Abstract
There is increasing concern that animal- and human reproduction may be adversely affected by exposure to xenoestrogens that activate estrogen receptors. There is evidence that one such compound, Bisphenol A (BPA), also induces meiotic and mitotic aneuploidy, suggesting that these kinds of molecules may also have effects on cell division. In an effort to understand how Bisphenol A might disrupt cell division, a phenotypic analysis was carried out using sea urchin eggs, whose early embryonic divisions are independent of zygotic transcription. Fertilized Lytechinus pictus eggs exposed to BPA formed multipolar spindles resulting in failed cytokinesis in a dose-dependent, transcriptionally independent manner. Using novel biotinylated BPA affinity probes to fractionate cell-free extracts, tubulin was identified as a candidate binding protein by mass spectroscopy, and BPA promoted microtubule polymerization and centrosome-based microtubule nucleationin vitro, but did not appear to display microtubule-stabilizing activity. Treatment of mammalian cells demonstrated that BPA-as well as a series of Bisphenol A derivatives induced ectopic spindle pole formation in the absence of centrosome overduplication. Together, these results suggest a novel mechanism by which Bisphenol A affects the nucleation of microtubules, disrupting the tight spatial control associated with normal chromosome segregation, resulting in aneuploidy.
Introduction
Xenoestrogens are a structurally diverse class of non-steroidal compounds that share structural features with steroid hormones and modulate the activity of nuclear hormone receptors (1). Although the potencies of xenoestrogens vary, their release into the environment has begun to have a measurable effect on the reproductive development of several species, and there is increasing concern that human reproduction is being affected as well (2, 3). One such weakly estrogenic compound that has increasingly become a cause for concern is Bisphenol A (4,4′ isopropylidenediphenol, BPA). Although less estrogenic than diethylstilbestrol (DES), Bisphenol A affects male and female reproductive development at low doses, and while much of the research to date has focused on BPA’s role as a potential endocrine disruptor (4, 5), non-genomic effects have been reported as well (1, 6). Nearly ubiquitous, BPA is found extensively in polycarbonate plastics, resins lining food containers, adhesives, and dental sealants, and leaching has been documented with many of these products (7-9). One particularly startling finding arose from a rodent colony accidentally exposed to Bisphenol A, where increases in synaptic abnormalities and meiotic aneuploidy were detected in mouse oocytes (10, 11). The detection of chromosome congression- and meiotic nondisjunction errors in exposed mice suggested that in addition to aberrantly activating the estrogen receptor, Bisphenol A may be directly interfering with the mechanics of cell division.
Like DES, Bisphenol A has been reported to transform cells in vitro and has been linked to tumor formation in animal models (12-16), although genotoxicity assays performed with Salmonella typhimurium indicated that BPA is not mutagenic (17). Studies in mammary tumor cell lines demonstrated that BPA is able to induce expression of estrogen responsive genes and promote proliferation (9), consistent with the notion that Bisphenol A promotes cellular proliferation though the estrogen receptor. In contrast, cell lines that lack measurable levels of estrogen receptors are also capable of BPA-induced cellular transformation (16). The same study as well as others reported an increase in aneuploidy with BPA exposure (18-21) although the concentrations required to induce aneuploidy in cultured cells were much higher than those reported for whole animal studies (11). However, alterations in spindle morphology were reported for both cultured somatic cells and oocytes (18, 21-24), suggesting that the reported congression failures of chromosomes at metaphase and non-disjunction at anaphase may due to BPA’s affect on microtubule assembly and organization. While mechanisms for non-disjunction might be based on BPA- and metabolite interactions with DNA (10, 16, 25-28), the appearance of altered spindle morphology suggest that BPA may also indirectly or directly target the mitotic apparatus to affect chromosome segregation and the maintenance of ploidy.
Studies concerning the endocrinology and developmental toxicology of Bisphenol A suggest that this compound is a potential threat to human health, but none of the studies to date have clearly established a molecular mechanism by which BPA increases aneuploidy through its alteration of the mitotic spindle. In an effort to understand how Bisphenol A disrupts the machinery of cell division, we undertook a multidisciplinary approach combining synthetic organic chemistry, imaging and biochemistry to identify tubulin as a direct target of BPA. In agreement with earlier findings, we find that BPA induces multipolar spindles in diverse cell types, and propose a model by which BPA produces multipolar spindles by promoting ectopic microtubule nucleation, disrupting spindle morphology and ultimately contributing to chromosome segregation defects and aneuploidy.
Results
Bisphenol A alters microtubule organization during early embryogenesis
Bisphenol A has been reported to disrupt mitotic and meiotic divisions, but the molecular mechanisms by which BPA induces aneuploidy remain elusive. Moreover, dramatic discrepancies have been reported between whole animal- and cultured cell models for the doses of BPA that induce aneuploidy and spindle disruption (10, 11, 18, 22). In an effort to better characterize non-genomic (ER-independent) effects of BPA during mitosis, we undertook a systematic examination of BPA effects on cell division in both embryonic- and somatic cells. As a first estimation of the effects of BPA on mitosis, we followed the first embryonic divisions of sea urchin embryos, which have been shown to be sensitive to estrogenic compounds (29), but whose early development is transcription-independent (30). Fertilized Lytechinus pictus eggs were exposed to BPA at concentrations ranging from 200 nM to 5 μM and followed through the first division by DIC timelapse microscopy (Figure 1 Panels A-D). Because exposure to BPA earlier than 20 minutes post-fertilization delayed pronuclear migration and fusion, experimental embryos were cultured in control seawater for 25 minutes prior to treatment with BPA. In comparison to DMSO controls (Figure 1, Panels A and B), BPA-treated embryos formed multiple, ectopic furrows and membrane blebs that later regressed to form spherical, binucleate eggs (Figure 1, Panels C and D), suggesting that there were defects in cleavage plane determination. BPA effects on cytokinesis were dose-dependent, with an IC50 of 3 μM, but effects were detected as low as 500 nM (Supplemental Figure 1). This dose range was comparable to levels previously described for rodents exposed to BPA (0.44 – 1.6 μM) (11) but lower than what was used on mouse oocytes matured in vitro (10-30 μM)(23). Early development of the sea urchin relies on maternal transcripts (30), and we found that pretreatment of eggs with 80 μM actinomycin D failed to suppress BPA-disruption of cleavage plane determination (not shown), suggesting that hormone receptor-mediated transcription could not account for the observed cell division defects.
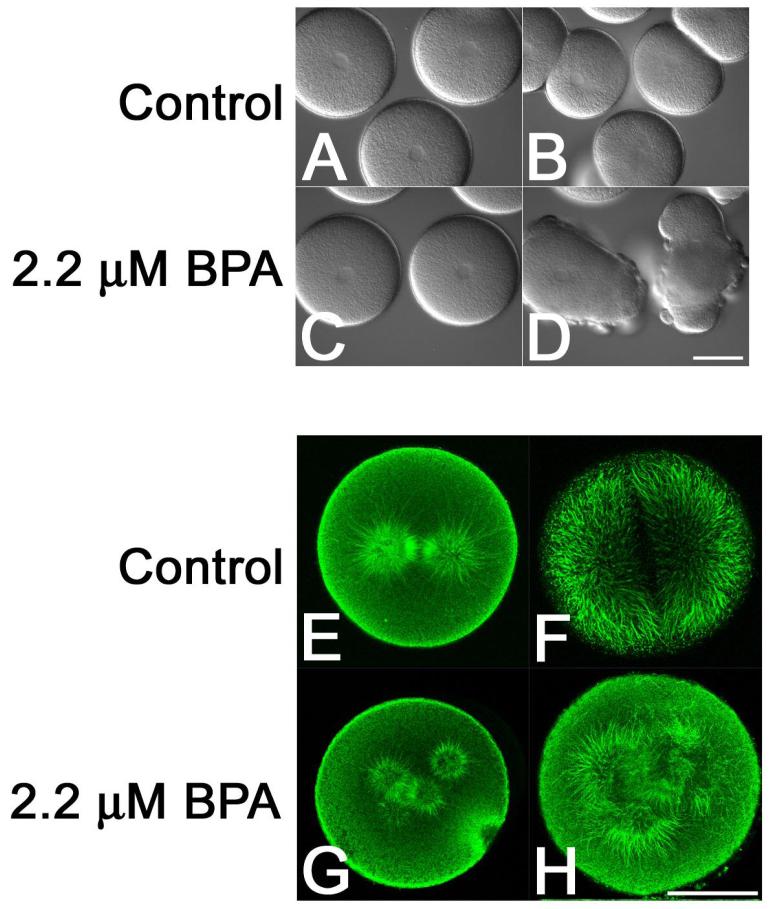
Figure 1. Dose-dependent effects of Bisphenol A on microtubule organization and cell division in sea urchin embryos.
L. pictus eggs were fertilized, stripped of their fertilization envelopes and cultured through the first division in the presence of O.1% DMSO (Panels A, B, E and F) or 2.2 μM BPA (Panels C, D G and H). Note that while the control embryos underwent normal cytokinesis (Panel B), BPA-treated embryos formed multiple, misplaced cleavage furrows (Panel D). Analysis of microtubule organization in metaphase (Panels E and G) and anaphase (F and H) embryos revealed the presence of normal metaphase and anaphase spindles in control embryos (Panels E and F) but supernumary spindle poles in BPA-treated embryos (Panels G and H). Bars for Panels D and H, 50 μm.
Microtubules of the mitotic apparatus are responsible for specifying the cleavage plane in all animal cells, and in echinoderm embryos, the explosive outgrowth of astral microtubules and their contact with the cortical actin cytoskeleton marks the position of the future contractile ring (31, 32). Because embryos exposed to BPA displayed failures in cleavage plane determination and cytokinesis, the organization of the mitotic spindle was examined in control- and BPA-treated embryos (Figure 1, Panels E-H). Whereas control eggs formed normal bipolar spindles that underwent a stereotypical transition from metaphase to anaphase (Figure 1, Panels E and F), eggs exposed to BPA displayed multiple spindle poles, that upon anaphase onset, resulted in a disorganized elongation of astral microtubules towards the cortex (Figure 1, Panels G and H). Monopolar spindles were occasionally observed, but unlike the appearance of multipolar spindles, there was no dose-dependent increase in the frequency of monopolar spindles. Because there are several possible mechanisms by which multipolar spindles may form (centrosome amplification, centriole splitting, de novo formation, etc), microtubule organization was followed in living cells using orientation-independent polarization microscopy, which allows for visualization of spindle formation without the use of fluorescent probes (33, 34). As shown in Figure 2, control embryos underwent normal spindle assembly, as evidenced by the presence of a birefringent bipolar spindle (Figure 2, Panels A-C, and Supplemental Movie 1). In Bisphenol A-treated cells (Figure 2, Panels D-I, Supplemental Movies 2 and 3), supernumery asters could be detected forming de novo (Figure 2, Panels E, F, H, and I, arrows). As mentioned above, monopolar spindles could also be detected forming near the nucleus following nuclear envelope breakdown (Figure 2, Panels E and F, asterisk), but in all 28 cells observed, we failed to observe spindle collapse (which would account for the presence of a monopole).
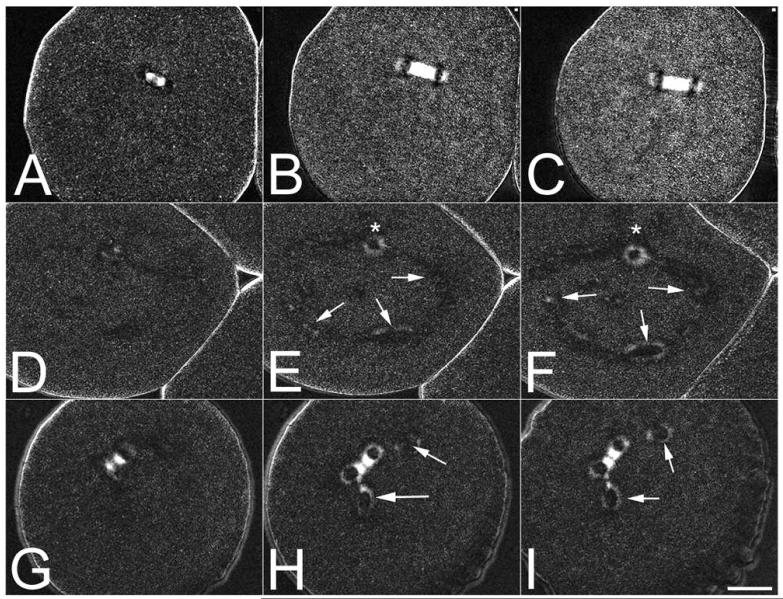
Figure 2. Visualization of ectopic spindle pole formation in Bisphenol A-treated sea urchin eggs.
L. pictus eggs exposed to either DMSO carrier (Panels A-C) or 2.2 μM BPA were compressed under fluorocarbon oil to aid in visualizing spindle formation, and followed by polarization microscopy. Whereas controls formed a normal, birefringent spindles (Panels A-C), embryos exposed to BPA could be observed forming monopolar spindles (*) as well as de novo microtubule organizing centers in the cytoplasm (Panels D-F, arrows). In other cells, asters could be observed splitting off the main spindle (Panels G-I, arrows). Bar 20 μM.
Design of BPA probes for affinity purification
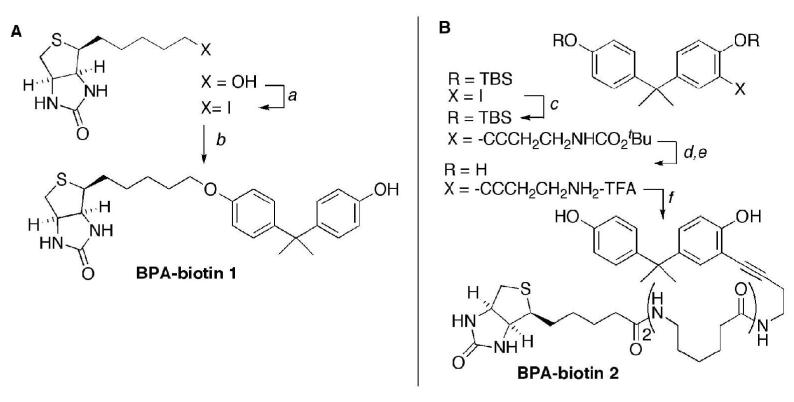
Bisphenol A treatment of sea urchin eggs produced defects in microtubule organization that resulted in cleavage failure (Figures 1 and 2), but had no other effects on cell cycle timing or progression. In order to identify cellular targets of BPA that may be involved in the observed dose-dependent effects on dividing cells, we initiated efforts to design a BPA affinity probe. The presence of a polar, acidic phenol group connected to a hydrophobic aliphatic backbone are key features of BPA that are recognized in structure activity models of estrogenicity (35-37), and we anticipated that these characteristics would also be essential for binding other proteins. The standard procedure for biotinylation involves coupling a nucleophilic residue on the substrate, typically an amine or thiol group, with an activated carboxylate or maleimide derivative of biotin respectively. BPA lacks suitable reactive functional groups of this type and possesses a non-polar backbone that likely interacts with hydrophobic protein binding sites. The design of small molecule affinity probes requires judicious introduction of functionality for conjugation to avoid adversely affecting the binding affinity due to unfavorable electrostatic interactions, modified solvation characteristics, altered lipophilicity, and increased steric interactions. We have recently described an alternative approach for the biotinylation of hydrophobic substrates that replaces the carboxylic group with a non-polar linkage to the 7-oxo-3-thia-6,8-diazabicyclo[3.3.0]oct-4-yl heterocycle that provides the major contribution to (strept)avidin binding affinity(38). Extending this approach, we designed two types of BPA probes (Figure 3). Compound 1 (BPA-biotin 1) possesses a phenolic ether connection to a reduced biotin fragment and provides minimal alteration of BPA’s hydrophobic backbone (Figure 3, Left panel). Compound 2 (BPA-biotin 2) incorporates a butynyl carboxamide as a spacer group attached at the ortho position of the aryl ring that preserves both of the phenol groups found in the original BPA (Figure 3, Right panel).
Figure 3. Synthesis of BPA-biotin affinity probes.
Panel A. The BPA-biotin derivative 1 was prepared by Williamson ether synthesis. The 5-hydroxy derivative was converted to 4-(5-iodopentyl)tetrahydro-1H-thieno[3,4-d]imidazol-2(3H)-one using I2, triphenylphosphine. Selective O-alkylation of BPA with the primary alkyl iodide gave the desired compound 1 as the major product.
Panel B. The BPA-biotin derivative 2 was prepared by sequential Sonogashira coupling and biotinylation. 4-(2-(4-Hydroxyphenyl)propan-2-yl)-2-iodophenol was protected as the bis-TBS ether using standard conditions, and was then coupled with tert-butyl but-3-ynylcarbamate to the alkyne product in excellent yield. The TBS protecting groups were removed with TBAF, and the tBoc group was cleaved with TFA to provide the corresponding ammonium salt. Biotinylation with biotin-L2-NHS in Et3N/DMF gave the desired compound 2. Both of the biotinylated probes 1 and 2 were purified by silica gel chromatography and structurally characterized by 1H- and 13C-NMR, and HPLC-MS.
Identification of tubulin as a BPA-binding protein
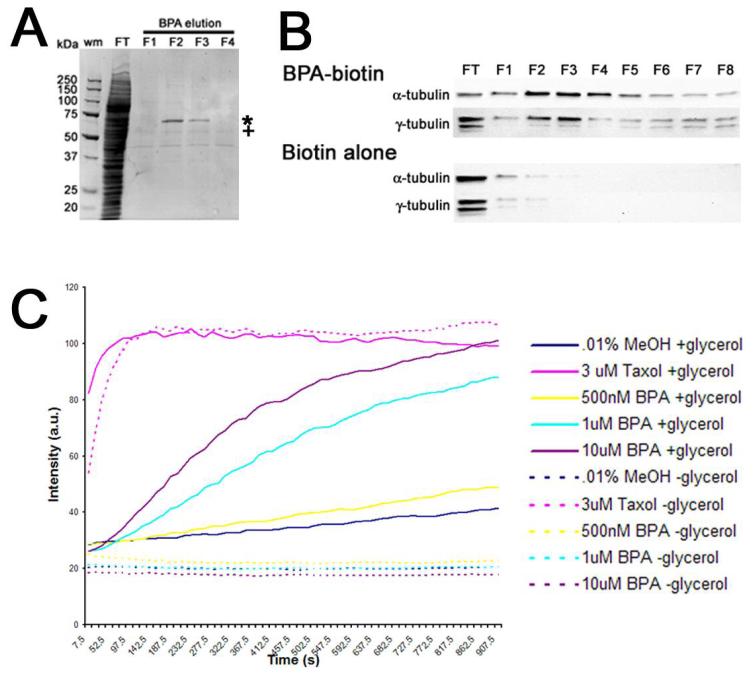
Biotin-linked analogs of BPA were used to fractionate cell-free extracts derived from Xenopus oocytes. The highly concentrated extracts of frog and clam oocytes are capable of replicating microtubule- and centrosomal dynamics that mirror the in vivo state, and have been used extensively for the study of cytoskeletal and cell cycle dynamics (39-41). Xenopus cytostatic factor (CSF)-arrested extracts were were incubated in the presence of BPA-biotin 1 or 2, and bound complexes were collected using streptavidin-agarose, and those proteins specifically interacting with Bisphenol A were eluted with 210 μM free BPA and resolved by SDS-PAGE (Figure 4, Panel A). Two bands of interest were identified that eluted with BPA from BPA-biotin matrices (Figure 4, Panel A, fractions F2 and F3, denoted by * and +) but not from biotin alone (not shown). MALDI-TOF analysis identified the upper band as aconitase, and the lower band (denoted with +) as α,β-tubulin. While the purification of aconitase was batch-dependent, α- as well as γ-tubulin consistently eluted from both of the BPA-biotin affinity probes as detected by Western blotting (Figure 4, Panel B: F2-F8), and was independent of the extract fractionated (Xenopus, sea urchin or surf clam). In contrast, neither α- nor γ-tubulin eluted from biotin control matrices (Figure 4, Panel B: F2-F8). Centrosomal components such as pericentrin and ε-tubulin, were not detected eluting from BPA affinity matrices, suggesting that centrosomes or centrosomal precursors were not associating with the matrix as a complex (not shown). To validate tubulin as a target for Bisphenol A, tubulin was polymerized in the presence of increasing concentrations of BPA, and followed by fluorimetry (Figure 4, Panel C). Because DMSO alone can promote microtubule polymerization, methanol was used as a solvent, which had no effect on microtubule polymerization (at 0.14%). As shown in Figure 4C, Bisphenol A promoted microtubule polymerization, although to a lesser extent than the potent microtubule stabilizer, taxol (Figure 4C, solid lines).BPA-induced microtubule polymerization could be detected in the presence of 20% glycerol, where glycerol acts as a general stabilizer of microtubules (42, 43). In contrast, microtubule polymerization in presence of ten-fold less glycerol was undetectable in either control- or BPA-treated samples (Figure 4C, dashed lines), whereas taxol-treated samples polymerized normally. The requirement of glycerol for BPA-induced microtubule polymerization suggested that BPA may not be stabilizing microtubules in the same manner as taxol, and this was further confirmed when BPA failed to protect microtubules from depolymerization by cold treatment or 4mM CaCl2 (not shown). Thus, while BPA was capable of promoting microtubule polymerization in vitro, it did not appear to act as a stabilizer.
Figure 4. Bisphenol A binds tubulin and promotes microtubule polymerization.
Panel A. CSF-arrested Xenopus extracts (XE) were incubated with biotinylated BPA (compound 1), and protein complexes were fractionated over strepavidin beads, and following the collection of the flow-through (FT) fraction and washing with BRB80 buffer, bound proteins were eluted (F1-F4) with 210 μM BPA. Bound proteins were then resolved on a 4-15% SDS PAGE gradient gel. Two bands were chosen for analysis and sequencing (* and +), and the band denoted by (+) was identified by mass spectroscopy as α-/β-tubulin. Panel B. Western blot confirmation of α-tubulin and γ-tubulin elution from BPA-biotin 1 affinity matrices in fractions 1-8 (FT corresponds to the flow-through or unbound fraction). In contrast, extract incubated with biotin alone showed that neither α-tubulin or γ-tubulin associated with the affinity matrix. Panel C. Tubulin was polymerized in the presence of 20% glycerol (Solid lines) either in the presence of carrier alone (0.1% MeOH), 3 μM Taxol, or increased concentrations of BPA.A parallel set of assays were performed in the presence of low glycerol (2%), where microtubules polymerize poorly unless in the presence of an additional stabilizing reagent (dashed lines). Samples were assembled on ice, warmed to 37 °C and readings were acquired every 15 sec. Note that while taxol-induced polymerization was independent of glycerol, BPA-induced polymerization did not occur in the absence of glycerol.
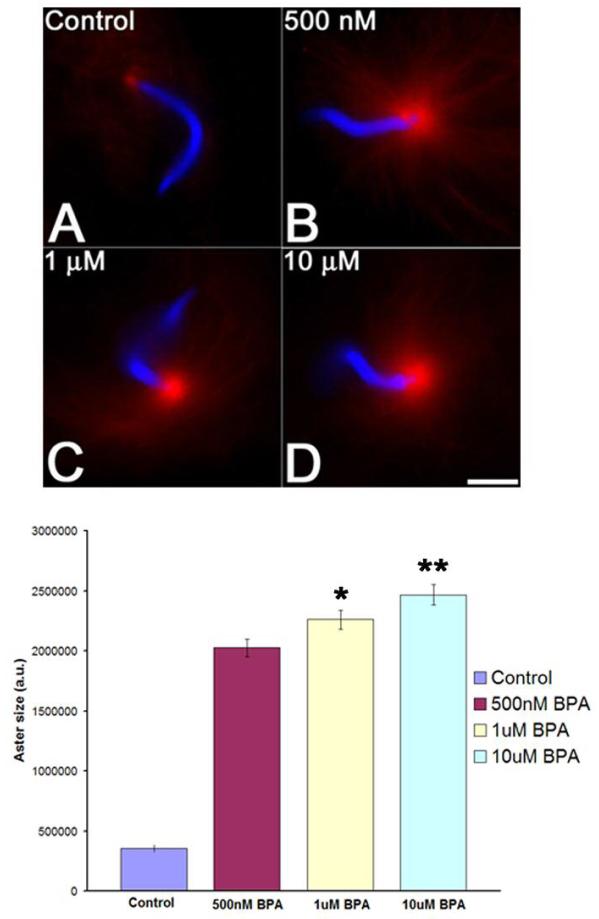
In vitro, BPA promoted microtubule polymerization but did not appear to act as a microtubule stabilizer in comparison with taxol. To better understand the action of BPA on microtubule polymerization, we followed centrosome-nucleated aster formation in the absence or presence of BPA.CSF extracts were supplemented with rhodamine-tubulin, sperm nuclei, andBisphenol A or carrier control, warmed to 15°C for ten minutes and fixed onto slides. While a small amount of nucleation at the centrosome was detected in controls under these conditions (Figure 5, Panel A), BPA-treated extracts (Figure 5, Top, Panels B-D) displayed a 5.7-fold increase in aster size at concentrations as low as 500 nM (Figure 5, Panels A and B). Small but statistically significant increases could be detected at higher doses (Figure 5B), and at concentrations above 10 μM, non-centrosomal microtubule nucleation appeared to predominate in the extracts, and sperm asters became more disorganized and difficult to quantify (not shown).The robust nucleation of microtubule asters observed in the presence of 500nM BPA contrasted sharply with the modest promotion of microtubule polymerization observed with purified tubulin at the same BPA concentration (Figure 4C) suggesting that BPA promoted microtubule nucleation.
Figure 5. Bisphenol A promotes microtubule nucleation from centrosomes in vitro.
Top: CSF-arrested Xenopus CSF extracts were incubated with sperm nuclei, rhodamine tubulin, and carrier control (O.8% MeOH) or BPA for 30 minutes on ice. Tubes were then warmed to 15°C for 10 minutes. Reactions were stopped by adding 1 μL of each reaction to 3 μL of fixative containing Hoescht 33342 and observed by wide-field epifluorescence. When compared with controls (Panel A), BPA-treated extracts displayed robust microtubule nucleation from centrosomes (Panels B-D). Bar, 10 μm.
Bottom: Quantification of BPA-enhanced aster formation in CSF Xenopus extracts. The area for single asters was calculated by first normalizing the background and then creating a threshold image. The areas were then calculated using ImageJ for 12 asters per condition. Small but significant differences could be detected between 500 nM and 1 μM BPA (p=0.03, asterisk) as well as between 1 and 10 μM (p=0.01, double asterisk). Error bars denote standard error.
Bisphenol A-induction of acentriolar spindle poles
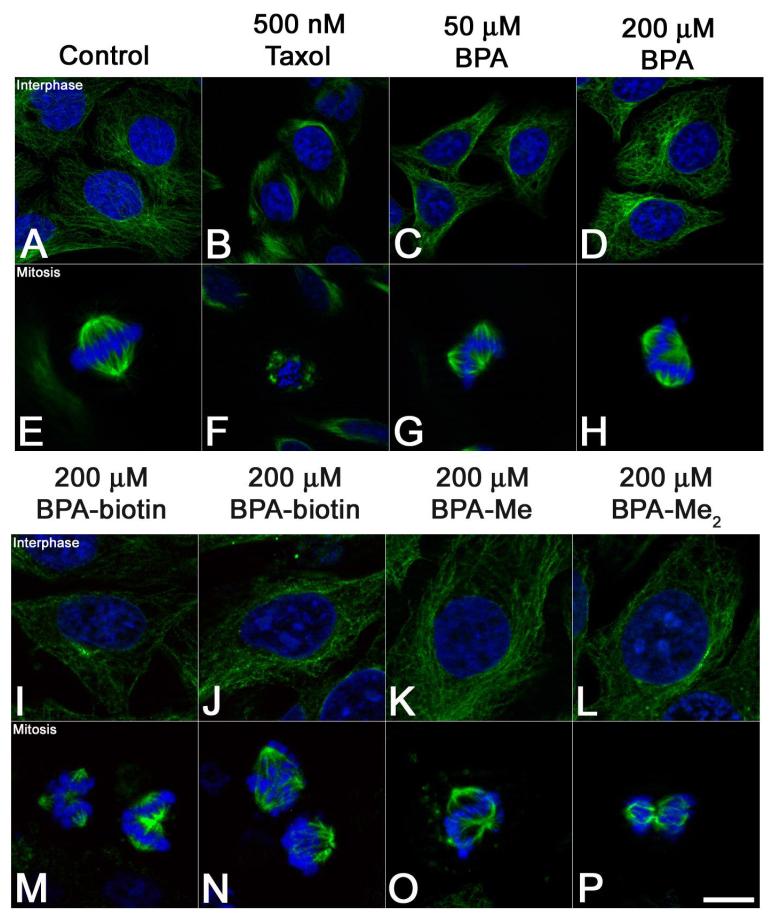
In animal cells, interphase- and mitotic microtubule arrays are organized by centrosomes, which serve as a scaffold for γ-tubulin-based microtubule nucleation and anchoring (44). Bisphenol A bound both α/β- and γ-tubulin in vitro (Figure 4), and altered microtubule organization in dividing sea urchin eggs (Figures 1 and 2), but the mechanisms by which these alterations occur were still not clear. We revisited BPA’s effects on microtubule organization in mammalian cells (Figure 6), where BPA has been shown to act as a disruptor of microtubule organization in cultured mammalian cells and oocytes (18, 22, 23), albeit at much higher concentrations than what we and others have observed for oocytes and early embryos (11, 23). In HeLa cells, microtubule stabilizers such as taxol suppress microtubule dynamics, resulting in microtubules of uniform length during both interphase and mitosis (Figure 6, Panels B and F). In contrast, BPA had no affect on interphase microtubule organization (Figure 6, panels C and D) at any concentration ranging from 500 nM to 200 μM, nor did BPA induce the small ectopic asters associated with taxol (Figure 6, Panel F). Instead, BPA induced the dose-dependent formation of ectopic spindle poles (Figure 6, Panels G and H; and Figure 7B) with an IC50 of 100 μM in a manner that was independent of the carrier used to solubilize BPA (DMSO or MeOH). The vast majority of cells observed (>90%, n=400) contained only one or two additional poles, and consistent with earlier reports (18, 21), these cells were able to progress through anaphase and initiate cytokinesis, cleaving into three or four daughter cells (not shown). Additionally, both biotin conjugates were capable of inducing ectopic poles (Figure 6, Panels M and N), as were Bisphenol A monomethyl ether (BPA-Me) and Bisphenol A dimethyl ether (BPA-Me2) (Figure 6, Panels O and P). These BPA ethers have 1.6- and 184-fold lower affinity for the estrogen receptor, respectively (45), yet there was no significant reduction in the ability of these analogs to induce multipolar spindles (Supplemental 3). Consistent with BPA, perturbations were only observed in mitotic cells (Figure 6, Panels M-P), with interphase microtubules remaining morphologically normal (Figure 6, Panels I-L). Together, these results indicated that both the biotin conjugates as well as analogs with lowered affinity for the estrogen receptor retained biological activity, suggesting that Bisphenol A’s effects on spindle morphology were extragenic.
Figure 6. Ectopic spindle pole formation in HeLa cells exposed to Bisphenol A.
Hela cells were exposed to 0.1% DMSO (Panels A and E), 500 nM Taxol (B and F), or Bisphenol A (Panels C-D, F-H), BPA-biotin 1 (I and M), BPA-biotin 2 (J and N), Bisphenol A monomethyl ether (BPA-Me) (K and O), and Bisphenol A dimethyl ether (BPA-Me2) (L and P) for four hours, processed for DNA (blue) and tubulin (green) localization, and representative images were acquired from cells in interphase (Panels A-D; I-L) and mitosis (Panels E-H; M-P). In contrast to Taxol-treated cells (Panel B and F) that displayed small, stellate asters, BPA-treated cells developed ectopic spindle poles (Panels G and H). Similarly, biotinylated-BPA analogs produced phenotypes consistent with the parent molecule (Panels M and N), as did methylated BPA analogs BPA-Me and BPA-Me2 (Panels O and P). Bar 10 μm.
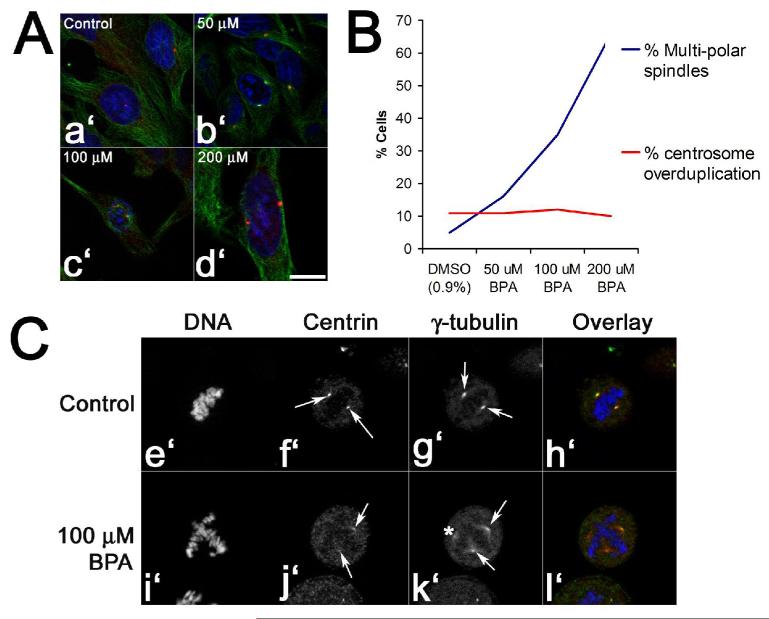
Figure 7. Bisphenol A does not drive centrosome amplification to generate ectopic spindle poles.
Panels A and B. Hela cells were exposed to either 0.1% DMSO or increasing doses of Bisphenol A for four hours, fixed and processed for DNA (blue), tubulin (green), and pericentrin (red) localization (Panel A, a’-d’; Bar, 10 μM). Cells were then scored for the presence of multipolar spindles or centrosomes at the G2/M transition (Panel B). Note that the number of pericentrin-positive centrosomes at G2/M does not increase even at 200 μM, where the majority of metaphase spindles are multipolar. Panel C. Centrin and γ-tubulin localization in control- (Panels e’ through h’) and BPA-treated eggs (Panels i’-l’). Compared to controls (Pictures f’ and g’, arrows), BPA-treated cells contained spindle poles that were positive for both γ-tubulin and centrin (Pictures j’ and k’, arrows), as well as diffuse poles that lacked centrin foci (asterisk k’). Bar, 10 μm.
Multipolar spindles are commonly found in tumor cell lines, and these defects typically arise due to uncoupling of the centrosome duplication cycle from normal cell cycle controls (46, 47). Our data (Figures 1, 2 and 6) as well as other reports have demonstrated the presence of multipolar spindles in cells exposed to Bisphenol A (18, 22), and the centrosomal component γ-tubulin was found to associate with BPA affinity matrices (Figure 4, Panel B), raising the possibility that BPA was inducing centrosome amplification. Given that spindle poles can self-organize in cell-free extracts or whole cells in the absence of centrosomes (48, 49), we sought to discriminate between those two possibilities by counting separated centrosomes at the G2/M transition, as well as the percentage of metaphase multipolar spindles in the absence or presence of Bisphenol A (Figure 7). Analysis of separated centrosomes revealed that even in the presence of 200 μM BPA, where nearly 70% of metaphase spindles are multipolar (Figure 7, Panel B), cells contained the normal complement of maturing centrosomes in late G2/early prophase (Figure 7, Panels A and B). Further, centrin and γ-tubulin localization revealed that metaphase cells containing supernumerary spindle poles, only two poles contained centrin foci (Figure 7, Panel C, i’-l’), and optical sectioning at 0.5 μm intervals through the cell failed to locate additional foci (not shown). Those spindle poles lacking centrin did contain γ-tubulin, but staining was diffuse (Figure 7, Panel k’) in comparison with centrin-positive controls (Figure 7, Panel g’). Those multipolar spindles that did contain more than two centrin foci (21%) were not significantly different than controls, suggesting that spindle pole splitting or centrosome amplification was not responsible for the aberrant pole formation in BPA-treated cells.
Discussion
There is an emerging body of evidence that estrogenic phenols such as diethylstilbestrol (DES) and Bisphenol A are carcinogenic, even though BPA fails to demonstrate mutagenicity by Ames testing (17, 50). BPA has been demonstrated to cause aneuploidy in both cell culture and in animal models (11, 15, 16, 18, 24), and while it is debatable whether aneuploidy is a causative- or aggravating factor in tumorigenesis (51, 52), the sensitivity of maturing mammalian oocytes to low concentrations of these compounds (10, 11) necessitates a mechanistic re-examination of Bisphenol A’s effects on mitosis. Using a chemical biological approach, we report here that Bisphenol A disrupts mitosis and cytokinesis by inducing the formation of ectopic microtubule organizing centers. Using biotinylated analogs of Bisphenol A, we have identified tubulin as a direct target of BPA, which was validated by demonstrating that BPA promoted microtubule polymerization and nucleation in vitro (Figures 4 and 5). And while our data does not definitively demonstrate the mechanism by which ectopic spindle poles are generated in vivo, these findings lend further credence to the notion that Bisphenol A is capable of disrupting normal cellular processes by mechanisms independent of the estrogen receptor.
Identification of BPA-binding proteins using biotinylated analogs
Bisphenol A is a structurally simple compound, which complicates the identification of critical functional groups as well as cellular targets. Because there are dozens of possible targets that participate in spindle assembly and cleavage plane determination (53-56), we chose a chemical biological approach to distinguish novel cellular targets using synthetic BPA-biotin conjugates as affinity agents. The biotinylation of proteins and nucleic acids is a mature technology, yet relatively few applications employing biotinylated analogs of small bioactive molecules to identify protein targets have been described. Several biotinylated steroid hormones conjugates have been generated, primarily as immunoassay probes (57-70), with relatively few examples using these reagents to study binding with cellular receptors (71-73). The potential value of this approach is evident in recent reports employing biotinylated analogs of small molecules to identify penicillin-binding proteins, targets of anti-inflammatory drugs, and investigate the actions of cancer drugs targeting DNA methyltransferases (74-76). We recently reported a new strategy for connecting biotin fragments to nonpolar substrates (38), and have adapted this approach for BPA to design two types of probes (Figure 3). Compound 1 (BPA-biotin 1) uses one of the phenolic oxygens for construction of a nonpolar ether linkage directly attached to a short biotin fragment, whereas Compound 2 (BPA-biotin 2) incorporates a butynyl spacer attached to the ortho-position of the phenol and connected to biotin using an extended linkage. BPA-biotin 1 retains one polar phenol and avoids modification of the nonpolar backbone of BPA, while BPA-biotin 2 exhibits both of the phenolic groups of BPA and uses the aryl backbone to attach an extended biotin appendage. We anticipated that both probes incorporated key structural elements of BPA and would be viable candidates for affinity purification of protein targets.
Using this approach, we were able to identify α/β– and γ-tubulin by affinity fractionation of Xenopus oocyte extracts (Figure 4), and both probes effectively bound tubulin with little observable difference using this qualitative assay (not shown). Additionally, these probes and simple methyl ether derivatives were efficacious in disrupting bipolar spindle organization in cultured cells (Figure 6). These results suggest that hydrophobic interactions of the BPA analogs are responsible for tubulin binding affinity. These observations may also indicate the likelihood that this contact occurs at surface accessible sites of the protein, since both BPA-biotin probes, incorporating short or extended linkages, exhibited affinity for α/β- and γ-tubulin. To date, this is the first report using a solid phase reagent for identifying Bisphenol A-binding proteins, and it is possible that this type of analog may be used for identifying additional targets. These findings also raise the possibility that probes can be designed to distinguish BPA-induced ER-activation from extragenic effects on tubulin nucleation.
BPA modulation of microtubule dynamics and organization
Affinity fractionation of Xenopus extracts identified tubulin as a BPA-associated protein, and in vitro analyses demonstrated that BPA directly affected microtubule assembly (Figures 4 and 5). Earlier studies had suggested that both DES and BPA disrupted microtubule organization by promoting microtubule disassembly (19, 20, 24, 77-80), which stands in contrast to both our in vitro and in vivo studies.). While BPA promoted microtubule assembly in vitro, it required glycerol, and in contrast to taxol, could not stabilize microtubules in its absence (Figure 4C). BPA also failed to stabilize microtubules against cold or calcium treatment, arguing further against its action as a microtubule stabilizer. BPA’s effects in vivo in echinoderm embryos and cultured cells was also not consistent with microtubule stabilizers such as taxol or hexylene glycol, with the most notable difference being that BPA had no effect on interphase microtubule organization even at concentrations as high as 0.2 mM (Figures 6 and 7). In contrast, at concentrations that had only a modest effect on the polymerization of purified tubulin, BPA robustly promoted microtubule nucleation from sperm centrosomes (Figure 5), suggesting that BPA’s was a facilitator of microtubule nucleation.
Bisphenol A’s affects on the microtubule cytoskeleton appear to be limited to mitosis. During the G2/M transition, microtubule dynamics undergo a dramatic increase in nucleation and turnover (81). In vitro, BPA promotes microtubule nucleation (Figure 5), raising the possibility that in mitotic cells, BPA may uncouple nucleation from the centrosome, where nucleation rates are already accelerated.
BPA has been reported to induce multipolar spindles (18, 22, 23), and exposure of maturing bovine oocytes with estradiol produces a very similar affect (82). Indeed, the estrogen receptor can directly induce the expression of Aurora A kinase (83-88), whose overexpression can drive centrosome amplification in many tumor types (86, 87, 89, 90). However, we found no evidence for centrosome amplification in cells at the G2/M boundary, nor did we find centrin localized to ectopic spindle poles (Figure 7). Lastly, methylated BPA analogs with diminished affinity for the estrogen receptor retained the capacity to induce spindle malformations (Figure 6 and Supplementary Figure 2), arguing against the involvement of the estrogen receptor as an indirect mediator of centrosome amplification in BPA-treated cells.
How does BPA induce ectopic spindle poles? Live cell analysis in sea urchin eggs suggests that ectopic asters may form de novo (Figure 2, Panels E, F, H and I). Alternatively, ectopic spindle poles may occur via spindle pole splitting, where the paired centrioles at one pole split and separate to form a new spindle pole. In sea urchin eggs, this is commonly observed during prolonged mitotic arrest (91, 92) or in response to β-mercaptoethanol (93). However, because BPA-induced ectopic poles in HeLa cells lacked centrin (and therefore centrioles) (Figure 7, Panel C), and the levels of centrin-positive supernumerary poles were no different than carrier controls (not shown), we find little evidence for spindle pole splitting. Furthermore, mammalian oocytes are acentriolar, so it is unlikely that the spindle defects observed in mammalian oocytes are due to centriole splitting per se (11, 23). However, given that γ-tubulin was also identified eluting from the BPA-biotin affinity matrices (Figure 4), its possible that centrosome fragmentation due to BPA’s effects on γ-tubulin could account for the generation of supernumerary spindle poles in both centriolar spindle poles in sea urchins and cultured cells, as well as in acentriolar spindles in mammalian cells. Moreover, centrosomes are not an absolute requirement for pole formation, and studies in Xenopus extracts, Drosophila embryos and cultured mammalian cells have demonstrated that centrosomes are dispensable for spindle formation (48, 94, 95). Indeed, both whole cells and Xenopus extracts are capable of organizing microtubule minus ends in the absence of centrosomes through a mechanism involving dynein and NUMA (49, 96-98). In such a self-organization model, kinetochore fibers nucleated and organized by active RanGTPase and further stabilized by BPA would become focused into ectopic spindle poles through a process of NUMA/dynein-mediated organization of microtubule minus ends. Ongoing efforts will determine whether this, indeed, is the case.
Methods
Embryo and mammalian cell culture
Lytechinus pictus sea urchins were obtained from Marinus Scientific (Garden Grove CA) and maintained in a chilled saltwater aquarium at 15°C. Eggs or sperm were obtained by injecting urchins with 0.5M KCl and collected gametes used immediately for all experiments. Eggs were fertilized with freshly diluted sperm, and fertilization envelopes were removed by passage through 105 μm Nytex several times before culturing in calcium-free seawater (CaFSW) at 15°C.
HeLa cells were cultured in DMEM supplemented with 10% fetal calf serum, sodium pyruvate, sodium bicarbonate and PSF. For BPA treatments, cells were treated with either carrier (0.1% DMSO) or BPA prediluted in media for 3 hours prior to fixation by immersion in methanol at −20°C.
General chemical methods
Unless noted otherwise, all chemicals were purchased from Sigma Co (St Louis, MO). BPA was dissolved fresh in DMSO or methanol at a concentration of 44 mM, and used immediately. Succinimidyl-6-(biotinamido)-6-hexanamido hexanoate (Biotin-L2-NHS) was purchased from Pierce. The Bisphenol A dimethyl ether (BPA-Me2) (1,1′-(1-methylethylidene)bis[4-methoxybenzene]) and Bisphenol A monomethyl ether (BPA-Me) (4-[1-(4-methoxyphenyl)-1-methylethyl]-phenol) were prepared by methylation of BPA using sodium hydride and methyl iodide in dimethylformamide. Detailed methods of the synthesis of the BPA affinity probes 1 and 2 are included in the Supplemental materials.
Live cell microscopy
Lytechinus pictus were fertilized and stripped of their fertilization membranes and incubated in calcium free sea water at 15°C for 25 minutes. After 25 minutes, embryos were treated with varying concentrations of DMSO or BPA and incubated for another 30 minutes. Cells were then followed by Nomarski/DIC or polarization microscopy. To better visualize the spindle in living cells, control- or BPA-treated eggs were settled onto protamine sulfate coated glass-bottomed 35 mm dishes (World Precision Instruments, Sarasota, FL), and compressed under Fluorinert FC-40 oil (99, 100). Time-lapse sequences were acquired using Zeiss Axiovert200M inverted microscopes configured for either standard Nomarski/DIC or orientation-independent polarization microscopy with a circular polarizer and 546 nm filter placed above the condenser, and a liquid crystal universal compensator (LC-Polscope, Cambridge Research Instruments, Woborn MA) placed below the reflector turret. An EXFO X-Cite 120 light source (Mississuaga, ONT) was used for transillumination, and images were acquired using a Q Imaging CCD camera controlled by PSJ software (Marine Biological Laboratory, Woods Hole, MA). Image stacks were acquired using a 3 nm retardance ceiling, exported as 8 bit tiffs to ImageJ, where movies and figures were then prepared.
Immunofluorescence
Eggs and early embryos were fixed and processed for tubulin localization according to previously described methods (100, 101). Hela cells were fixed by immersion in cold methanol for thirty minutes at −20°C before rehydration in phosphate-buffered saline (PBS). Both sea urchin eggs and HeLa cells were blocked by incubation in phosphate-buffered saline containing 5% Bovine Serum Albumin (blocking buffer), for one hour at room temperature. Cells were then placed into 1:1000 dilutions of mouse anti-tubulin (Sigma, Co, St. Louis, MO) in blocking buffer overnight at 4°C. In experiments with cultured cells, cells were also counterstained with 1:100 Rabbit anti-centrin (Sigma) or 1:500 Rabbit anti-pericentrin (Covance, Berkeley, CA). Primary antibodies were detected using Alexafluor-conjugated secondary antibodies (Molecular Probes, Eugene OR). After washing, cells were mounted in 90% glycerol/1× PBS and stored at −20°C. Sea urchin embryos were imaged using an Olympus Fluoview confocal microscope at the Central Microscopy Facility at the Marine Biological Laboratory, and HeLa cell images were acquired using a Zeiss Axiovert 200M inverted microscope equipped with epifluorescence optics and an Apotome structured illumination module (Carl Zeiss, Thornwood, NY). All acquired mages were exported into eight bit tiff files and figures were prepared using Image J and Adobe Photoshop software.
Cell-free extract preparation
Cytostatic factor-arrested Xenopus oocyte cell free extracts were prepared according to previously published protocols (102). Extracts were clarified by spinning at 16,000 rpms in 4°C for 10 minutes. After separating debris and lipids, proteinase inhibitors (20 ug/mL), cytochalasin D (0.4 ug/mL), and 20× energy mix (3M creatine phosphate, 0.4 M ATP, pH 7.4, 40mM EGTA, ph 7.7, and 0.4M MgCl2) were added to the extract, which was used immediately or supplemented with 150 mM sucrose and snap frozen in liquid nature for later use.
Cell-free extracts were prepared from fertilized sea urchin eggs 30 minutes post-insemination according to (103) and stored at −80°C until use. Extracts were also prepared from activated Spisula solidissma surf clam oocytes according to previously published protocols (104, 105) snap frozen, and stored at −80°C until use.
Affinity fractionation of BPA-binding proteins
Xenopus cytoplasmic extracts were thawed and then clarified by centrifugation at 16,000 rpms in 4°C for 10 minutes. 50 μl of 1 mg/ml BPA-biotin 1 or 2 or biotin alone was added to 500μl of clarified extract and incubated at 4°C for twenty minutes. 150 μl Strepavidin beads in a 30% slurry in wash buffer (BRB80 + proteinase inhibitors + 0.2M ATP) were added to the extracts and incubated at 4°C for an additional 20 minutes. The suspension was allowed to settle in a column, the flow-through fraction collected and washed with 10 ml of cold wash buffer. To specifically elute BPA-binding proteins, the column was eluted with 210 μM BPA in wash buffer, and 300 μl fractions were collected, snap frozen with liquid nitrogen and stored at −80°C. Fractions were resolved on 4-15% SDS-PAGE gradient gels (Bio-Rad, Hercules, CA), and bands were visualized using SYPRO-RUBY (Invitrogen, Carlsbad, CA). For proteomic analysis, unstained gels were washed twice for 10 minutes in deionized water, incubated in Bio-safe Coomassie Stain (Biorad) for 1 hour, and then destained with deionized water overnight. Under a clean hood, bands of interest were carefully excised and placed into a methanol-washed microcentrifuge tube. Proteolysis, peptide recovery and MALDI-TOF analysis were performed at the Baylor College of Medicine Protein Chemistry Facility.
Measuring effects of Bisphenol A on microtubule polymerization in vitro
Microtubule polymerization in the presence of taxol or BPA was monitored using a fluorescence-based, commercially-available assay from Cytoskeleton (Denver, CO). Because DMSO alone is capable of promoting microtubule polymerization at concentrations above 0.2%, BPA was reconstituted in methanol and further diluted in deionized water for the assay. In some conditions, glycerol in the polymerization buffer (20% glycerol, 80 mM PIPES, 0.5 mM EGTA, 2mM MgCl2, pH 6.9) was reduced to 2% to determine whether BPA acted as a microtubule stabilizer. Reactions were kept on ice until added to a 50 μl cuvette and warmed to 37°C. Microtubule polymerization was monitored in a temperature-controlled Carey Eclipse Fluorescence Spectrophotometer (Varian, Inc, Walnut Creek, CA) for fifteen minutes, with readings acquired every 15 seconds. Data was imported into a Microsoft Excel spreadsheet for further analysis and graph generation.
Microtubule nucleation was followed in Xenopus CSF-extracts by supplementing thawed extracts with 20× energy mix, 60 μg/ml Rhodamine-tubulin (Cytoskeleton, Denver, CO), and demembranated sperm nuclei and stored on ice. Bisphenol A was solubilized in methanol and further diluted to 50× stocks in BRB80 buffer. Extracts were supplemented with BPA, pre-incubated on ice for 30 minutes, and warmed to 15°C for 10 minutes. 1 μL of each reaction were mixed with 3 μL of fixative containing Hoescht 33342 and mounted for imaging. Images were acquired using a Zeiss Axiovert 200M inverted microscope equipped with a 63× NA 1.4 planapo objective and exported into eight bit tiff files and figures were prepared using Image J and Adobe Photoshop software. For quantification of BPA-induced asters, 8 bit images of single asters were normalized for background, and aster size was quantified using Image J software, and statistical analyses were performed using a pairwise student’s t-test.
Supplementary Material
Supplemental Data
Supplemental Methods (PDF file). Detailed materials and methods for the synthesis of BPA-biotin affinity probes 1 and 2.
Supplementary Figure 1. (PDF file) Bisphenol A disrupts sea urchin cell divisions in a dose-dependent manner.
Dose response curve of sea urchin embryos exposed to BPA. L. pictus eggs exposed to either DMSO or increasing concentrations of Bisphenol A 20 minutes post fertilization and cultured for 2 hours. Each condition was performed in triplicate, and following the second division, 100 embryos were scored per well for successful cleavage.
Supplementary Figure 2. (PDF file) Methylated analogs of Bisphenol A disrupt mitotic spindle formation in a manner similar to the parent molecule.
Panel A. Chemical structures of the parent molecule (BPA) and methylated analogs (BPA-Me and BPA-Me2). Panel B. Hela cells were exposed to DMSO (either at 0.1% or 0.9%, the latter of which matches the carrier concentration in experimental treatments) or 200 μM BPA, BPA-Me, or BPA-Me2 for four hours, fixed and processed. Cells were then scored for alterations in bipolar spindle formation. When compared to the parent molecule, the methylated BPA analogs displayed similar capacities to induce ectopic spindle pole formation.
Supplemental Movie 1. (quicktime MOV file) Spindle assembly in a flattened sea urchin egg. L. pictus eggs were compressed under fluorocarbon oil to better image spindle assembly, and spindle assembly was followed using orientation-independent polarization microscopy. Images were acquired every 30 seconds, and played back at 10 frames/sec. Movie corresponds to Figure 2, Panels A-C.
Supplemental Movie 2. (Quicktime MOV file). De novo formation of ectopic asters in a BPA-treated sea urchin egg. L. pictus eggs were treated with 2.2 μM BPA Bisphenol A for twenty minutes before being compressed under fluorocarbon oil, and imaged using orientation-independent polarization microscopy. Images were acquired every 30 seconds, and played back at 10 frames/sec. Movie corresponds to Figure 2, Panels D-F.
Supplemental Movie 3. (Quicktime MOV file). De novo formation of ectopic asters in a BPA-treated sea urchin egg. L. pictus eggs were treated with 2.2 μM BPA Bisphenol A for twenty minutes before being compressed under fluorocarbon oil, and imaged using orientation-independent polarization microscopy. Images were acquired every 30 seconds, and played back at 10 frames/sec. Movie corresponds to Figure 2, Panels G-I.
Acknowledgements
The authors would like to thank Snezna Rogelj and Tim Mitchison for their comments and suggestions, Bob Morris for assistance in image quantification, and Rudolf Oldenbourg and Grant Harris for their assistance with LC-Polscope imaging at the Marine Biological Laboratory. This work is supported by S06 GM08136, GM61222, P20 RR16480, and the Robert Day Allen- and Baxter Fellowships at the Marine Biological Laboratory.
Literature Cited
- 1.Watson CS, Bulayeva NN, Wozniak AL, Alyea RA. Steroids. 2006 doi: 10.1016/j.steroids.2006.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colborn T, vom Saal FS, Soto AM. Environ Health Perspect. 1993;101:378–84. doi: 10.1289/ehp.93101378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hawley RS, Warburton D. PLoS Genet. 2007;3:e6. doi: 10.1371/journal.pgen.0030006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.vom Saal FS, Hughes C. Environ Health Perspect. 2005;113:926–33. doi: 10.1289/ehp.7713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maffini MV, Rubin BS, Sonnenschein C, Soto AM. Mol Cell Endocrinol. 2006;254-255:179–86. doi: 10.1016/j.mce.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 6.Roy D, Palangat M, Chen CW, Thomas RD, Colerangle J, Atkinson A, Yan ZJ. J Toxicol Environ Health. 1997;50:1–29. doi: 10.1080/009841097160573. [DOI] [PubMed] [Google Scholar]
- 7.Brotons JA, Olea-Serrano MF, Villalobos M, Pedraza V, Olea N. Environ Health Perspect. 1995;103:608–12. doi: 10.1289/ehp.95103608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fung EY, Ewoldsen NO, St Germain HA, Jr., Marx DB, Miaw CL, Siew C, Chou HN, Gruninger SE, Meyer DM. J Am Dent Assoc. 2000;131:51–8. doi: 10.14219/jada.archive.2000.0019. [DOI] [PubMed] [Google Scholar]
- 9.Krishnan AV, Stathis P, Permuth SF, Tokes L, Feldman D. Endocrinology. 1993;132:2279–86. doi: 10.1210/endo.132.6.8504731. [DOI] [PubMed] [Google Scholar]
- 10.Susiarjo M, Hassold TJ, Freeman E, Hunt PA. PLoS Genet. 2007;3:e5. doi: 10.1371/journal.pgen.0030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hunt PA, Koehler KE, Susiarjo M, Hodges CA, Ilagan A, Voigt RC, Thomas S, Thomas BF, Hassold TJ. Curr Biol. 2003;13:546–53. doi: 10.1016/s0960-9822(03)00189-1. [DOI] [PubMed] [Google Scholar]
- 12.Ho SM, Tang WY, Belmonte de Frausto J, Prins GS. Cancer Res. 2006;66:5624–32. doi: 10.1158/0008-5472.CAN-06-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prins GS, Birch L, Tang WY, Ho SM. Reprod Toxicol. 2007;23:374–82. doi: 10.1016/j.reprotox.2006.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsutsui T, Barrett JC. Environ Health Perspect. 1997;105(Suppl 3):619–24. doi: 10.1289/ehp.97105s3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsutsui T, Tamura Y, Suzuki A, Hirose Y, Kobayashi M, Nishimura H, Metzler M, Barrett JC. Int J Cancer. 2000;86:151–4. doi: 10.1002/(sici)1097-0215(20000415)86:2<151::aid-ijc1>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 16.Tsutsui T, Tamura Y, Yagi E, Hasegawa K, Takahashi M, Maizumi N, Yamaguchi F, Barrett JC. Int J Cancer. 1998;75:290–4. doi: 10.1002/(sici)1097-0215(19980119)75:2<290::aid-ijc19>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 17.Andersen M, Kiel P, Larsen H, Maxild J. Nature. 1978;276:391–2. doi: 10.1038/276391a0. [DOI] [PubMed] [Google Scholar]
- 18.Ochi T. Mutat Res. 1999;431:105–21. doi: 10.1016/s0027-5107(99)00190-6. [DOI] [PubMed] [Google Scholar]
- 19.Lehmann L, Metzler M. Chem Biol Interact. 2004;147:273–85. doi: 10.1016/j.cbi.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 20.Metzler M, Pfeiffer E. Environ Health Perspect. 1995;103(Suppl 7):21–2. doi: 10.1289/ehp.95103s721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parry EM, Parry JM, Corso C, Doherty A, Haddad F, Hermine TF, Johnson G, Kayani M, Quick E, Warr T, Williamson J. Mutagenesis. 2002;17:509–21. doi: 10.1093/mutage/17.6.509. [DOI] [PubMed] [Google Scholar]
- 22.Nakagomi M, Suzuki E, Usumi K, Saitoh Y, Yoshimura S, Nagao T, Ono H. Teratog Carcinog Mutagen. 2001;21:453–62. doi: 10.1002/tcm.1032. [DOI] [PubMed] [Google Scholar]
- 23.Can A, Semiz O, Cinar O. Mol Hum Reprod. 2005;11:389–96. doi: 10.1093/molehr/gah179. [DOI] [PubMed] [Google Scholar]
- 24.Pfeiffer E, Rosenberg B, Deuschel S, Metzler M. Mutat Res. 1997;390:21–31. doi: 10.1016/s0165-1218(96)00161-9. [DOI] [PubMed] [Google Scholar]
- 25.Steiner S, Honger G, Sagelsdorff P. Carcinogenesis. 1992;13:969–72. doi: 10.1093/carcin/13.6.969. [DOI] [PubMed] [Google Scholar]
- 26.Iso T, Watanabe T, Iwamoto T, Shimamoto A, Furuichi Y. Biol Pharm Bull. 2006;29:206–10. doi: 10.1248/bpb.29.206. [DOI] [PubMed] [Google Scholar]
- 27.Edmonds JS, Nomachi M, Terasaki M, Morita M, Skelton BW, White AH. Biochem Biophys Res Commun. 2004;319:556–61. doi: 10.1016/j.bbrc.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 28.Atkinson A, Roy D. Biochem Biophys Res Commun. 1995;210:424–33. doi: 10.1006/bbrc.1995.1678. [DOI] [PubMed] [Google Scholar]
- 29.Roepke TA, Snyder MJ, Cherr GN. Aquat Toxicol. 2005;71:155–73. doi: 10.1016/j.aquatox.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 30.Gross PR. J Exp Zool. 1964;157:21–38. doi: 10.1002/jez.1401570107. [DOI] [PubMed] [Google Scholar]
- 31.Rappaport R. Cytokinesis in Animal Cells. Cambridge University Press; Cambridge: 1996. [Google Scholar]
- 32.Burgess DR, Chang F. Trends Cell Biol. 2005;15:156–62. doi: 10.1016/j.tcb.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 33.Oldenbourg R. Methods Cell Biol. 1999;61:175–208. doi: 10.1016/s0091-679x(08)61981-0. [DOI] [PubMed] [Google Scholar]
- 34.Inoue S, Oldenbourg R. Mol Biol Cell. 1998;9:1603–7. doi: 10.1091/mbc.9.7.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao H, Katzenellenbogen JA, Garg R, Hansch C. Chem Rev. 1999;99:723–44. doi: 10.1021/cr980018g. [DOI] [PubMed] [Google Scholar]
- 36.Fang H, Tong W, Shi LM, Blair R, Perkins R, Branham W, Hass BS, Xie Q, Dial SL, Moland CL, Sheehan DM. Chem Res Toxicol. 2001;14:280–94. doi: 10.1021/tx000208y. [DOI] [PubMed] [Google Scholar]
- 37.Anstead GM, Carlson KE, Katzenellenbogen JA. Steroids. 1997;62:268–303. doi: 10.1016/s0039-128x(96)00242-5. [DOI] [PubMed] [Google Scholar]
- 38.Corona C, Bryant BK, Arterburn JB. Org Lett. 2006;8:1883–6. doi: 10.1021/ol060458r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Longo FJ, Mathews L, Palazzo RE. Dev Biol. 1994;162:245–58. doi: 10.1006/dbio.1994.1082. [DOI] [PubMed] [Google Scholar]
- 40.Palazzo RE, Vaisberg E, Cole RW, Rieder CL. Science. 1992;256:219–21. doi: 10.1126/science.1566068. [DOI] [PubMed] [Google Scholar]
- 41.Desai A, Murray A, Mitchison TJ, Walczak CE. Methods Cell Biol. 1999;61:385–412. doi: 10.1016/s0091-679x(08)61991-3. [DOI] [PubMed] [Google Scholar]
- 42.Herzog W, Weber K. Proc Natl Acad Sci U S A. 1977;74:1860–4. doi: 10.1073/pnas.74.5.1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shelanski ML, Gaskin F, Cantor CR. Proc Natl Acad Sci U S A. 1973;70:765–8. doi: 10.1073/pnas.70.3.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zimmerman WC, Sillibourne J, Rosa J, Doxsey SJ. Mol Biol Cell. 2004;15:3642–57. doi: 10.1091/mbc.E03-11-0796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coleman KP, Toscano WA, Wiese TE. QSAR Comb. Sci. 2003;22:78–88. [Google Scholar]
- 46.Doxsey S. Mol Cell. 2002;10:439–40. doi: 10.1016/s1097-2765(02)00654-8. [DOI] [PubMed] [Google Scholar]
- 47.Kramer A, Neben K, Ho AD. Leukemia. 2002;16:767–75. doi: 10.1038/sj.leu.2402454. [DOI] [PubMed] [Google Scholar]
- 48.Khodjakov A, Cole RW, Oakley BR, Rieder CL. Curr Biol. 2000;10:59–67. doi: 10.1016/s0960-9822(99)00276-6. [DOI] [PubMed] [Google Scholar]
- 49.Merdes A, Ramyar K, Vechio JD, Cleveland DW. Cell. 1996;87:447–58. doi: 10.1016/s0092-8674(00)81365-3. [DOI] [PubMed] [Google Scholar]
- 50.Ivett JL, Brown BM, Rodgers C, Anderson BE, Resnick MA, Zeiger E. Environ Mol Mutagen. 1989;14:165–87. doi: 10.1002/em.2850140306. [DOI] [PubMed] [Google Scholar]
- 51.Giet R, Petretti C, Prigent C. Trends Cell Biol. 2005;15:241–50. doi: 10.1016/j.tcb.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 52.Weaver BA, Cleveland DW. Curr Opin Cell Biol. 2006;18:658–67. doi: 10.1016/j.ceb.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 53.Echard A, Hickson GR, Foley E, O’Farrell PH. Curr Biol. 2004;14:1685–93. doi: 10.1016/j.cub.2004.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goshima G, Vale RD. J Cell Biol. 2003;162:1003–16. doi: 10.1083/jcb.200303022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goshima G, Wollman R, Goodwin SS, Zhang N, Scholey JM, Vale RD, Stuurman N. Science. 2007;316:417–21. doi: 10.1126/science.1141314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Skop AR, Liu H, Yates J, 3rd, Meyer BJ, Heald R. Science. 2004;305:61–6. doi: 10.1126/science.1097931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bodmer DM, Tiefenauer LX, Andres RY. J Steroid Biochem. 1989;33:1161–6. doi: 10.1016/0022-4731(89)90425-1. [DOI] [PubMed] [Google Scholar]
- 58.Hauptmann H, Metzger J, Schnitzbauer A, Cuilleron CY, Mappus E, Luppa PB. Steroids. 2003;68:629–39. doi: 10.1016/s0039-128x(03)00092-8. [DOI] [PubMed] [Google Scholar]
- 59.Hauptmann H, Paulus B, Kaiser T, Herdtweck E, Huber E, Luppa PB. Bioconjug Chem. 2000;11:239–52. doi: 10.1021/bc9901402. [DOI] [PubMed] [Google Scholar]
- 60.Hauptmann H, Paulus B, Kaiser T, Luppa PB. Bioconjug Chem. 2000;11:537–48. doi: 10.1021/bc9901651. [DOI] [PubMed] [Google Scholar]
- 61.Hussey SL, Muddana SS, Peterson BR. J Am Chem Soc. 2003;125:3692–3. doi: 10.1021/ja0293305. [DOI] [PubMed] [Google Scholar]
- 62.Kaiser T, Gudat P, Stock W, Pappert G, Grol M, Neumeier D, Luppa PB. Anal Biochem. 2000;282:173–85. doi: 10.1006/abio.2000.4596. [DOI] [PubMed] [Google Scholar]
- 63.Lacorn M, Fleischer K, Willig S, Gremmel S, Steinhart H, Claus R. J Immunol Methods. 2005;297:225–36. doi: 10.1016/j.jim.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 64.Luppa P, Birkmayer C, Hauptmann H. Bioconjug Chem. 1994;5:167–71. doi: 10.1021/bc00026a009. [DOI] [PubMed] [Google Scholar]
- 65.Luppa P, Hauck S, Schwab I, Birkmayer C, Hauptmann H. Bioconjug Chem. 1996;7:332–7. doi: 10.1021/bc960015f. [DOI] [PubMed] [Google Scholar]
- 66.Mares A, DeBoever J, Stans G, Bosmans E, Kohen F. J Immunol Methods. 1995;183:211–9. doi: 10.1016/0022-1759(95)00057-h. [DOI] [PubMed] [Google Scholar]
- 67.Muddana SS, Peterson BR. Org Lett. 2004;6:1409–12. doi: 10.1021/ol0497537. [DOI] [PubMed] [Google Scholar]
- 68.Tiefenauer LX, Andres RY. J Steroid Biochem. 1990;35:633–9. doi: 10.1016/0022-4731(90)90302-9. [DOI] [PubMed] [Google Scholar]
- 69.Wang S, Lin S, Du L, Zhuang H. Anal Bioanal Chem. 2006;384:1186–90. doi: 10.1007/s00216-005-0268-2. [DOI] [PubMed] [Google Scholar]
- 70.Zhao J, Wang Y, Mi J, Li Y, Chang W. J Immunoassay Immunochem. 2003;24:369–82. doi: 10.1081/IAS-120025775. [DOI] [PubMed] [Google Scholar]
- 71.Sato S, Kwon Y, Kamisuki S, Srivastava N, Mao Q, Kawazoe Y, Uesugi M. J Am Chem Soc. 2007;129:873–80. doi: 10.1021/ja0655643. [DOI] [PubMed] [Google Scholar]
- 72.Redeuilh G, Secco C, Baulieu EE. J Biol Chem. 1985;260:3996–4002. [PubMed] [Google Scholar]
- 73.Eisen C, Meyer C, Dressendorfer R, Strasburger C, Decker H, Wehling M. Eur J Biochem. 1996;237:514–8. doi: 10.1111/j.1432-1033.1996.0514k.x. [DOI] [PubMed] [Google Scholar]
- 74.Schirrmacher E, Beck C, Brueckner B, Schmitges F, Siedlecki P, Bartenstein P, Lyko F, Schirrmacher R. Bioconjug Chem. 2006;17:261–6. doi: 10.1021/bc050300b. [DOI] [PubMed] [Google Scholar]
- 75.Honda T, Janosik T, Honda Y, Han J, Liby KT, Williams CR, Couch RD, Anderson AC, Sporn MB, Gribble GW. J Med Chem. 2004;47:4923–32. doi: 10.1021/jm049727e. [DOI] [PubMed] [Google Scholar]
- 76.Dargis M, Malouin F. Antimicrob Agents Chemother. 1994;38:973–80. doi: 10.1128/aac.38.5.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sharp DC, Parry JM. Carcinogenesis. 1985;6:865–71. doi: 10.1093/carcin/6.6.865. [DOI] [PubMed] [Google Scholar]
- 78.Hartley-Asp B, Deinum J, Wallin M. Mutat Res. 1985;143:231–5. doi: 10.1016/0165-7992(85)90086-7. [DOI] [PubMed] [Google Scholar]
- 79.Wheeler WJ, Cherry LM, Downs T, Hsu TC. Mutat Res. 1986;171:31–41. doi: 10.1016/0165-1218(86)90006-6. [DOI] [PubMed] [Google Scholar]
- 80.Tucker RW, Barrett JC. Cancer Res. 1986;46:2088–95. [PubMed] [Google Scholar]
- 81.Desai A, Mitchison TJ. Annu Rev Cell Dev Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- 82.Beker-van Woudenberg AR, van Tol HT, Roelen BA, Colenbrander B, Bevers MM. Biol Reprod. 2004;70:1465–74. doi: 10.1095/biolreprod.103.025684. [DOI] [PubMed] [Google Scholar]
- 83.Miyoshi Y, Iwao K, Egawa C, Noguchi S. Int J Cancer. 2001;92:370–3. doi: 10.1002/ijc.1200. [DOI] [PubMed] [Google Scholar]
- 84.Lo YL, Yu JC, Chen ST, Yang HC, Fann CS, Mau YC, Shen CY. Int J Cancer. 2005;115:276–83. doi: 10.1002/ijc.20855. [DOI] [PubMed] [Google Scholar]
- 85.Li JJ, Weroha SJ, Lingle WL, Papa D, Salisbury JL, Li SA. Proc Natl Acad Sci U S A. 2004;101:18123–8. doi: 10.1073/pnas.0408273101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hontz AE, Li SA, Lingle WL, Negron V, Bruzek A, Salisbury JL, Li JJ. Cancer Res. 2007;67:2957–63. doi: 10.1158/0008-5472.CAN-06-3296. [DOI] [PubMed] [Google Scholar]
- 87.Goepfert TM, Adigun YE, Zhong L, Gay J, Medina D, Brinkley WR. Cancer Res. 2002;62:4115–22. [PubMed] [Google Scholar]
- 88.Dai Q, Cai QY, Shu XO, Ewart-Toland A, Wen WQ, Balmain A, Gao YT, Zheng W. Cancer Epidemiol Biomarkers Prev. 2004;13:2065–70. [PubMed] [Google Scholar]
- 89.Li D, Zhu J, Firozi PF, Abbruzzese JL, Evans DB, Cleary K, Friess H, Sen S. Clin Cancer Res. 2003;9:991–7. [PubMed] [Google Scholar]
- 90.Meraldi P, Honda R, Nigg EA. Curr Opin Genet Dev. 2004;14:29–36. doi: 10.1016/j.gde.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 91.Shuster CB, Burgess DR. Curr Biol. 2002;12 doi: 10.1016/s0960-9822(02)00838-2. [DOI] [PubMed] [Google Scholar]
- 92.Hinchcliffe EH, Cassels GO, Rieder CL, Sluder G. J Cell Biol. 1998;140:1417–26. doi: 10.1083/jcb.140.6.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sluder G, Rieder CL. J Cell Biol. 1985;100:887–96. doi: 10.1083/jcb.100.3.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Megraw TL, Kao LR, Kaufman TC. 2001;11:116–20. doi: 10.1016/s0960-9822(01)00017-3. [DOI] [PubMed] [Google Scholar]
- 95.Heald R, Tournebize R, Blank T, Sandaltzopoulos R, Becker P, Hyman A, Karsenti E. Nature. 1996;382:420–5. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- 96.Heald R, Tournebize R, Habermann A, Karsenti E, Hyman A. J Cell Biol. 1997;138:615–28. doi: 10.1083/jcb.138.3.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Merdes A, Heald R, Samejima K, Earnshaw WC, Cleveland DW. J Cell Biol. 2000;149:851–62. doi: 10.1083/jcb.149.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Manning AL, Compton DA. Curr Biol. 2007;17:260–5. doi: 10.1016/j.cub.2006.11.071. [DOI] [PubMed] [Google Scholar]
- 99.Stack C, Lucero AJ, Shuster CB. Dev Dyn. 2006;235:1042–52. doi: 10.1002/dvdy.20695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lucero A, Stack C, Bresnick AR, Shuster CB. Mol Biol Cell. 2006;17:4093–104. doi: 10.1091/mbc.E06-02-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wong GK, Allen PG, Begg DA. Cell Motil Cytoskeleton. 1997;36:30–42. doi: 10.1002/(SICI)1097-0169(1997)36:1<30::AID-CM3>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 102.Murray AW. Methods Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- 103.Gliksman NR, Parsons SF, Salmon ED. Methods Cell Biol. 1993;39:237–51. doi: 10.1016/s0091-679x(08)60174-0. [DOI] [PubMed] [Google Scholar]
- 104.George O, Johnston MA, Shuster CB. Cell Cycle. 2006;5:2648–56. doi: 10.4161/cc.5.22.3444. [DOI] [PubMed] [Google Scholar]
- 105.Palazzo RE, Vogel JM. Methods Cell Biol. 1999;61:35–56. doi: 10.1016/s0091-679x(08)61974-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data
Supplemental Methods (PDF file). Detailed materials and methods for the synthesis of BPA-biotin affinity probes 1 and 2.
Supplementary Figure 1. (PDF file) Bisphenol A disrupts sea urchin cell divisions in a dose-dependent manner.
Dose response curve of sea urchin embryos exposed to BPA. L. pictus eggs exposed to either DMSO or increasing concentrations of Bisphenol A 20 minutes post fertilization and cultured for 2 hours. Each condition was performed in triplicate, and following the second division, 100 embryos were scored per well for successful cleavage.
Supplementary Figure 2. (PDF file) Methylated analogs of Bisphenol A disrupt mitotic spindle formation in a manner similar to the parent molecule.
Panel A. Chemical structures of the parent molecule (BPA) and methylated analogs (BPA-Me and BPA-Me2). Panel B. Hela cells were exposed to DMSO (either at 0.1% or 0.9%, the latter of which matches the carrier concentration in experimental treatments) or 200 μM BPA, BPA-Me, or BPA-Me2 for four hours, fixed and processed. Cells were then scored for alterations in bipolar spindle formation. When compared to the parent molecule, the methylated BPA analogs displayed similar capacities to induce ectopic spindle pole formation.
Supplemental Movie 1. (quicktime MOV file) Spindle assembly in a flattened sea urchin egg. L. pictus eggs were compressed under fluorocarbon oil to better image spindle assembly, and spindle assembly was followed using orientation-independent polarization microscopy. Images were acquired every 30 seconds, and played back at 10 frames/sec. Movie corresponds to Figure 2, Panels A-C.
Supplemental Movie 2. (Quicktime MOV file). De novo formation of ectopic asters in a BPA-treated sea urchin egg. L. pictus eggs were treated with 2.2 μM BPA Bisphenol A for twenty minutes before being compressed under fluorocarbon oil, and imaged using orientation-independent polarization microscopy. Images were acquired every 30 seconds, and played back at 10 frames/sec. Movie corresponds to Figure 2, Panels D-F.
Supplemental Movie 3. (Quicktime MOV file). De novo formation of ectopic asters in a BPA-treated sea urchin egg. L. pictus eggs were treated with 2.2 μM BPA Bisphenol A for twenty minutes before being compressed under fluorocarbon oil, and imaged using orientation-independent polarization microscopy. Images were acquired every 30 seconds, and played back at 10 frames/sec. Movie corresponds to Figure 2, Panels G-I.