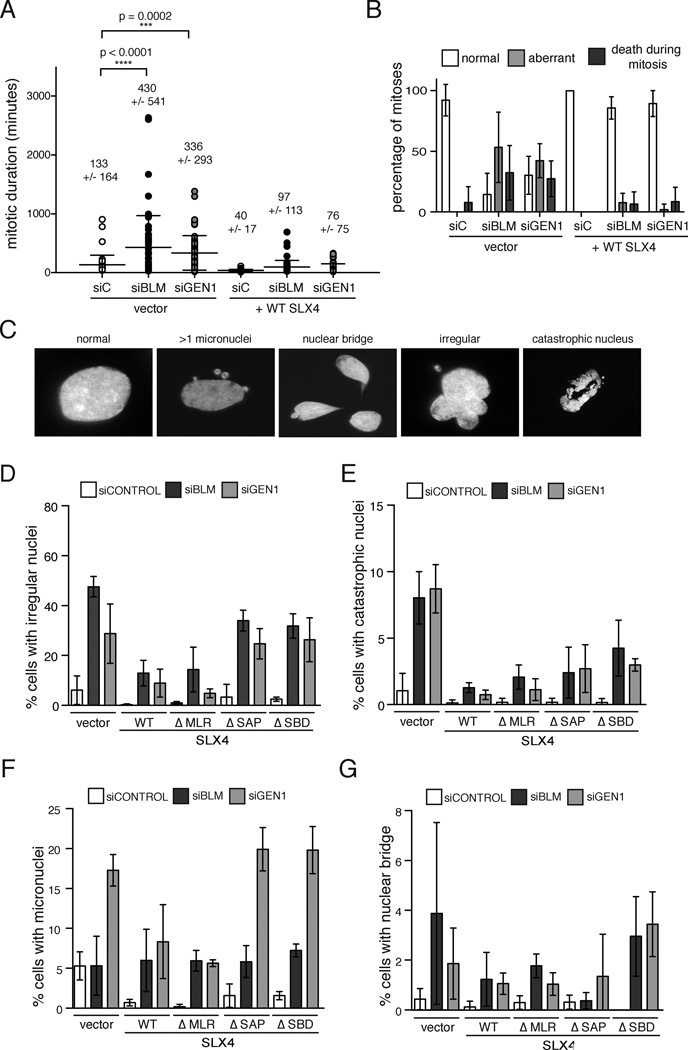
To identify the mode of cell death in cells deficient for SLX4 and BLM, or SLX4 and GEN1, we employed time-lapse microscopy of GFP-H2B expressing SLX4 null and complemented cells depleted of BLM or GEN1. Analysis of mitotic duration showed that the length of mitosis was significantly prolonged in SLX4-null cells depleted of either BLM or GEN1 (Figure 2A) and that the defect was suppressed by expression of WT SLX4. We observed that synthetic lethality in human cells deficient for these proteins predominantly occurs as a consequence of dysfunctional mitosis. 89% of mitoses in SLX4 and BLM doubly deficient cells and 68% of mitoses in SLX4 and GEN1 deficient cells were aberrant (failed to segregate, had lagging chromosomes, or gave rise to more than two daughters) or resulted in cell death (Figure 2B). Cleavage of caspase 3 was increased in SLX4 null cells depleted of GEN1 or BLM suggesting that some of the cells were dying by apoptosis (Figure S2A). Once more, expression of WT-SLX4 rescued mitotic dysfunction and caspase activation in the absence of GEN1 or BLM (Figure 2B, Figure S2A). The prominent feature of aberrant mitoses in both SLX4-GEN1 and SLX4-BLM deficient backgrounds was mitotic failure resulting in binucleated and multinucleated cells (Supplementary table S1). We next assessed the nuclear morphology in doubly deficient cells by quantifying a number of distinct nuclear abnormalities (Figure 2C). Depletion of BLM or GEN1 in SLX4 null cells led to an increase in the number of cells with irregular nuclei and catastrophic nuclei and the defect was suppressed by WT or SLX4ΔMLR and partially suppressed by SLX4ΔSAP or SLX4ΔSBD (Figure 2D and 2E). An increase in micronuclei (Figure 2F) was specifically associated with GEN1 depletion while nuclear bridges were increased in SLX4 null cells with both GEN1 and BLM depletion (Figure 2G). The nuclear abnormalities present in SLX4 null cells depleted of either GEN1 or BLM suggested increased levels of genome instability, which was confirmed by the increased staining of phosphorylated H2AX (γH2AX) in SLX4-null cells depleted of GEN1 or BLM (Figure S2B, S2C). The majority of micronuclei were γH2AX positive suggesting that they were derived from broken chromosomes.
Figure 2. Mitotic duration is greatly increased and the majority of mitoses are abnormal in SLX4 deficient cells depleted of GEN1 or BLM.
(A). Duration of mitosis (in minutes) assessed by live-cell imaging of GFP-H2B-positive SLX4-null cells complemented with vector or WT SLX4 and also transfected with the indicated siRNAs. Each value plotted represents a single cell undergoing mitosis. Cells were scored from 5 independent movies for each experimental condition. p values were determined by one-way ANOVA. (B). Outcome of mitoses in the experiment described in (A). Cells entering mitosis and segregating to yield two daughter cells were scored as ‘normal’. Cells that failed to segregate or did so with lagging chromosomes or multiple daughters were scored as ‘aberrant’. Cells that died during mitosis or those with daughter cells that died during mitotic exit were scored as ‘death during mitosis’. Values plotted represent the mean percentages of scores from 5 independent movies for each experimental condition. Error bars represent the standard deviation between the 5 replicates. See also Table S1. (C). to (G). Nuclear morphology of SLX4-null cells depleted of GEN1 or BLM. SLX4-null and complemented cells transfected with indicated siRNAs were stained with DAPI 96 hrs post second siRNA transfection. Nuclear morphology was scored according to the example panel shown in (C). Cells with irregular nuclei (D), with catastrophic nuclei (E), with micronuclei (F), and with nuclear bridges (G) were counted. Note that cells that die during mitosis were not counted since they were not attached to a coverslip. Values plotted represent the mean percentage from three experimental replicates. Error bars represent the standard deviation of the replicate means. See also Figure S2.
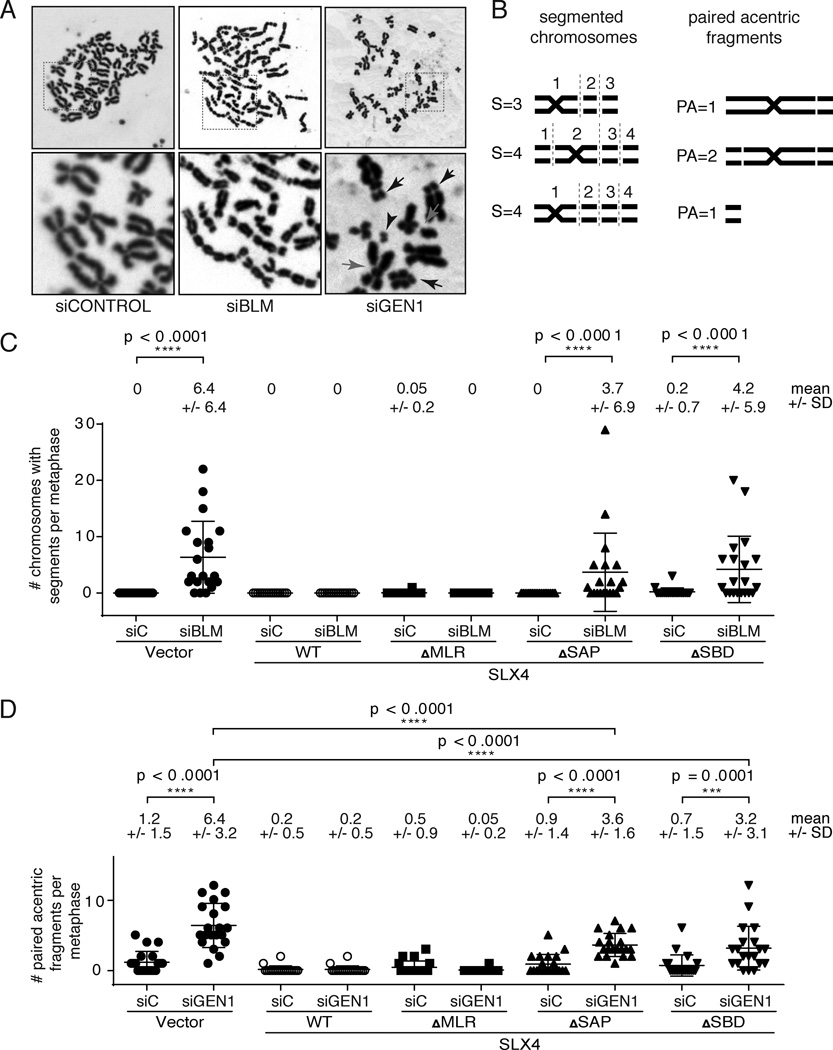
In order to directly evaluate the type and level of genomic instability, we assessed chromosomal morphology on metaphase spreads in SLX4-null cells depleted of BLM or GEN1. Depletion of BLM from SLX4-null cells led to the appearance of a striking phenotype of chromosomal segmentation (Figure 3A and 3B). This phenotype was previously observed when different combinations of nucleases capable of processing HJs were depleted from cells deficient for BLM protein (Wechsler et al., 2011). The authors attributed the chromosome segmentation phenotype to lack of proper condensation at the sites of chromatid entanglements that could not be properly processed in the absence of HJ dissolution or resolution (Wechsler et al., 2011). Cells expressing SLX4ΔSBD lacking the SLX1 interaction or SLX4ΔSAP lacking MUS81 interaction also showed very high levels of segmentation indicating that the SLX1 and MUS81 activities associated with SLX4 are essential for the suppression of segmentation (Figure 3C).
Figure 3. Depletion of BLM or GEN1 in the absence of SLX4 results in distinct chromosomal phenotypes that are dependent on MUS81 and SLX1.
(A) Metaphase spreads from SLX4-null cells depleted of BLM or GEN1 showing presence of segmented chromosomes (siBLM) or paired acentric fragments (siGEN1). Upper panels display whole metaphases. The boxed area is shown below at higher magnification. Paired acentric fragments (arrowhead) appear to originate from telomere-proximal fragments that affect both sister chromatids (black arrow. Chromatid breaks are also seen (red arrows). See Figure S4 for staining for telomeric DNA. (B) Examples of scoring of sectors and paired acentric fragments used for quantification. (C) Quantification of segmented chromosomes in SLX4-null and complemented cells depleted of BLM. Chromosomes with 3 or more segments were scored in the indicated cell lines. There had to be at least two paired fragments, on the p and/or q arm, extending from the centric fragment, separated by gaps or breaks, for a total of at least three segments. SLX4-null cells depleted of GEN1 showed no segmented chromosomes. (D) The acentric fragments (distinct pairs only) were quantified in SLX4-null and complemented cells depleted of GEN1. 20 metaphases were scored for each experiment shown in this figure and the analysis was blinded. Means and standard deviations are shown above each graph. Significance was determined by one-way ANOVA. p values are indicated for the statistically significant comparisons of interest.
Depletion of GEN1 from SLX4 cells does not result in chromosomal segmentation (Figure 3A). However, metaphases from SLX4 and GEN1 deficient cells showed the presence of paired acentric telomere-proximal DNA fragments (Figure 3A and 3D, black arrows) that often become physically separated from the originating chromosome (Figure 3A, black arrowhead). These fragments are positive for telomeric DNA (Figure S3A) indicating that they arise from the tips of chromosomes. Similar to the segmentation phenotype, the fragmentation is present on both chromatids and appears to be symmetrical. Once again, complementation of the SLX4-null cells with SLX4 that lacks the MUS81 or SLX1-association were not able to fully suppress the formation of these paired acentric fragments although some suppression was evident.
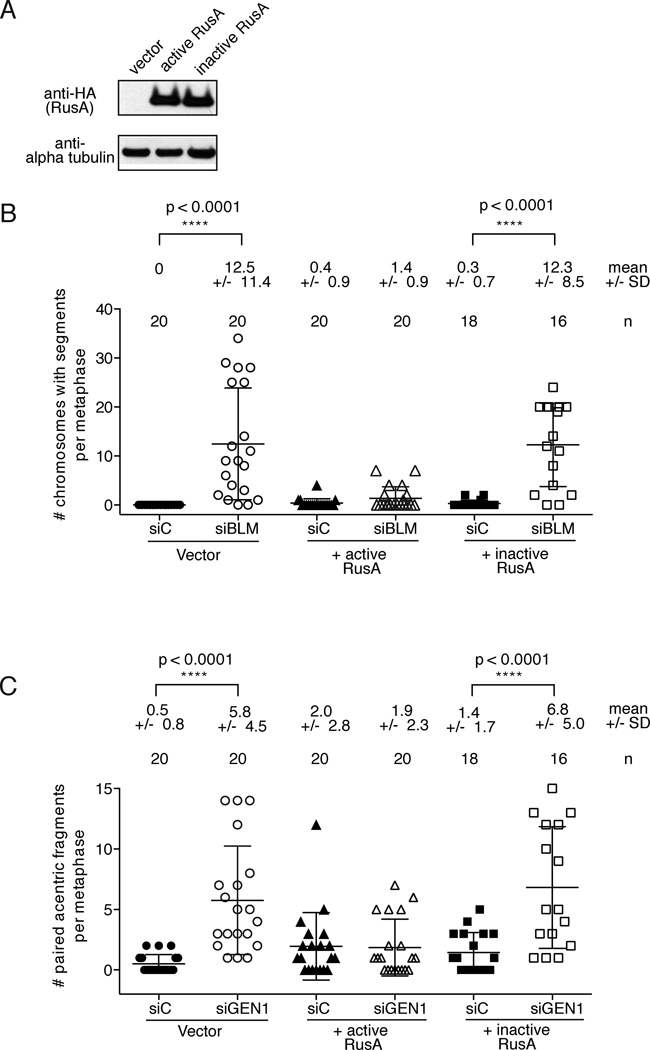
We hypothesized that the segmentation and formation of the acentric fragments was due to persistence of unresolved HJs in cells deficient in SLX4 and BLM or GEN1. To test that assertion we expressed one of the bacterial HJ resolvases, RusA (Doe et al., 2000; Saintigny et al., 2002 and Figure 4A) in SLX4 deficient cells and assessed segmentation and formation of acentric fragments after depletion of BLM or GEN1 (Figure 4B and C). RusA cleaves Holliday junctions in a symmetrical manner to yield nicked duplex products (Chan et al., 1997, reviewed in Sharples et al., 1999). Exogenous expression of bacterial RusA has been used to pinpoint Holliday junction resolution defects in yeast and human cells (Boddy et al., 2001; Doe et al., 2000; Saintigny et al., 2002). Expression of wild type RusA in SLX4-null cells depleted of BLM or GEN1 was able to suppress both segmentation and acentric fragment formation respectively. In contrast, a RusA D70N mutant deficient for in vitro HJ resolvase activity was able to suppress neither the segmentation nor the acentric fragment formation. This suggests that the observed chromosomal abnormalities are the outcome of inefficient HJ resolution in cells that are deficient for SLX4 and BLM or SLX4 and GEN1.
Figure 4. Exogenous expression of RusA rescues chromosomal segmentation or paired acentric fragmentation following depletion of BLM or GEN1 in SLX4-null cells.
(A). SLX4-null cells were stably transduced with either empty vector, HA-tagged active RusA or HA-tagged inactive RusA (D70N). HA-tagged RusA expression levels are shown by anti-HA western analysis. (B). Quantification of segmented chromosomes in RusA expressing SLX4-null cells depleted of BLM. Chromosomes with 3 or more segments were scored in the indicated cell lines. (C). Quantification of paired acentric fragments in RusA expressing SLX4-null cells depleted of GEN1. Acentric fragments in distinct pairs only were scored in the indicated cell lines. (B) and (C) The number of metaphases scored, means and standard deviations are shown above each graph. Analysis was blinded for both experiments in which SLX4-null cells expressing vector control or active RusA were depleted of control or BLM protein. Significance was determined by one-way ANOVA. p values are indicated for the statistically significant comparisons of interest.
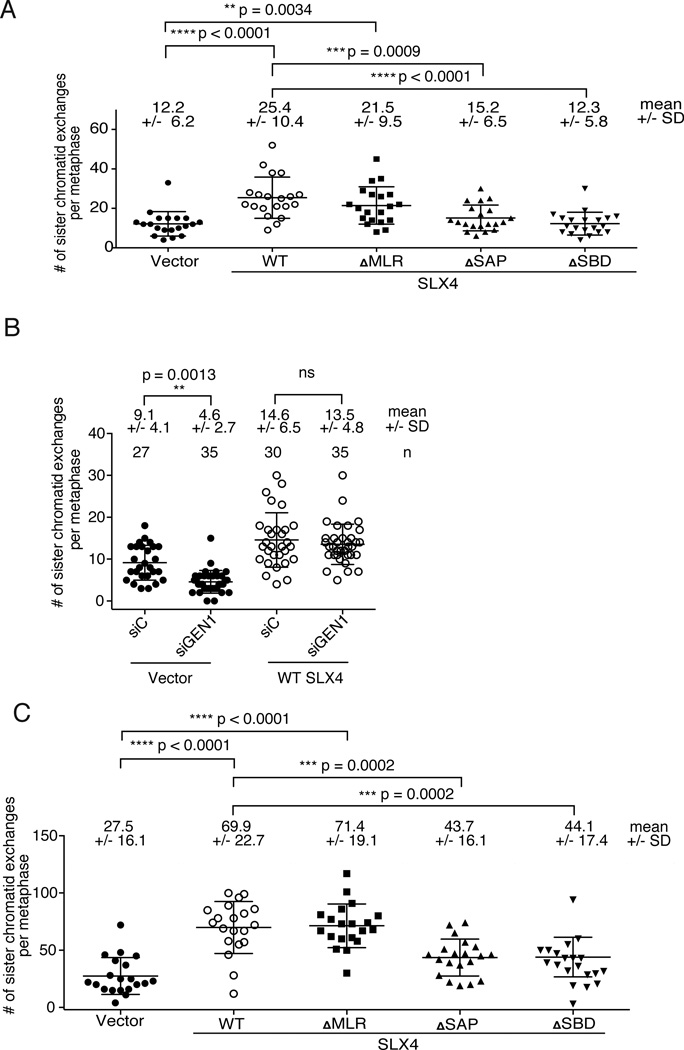
To test directly the contribution of SLX4-associated nucleases and GEN1 to HJ resolution activity, we have assessed the formation of sister chromatid exchanges (SCE) which arise from crossover events and are therefore markers for nucleolytic processing of HJs (Chaganti et al., 1974; Latt et al., 1975). As expected, mitomycin C (MMC)-induced SCEs were low in SLX4 null cells and were increased when the cells are complemented with WT SLX4 or SLX4 that is unable to interact with XPF (Figure 5A). However, MMC-induced SCEs remained low in cells that lacked SLX4-associated MUS81 or SLX1. Depletion of GEN1 in SLX4-null cells, but not in the SLX4-complemented cells led to further suppression of SCEs (Figure 5B). A different way of inducing SCEs is to deplete BLM from cells (Chaganti et al., 1974). Low levels of SCEs in SLX4 null cells depleted of BLM were greatly increased when cells were complemented with WT SLX4 (Figure 5C and Figure S4). Once again, cells expressing SLX4ΔSAP or SLX4ΔSBD were not able to complement the SCE formation suggesting that the SLX4-associated nucleolytic complex forms an in vivo HJ resolvase with both MUS81 and SLX1 participating in HJ resolution.
Figure 5. Both MUS81 and SLX1 interactions with SLX4 are necessary for the formation of SLX4-dependent sister chromatid exchanges.
(A). Quantification of sister chromatid exchanges (SCEs) after SLX4-null and complemented cells were treated with low levels of interstrand crosslinking agents (one hour treatment with 0.1 micrograms of MMC per mL of media) (B). Quantification of SCEs after siRNA-mediated depletion of GEN1 from SLX4-null or WT complemented cells and treatment with low levels of interstrand crosslinking agents. (C). Quantification of SCEs after siRNA mediated depletion of BLM from SLX4-null and complemented cells. SCEs were scored blindly (A and C) and not blindly (B) on 20 metaphases from each cell line. Means and standard deviations are given above the graphs. Significance was determined by one-way ANOVA. p values are indicated for the statistically significant comparisons. See also Figure S4.
Discussion
Taken together, we show that mitotically growing cells rely on HJ dissolution provided by BLM-TOP3α-RMI1-RMI2 or HJ resolution supplied by MUS81-EME1 and SLX1 working together in the context of SLX4. Since GEN1 is unable to prevent the synthetic lethality of SLX4 and BLM deficiencies, we conclude that GEN1 cannot substitute for SLX4-dependent activity during HJ processing in vivo. Our findings in human cells have striking parallels to the work from the budding yeast, in which the Slx4-Slx1 complex as well as the Mus81-Mms4 were identified as being synthetically lethal with the yeast RecQ helicase Sgs1, a BLM ortholog, and also with Top3, SGS1-interacting topoisomerase (Mullen et al., 2001, reviewed in Rouse, 2009). These parallels emphasize the evolutionarily conserved redundancy in the processing of replication-induced DNA intermediates.
Synthetic lethality between human SLX4 and GEN1 loss indicates that GEN1 activity is necessary in the setting of SLX4 deficiency even in the presence of BLM complex, showing for the first time an important function of human GEN1 in vivo. This also highlights that nucleolytic HJ processing is essential for cellular viability in human cells.
The strict requirement of both MUS81 and SLX1 for the suppression of synthetic lethality in cases of BLM and GEN1 depletion suggests that SLX4-MUS81-EME1-SLX1 forms an in vivo HJ resolvase as has been previously proposed (Munoz et al., 2009; Svendsen et al., 2009). We corroborated this finding by showing that MMC-induced as well as BLM depletion-induced SCEs were dependent on both MUS81 and SLX1 association with SLX4. The data presented here suggest a concerted action of the SLX4-MUS81-EME1-SLX1 complex. It is important, however, to emphasize that MUS81-EME1 also has functions that are dependent on SLX4 but that are independent of SLX1. This notion is supported by previous studies that showed that SLX4ΔSAP was unable to confer resistance to Topoisomerase I or to PARP inhibitors and it stresses that SLX4 engages different nucleases in different lesion contexts (Figure S5). Determining how SLX4 is regulated will be crucial to the understanding of the in vivo functions of these nucleases.
GEN1 depletion in SLX4 null cells led to further suppression of SCEs but such suppression was not seen in the SLX4-complemented cells. This suggests that although GEN1 is capable of resolving the same structures as SLX4 complexes, SLX4 dependent processes are very efficient at HJ resolution and GEN1 may act as a backup pathway. The observation that GFP-tagged GEN1 protein expressed from an integrated BAC is cytoplasmic and may only gain access to the DNA after nuclear envelope breakdown (Matos et al., 2011) suggests that GEN1 may act in mitosis on structures that have not been processed by SLX4-dependent nucleases earlier in the cell cycle. What emerges is a hierarchy of HJ processing with dissolution using the BLM complexes being favored since it avoids crossover product formation with the potential loss of heterozygosity or unequal sister chromatid exchanges. The second in line during HJ processing is the SLX4-dependent resolution and finally GEN1-dependent resolution is third (Figure 6). Importantly, we show that there also exist HJ structures that cannot be processed by BLM complex and these absolutely require nucleolytic resolution provided by SLX4 complex or GEN1. It is also possible that other nucleases capable of action on HJs are still present in the mitotic cells; however, our synthetic lethality data indicate that they are unable to substitute for the three activities associated with BLM, SLX4, and GEN1.
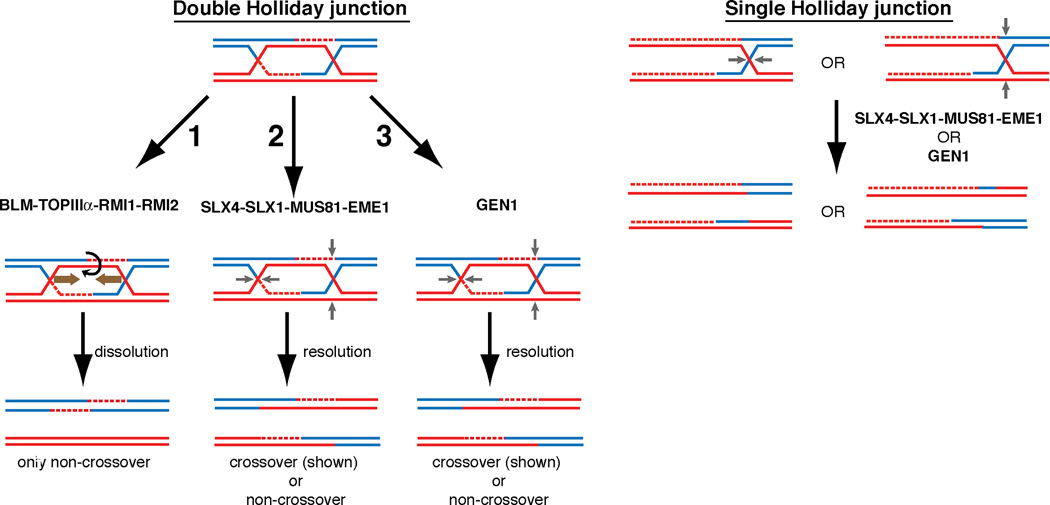
Figure 6. Model of Holliday junction processing in mitotically growing human cells.
(1) The BLM-TOP3α-RMI1-RMI2 Holliday junction dissolvase complex facilitates branch migration of dHJs towards one another and decatenation of the DNA strands without the use of structure specific endonucleases. This process yields entirely non-crossover reaction products. (2) SLX4 associated nucleases provide essential nucleolytic processing of dHJs that are not dissolved in the absence of the BLM complex. The strict requirement of both MUS81 and SLX1 interaction with SLX4 for the suppression of synthetic lethality in the absence of BLM as well as the increase of SCEs after MMC treatment or BLM depletion suggests that they together form an in vivo HJ resolvase. (3) GEN1 is able to process HJs but is unable to prevent the synthetic lethality of SLX4-BLM deficiency. Both SLX4-dependent or GEN1 mediated resolution of dHJs may yield both cross-over and non-crossover products. A single Holliday junction or another substrate unsuitable for BLM complex mediated activity requires SLX4-complexed nucleases or GEN1. GEN1-SLX4 synthetic lethality indicates that GEN1 activity is necessary in the setting of SLX4 deficiency even in the presence of the BLM complex. In this setting, SLX4-interacting MUS81-EME1 and SLX1 are again required for the suppression of GEN1-SLX4 synthetic lethality.
Lack of BLM-dependent dissolution or SLX4-dependent resolution in mitotically dividing cells results in chromosomal segmentation that has been previously seen when various combinations of HJ resolving nucleases were depleted from BLM deficient cells (Wechsler et al., 2011). In our experiments, the absence of both BLM-mediated dissolution and the SLX4-associated nucleases was sufficient to reveal a robust chromosome segmentation phenotype despite the presence of GEN1. Disappearance of this chromosomal phenotype upon expression of RusA suggests that segmentation arises due to the inability to properly process HJs. It is indeed possible that the sites between segments correspond to the unresolved HJs.
The presence of chromosomal segmentation even when GEN1 is expressed highlights that GEN1 is unable to process the apparent high levels of HJs that arise spontaneously when BLM and SLX4 are absent. Wechsler et al. observed robust chromosomal segmentation only after SLX4 (or MUS81) and GEN1 were depleted. We speculate that the discrepancy comes from the incomplete siRNA-mediated depletion of SLX4 (or MUS81) in the previous work thus requiring co-depletion of GEN1 in order to observe a robust segmentation phenotype.
Acentric fragments appearing in the absence of SLX4 and GEN1 were also efficiently removed when RusA was overexpressed suggesting that they also represent HJs that can be resolved by RusA. Since the acentric fragments are present in cells that have robust BLM activity, they have to arise from intermediates that cannot be processed by the BLM complex. Thus, it is possible that these sites may represent unresolved single HJs. These would not be suitable substrates for dissolution and would have to be resolved by SLX4 complex or by GEN1.
Appearance of chromosomal segmentation and acentric fragment formation in cells untreated with any DNA damaging agents indicates that proper HJ processing is necessary during unperturbed replication. Without the HJ processing enzymes, the unprocessed HJs link the two sister chromatids and hinder the ability of the mitotic spindle to segregate chromosomes causing the apparent mitotic demise in cells deficient for SLX4 and BLM protein or SLX4 and GEN1. The appropriate processing of HR intermediates in mitotic mammalian cells is an essential process for cell viability and the suppression of genomic instability. Essential processes like these are difficult to dissect as organisms evolve complex back-up mechanisms to avert the detrimental effects of genome instability such as cell death and cancer. Our model system has allowed us to address the essential aspects of HJ processing and we conclude that the SLX4-MUS81-EME1-SLX1 complex is an in vivo HJ resolvase in human cells, supplementing BLM dependent double HJ dissolvase activity. Even when the BLM complex is active, nucleolytic HJ resolution is still essential for viability and depends on SLX4 and GEN1 functions.
Experimental procedures
Cell lines and cell culture
SLX4-null (RA3331/E6E7/hTERT) and complemented cell lines were previously described (Kim et al., 2013). Cells were grown in Dulbecco Modified Eagle medium (DMEM) supplemented with 15% (v/v) FBS, 100 units of penicillin per mL and 0.1 mg streptomycin per mL (all from Invitrogen). Active RusA and inactive RusA (D70N) expression vectors were a kind gift from the Monnat lab (University of Washington). The ORFs and N terminal NLS sequences were subcloned into lentiviral vectors. Active RusA and inactive rusA stably expressing cell lines were generated by lentiviral transduction of SLX4-null (RA3331/E6E7/hTERT) cells.
RNAi
siRNA (Ambion) were transfected using Lipofectamine RNAiMAX (Invitrogen) with single siRNAs or pools of 3 siRNAs as indictated with a final siRNA concentration of 25 nM. Cells were transfected twice, first by reverse transfection and 24 hours later by forward transfection as outlined in the manufacturer’s instructions.
Cell growth and survival assays
Cells transfected twice with siRNAs, were collected 72 hours after the first siRNA transfection and plated in equal number (1,000 cells per well) in a 96-well transparent plates for observation and light microsopy or in 96-well Opaque White Microtest plate (BD Biosciences, 353296). After 5 days in culture, cell survival was determined using the Cell Titer-Glo reagent (Promega) according to the manufacturer's instruction. Survival assay was assessed at least in triplicate and error bars depicted represent standard deviations.
Metaphase spreading and sister chromatid exchange assay
Cells were arrested with colcemid (0.17 µg per mL of media) for 90 minutes, harvested, incubated for 10 min at 37 °C in 0.075 M KCl and fixed in freshly prepared methanol:glacial acidic acid (3:1 vol/vol). Metaphase spreads were prepared by dropping the cell suspension onto slides pre-wetted with ddH20. Slides were dried at 42°C for 60 min before staining with KaryoMAX Giemsa (Invitrogen) in Gurr Buffer for 3 min. After rinsing with fresh Gurr Buffer followed by distilled water, the slides were fully dried then scanned unmounted using the Metasystems Metafer application.
Sister chromatid exchange protocol was adapted from (Chu et al., 2010). Cells were cultured for 24 hours in the presence of 10 ug/mL BrdU (BD Pharmingen). Cells were then treated with 0.1 ug/mL MMC for one hour (Sigma Aldrich), washed once with PBS and replenished with fresh culture media containing a final concentration of 10 ug/mL BrdU and cultured for a further 24 hours. For sister chromatid exchange analysis of siRNA depleted cells, cells were transfected with siRNA twice as described above. Media was supplemented with 10ug/mL BrdU during the second (forward) transfection. BrdU labeling, MMC treatment (where appropriate, as indicated in figure legends) and metaphase preparation were as described above. Slides were differentially stained for sister chromatid exchange analysis as described fully in supplemental material.
Live cell imaging
For time-lapse microscopy cells were plated on 35mm glass bottom dishes (MatTek) and transferred to an LCV110 VivaView Incubator Microscope (Olympus) 48 hours following forward transfection of siRNAs. Cells were maintained at 37 degrees with 5% CO2 for the duration of imaging (48 hours). Each stage position was imaged every 5 minutes using a 20× DIC objective and 0.5× magnification changer. Images were obtained for DIC and GFP and movies were prepared using Metamorph software.
Immunofluorescence
Cells were fixed in 3.7% formaldehyde, and permeabilized with NP-40, blocked with PBG (0.2% [weight/volume] cold fish gelatin, 0.5% [weight/volume] BSA in PBS), and incubated with anti-γH2AX (Millipore #05-636) in blocking buffer. Cells were washed and incubated with the appropriate Alexa fluor secondary antibody. After 3 additional washes in PBG, the coverslips were embedded in Vectashield (Vector Laboratories) supplemented with 4,6-diamidino-2-phenylindole (DAPI).
Western blotting and immunoprecipitation
For western blotting, cells were lysed directly in Laemmli sample buffer (BioRad) and separated by 8 or 12% SDS-PAGE prepared using Protogel (National Diagnostics) as per manufacturers instructions. Proteins were transferred to PVDF membrane (Immobilon, Millipore) and incubated with primary antibodies as indicated. For a full list of antibodies see supplemental material. For immunoprecipitations, cells were lysed in MCLB (50mM Tris-HCl, pH 7.5, 150mM NaCl, and 0.5% Nonidet P-40) supplemented with protease inhibitors (Roche Diagnostics). A total of 1 mg of protein extract was incubated with 10 µL of anti-HA agarose (Sigma-Aldrich, A 2095). Immunoprecipitates were eluted in Tris-glycine SDS sample buffer and size fractionated on Novex 3%-8% Tris-Acetate gel (Invitrogen).
Quantitative RT-PCR
Total RNA was prepared using RNeasy kit (Qiagen) as suggested by the manufacturer. 1 microgram of total RNA was reverse transcribed with SuperScript III First-Strand Synthesis Supermix (Invitrogen) and used for quantitative RT-PCR using Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen) according to the manufacturer's instruction. Primers are listed in supplemental material.
Supplementary Material
Highlights.
Human SLX4-MUS81-EME1 and SLX4-SLX1 are both required for in vivo HJ resolution
SLX4-dependent HJ resolution is essential in the absence of HJ dissolution by BLM
HJ resolvase GEN1 is essential in the absence of SLX4
HJ resolution is required for chromosomal integrity during unperturbed S phase
Acknowledgments
This work was supported in part by the Anderson Cancer Center at Rockefeller University, Burroughs Wellcome Fund Career Award for Medical Scientists, Starr Center Consortium grant, the Doris Duke Clinical Scientist Development Award, and the National Center for Research Resources (grant UL1RR024143). A.S. is a Rita Allen Foundation scholar, an Irma T. Hirschl scholar, and an Alexandrine and Alexander Sinsheimer Foundation scholar. Y.K. was partially supported by the Sookmyung Women's University research grant 1-1303-0055. M.C.K. is supported by an American Cancer Society-J.T.Tai postdoctoral fellowship. We are grateful to Jayanta Chaudhuri (Memorial Sloan Kettering Cancer Center) for the kind generosity with anti-GEN1 antibody and to Ray Monnat Jr. (University of Washington) and Matthew Whitby (University of Oxford) for sharing RusA expression vectors. We also thank Alison North and members of the bio-imaging resource center at the Rockefeller University for expert advice and encouragement and Mayte Suarez-Farinas and Joel Correa da Rosa for assistance with statistical analysis. We are grateful to members of the Smogorzewska lab especially Anderson Wang for helpful discussion and critical reading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andersen SL, et al. Drosophila MUS312 and the vertebrate ortholog BTBD12 interact with DNA structure-specific endonucleases in DNA repair and recombination. Mol Cell. 2009;35:128–135. doi: 10.1016/j.molcel.2009.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy MN, et al. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell. 2001;107:537–548. doi: 10.1016/s0092-8674(01)00536-0. [DOI] [PubMed] [Google Scholar]
- Cejka P, et al. Rmi1 stimulates decatenation of double Holliday junctions during dissolution by Sgs1-Top3. Nat Struct Mol Biol. 2010;17:1377–1382. doi: 10.1038/nsmb.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaganti RS, et al. A manyfold increase in sister chromatid exchanges in Bloom's syndrome lymphocytes. Proc Natl Acad Sci U S A. 1974;71:4508–4512. doi: 10.1073/pnas.71.11.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SN, et al. Sequence specificity and biochemical characterization of the RusA Holliday junction resolvase of Escherichia coli. J Biol Chem. 1997;272:14873–14882. doi: 10.1074/jbc.272.23.14873. [DOI] [PubMed] [Google Scholar]
- Chen XB, et al. Human Mus81-associated endonuclease cleaves Holliday junctions in vitro. Mol Cell. 2001;8:1117–1127. doi: 10.1016/s1097-2765(01)00375-6. [DOI] [PubMed] [Google Scholar]
- Chu WK, et al. BLM has early and late functions in homologous recombination repair in mouse embryonic stem cells. Oncogene. 2010;29:4705–4714. doi: 10.1038/onc.2010.214. [DOI] [PubMed] [Google Scholar]
- Constantinou A, et al. Holliday junction resolution in human cells: two junction endonucleases with distinct substrate specificities. EMBO J. 2002;21:5577–5585. doi: 10.1093/emboj/cdf554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe CL, et al. Partial suppression of the fission yeast rqh1(−) phenotype by expression of a bacterial Holliday junction resolvase. EMBO J. 2000;19:2751–2762. doi: 10.1093/emboj/19.11.2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekairi S, et al. Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell. 2009;138:78–89. doi: 10.1016/j.cell.2009.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R. Mechanism for Gene Conversion in Fungi. Genet Res. 1964;5 doi: 10.1017/S0016672308009476. 282-&. [DOI] [PubMed] [Google Scholar]
- Ip SC, et al. Identification of Holliday junction resolvases from humans and yeast. Nature. 2008;456:357–361. doi: 10.1038/nature07470. [DOI] [PubMed] [Google Scholar]
- Kim Y, et al. Mutations of the SLX4 gene in Fanconi anemia. Nat Genet. 2011;43:142–146. doi: 10.1038/ng.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, et al. Regulation of multiple DNA repair pathways by the Fanconi anemia protein SLX4. Blood. 2013;121:54–63. doi: 10.1182/blood-2012-07-441212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottemann MC, Smogorzewska A. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature. 2013;493:356–363. doi: 10.1038/nature11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuraoka I, et al. Repair of an interstrand DNA cross-link initiated by ERCC1-XPF repair/recombination nuclease. J Biol Chem. 2000;275:26632–26636. doi: 10.1074/jbc.C000337200. [DOI] [PubMed] [Google Scholar]
- Latt SA, et al. Induction by alkylating agents of sister chromatid exchanges and chromatid breaks in Fanconi's anemia. Proc Natl Acad Sci U S A. 1975;72:4066–4070. doi: 10.1073/pnas.72.10.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Heyer WD. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 2008;18:99–113. doi: 10.1038/cr.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos J, et al. Regulatory control of the resolution of DNA recombination intermediates during meiosis and mitosis. Cell. 2011;147:158–172. doi: 10.1016/j.cell.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen, et al. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics. 2001;157:103–118. doi: 10.1093/genetics/157.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz IM, et al. Coordination of structure-specific nucleases by human SLX4/BTBD12 is required for DNA repair. Mol Cell. 2009;35:116–127. doi: 10.1016/j.molcel.2009.06.020. [DOI] [PubMed] [Google Scholar]
- Ray Chaudhuri A, et al. Topoisomerase I poisoning results in PARP-mediated replication fork reversal. Nat Struct Mol Biol. 2012;19:417–423. doi: 10.1038/nsmb.2258. [DOI] [PubMed] [Google Scholar]
- Rouse J. Control of genome stability by SLX protein complexes. Biochem Soc Trans. 2009;37:495–510. doi: 10.1042/BST0370495. [DOI] [PubMed] [Google Scholar]
- Saintigny Y, et al. Homologous recombination resolution defect in werner syndrome. Mol Cell Biol. 2002;22:6971–6978. doi: 10.1128/MCB.22.20.6971-6978.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz EK, Heyer WD. Processing of joint molecule intermediates by structure-selective endonucleases during homologous recombination in eukaryotes. Chromosoma. 2011;120:109–127. doi: 10.1007/s00412-010-0304-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharples GJ, et al. Holliday junction processing in bacteria: insights from the evolutionary conservation of RuvABC, RecG, and RusA. J Bacteriol. 1999;181:5543–5550. doi: 10.1128/jb.181.18.5543-5550.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoepker C, et al. SLX4, a coordinator of structure-specific endonucleases, is mutated in a new Fanconi anemia subtype. Nat Genet. 2011;43:138–141. doi: 10.1038/ng.751. [DOI] [PubMed] [Google Scholar]
- Svendsen JM, et al. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell. 2009;138:63–77. doi: 10.1016/j.cell.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak JW, et al. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Taylor ER, McGowan CH. Cleavage mechanism of human Mus81-Eme1 acting on Holliday-junction structures. Proc Natl Acad Sci U S A. 2008;105:3757–3762. doi: 10.1073/pnas.0710291105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler, et al. Aberrant chromosome morphology in human cells defective for Holliday junction resolution. Nature. 2011;471:642–646. doi: 10.1038/nature09790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Hickson ID. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.