Abstract
Misfolding and subsequent aggregation of alpha-synuclein (α-Syn) protein are critically involved in the development of several neurodegenerative diseases, including Parkinson’s disease (PD). Three familial single point mutations, A30P, E46K, and A53T, correlate with early-onset PD; however the molecular mechanism of the effects of these mutations on the structural properties of α-Syn and its propensity to misfold remains unclear. Here, we address this issue utilizing a single molecule AFM force spectroscopy approach in which structural details of dimers formed by all four variants of α-Syn are characterized. Analysis of the force spectroscopy data reflecting contour length distribution for α-Syn dimer dissociation suggests that multiple segments are involved in the assembly of the dimer. The interactions are not limited to the central non-amyloid-beta component (NAC) of the protein, but rather expand beyond this segment. All three mutations alter the protein’s folding and interaction patterns affecting interactions far beyond their immediate locations. Implementation of these findings to our understanding of α-Syn aggregation pathways is discussed.
The aggregation of alpha-synuclein (α-Syn) is a hallmark of several neurodegenerative diseases including Parkinson’s disease. α-Syn is an abundant presynaptic protein that belongs to a group of natively unfolded proteins lacking secondary/tertiary structure in aqueous solutions.1, 2 Although unstructured, some preferences for adopting α-helical structure have been observed in alpha-synuclein primarily within the N-terminal domain and the non-amyloid-beta component (NAC) region when the protein binds to lipids of membranes and vesicles.3 Structural transitions of α-Syn are also involved in the formation of amyloid-like fibrils with a high content of β-sheet secondary structure in vivo and under a variety of conditions in vitro.4–7
Various studies established that the aggregation rate of α-Syn depends on environmental conditions and is accelerated by various factors such as low pH and the presence of biogenic polyamines.8–14 Changes in primary sequence associated with early-onset familial Parkinson’s (A30P,10 E46K12 and A53T11 single point mutants) also enhance the rate of protein aggregation compared to the wild-type. The pathway of aggregation, however, was found to differ for each mutant. Whereas, E46K and A53T aggregate more rapidly than WT with the formation of fibrils,15, 16 A30P assembles primarily in oligomeric forms.17, 18 Unraveling the effects of these factors on misfolding of α-Syn is important for understanding the molecular basis of the disease development.
The main challenge in studying protein misfolding phenomena is that misfolded conformations that lead to the pathological aggregation of a protein are only transiently populated. The ensemble for some proteins consists of rapidly interconverting compact conformations indicating the highly dynamic nature of protein conformational space.19 It was recently demonstrated that the aggregation propensity of α-Syn depends on how fast the protein accesses aggregation-prone conformations – a process termed reconfiguration rate.20–22 A monomeric protein must undergo structural reorganization to adopt transient conformational states (misfolded conformations) in order to initiate aggregation.23 Interactions between misfolded proteins facilitate the very first step in the aggregation process – dimerization. Subpopulation of misfolded states in the overall ensemble can be very small. More importantly it is not obvious which of the many species in the conformational space lead to pathological aggregation. Increasing evidence suggests that α-Syn populates an ensemble of conformers ranging from extended to highly collapsed forms in aqueous solutions.24, 25 Conditions that favor more compact structures have been reported to significantly increase aggregation rates of α-Syn.26–28 Ensemble-based (bulk) methods, however, cannot access a wealth of information that a heterogeneous ensemble of alpha-synuclein conformations might offer with respect to the aggregation process.
Single-molecule techniques can characterize properties of these individual molecules without contribution from the rest of the ensemble. Recently, AFM single-molecule force spectroscopy (SMFS) has been applied to study the conformational diversity of monomeric α-Syn.27, 28 The study identified three main distinct conformations that exist within monomeric alpha-synuclein based on their resistance to mechanical pulling. These conformations were classified as random coil, mechanically weak, and beta-like structures. The latter were proposed to be directly related to the aggregation of α-Syn.28 Interestingly, pathological conditions which are known to increase the propensity of alpha-synuclein to aggregate, such as high ionic strength and the presence of Cu2+, as well as mutations A30P, A53T and E46K,27 shifted the conformational equilibrium towards these beta-like structures. We have approached the misfolding problem differently. We have used a recently proposed novel method to characterize misfolded states in dimeric forms of amyloidogenic proteins rather than monomeric proteins (reviewed in 29). In this approach, interactions between proteins immobilized on an AFM tip and the probed surface are measured with force spectroscopy with significant rupture forces, suggesting that misfolded proteins form dimeric states. Therefore, instead of probing the whole conformational space of a protein we probe only conformers that are capable of forming dimers – the simplest aggregated species. The advantage of this methodology is, therefore, that it probes conformers that associated with one another at the single-molecule level. We have recently applied this methodology to characterize the effect of spermidine on α-Syn misfolding.14
Here we report an AFM force spectroscopy study of α-Syn folding and the effect of single point mutations of α-Syn on the structure of dimeric contact. Pair-wise interactions were probed between individual α-Syn molecules at low pH conditions that induce conformational transitions associated with enhanced aggregation. We applied the rupture length analysis to identify segments of α-Syn involved in the formation of dimers. This analysis showed that only a limited span of the C-terminal region of wild-type α-Syn is involved in the stabilization of dimers. Importantly, the single-point mutations located within the N-terminal region of the protein dramatically change the pattern of protein misfolding by extending interactions into the C-terminal region. These results suggest that the single point mutations affect distantly located interacting segments of the misfolded α-Syn. A model of the protein assembly involving misfolded dimers is proposed.
MATERIALS AND METHODS
Materials
The 20 mM stock solution of β-mercaptoethanol was prepared in water. The 2.92 mM stock solution of maleimide-polyethylene glycol-silatrane (MAS)30 was dissolved in water and stored at −20 °C. The buffers used in force measurement experiments were: pH 3.7 (10 mM sodium acetate/acetic acid) and pH 7.0 (PBS buffer). All buffers were adjusted by sodium chloride to a final ionic strength of 150 mM. All other reagents used were of analytical grade from Sigma-Aldrich unless specified otherwise. Deionized water (18.2 MΩ, 0.22 μm pore size filter, APS Water Services Corp., Van Nuys, CA) was used for all experiments.
Preparation of recombinant α-Syn proteins
Wild type A140C α-synuclein in which the C-terminal alanine was replaced with a cysteine and the double mutant A30P-A140C were prepared as described in 14. A cDNA encoding human α-Syn E46K or A53T was amplified by PCR and subcloned as an Nde I - Hind III fragment into the vector pT7-7, yielding the constructs pT7-E46K and pT7-A53T, respectively. The double mutants E46K-A140C A53T-A140C were generated by replacing a 354-base pair fragment from pT7-7 A140C (excised by digesting with Nde I and BamH I) with the equivalent DNA fragment from pT7-E46K or pT7-A53T, respectively. The sequence of the α-Syn-encoding insert in each construct was verified using an Applied Biosystems (ABI 3730 XL) DNA sequencer. Each α-Syn variant was expressed in the E. coli strain BL21 (DE3) and purified as described in 14, 31. To ensure that the alpha-synuclein variants were purified as full-length, intact proteins, each lot of protein was characterized using two methods.14 As one approach, each protein was analyzed by reacting with 5,5′-dithiobis-(2-nitrobenzoic acid) to determine levels of reduced cysteine. From measurements of the absorbance of protein-bound 2-nitro-5-thiobenzoate at 412 nm, it was determined that the ratio of cysteine residues to protein molecules was approximately 1:1. As a second approach, each α-Syn variant was further purified by reverse phase high performance liquid chromatography and analyzed by matrix assisted laser desorption ionization (MALDI) mass spectrometry. The mass-to-charge (m/z) values obtained from the MALDI analysis corresponded to the predicted values for full-length α-Syn.
Tip modification
Silicon nitride (Si3N4) AFM tips (SNL-10, Bruker AFM Probes) were immersed in ethanol for 30 min, rinsed thoroughly with water and dried followed by UV treatment for 30 min (CL-1000 Ultraviolet Crosslinker, UVP, Upland, CA). After the UV treatment, AFM tips were immersed in an aqueous solution of 167 μM maleimide silatrane (MAS) for 3 h followed by multiple rinses in water. For covalent attachment of α-Syn to MAS functionalized tips, a 19 nM protein solution in PBS buffer, pH 7.0, was treated with 1 mM DTT (to reduce any disulfide bonds formed between α-Syn molecules) for 10 min and filtered through 10 kDa MWCO centrifuge filter devices (Amicon Ultra-0.5, Millipore) 3 times. MAS functionalized tips were immersed in a 19 nM solution of α-Syn for 1 h. After rinsing with PBS buffer, unreacted maleimide moieties were quenched with 10 mM β-mercaptoethanol in PBS for 10 min. These functionalized probes were washed with PBS and immediately used for force measurements.
Surface modification
Mica sheets (Asheville-Schoonmaker Mica Co., Newport News, VA) were cut into ~1.5 cm×1.5 cm squares and glued onto glass slides using epoxy glue EPO-TEK 353ND (Epoxy Technology, Inc., Billerica, MA). Freshly cleaved mica surfaces were treated with MAS for 3 hours. MAS modified mica squares were rinsed with water several times to remove non-bound MAS, incubated with a 19 nM α-Syn solution (in PBS, prepared as described in the previous section) for 1 h, and rinsed with PBS buffer. Unreacted maleimide moieties were quenched with 10 mM β-mercaptoethanol in PBS buffer for 10 min. The functionalized mica surfaces were washed with PBS buffer and immediately used for force measurements. The low concentration of alpha-synuclein (19 nM) for surface modification was previously identified to provide a sparse surface coverage of monomeric protein. AFM imaging was used to verify that there was only one pair of molecules interacting within the contact area in the force measurements.40
Force measurements
Force–distance measurements were performed in either PBS buffer or acetate buffer (pH 3.7) at room temperature using the Force Robot 300 (JPK Instruments, Berlin, Germany). Silicon nitride cantilevers (SNL-10, Bruker AFM probes) with nominal values of spring constants of ~0.1 N/m were used. Spring constants for each cantilever were obtained using a thermal method with the Force Robot 300 instrument. The ramp size was 200 nm with approach and retraction velocities kept constant at 600 nm/s. A low trigger force (200 pN) was applied. Force-distance curves were acquired systematically at different points on the surface set by a grid (5 by 5 μm) where points were separated from each other by 50 nm. The dwell time was set at 0.5 s.
Data analysis
Thousands of force-distance curves were collected for each α-Syn variant. The curves were filtered using the Data Processing (DP) software of the JPK Force Robot 300 instrument selecting those that contain rupture events. The curves were then fitted with a Worm Like Chain (WLC) model incorporated into the DP software of JPK. The WLC model was chosen because it describes well the elasticity of a polypeptide chain.32–34 Rupture force value was measured at the rupture point. The contour length (LC) was evaluated as a variable parameter for the best fit of the curve while keeping the persistence length constant at 0.4 nm. This value of persistence length was found to adequately describe a variety of proteins.32, 35, 36 Measured values of contour length were assembled into statistical histograms and fitted in MagicPlot Pro 2.0 software with Gaussian functions to determine maxima of LC distributions (LC,MAX ± SD). The length of the alpha-synuclein segment that undergoes stretching upon applied force was estimated following an approach proposed earlier.37 The length of this segment was calculated by subtracting the length of the MAS linker (3 nm) from the total measured contour length and taking into account that one amino acid residue contributes 0.34 nm to the total contour length.38
RESULTS
Experimental system and approach
α-Syn molecules were covalently attached to an AFM probe and the mica surface via maleimide – cysteine coupling for the force spectroscopy experiments. The wild-type α-Syn sequence and its mutants (A30P, A53T and E46K) do not contain cysteine residues except the one we introduced at the very end of the C-terminus (A140C). The cysteine-maleimide coupling chemistry ensured very site-specific immobilization of the protein on the AFM substrate and tip surfaces required for quantitative analysis of the rupture lengths.14, 29, 30 The following structural properties of α-Syn were considered for using the C-terminus for immobilization. There are three distinct domains in the primary structure of α-Syn: N-terminus (1–60 aa), NAC region (61–95 aa), and C-terminus (96–140 aa). We chose the end of the C-terminus as the point of α-Syn covalent attachment because structural changes in the protein were suggested to occur only within the N-terminal part and the NAC region of the protein.3 The N-terminal and NAC regions are responsible for the transition to alpha-helical structure and association with lipids.3 The NAC region was also found in fibrils and accommodates β-sheet secondary structure.39 On the other hand, the C-terminus of α-Syn is a highly acidic and proline-rich region that has been suggested to be in a fully random conformation in both free and lipid bound forms of α-Syn.3 The C-terminal region was expected to act as a flexible linker for probing interactions between α-Syn molecules in force measurements. Therefore, we have used only short linker within MAS tether (4 units of ethylene glycol) for covalent attachment of α-Syn molecules.
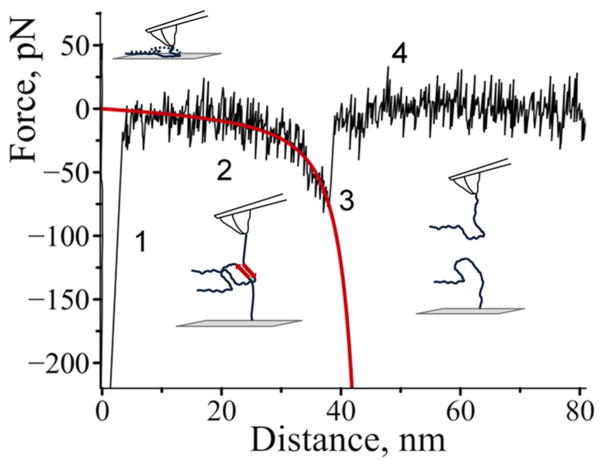
Interactions between covalently attached α-Syn molecules were probed in multiple approach-retraction cycles at various positions on the surface. If no stable dimeric contact is formed the retraction of the tip does not detect any rupture event. However, a stable dimeric contact produces a specific rupture signature in the force-distance curve. The beginning of such a force curve usually contains an adhesion peak (1) due to short-range non-specific interactions between the tip and the surface (illustrated in Figure 1). The adhesion peak is followed by a complete rupture event of dimeric contact (3) where the tip comes free from the surface (4). The rupture event is preceded by a part characteristic of the stretching of polymer linkers (2).
Figure 1.
Typical regions on the force-distance curves: (1) an adhesion peak due to short-range non-specific interactions between the tip and the surface, (2) gradual increase in force characteristic of polymer stretching, (3) complete rupture and (4) region where tip comes free from the surface.
The control experiments carried out with only one of the surfaces carrying protein (modified tip/unmodified surface or modified surface/unmodified tip) show no or only a few rupture events in force-distance curves (Fig. S1). These control experiments suggest that the ruptures we observed are not due to non-specific interactions of α-Syn molecules with the surface or AFM tip but rather specific interactions which are probed only when a contact between α-Syn molecules is established. The formation of a dimer makes it possible to observe subtle features of α-Syn dimeric structure and conformational characteristics of misfolded proteins that normally would not be detectable using ensemble measurements. Notably, detectable interactions were also observed between α-Syn at neutral pH in the presence of spermidine.14
Effect of pH on misfolding of α-Syn
We have shown recently 14, 40 that the interactions between α-Syn molecules are very weak at neutral pH, and this finding correlates with the low aggregation propensity of α-Syn at this pH.41 Several recent studies have utilized low pH to convert α-Syn to its aggregation-prone state.8, 9, 20, 42–45 We also used low pH to stimulate misfolding of α-Syn in our experiments. The effect of pH is evident from Fig. 2A which shows a set of representative force-distance curves measured for wild type α-Syn at neutral pH. No rupture events were observed beyond the short range adhesion peak. Next, we continued to probe interactions using the same tip and the same substrate but replaced the buffer in the cell to perform probing at a lower pH value − 3.7. Fig. 2B demonstrates that at low pH specific interactions appear in the force-distance curves. In addition to curve (i) with a shape similar to the one detected at neutral pH, clear rupture events were detected (curves (ii and iii). We assembled the rupture events into three major groups: NR - force curves with no rupture events (Fig. 2B-i), SR - curves that feature a single rupture event (Fig. 2B-ii) and MR - curves with multiple ruptures (Fig. 2B-iii).
Figure 2.
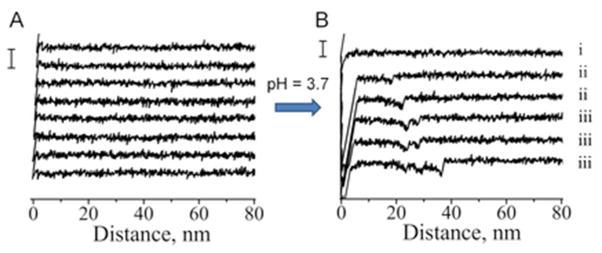
Representative force-distance curves for wild type α-Syn. A) measured at neutral pH (scale bar 100 pN) and B) measured at low pH = 3.7 (scale bar 100 pN): i) with no rupture events (group NR), ii) single ruptures (group SR) and iii) multiple ruptures (group MR).
Force distance curves with single rupture: Wild type protein
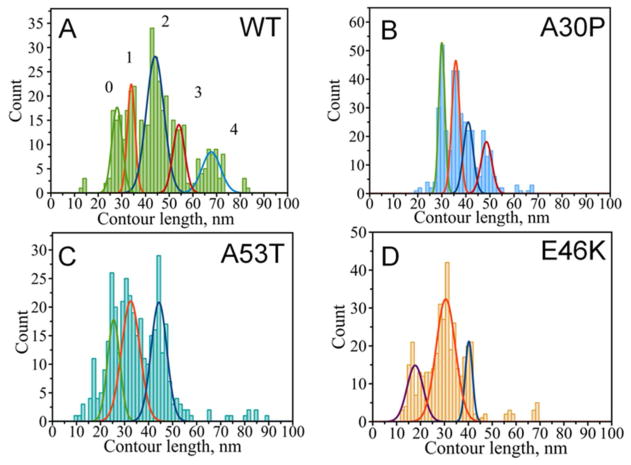
First, we analyzed force distance curves that belong to group SR with a single rupture peak. We followed an approach recently developed in our lab that allows localizing segments of the protein involved in dimer formation.37, 40, 46 Using this approach we characterized structural differences of misfolded α-Syn variants detected at low pH. Each force-distance curve with a specific rupture event was fitted with a worm-like chain model providing a contour length (LC) that characterizes the length of stretchable parts of α-Syn molecules not involved in dimeric interaction. A statistical histogram of LC values measured this way for SR force distance curves of WT α-Syn is shown in Fig. 3A. The distribution has a set of clear features and was approximated with five peaks representing the most reproducible positions of the rupture events. A Gaussian fit of the histogram resulted in maxima of the distribution at 28 nm (peak 0), 34 nm (peak 1), 44 nm (peak 2), 54 nm (peak 3) and 68 nm (peak 4). The most prominent peak appears at 44 nm which contributes 42% of all detected single rupture curves for WT α-Syn. The fact that the distribution reveals several peaks suggests that there is more than one conformation of WT α-Syn capable of forming the dimer.
Figure 3.
Contour length distribution for alpha-synuclein variants – group B force-distance curves with a single rupture. A) wild type (with maxima at 28±2 nm (peak 0), 34±2 nm (peak 1), 44±4 nm (peak 2), 54±3 nm (peak 3) and 68±4 nm (peak 4)), B) A30P mutant (with maxima at 30±1, 36±2, 41±2 and 49±2 nm), C) A53T mutant (with maxima at 26±3, 33±4 and 44±3 nm), D) E46K mutant (with maxima at 17±3, 31±4 and 40±2 nm).
Force distance curves with single rupture: Mutants
Next we probed interactions involving pathological single point mutants which correlate with familial early onset Parkinson’s disease, A30P, A53T and E46K.10–12 Again, similar to wild type α-Syn, probing interactions involving mutants at pH 7.0 did not reveal specific rupture events. However, specific rupture events appeared on the force-distance curves measured at pH = 3.7. Similar to WT α-Syn, all three mutants had three major groups of force-distance curves: NR (no rupture), SR (single rupture) and MR (multiple ruptures). Results of force-distance curve analysis are shown in Figs. 3B–D. Similar to the data for WT α-Syn, the distributions of contour lengths have specific features, although they appear simpler with fewer peaks present and slightly narrower than for WT α-Syn (Figs. 3, A–D). The distributions reveal different patterns for each α-Syn variant suggesting the existence of more than one conformation within the dimer. The major effect of the contour length measurements is a dramatic sensitivity of the contour length profiles upon introducing mutations that can be seen without any additional analysis (Fig. 3). This effect reflects the change of the interprotein interaction and we used fitting of the overall contour lengths distribution profiles with a set of peaks to facilitate the data interpretation. Four prominent peaks are clearly seen for the A30P mutant with maxima of Gaussians at 30, 36, 41 and 49 nm (Fig. 3, B). Both A53T and E46K mutants had their contour length distributions skewed to shorter LC values as compared to both WT and A30P (Figs. 3, C,D). The majority of the rupture events for all α-Syn variants appear within the range of ~ 26 nm to ~ 50 nm. Although, there were other interactions observed at longer and shorter distances, these had a lower probability than the peaks in the 26–50 nm range. Moreover, other peaks outside this region are not present for all the variants. For example, the shortest peak observed for E46K at 17 nm is not present in the histogram for other variants. Also the longest peak at 68 nm does not have a counterpart in histograms for the mutants suggesting that the interaction producing this long rupture event is specific only for WT.
The comparison of contour length distributions shown in Fig. 3 and Table 1 demonstrates the discrete distribution of the rupture events as revealed by contour lengths. However two major peaks, peak 1 at ~ 30–34 nm and peak 2 at ~40–44 nm appear in all the mutants and WT protein. All mutations remove distant interactions (peak ~ 68 nm for the WT α-Syn sample), but short interactions appear in the A53T and E46K distributions. Also, similar interactions (peak 1 and peak 2) have different contributions to the overall interaction pattern. For example, interactions of WT are dominated by peak 2 which comprises 42% of all the detected interactions. Peak 1 contributes only 15%. In contrast, the A53T mutant has equal contributions of these two peaks at 31% each, and A30P has almost equal contributions of peak 1 and peak 2, 30% and 24% respectively. The E46K profile is the opposite of the WT α-Syn profile with peak 1 dominating at 61% compared to 15% for peak 2. These data show preferences of conformations for these variants. E46K has shorter conformations while WT has longer conformations and A53T and A30P are somewhere in the middle.
Table 1.
Relative contribution of single (SR) and multiple rupture (MR) curves for different variants. Corresponding contour lengths are also shown with relative contribution of each peak.
| SR # of curves | LC | MR # of curves | LC (last) | LC (internal) | |
|---|---|---|---|---|---|
|
| |||||
| A30P | 398 | 30±1 (28 %) | 53 | 25±3 (24 %) | 17±5 (27 %) |
| 36±2 (33 %) | - | 34±5 (73 %) | |||
| 41±2 (22 %) | 38±3 (62 %) | ||||
| 49±2 (17 %) | 48±1 (14 %) | ||||
|
| |||||
| WT | 421 | 28±2 (16 %) | 233 | 25±3 (25 %) | 15±2 (20 %) |
| 34±2 (15 %) | - | 23±4 (50 %) | |||
| 44±4 (42 %) | 40±5 (55 %) | 33±3 (17 %) | |||
| 54±3 (14 %) | 50±1 (11 %) | 43±5 (13 %) | |||
| 68±4 (13 %) | 63±3 (9 %) | ||||
|
| |||||
| A53T | 372 | 26±3 (24 %) | 674 | 28±6 (28 %) | 13±2 (19 %) |
| 33±4 (41 %) | - | 21±4 (32 %) | |||
| 44±3 (35 %) | 38±2 (30 %) | 30±2 (27 %) | |||
| 48±4 (42 %) | 39±5 (22 %) | ||||
|
| |||||
| E46K | 317 | 17±3 (24 %) | 498 | - | 19±3 (39 %) |
| 31±4 (61 %) | - | 26±3 (31 %) | |||
| 40±2 (15 %) | 41±3 (100 %) | 34±4 (24 %) | |||
| 40±2 (6 %) | |||||
Force distance curves with a multiple-rupture pattern
The available set of data allowed us to analyze group MR force distance curves characterized by a multiple-rupture pattern similar to the one shown in Fig. 2B-iii. We have compared the relative contribution of group SR vs. group MR force-distance curves in the overall set of the force distance curves with specific rupture events. The A30P mutant had a majority of the force curves with single rupture peaks, which contributed 81% of all force-distance curves with specific rupture events. The relative contribution of SR curves for all the variants decreased in the following order A30P>WT>A53T>E46K (81%>63%>37%>24%). We also characterized the force curves by the number of multiple events termed the curve complexity. Complexity of the curves, defined by the number of rupture peaks in a single force distance curve, increased in the following order: A30P<WT<A53T<E46K. The E46K mutant had the highest degree of force curve complexity among all the variants.
Force distance curves with a multiple-rupture pattern: Wild type protein
Here we analyzed force distance curves with a multiple-rupture pattern. A representative curve of this kind is shown on Fig. 2B-iii. We interpreted the last rupture peak as the one leading to dimer dissociation and the rest of the rupture peaks were termed internal ruptures. They correspond to the rupture of interactions between protein segments involved in intramolecular or intermolecular α-Syn folding. Both the last rupture in MR curves and the single rupture in SR curves correspond to force-induced dissociation of dimers, and, therefore, it is instructive to compare these rupture events with each other. Analysis of contour length for both the last rupture peaks in group MR and the single rupture peaks in group SR is shown on Fig. 4A and Fig. 4B. The histograms are very similar with minor variations. These variations are manifested in slight shift of peak maxima to smaller values in the case of the group MR distribution. It appears that the relative contribution of any particular peak is similar for the last rupture histogram (group MR) and the histogram for group SR. For example, a peak at 44 nm (corresponding to the peak 2 of the SR distribution) is dominant in both histograms. Interestingly, a peak at 34 nm (corresponding to the peak 1 of the SR distribution) is not present in the histogram of contour lengths measured for the last ruptures (Fig. 4, B). Although, there might be some events registered at that distance, it is obvious that they do not make up a clear peak leaving a gap between the peak at 25 nm and the peak at 40 nm.
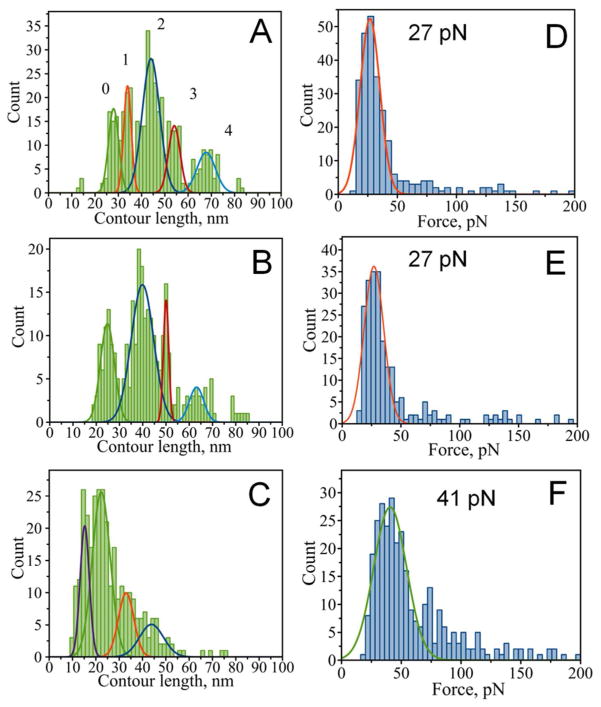
Figure 4.
Rupture contour length histograms (A,B,C) and rupture force histograms (D,E,F) for wild type α-Syn. A and D) single rupture event group (with contour length maxima at 28±2, 34±2, 44±4, 54±3 and 68±4 nm and a maximal rupture force of 27±8 pN), B and E) last events from multiple rupture group (with contour length maxima at 25±3, 40±5, 50±1 and 63±3 nm and a maximal rupture force of 27±8 pN), C and F) internal ruptures from multiple rupture group (with contour length maxima at 15±2, 23±4, 33±3, and 43±5 nm and a maximal rupture force of 41±14 pN).
Next we analyzed the contour length for internal ruptures of the MR force distance curves. The LC distribution for these ruptures is shown in Fig. 4, C. In general, the distribution of contour lengths is shifted to smaller values as compared to both the last event of MR curves (Fig. 4, B) and the single event in SR curves (Fig. 4, A). Such behavior of the contour length distributions is expected for the sequential rupture of multiple contacts established between polypeptide chains of α-Syn. A peak corresponding to the peak 1 of the group SR distribution (with a maximum contour length of 33 nm) appeared again when we plotted the contour length distribution for internal ruptures as illustrated by Fig. 4, C. A peak corresponding to peak 2 of the SR distribution (with a maximum contour length of 43 nm) is also present; however, it no longer dominates the distribution.
Force distance curves with a multiple-rupture pattern: Single point mutants
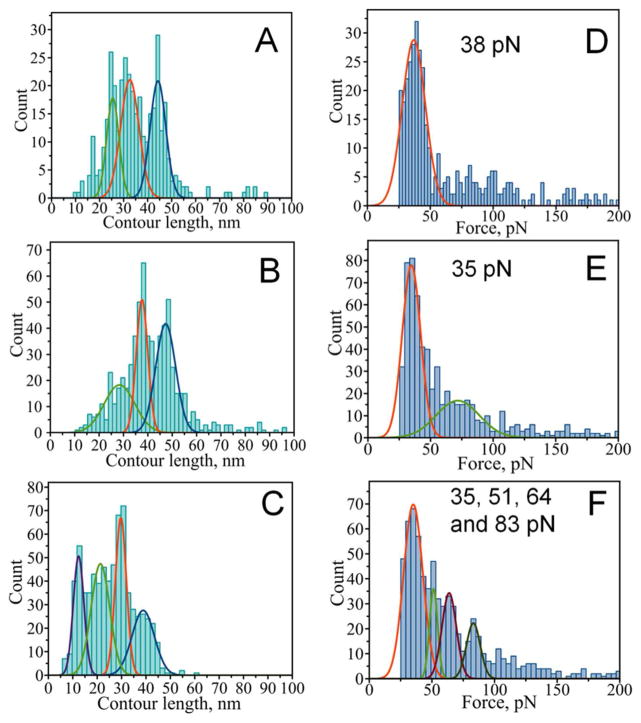
We have also analyzed force distance curves with a multiple-rupture pattern for the pathological mutants of α-Syn. Comparison of contour lengths for both the last rupture peaks in group MR and the single rupture peaks in group SR revealed that A30P and A53T mutants behave similarly to WT. Figures 5, A and B show histograms for single rupture peaks (SR curves) and last rupture peaks (MR curves) for A53T mutant. Again, peak 4 (at 63 nm) characteristic for WT is not present in the histogram for the mutant. Peak 1 with maximum at ~30 nm does not completely disappear for A53T as it does for WT but its contribution is smaller than in the histogram for single rupture curves (Fig. 5, A and B). A30P mutant has very similar to A53T histograms of contour length for both types of analyzed rupture events with only minor variations (Fig. S2). Similar to WT, a peak with a maximum contour length of ~30 nm vanishes in the histogram of last rupture peaks for A30P (Fig. S2, A and B). Also, despite much complexity in the number of curves with multiple ruptures and the number of ruptures per curve, the contour length distribution for the E46K mutant features only a single peak at 40 nm when the last rupture is analyzed (Fig. S3, B). Interestingly, while group SR events for all the mutants (particularly E46K) are dominated by a peak at ~30 nm, a peak at ~40 nm becomes dominant in the last rupture events of MR curves for all the mutants.
Figure 5.
Rupture contour length histograms (A,B,C) and rupture force histograms (D,E,F) for the A53T mutant of α-Syn. A and D single rupture event group (with contour length maxima at 26±3, 33±4 and 44±3 nm and a maximal rupture force of 38±9 pN), B) last events from multiple rupture group (with contour length maxima at 28±6, 38±2 and 48±4 nm and a maximal rupture force of 35±7 pN), C) internal ruptures from multiple rupture group (with contour length maxima at 13±2, 21±4, 30±2 and 39±5 nm and rupture force maxima of 35±8, 51±3, 64±6, and 83±6 pN).
The internal rupture events of the group MR force distance curves were also analyzed. The LC distribution for these ruptures is shown in Figures 5-C, S2-C and S3-C. In general, similar to WT protein, the distributions are shifted to smaller values as compared to both single events in simple curves (Figs. 5-A, S2-A and S3-A) and the last event of complex curves (Figs. 5-B, S2-B and S3-B).
Rupture forces measured for all the mutants are also similar to WT (Figs. 5, S2 and S3). Although, generally similar there are differences among the variants. For example, the E46K mutant had a double distribution of rupture forces at 27 and 50 pN for the last event of MR curves compared to a single peaked distribution for SR curves (Fig. S3, D and E). The second peak at 50 pN resembles forces detected for internal events of E46K, which peaked at 49 pN (Fig. S3, F). This suggests that last events in MR curves for E46K have a double nature: one that resembles single events and another one resembling internal ruptures with higher force. The rupture forces of the A53T mutant also had a higher contribution of large forces for internal ruptures. Figure 5 shows that in addition to small forces (35 pN; Fig 5, E first peak) similar to the forces of the single rupture events we have also observed a contribution from larger forces (72 pN; Fig. 5, E second peak) for the A53T mutant. This situation is opposite to WT (Fig. 4) and A30P (Fig. S2) but similar to E46K mutant (Fig. S3).
To correlate the rupture force values with the contour lengths we made 2D scatter plots and histograms of the rupture forces and contour lengths in one combined plot shown in Figures S7–S10. Such plots illustrate graphically that the majority of ruptures take place at low force and this dependence is common for all LC peaks.
DISCUSSION
Single molecule force spectroscopy probing of interactions between α-Syn molecules within a dimeric contact provided new insight into the pattern of α-Syn assembly within the dimers. Reproducible interactions suggest that α-Syn structure within the dimers is characterized by a set of well-defined conformations. We used the approach described in 37, 46 to characterize the protein segments involved in the α-Syn folding and interactions. In our immobilization procedure, the proteins are covalently bound to the tip and surface at the C-terminus. Therefore, the position of the peak on the force-distance curve is defined by the protein length between the C-terminus and the interacting segment. The closer the segment to C-terminus, the shorter the contour length measured with AFM. This is schematically shown in Fig. 6, A where contour length LC1 is defined by the rupture between the segments shown with red arrows. Longer contour length, LC2, corresponds to the dissociation of distantly located segments shown with blue arrows. Using 0.34 nm as a contribution of one amino acid residue to the total contour length 38 we estimated that LC1 and LC2 correspond to the following C-terminal parts of α-Syn molecules not participating in interactions: ~94–140 and ~79–140 residues respectively matching values of 34 nm (peak1) and 44 nm (peak2) (Fig. S4).
Figure 6.
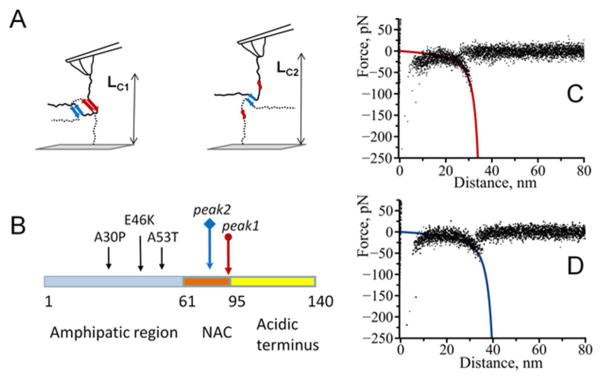
Interaction model of alpha-synuclein molecules. The model describes two (observed for all the variants of alpha-synuclein) major peaks in the contour length histograms as sites of same identity interacting with each other with the formation of a dimer. A) Position of the interacting site further from C-end (point of attachment) results in longer contour length value. B) Positions of the beginning of detected interaction sites. Colored arrows correspond to two detected major interaction sites schematically shown in A), black arrows show the positions of A30P, E46K and A53T mutations in alpha-synuclein. C) and D) superposition of representative force-distance curves for the detected rupture events corresponding to LC1 and LC2.
Structure of WT α-Syn in dimers
According to Fig. 4A, the contour lengths distribution of the SR force-distance curves features five peaks occurring at distinct distances. Peak 1 (34 nm) and peak 2 (44 nm) comprise the majority of the contour lengths; other peaks are located at 54 nm, 68 nm and 28 nm suggesting that protein segments located at these positions are involved in protein folding within dimers. We compared our data on the location of these interacting segments with known structure of α-Syn within fibrils. A recent solid state NMR study of full-length α-Syn fibrils showed an extended core with multiple beta-strands that span residues 38–96.47, 48 Assuming that the C-terminus is a stretched polypeptide chain acting as a linker, residues 38–96 correspond to the contour length values between 72 and 32 nm. Our observed range of contour length values vary between 23 to 73 nm which is close to the estimated LC range based on the NMR data. Figure S4 compares secondary structure of WT α-Syn taken from 48 with the positions of interacting segments estimated based on our contour length analysis. Remarkably, the peaks in the contour length distribution coincide very well with the location of β-strands in the amyloid fibrils of WT α-Syn determined from ssNMR (see table in Figure S4).48 Despite this similarity, there are some differences: 1) a peak at 59 nm predicted from the NMR structure is missing in our measurements, and 2) we observe a single peak at 44 nm instead of the predicted two peaks at 42 nm and 48 nm. The majority of events were observed at this contour length (peak 2 = 44 nm) in the LC distribution of WT α-Syn. Interestingly, this interaction coincides with the position of an 11 aa long segment of the NAC region. This segment comprising residues 73–83 has been suggested to determine α-Syn’s ability to form fibrils.39, 49
The multiple rupture events provide additional insight into the α-Syn structure within dimers. Force-induced dissociation of the dimer results in a complete separation of two monomers and thus AFM tip comes free from the surface (segment 4 in Fig. 1). This complete dissociation appears as the last rupture in the force distance curves with multiple rupture peaks. Comparison of the last rupture in MR curves with SR curves suggested these two are very similar. For example, peak 2 at 44 nm is dominant in both histograms. This peak corresponds to the 73–83 aa stretch which is critically involved in α-Syn folding within fibrils.39, 49 Three other peaks at 25, 50 and 63 nm have similar positions in the histogram for group MR events compared to the LC histogram for group SR events. One major difference is the absence of peak 1 at 34 nm in the histogram of contour lengths of the last ruptures (Fig. 4, B).
While the LC distributions of the SR curves and the last rupture peaks of the MR curves are similar, the LC histogram for internal ruptures of the MR curves is quite different. The contour length distribution is skewed to shorter contour length values (Fig. 4, C) with the first peak in the histogram being at LC = 15 nm. This peak corresponds to a very short non-interacting segment ~122–140 aa which is further in the C-terminal region than expected based on NMR studies of fibrillar structures.47, 48 The results of the LC analysis lead to the following assumptions of the structural features of α-Syn within misfolded dimers: 1) the interacting regions which are capable of forming dimers extend beyond the NAC region into the C-terminal part, and 2) the C-terminal part forms a compact configuration which resists external force applied in the pulling experiments.
The first assumption suggests a simplified model where the observed short interactions occur between the segments beyond the NAC region and this model is supported by the recent NMR studies of α-Syn in the free state.44, 50 According to these data, the segment spanning residues 110–120 has a propensity for the formation of β structure. The neutralization of negative charges within the C-terminus at low pH reduces intermolecular repulsion. This effect along with the preference for β structure formation might effectively induce interactions between regions located outside the NAC.
Our second assumption is based on the possibility of the C-terminal region to fold at low pH. Indeed, several studies observed that low pH values induce a collapse of the C-terminal region due to the shielding of negative charges.8, 43, 51, 52 The existence of contacts between different segments due to the compactness of the C-terminal region was observed in AFM single-molecule unfolding of monomeric α-Syn.27, 28 These studies showed that the intramolecular contacts are strong enough to resist mechanical pulling.
Effect of mutations on the structure of α-Syn folding in dimers
Three single point mutations A30P, E46K and A53T of α-Syn identified in familial early-onset PD alter the interaction pattern between molecules characteristic of the SMFS experiments. The following differences between mutants and WT were found: 1) contour length distributions for all three mutants lack the peak at LC = 68 nm (peak 4) and 2) interactions at shorter contour lengths become more dominant.
According to our estimates peak 4 corresponds to the interaction segment starting at around the 45th residue. All three mutations are located in the proximity of this interacting site. Thus, we hypothesize that the absence of the peak at this LC indicates a direct influence of a mutation on the interacting propensity of this segment, thereby reducing its ability to form a stable dimeric contact. This finding suggests that the protein’s segment around the 45th residue is structured and involved in the protein folding, but mutations destabilize this structure. Therefore, weakening of the protein structure in this region facilitates α-Syn misfolding and aggregation.
The appearance of short interactions in mutants is an indication of the stabilization of interactions within the C-terminal region of the protein. This effect is more pronounced for the E46K and A53T mutants as demonstrated by Fig. 3. We hypothesize that pathological mutants, especially A53T and E46K, in the misfolded state have a higher propensity for β structure in the C-terminal part. This assumption is in agreement with ssNMR analysis of fibrils formed by A53T mutant that detected the formation of an extended β-sheet core in the C-terminal region as compared to WT α-Syn.48
Despite the variability in positions of the short-range interactions, all four variants of α-Syn have two common peaks in contour length distributions: peak 1 at around ~30 nm and peak 2 at ~40 nm. Peak 2 corresponding to the 73–83 aa segment of the NAC region was expected to dominate the LC distributions for mutants as it did for WT. Although, all three pathological mutants have this peak, its contribution becomes smaller and decreases in the following order A53T>A30P>E46K. Such behavior is consistent with observations made in NMR studies that fibrils of mutants are structurally perturbed and the effects of the mutations increase in the order A30P<A53T<E46K.47 The decrease in the intensity of peak 2 is accompanied by an increase in intensity of another peak common to all variants, peak 1 at ~30 nm, which becomes dominant for the mutants. For example, this interaction comprises 61% of all events for the E46K mutant while peak 2 contributes only 15%. This indicates that the mutants prefer to assemble via interactions of segment 2 (segment s2 in Fig. S4) in contrast to WT whose preference is segment 3 (segment s3 in Fig. S4).
Similar to WT, the MR force-distance curves with multiple rupture events were also present for the mutants. The A30P mutant has a lower contribution of the MR curves in comparison to WT (19% vs 37%) while A53T and E46K mutants have a higher contribution of these force-distance curves, 63% and 76% respectively. Internal ruptures of the MR force curves for the mutants appear shifted to short contour length values suggesting that the observed multiple ruptures promote interactions at short distances. Both assumptions described above for the observation of short rupture distances in the case of WT α-Syn are also valid for the mutants. Collapse and compactness of the C-terminal region at low pH may result in LC shortening. Indeed, pathological mutants were found to establish stronger contacts than WT27 thus increasing compactness of C-terminus. Also, we cannot exclude that the observed short interactions appear due to the expansion of interacting modalities beyond the NAC region. According to our model of intermolecular interactions, the shortest interaction observed (LC = 13–15 nm) corresponds to contacts between monomers upon dimerization extending into the C-terminal region as far as the 125th residue (Fig. S5, A). These results suggest a critical involvement of the C-terminal region in α-Syn misfolding and interactions within the dimer.
Possible pathways for α-Syn self-assembly
Comparison of the aggregation pathways suggests that the A53T and E46K mutations accelerate fibrillization rates compared to WT15, 16 while A30P prefers an oligomerization route.17, 18 Our SMFS data also show differences in interactions between monomeric molecules of α-Syn. It is therefore tempting to correlate our data with the known aggregation patterns for WT and mutants of α-Syn. According to our data, the major difference for all the variants of α-Syn is in the number of the SR force distance curves vs the MR curves. The elevated relative contribution of multiple ruptures correlates very well with the higher rate of amyloid fibril formation for E46K and A53T mutants compared to WT.17, 18, 53 The A30P mutant shows a slower fibrillization rate17, 18 correlating with the fact that the SR curves are mostly detected for this mutant. Multiple contacts between monomers may also significantly slow down the internal reconfiguration rate of α-Syn which in turn would promote further association steps. Notably, a slow reconfiguration rate of monomers under conditions when aggregation is enhanced has been proposed as a key factor in determining how fast α-Syn molecules associate with each other.20–22 Our data suggest that A53T and E46K mutations increase the possibility for monomers to establish multiple contacts which may stabilize misfolded states of the protein in the dimer and allow for further growth of aggregated species. A similar hypothesis has been recently proposed based on measurements of dimer stability (lifetime) for several amyoidogenic proteins/peptides including α-Syn.29
Although the model shown in Fig. 6 depicts interacting regions in shearing geometry which rupture sequentially upon pulling, a more complex situation can be envisioned. For example, multiple contacts between monomers may promote the formation of a folded dimeric structure as illustrated in Fig. S6. The structure resembles a fold of alpha-synuclein within fibrils with several layers of parallel in-register β-sheets as recently proposed.54, 55 Such folded structure establishes inter- and intramolecular contacts between β-sheets.
CONCLUSIONS
In summary, we report novel findings on the pattern of misfolding of α-Syn and the effects of single point mutations on the protein’s dimerization, the very first step of its self-assembly. Single molecule force spectroscopy identified local interacting segments within monomeric α-Syn revealing a great deal of heterogeneity in the dimer conformations. Several segments involved in dimer formation are found in the core of amyloid fibrils suggesting that a link exists between structures of α-Syn at first steps of self-assembly and in mature fibrils. Our data demonstrate that the pathogenic mutations A30P, E46K and A53T do not increase the propensity of α-Syn to misfold but rather change conformational preferences of α-Syn. There is a direct effect of mutations through destabilization of interactions involving regions located close to the position of the mutations. Additionally, indirect contribution of mutations to interactions between segments located within the C-terminal region distant from the mutation sites is identified. Importantly, E46K and A53T mutations increase dimerization propensity by promoting simultaneous interactions of multiple segments, while the A30P mutant prefers single type interactions compared to WT. We hypothesize that such differences in interacting patterns can define the aggregation pathway of the protein and future studies are needed to clarify this hypothesis. The single molecule force spectroscopy approach described here was used to study the formation of a dimer. This method can be extended to analyze further steps of aggregation process if modeled dimer structure is probed by monomer or/and designed dimers.
Supplementary Material
Acknowledgments
We thank members of the Lyubchenko lab for the discussion of the results and critical comments.
Funding Information
The work was supported by grants from the National Institutes of Health (5P01GM091743-02 and 5R01 GM096039-02) and National Science Foundation (EPS – 1004094).
ABBREVIATIONS
- α-Syn
alpha-synuclein
- AFM
atomic force microscopy
- NAC
non-amyloid-beta component
- NR
no rupture
- SR
single rupture
- MR
multiple rupture
- SMFS
single molecule force spectroscopy
- WT
wild type
- PD
Parkinson’s disease
Footnotes
Supporting information includes Figures S1–S10. Supporting materials may be accessed free of charge online at http://pubs.acs.org.
References
- 1.Bisaglia M, Mammi S, Bubacco L. Structural insights on physiological functions and pathological effects of alpha-synuclein. Faseb J. 2009;23:329–340. doi: 10.1096/fj.08-119784. [DOI] [PubMed] [Google Scholar]
- 2.Weinreb PH, Zhen W, Poon AW, Conway KA, Lansbury PT., Jr NACP, a protein implicated in Alzheimer’s disease and learning, is natively unfolded. Biochemistry. 1996;35:13709–13715. doi: 10.1021/bi961799n. [DOI] [PubMed] [Google Scholar]
- 3.Eliezer D, Kutluay E, Bussell R, Jr, Browne G. Conformational properties of alpha-synuclein in its free and lipid-associated states. J Mol Biol. 2001;307:1061–1073. doi: 10.1006/jmbi.2001.4538. [DOI] [PubMed] [Google Scholar]
- 4.Conway KA, Harper JD, Lansbury PT., Jr Fibrils formed in vitro from alpha-synuclein and two mutant forms linked to Parkinson’s disease are typical amyloid. Biochemistry. 2000;39:2552–2563. doi: 10.1021/bi991447r. [DOI] [PubMed] [Google Scholar]
- 5.Iwai A, Yoshimoto M, Masliah E, Saitoh T. Non-A beta component of Alzheimer’s disease amyloid (NAC) is amyloidogenic. Biochemistry. 1995;34:10139–10145. doi: 10.1021/bi00032a006. [DOI] [PubMed] [Google Scholar]
- 6.Spillantini MG, Schmidt ML, Lee VMY, Trojanowski JQ, Jakes R, Goedert M. [alpha]-Synuclein in Lewy bodies. Nature. 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 7.Baba M, Nakajo S, Tu PH, Tomita T, Nakaya K, Lee VM, Trojanowski JQ, Iwatsubo T. Aggregation of alpha-synuclein in Lewy bodies of sporadic Parkinson’s disease and dementia with Lewy bodies. Am J Pathol. 1998;152:879–884. [PMC free article] [PubMed] [Google Scholar]
- 8.McClendon S, Rospigliosi CC, Eliezer D. Charge neutralization and collapse of the C-terminal tail of alpha-synuclein at low pH. Protein Sci. 2009;18:1531–1540. doi: 10.1002/pro.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uversky VN, Li J, Fink AL. Evidence for a partially folded intermediate in alpha-synuclein fibril formation. J Biol Chem. 2001;276:10737–10744. doi: 10.1074/jbc.M010907200. [DOI] [PubMed] [Google Scholar]
- 10.Kruger R, Kuhn W, Muller T, Woitalla D, Graeber M, Kosel S, Przuntek H, Epplen JT, Schols L, Riess O. Ala30Pro mutation in the gene encoding alpha-synuclein in Parkinson’s disease. Nat Genet. 1998;18:106–108. doi: 10.1038/ng0298-106. [DOI] [PubMed] [Google Scholar]
- 11.Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, Pike B, Root H, Rubenstein J, Boyer R, Stenroos ES, Chandrasekharappa S, Athanassiadou A, Papapetropoulos T, Johnson WG, Lazzarini AM, Duvoisin RC, Di Iorio G, Golbe LI, Nussbaum RL. Mutation in the α-Synuclein Gene Identified in Families with Parkinson’s Disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 12.Zarranz JJ, Alegre J, Gómez-Esteban JC, Lezcano E, Ros R, Ampuero I, Vidal L, Hoenicka J, Rodriguez O, Atarés B, Llorens V, Tortosa EG, del Ser T, Muñoz DG, de Yebenes JG. The new mutation, E46K, of α-synuclein causes parkinson and Lewy body dementia. Annals of Neurology. 2004;55:164–173. doi: 10.1002/ana.10795. [DOI] [PubMed] [Google Scholar]
- 13.Uversky VN, Li J, Fink AL. Metal-triggered structural transformations, aggregation, and fibrillation of human alpha-synuclein. A possible molecular link between Parkinson’s disease and heavy metal exposure. J Biol Chem. 2001;276:44284–44296. doi: 10.1074/jbc.M105343200. [DOI] [PubMed] [Google Scholar]
- 14.Krasnoslobodtsev AV, Peng J, Asiago JM, Hindupur J, Rochet JC, Lyubchenko YL. Effect of spermidine on misfolding and interactions of alpha-synuclein. PLoS One. 2012;7:e38099. doi: 10.1371/journal.pone.0038099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greenbaum EA, Graves CL, Mishizen-Eberz AJ, Lupoli MA, Lynch DR, Englander SW, Axelsen PH, Giasson BI. The E46K mutation in alpha-synuclein increases amyloid fibril formation. J Biol Chem. 2005;280:7800–7807. doi: 10.1074/jbc.M411638200. [DOI] [PubMed] [Google Scholar]
- 16.Conway KA, Harper JD, Lansbury PT. Accelerated in vitro fibril formation by a mutant alpha-synuclein linked to early-onset Parkinson disease. Nat Med. 1998;4:1318–1320. doi: 10.1038/3311. [DOI] [PubMed] [Google Scholar]
- 17.Conway KA, Lee SJ, Rochet JC, Ding TT, Williamson RE, Lansbury PT., Jr Acceleration of oligomerization, not fibrillization, is a shared property of both alpha-synuclein mutations linked to early-onset Parkinson’s disease: implications for pathogenesis and therapy. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:571–576. doi: 10.1073/pnas.97.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J, Uversky VN, Fink AL. Effect of familial Parkinson’s disease point mutations A30P and A53T on the structural properties, aggregation, and fibrillation of human alpha-synuclein. Biochemistry. 2001;40:11604–11613. doi: 10.1021/bi010616g. [DOI] [PubMed] [Google Scholar]
- 19.Mukhopadhyay S, Krishnan R, Lemke EA, Lindquist S, Deniz AA. A natively unfolded yeast prion monomer adopts an ensemble of collapsed and rapidly fluctuating structures. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:2649–2654. doi: 10.1073/pnas.0611503104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahmad B, Chen Y, Lapidus LJ. Aggregation of alpha-synuclein is kinetically controlled by intramolecular diffusion. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:2336–2341. doi: 10.1073/pnas.1109526109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahmad B, Lapidus LJ. Curcumin prevents aggregation in alpha-synuclein by increasing reconfiguration rate. J Biol Chem. 2012;287:9193–9199. doi: 10.1074/jbc.M111.325548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lapidus LJ. Understanding protein aggregation from the view of monomer dynamics. Mol Biosyst. 2013;9:29–35. doi: 10.1039/c2mb25334h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rubinstein A, Lyubchenko YL, Sherman S. Dynamic properties of pH-dependent structural organization of the amyloidogenic beta-protein (1–40) Prion. 2009;3:31–43. doi: 10.4161/pri.3.1.8388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang A, Stultz CM. Finding order within disorder: elucidating the structure of proteins associated with neurodegenerative disease. Future Medicinal Chemistry. 2009;1:467–482. doi: 10.4155/fmc.09.40. [DOI] [PubMed] [Google Scholar]
- 25.Ullman O, Fisher CK, Stultz CM. Explaining the structural plasticity of alpha-synuclein. Journal of the American Chemical Society. 2011;133:19536–19546. doi: 10.1021/ja208657z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uversky VN. A protein-chameleon: conformational plasticity of alpha-synuclein, a disordered protein involved in neurodegenerative disorders. J Biomol Struct Dyn. 2003;21:211–234. doi: 10.1080/07391102.2003.10506918. [DOI] [PubMed] [Google Scholar]
- 27.Brucale M, Sandal M, Di Maio S, Rampioni A, Tessari I, Tosatto L, Bisaglia M, Bubacco L, Samori B. Pathogenic mutations shift the equilibria of alpha-synuclein single molecules towards structured conformers. Chembiochem. 2009;10:176–183. doi: 10.1002/cbic.200800581. [DOI] [PubMed] [Google Scholar]
- 28.Sandal M, Valle F, Tessari I, Mammi S, Bergantino E, Musiani F, Brucale M, Bubacco L, Samori B. Conformational equilibria in monomeric alpha-synuclein at the single-molecule level. PLoS Biol. 2008;6:e6. doi: 10.1371/journal.pbio.0060006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lyubchenko YL, Kim BH, Krasnoslobodtsev AV, Yu J. Nanoimaging for protein misfolding diseases. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2010;2:526–543. doi: 10.1002/wnan.102. [DOI] [PubMed] [Google Scholar]
- 30.Kransnoslobodtsev AV, Shlyakhtenko LS, Ukraintsev E, Zaikova TO, Keana JF, Lyubchenko YL. Nanomedicine and protein misfolding diseases. Nanomedicine. 2005;1:300–305. doi: 10.1016/j.nano.2005.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rochet J-C, Lansbury PT., Jr Amyloid fibrillogenesis: themes and variations. Curr Op Struct Biol. 2000;10:60–68. doi: 10.1016/s0959-440x(99)00049-4. [DOI] [PubMed] [Google Scholar]
- 32.Rief M, Gautel M, Oesterhelt F, Fernandez JM, Gaub HE. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 33.Tskhovrebova L, Trinick J, Sleep JA, Simmons RM. Elasticity and unfolding of single molecules of the giant muscle protein titin. Nature. 1997;387:308–312. doi: 10.1038/387308a0. [DOI] [PubMed] [Google Scholar]
- 34.Stirnemann G, Giganti D, Fernandez JM, Berne BJ. Elasticity, structure, and relaxation of extended proteins under force. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:3847–3852. doi: 10.1073/pnas.1300596110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sarkar A, Caamano S, Fernandez JM. The mechanical fingerprint of a parallel polyprotein dimer. Biophysical journal. 2007;92:L36–38. doi: 10.1529/biophysj.106.097741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oesterhelt F, Oesterhelt D, Pfeiffer M, Engel A, Gaub HE, Müller DJ. Unfolding Pathways of Individual Bacteriorhodopsins. Science. 2000;288:143–146. doi: 10.1126/science.288.5463.143. [DOI] [PubMed] [Google Scholar]
- 37.Yu J, Malkova S, Lyubchenko YL. alpha-Synuclein misfolding: single molecule AFM force spectroscopy study. J Mol Biol. 2008;384:992–1001. doi: 10.1016/j.jmb.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 38.Yang G, Cecconi C, Baase WA, Vetter IR, Breyer WA, Haack JA, Matthews BW, Dahlquist FW, Bustamante C. Solid-state synthesis and mechanical unfolding of polymers of T4 lysozyme. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:139–144. doi: 10.1073/pnas.97.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.El-Agnaf OM, Bodles AM, Guthrie DJ, Harriott P, Irvine GB. The N-terminal region of non-A beta component of Alzheimer’s disease amyloid is responsible for its tendency to assume beta-sheet and aggregate to form fibrils. Eur J Biochem. 1998;258:157–163. doi: 10.1046/j.1432-1327.1998.2580157.x. [DOI] [PubMed] [Google Scholar]
- 40.Yu J, Warnke J, Lyubchenko YL. Nanoprobing of alpha-synuclein misfolding and aggregation with atomic force microscopy. Nanomedicine : nanotechnology, biology, and medicine. 2011;7:146–152. doi: 10.1016/j.nano.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 41.McAllister C, Karymov MA, Kawano Y, Lushnikov AY, Mikheikin A, Uversky VN, Lyubchenko YL. Protein interactions and misfolding analyzed by AFM force spectroscopy. J Mol Biol. 2005;354:1028–1042. doi: 10.1016/j.jmb.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 42.Bernstein SL, Liu D, Wyttenbach T, Bowers MT, Lee JC, Gray HB, Winkler JR. α-Synuclein: Stable compact and extended monomeric structures and pH dependence of dimer formation. Journal of the American Society for Mass Spectrometry. 2004;15:1435–1443. doi: 10.1016/j.jasms.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 43.Trexler AJ, Rhoades E. Single molecule characterization of alpha-synuclein in aggregation-prone states. Biophysical journal. 2010;99:3048–3055. doi: 10.1016/j.bpj.2010.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu KP, Weinstock DS, Narayanan C, Levy RM, Baum J. Structural reorganization of alpha-synuclein at low pH observed by NMR and REMD simulations. J Mol Biol. 2009;391:784–796. doi: 10.1016/j.jmb.2009.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoyer W, Antony T, Cherny D, Heim G, Jovin TM, Subramaniam V. Dependence of alpha-synuclein aggregate morphology on solution conditions. Journal of molecular biology. 2002;322:383–393. doi: 10.1016/s0022-2836(02)00775-1. [DOI] [PubMed] [Google Scholar]
- 46.Kim BH, Palermo NY, Lovas S, Zaikova T, Keana JF, Lyubchenko YL. Single-molecule atomic force microscopy force spectroscopy study of Abeta-40 interactions. Biochemistry. 2011;50:5154–5162. doi: 10.1021/bi200147a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Comellas G, Lemkau LR, Nieuwkoop AJ, Kloepper KD, Ladror DT, Ebisu R, Woods WS, Lipton AS, George JM, Rienstra CM. Structured Regions of α-Synuclein Fibrils Include the Early-Onset Parkinson’s Disease Mutation Sites. Journal of Molecular Biology. 2011;411:881–895. doi: 10.1016/j.jmb.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heise H, Celej MS, Becker S, Riedel D, Pelah A, Kumar A, Jovin TM, Baldus M. Solid-state NMR reveals structural differences between fibrils of wild-type and disease-related A53T mutant alpha-synuclein. Journal of molecular biology. 2008;380:444–450. doi: 10.1016/j.jmb.2008.05.026. [DOI] [PubMed] [Google Scholar]
- 49.Giasson BI, Murray IV, Trojanowski JQ, Lee VM. A hydrophobic stretch of 12 amino acid residues in the middle of alpha-synuclein is essential for filament assembly. J Biol Chem. 2001;276:2380–2386. doi: 10.1074/jbc.M008919200. [DOI] [PubMed] [Google Scholar]
- 50.Rospigliosi CC, McClendon S, Schmid AW, Ramlall TF, Barre P, Lashuel HA, Eliezer D. E46K Parkinson’s-linked mutation enhances C-terminal-to-N-terminal contacts in alpha-synuclein. Journal of molecular biology. 2009;388:1022–1032. doi: 10.1016/j.jmb.2009.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cho M-K, Nodet G, Kim H-Y, Jensen MR, Bernado P, Fernandez CO, Becker S, Blackledge M, Zweckstetter M. Structural characterization of α-synuclein in an aggregation prone state. Protein Science. 2009;18:1840–1846. doi: 10.1002/pro.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fernandez CO, Hoyer W, Zweckstetter M, Jares-Erijman EA, Subramaniam V, Griesinger C, Jovin TM. NMR of alpha-synuclein-polyamine complexes elucidates the mechanism and kinetics of induced aggregation. Embo J. 2004;23:2039–2046. doi: 10.1038/sj.emboj.7600211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Choi W, Zibaee S, Jakes R, Serpell LC, Davletov B, Crowther RA, Goedert M. Mutation E46K increases phospholipid binding and assembly into filaments of human alpha-synuclein. FEBS Lett. 2004;576:363–368. doi: 10.1016/j.febslet.2004.09.038. [DOI] [PubMed] [Google Scholar]
- 54.Chen M, Margittai M, Chen J, Langen R. Investigation of alpha-synuclein fibril structure by site-directed spin labeling. J Biol Chem. 2007;282:24970–24979. doi: 10.1074/jbc.M700368200. [DOI] [PubMed] [Google Scholar]
- 55.Vilar M, Chou HT, Luhrs T, Maji SK, Riek-Loher D, Verel R, Manning G, Stahlberg H, Riek R. The fold of alpha-synuclein fibrils. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:8637–8642. doi: 10.1073/pnas.0712179105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.