Abstract
Background
The relevance of recurrent molecular abnormalities in cytogenetically normal (CN) acute myeloid leukemia (AML) was recently acknowledged by the inclusion of molecular markers such as NPM1, FLT3, and CEBPA as a complement to cytogenetic information within both the World Health Organization and the European Leukemia Net classifications. Mitochondrial metabolism is different in cancer and normal cells. A novel cytosolic type 2-hydroxybutyrate dehydrogenase, BDH2, originally named DHRS6, plays a physiological role in the cytosolic utilization of ketone bodies, which can subsequently enter mitochondria and the tricarboxylic acid cycle. Moreover, BDH2 catalyzes the production of 2, 3-DHBA during enterobactin biosynthesis and participates in 24p3 (LCN2)-mediated iron transport and apoptosis.
Results
We observed that BDH2 expression is an independent poor prognostic factor for CN-AML, with an anti-apoptotic role. Patients with high BDH2 expression have relatively shorter overall survival (P = 0.007) and a low complete response rate (P = 0.032). BDH2-knockdown (BDH2-KD) in THP1 and HL60 cells increased the apoptosis rate under reactive oxygen species stimulation. Decrease inducible survivin, a member of the inhibitors of apoptosis family, but not members of the Bcl-2 family, induced apoptosis via a caspase-3-independent pathway upon BDH2-KD.
Conclusions
BDH2 is a novel independent poor prognostic marker for CN-AML, with the role of anti-apoptosis, through surviving.
Keywords: BDH2, Survivin, Cytogenetically normal acute myeloid leukemia, Prognosis, Survival, Apoptosis
Background
While morphological evaluation of bone marrow (BM) and blood remains a cornerstone for the diagnosis of acute myeloid leukemia (AML), the presence or absence of specific cytogenetic and molecular abnormalities is useful not only for determining overall prognosis but also for guiding treatment. Clonal chromosomal aberrations are not detected in 40–50% of patients [1]. Both the World Health Organization and the European Leukemia Net classifications have included molecular markers such as NPM1, FLT3 and CEBPA as a prognostic factor for cytogenetically normal-acute myeloid leukemia (CN-AML) reinforcing their importance in cytogenetics [2,3]. Other mutated genes (e.g., WT1, IDH1/IDH2, TET2, RUNX1, and MLL) or aberrantly expressed ones (e.g., BAALC, ERG, EVI1, and miR-181a) will likely become useful in refining molecular risk in CN-AML [4-16]. Mithat Gönen et al. reported a panel of genetic profile that related to prognosis of AML [17].
In 1926, Otto Warburg initiated research on mitochondrial alterations in cancer and proposed a mechanism to explain the differences in energy metabolism between normal and cancer cells, suggesting that mitochondrial alterations could provide unique therapeutic targets in various cancer types [18]. Understanding the existing cross-talk between cellular metabolism and growth control has resulted in a better understanding of normal and disease processes and has facilitated the discovery of new treatment modalities in oncology, for example, trastuzumab and imatinib [19,20].
Guo et al. identified BDH2 as a short-chain dehydrogenase/reductase family member, originally named as DHRS6[21] BDH2 is a novel cytosolic-type 2-hydroxybutyrate dehydrogenase and has a physiological role in the utilization of cytosolic ketone bodies, which can subsequently enter mitochondria and the tricarboxylic acid cycle [21]. Recently, Devireddy et al. reported that BDH2 catalyzes the production of 2,3-DHBA during enterobactin biosynthesis and participates in 24p3 (LCN2)-mediated iron transport and apoptosis [22]. We reported that LCN2 expression is associated with prognosis in CN-AML previously [23]. While BDH2 is a gene related to mitochondria metabolism and participates in LCN2 mediated iron transport and apoptosis, we have interest to know if BDH2 has prognostic impact on CN-AML.
Here, we analyzed the relationship between BDH2, an enzyme related to the lipid metabolic pathway and iron transport, and CN-AML. We assessed whether BDH2 is a prognostic factor for patient survival. Additionally, we investigated the mechanism underlying the prognostic ability of BDH2 by using RNA interference-mediated knockdown of BDH2 (BDH2-KD) in cell lines.
Methods
Ethics statement
This research was approved by Kaohsiung-Medical University Chung-Ho Memorial Hospital institutional review boards and ethics committees. All human participants provided written informed consent.
Patients
We enrolled 130 patients newly diagnosed with CN-AML from August 2001 to February 2012, in a single medical center for retrospective analysis. Only 113 patients (including 49 female and 64 male patients) could be analyzed because of poor RNA quality in the remaining 17 samples. The average patient age was 54.5 years (age range, 22–86 years), with 47 patients more than 60 y/o. We also collected 43 normal BM samples, defined by normal BM findings in both BM aspiration smears and biopsy pathology reports and with no cytogenetic abnormalities, as a control group. Most patients were lymphoma without BM involvement. Moreover, we random collected bone marrow samples with good RNA quality from 10 newly diagnosed AML patients with AML-ETO fusion gene (translocation the AML1 [CBFA2, RUNX1] gene in the 21q22 region is fused to the ETO [MTG8 , RUNX1T] gene in the 8q22 region), 3 patients with cytogenetic finding of inv (16), as a good risk group, and 25 patients with multiple chromosomes abnormalities (more than 3 abnormalities) as a poor prognostic group from the samples in the past 10 years. Patients with chromosome 8 abnormality was excluded to avoid contaminated by myelodysplasia syndrome transformed AML.
Eighty-six CN-AML patients received conventional intensive induction chemotherapy consisting of 7 days of cytarabine at 200 mg·m−2·day−1 and 3 days of daunorubicin at 45 mg·m−2·day−1 (I3A7). Patients who failed to achieve complete remission (CR) but attained partial remission received the second induction chemotherapy with 70% doses of I3A7 under nadir status (between 7 and 10 days after first intensive chemotherapy). Patients achieving CR received consolidation chemotherapy with high dose AraC (cytarabine at 1 to 2 g·m−2·day−1 on day 1, 3 and 5). Based on the findings of these patients, we analyzed clinical outcomes, including overall survival (OS) and leukemia-free survival (LFS). Patients without CR after 2 rounds of intensive chemotherapy with good performance status (Eastern Cooperative Oncology Group, ECOG 0 or 1) and those with CR with a poor prognostic factor, including patients with delayed CR and FLT3 internal tandem duplication (FLT3-ITD) mutation detected after 2011, received hematopoietic stem cell transplantation (HSCT).
Clinical end points
CR was defined as recovery of morphologically normal BM and blood counts (i.e., neutrophil count ≥ 1.5 × 109/L and platelet count ≥ 100 × 109/L) and no circulating leukemic blasts or evidence of extramedullary leukemia. Relapse was defined by ≥5% BM blasts, circulating leukemic blasts, or development of extramedullary leukemia. OS was measured from the date of initial diagnosis until the date of death, censoring for patients alive at the last follow-up. LFS was measured from the date of CR until the date of relapse or death, regardless of the cause of death, censoring for patients alive at the last follow-up. Relapse-free survival (RFS) was measured from the date of diagnosis until the date of relapse or death.
Quantitative real-time reverse transcriptase-polymerase chain reaction (qRT-PCR) for mRNA expression analysis of BDH2, LCN2, Meningionma1 (MN1), ETS-related gene (ERG), micro-RNA-181a (miR-181a), and micro-RNA-3151 (miR-3151).
BM samples were collected at first diagnosis, and total RNA was extracted using the Trizol method (Invitrogen, Carlsbad, CA, USA). Furthermore, the same method was used to extract total RNA for RNA interference-mediated BDH2-KD in THP1 and HL60 cell lines. The RNA input (2 μg) for cDNA synthesis was determined by OD260 measurement, and cDNA was reverse transcribed using a TaqMan® High Capacity Reverse Transcription Kit (Applied Biosystems, Carlsbad, CA, USA) according to the manufacturer’s protocols. The cDNA sequences of BDH2 and LCN2 were evaluated, and the specific forward and reverse primers and TaqMan® probe were designed using Primer Express software version 1.5 (Applied Biosystems). The TaqMan® MGB probe designed by the software was synthesized and labeled with FAM fluorescent dye (Applied Biosystems). The mRNA expression levels of BDH2 and LCN2 were analyzed by qRT-PCR with the following primer sets and probes. BDH2: forward primer 5′-TTC CAG CGT CAA AGG AGT TGT-3′, reverse primer 5′-TTC CTG GGC ACA CAC AGT TG-3′, TaqMan® MGB probe 5′-ACA GAT GTG TGT GTA CAG CAC-3′ and LCN2: forward primer 5′-GGT ATG TGG TAG GCC TGG CA-3′, reverse primer 5′-AAC AGG ACG GAG GTG ACA TTG T-3′, TaqMan® MGB probe 5′-ACC CGC AAA AGA TGT A-3′. Expression of human β-actin was used to normalize BDH2, LCN2, ERG and MN1gene expression in qRT-PCR. Expression of human U6 snRNA was used to normalized miRNA181a and miRNA3151 gene expression in qRT-PCR. This TaqMan® endogenous control and primers and TaqMan® probes of ERG1, MN1, miRNA-181a and miRNA-3151 were purchased from Applied Biosystems. All reactions were carried out in a 25-μL final volume containing 200 ng of cDNA (as total input RNA), 400 nM of each primer, 200 nM of probe, and 12.5 μL of 2X TaqMan® Universal PCR Master Mix (Applied Biosystems).
For miRNA detection, RT reactions were performedwith 10 ng of total RNA, 50 nM stem–loop microRNA-specific RT primers, 1× RT buffer, 0.25 mM of dNTPs, 3.33 U/μl MultiScribe RTase and 0.25 U/μl RNase inhibitor. The reaction mixture was incubated for 30 min at 16°C and 30 min at 42°C, followed by 5 min incubation at 85°C to inactivate the RTase enzyme. RT products were subjected to microRNA expression assay for real-time quantitative PCR in a 20-μl final volume containing 2 μl of RT product, 1 μl of 20× TaqMan® micro-RNA Assay (Applied Biosystems), and 10 μl of 2× TaqMan® Universal PCR Master Mix (Applied Biosystems).
qRT-PCR was performed in an ABI Villi 7 Sequence Detector (Applied Biosystems), and the PCR cycling parameters were set as follows: 95°C for 10 min followed by 40 cycles of PCR reactions at 95°C for 20 seconds and 60°C for 1 min. The expression levels of the BDH2 and LCN2 genes were normalized to the internal control β-actin to obtain the relative threshold cycle (ΔCT). The relative expression between CN-AML and controls was calculated by the comparative CT (ΔΔCT) method. The CT values of β-actin were controlled between 18 and 22.
Mutation analysis of NPM1, FLT3, CEBPA, mixed lineage leukemia gene (MLL), IDH1/2 and DNMT3A
BM samples that were collected at diagnosis were retrospectively analyzed for gene mutations. Genomic DNA was extracted from mononuclear cell preparations using an Illustra™ blood genomicPrep Mini Spin Kit (GE Healthcare UK Limited, UK). The additional molecular markers associated with AML with normal karyotype, i.e., FLT3-ITD, FLT3 tyrosine kinase domain (FLT3-TKD) mutation, NPM1 mutation, CEBPA mutation, isocitrate dehydrogenase 1/2 (IDH1/2), DNA (cytosine-5)-methyltransferase 3A (DNMT3A) and mixed lineage leukemia gene (MLL) were screened as previously described [17,24-27].
PCR products were analyzed by agarose gel electrophoresis and purified using a QIAquick PCR-purification kit (Qiagen, Chatsworth, CA, USA). Purified PCR products were directly sequenced with the forward or reverse primers of each gene using an ABI BigDye Terminator Cycle Sequencing Kit (Applied Biosystems) in an ABI Prism 310 DNA sequencer (Applied Biosystems).
Cell culture
The THP1 cell line, an acute myelomonocytic leukemia cell line, was cultured in RPMI medium (GIBCO, Life Technologies, Hong Kong) supplemented with 10% fetal bovine serum (FBS) (GIBCO), 1% penicillin/streptomycin (P/S), 4.5 g/L glucose, 10 mM HEPES (4-(2-hydroxyethyl)-1-piperazineethane-sulfonic acid), 1 mM sodium pyruvate, and 1% beta-mercaptoethanol. For THP1 cells infected with shRNA empty vector and shRNA-BDH2 lentivirus, 1 μg/mL puromycin (Sigma-Aldrich, USA) was added as a stress selector.
HL60 cells, an acute promyelocytic leukemia cell line with t(15;17), were cultured in IMDM medium (GIBCO) supplemented with 1.5 g/L sodium bicarbonate, 20% FBS, and 1% P/S. Puromycin (1 μg/mL) was added to select cells with RNA interference-mediated BDH2-KD.
All cells were cultured at 37°C in a humidified atmosphere containing 5% CO2. All native cell lines were purchased from Food Industry Research and Development Institute, Taiwan.
RNA interference-mediated BDH2-KD in THP1 and HL60 cells
The shRNA-BDH2 lentivirus particle was purchased from Sigma. The clones TRCN0000036735, 0000036736, 0000036738, and 00000244979 were identified as shRNA-BDH2-1, shRNA-BDH2-2, shRNA-BDH2-3, and shRNA-BDH2-4, respectively. Naive THP1 and HL60 cells were transduced with lentiviruses expressing shRNAs and selected for puromycin resistance (1 μg/mL). The knockdown efficiency was assessed by qRT-PCR and western blot analyses.
Statistical analysis
We used software SPSS 17.0 for statistical analysis. ANOVA was used to compare the differences in BDH2 mRNA expression and LCN2 mRNA expression between patients with CN-AML, AML-ETO (+) under molecular studies, inv (16) under cytogenetic study and those with normal BM. Correlation regression was used for analysis if there was a correlation between BDH2 and LCN2 expression. We used the receiver operating characteristic (ROC) curve to estimate the cutoff point for BDH2 to predict death in CN-AML patients.
Time-to-event analysis involved estimating the probability that an event would occur at different time points. The end-point of follow-up for patients who developed AML was the date of death and for those who were lost to follow-up was the date of the last visit, to arrive at “censored” data. Two-sample t-tests and X2 square tests were used to analyze the differences in age, sex, peripheral white blood cells (WBCs), hemoglobin (Hb), platelet and blast counts, CD34 and blasts percentage in BM, percentage of FLT3-ITD, FLT3-TKD, NPM1 mutations, CEBPA mutation, IDH1/2, DNMT3A and MLL mutations in the BDH2 low and high expression groups (BDH2low and BDH2high, respectively) and younger and older patients group (separate at 60 year-old). ANOVA were used to analyze ERG, NM1, miR-181a and miR-3151 in different groups of BDH2 expression and age. We divided low and high expression of BDH2 using the median BDH2 expression level (ΔCT-BDH2 = 9.0060), which was similar to the cutoff point from the ROC curve prediction (ΔCT-BDH2 = 9.0113). The Kaplan–Meier estimates were computed for the risk among different categories, were compared by Log-rank tests. The analysis was performed to estimate the differences in OS among patients with different BDH levels, and FLT3-ITD mutation as well as the differences in LFS and RFS between BDH2high and BDH2low groups.
Two sets of hazard rate ratios (HRR) were computed for analysis factors by cox regression analyses. The univariate HRRs were estimated from separate Cox regressions with one analysis factor at a time. The factors included all molecular analyses and other parameters. The multivariable-adjusted HRRs were computed from Cox regression with additional variables of the factors that were identified as being statistically significant in the univariate analysis.
Apoptosis assay and flow cytometry
We treated THP1 and HL60 cells lines, including shRNA-BDH2 infected, shRNA empty vector infected, and parental cells, with 50 μM H2O2 for 30 min and with 15 μM H2O2 for 30 min, respectively. Then apoptosis was examined using the ApoScreen™ Annexin V-FITC Apoptosis Kit (Beckman Coulter, CA, USA). Meanwhile, caspase-3 was also analyzed by flow cytometry in cells treated with H2O2 using FITC rabbit anti-active caspase-3 (BD Pharmingen, CA, USA).
Western blot analysis
Cells were treated with 15 and 50 μM H2O2 for 2 h and were then lysed in cold lysis buffer supplemented with protease inhibitor cocktails (BioExpress, UT, USA, added at a 1:100 dilution). Cell lysates were separated by SDS-PAGE and transferred to a polyvinylidene fluoride membrane (PerkinElmer, Taiwan). Blots were probed with primary antibodies specific for the following proteins: PARP, caspase-3, survivin (Cell Signaling Technology, MA, USA), Bcl-xL, Bax, XIAP (Santa Cruz Biotechnology, Texas, USA), BDH (Sigma), and β-actin (Millipore Corporation, MA, USA). After incubation with the antibody, the proteins were detected with enhanced chemiluminescence (ECL, PerkinElmer).
JC-1 mitochondrial membrane potential detection
The mitochondrial membrane potential was analyzed using a MitoProbe™ JC-1 Assay Kit (Life technologies, CA, USA). The collapse of the electrochemical gradient across the mitochondrial membrane was measured using a fluorescent cationic dye, JC-1. This dye exhibits potential-dependent accumulation in the mitochondrial matrix. Further, 1 × 106 cells were incubated with 2 mM JC-1 at 37°C and 5% CO2 for 30 min. Cells were washed twice with PBS at 4°C, resuspended in 1 mL PBS, and assessed by a Beckman Coulter Epics XL.
Results
Association of BDH2 expression with molecular markers and clinical characteristics
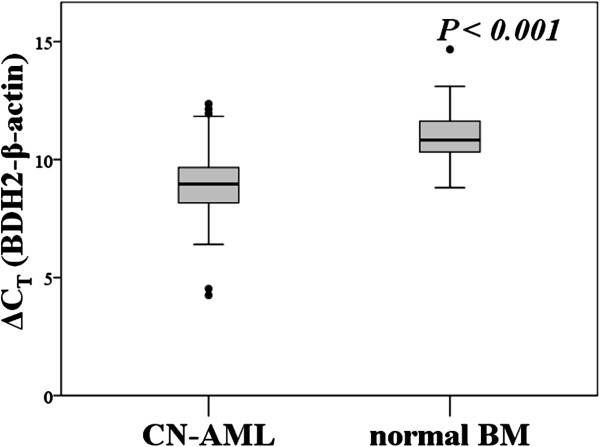
The mRNA expression of BDH2 was higher in patients with CN-AML and poor risk than in those with normal BM (P < 0.001, Figure 1 and Additional file 1: Figure S1). The mRNA expression of LCN2 was lower in patients with CN-AML, inv (16) and poor risk group that we already reported [23]. However, we did not see difference of BDH2 mRNA expression between good risk groups and normal bone marrow or CN-AML patients. There was no correlation between BDH2 and LCN2 expression (Additional file 2: Figure S2).
Figure 1.
mRNA expression levels of BDH2 in bone marrow.BDH2mRNA expression levels in bone marrow from patients with normal bone marrow and cytogenetically normal AML (CN-AML). P<0.001. Higher ΔCT-BDH2 means lower mRNA expression levels. BM: bone marrow. Sample number of bone marrows from normal bone marrow and CN-AML are 43 and 113.
We focused on CN-AML patients and used the median BDH2 mRNA expression level, ΔCT-BDH2 = 9.0060, as the cutoff point. This value is the same as the cutoff point predicted by the ROC curve. Among the 113 patients newly diagnosed with CN-AML, no differences were observed between BDH2high and BDH2low groups with regard to clinical features or biological characteristics such as age, sex, WBCs, Hb, platelets, blasts in peripheral blood, blasts in BM, amount of CD34 expression in BM myeloblasts, and French-American-British (FAB) classification subtypes (Table 1). Moreover, no differences were observed with regard to these clinical features between the 2 groups, among the 86 patients with CN-AML with intensive induction chemotherapy (data not shown).
Table 1.
Comparison of clinical manifestations between patients with AML in low and high BDH2 expression groupsa
| Variables | Total (n = 113) | Low BDH2, expression, (n = 57) | High BDH2, expression, (n = 56) | P |
|---|---|---|---|---|
| Sexb |
113 |
57 |
56 |
0.467 |
| Male |
64 |
23 |
26 |
|
| Female |
49 |
34 |
30 |
|
| Age (years)c |
54.5 (21–86) |
48 (22–86) |
57 (21–85) |
0.193 |
| Laboratory datac |
|
|
|
|
| WBC, ìL−1 |
30,830 |
37,615 |
24,370 (600–243,290) |
0.452 |
| (300–296,300) |
(300–216,820) |
|||
| Hb, g/dL |
8.3 |
8.3 |
8.4 |
0.365 |
| (4.2–15.6) |
(4.4–14.3) |
(4.2–15.6) |
||
| Platelet, ×1,000/ìL |
42 |
44.5 |
41 |
0.582 |
| (3–369) |
(4–175) |
(3–369) |
||
| Blast, ìL−1 |
20,188.5 |
13,074.15 |
26,001 |
0.172 |
| (0–287,411) |
(126–145,572.7) |
(0–287,411) |
||
| Blast in BM, % |
74.8% (24.8%–97.4%) |
70.8% (24.8%–94.7%) |
79% (27.2%–94.4%) |
0.424 |
| CD34 (+) in BM, % |
31.1% |
29.5% |
33.25% (0%–99.5%) |
0.272 |
| (0%–99.9%) |
(0%–99.9%) |
|||
| FABb |
|
|
|
|
| M0 |
3 |
1 (1.75%) |
2 (2.44%) |
0.057 |
| M1 |
28 |
8 (13.89%) |
20 (43.90%) |
|
| M2 |
47 |
25 (47.23%) |
22 (34.15%) |
|
| M3 |
0 |
0 (0%) |
0 (0%) |
|
| M4 |
23 |
15 (23.61%) |
9 (14.63%) |
|
| M5 |
7 |
6 (9.72%) |
1 (0%) |
|
| M6 |
0 |
0 (0%) |
0 (0%) |
|
| M7 |
4 |
2 (2.78%) |
2 (4.88%) |
|
| Undetermined |
0 |
0 (0%) |
0 (0%) |
|
| Induction responsebd |
|
|
|
|
| CR |
61 (70.93%) |
37 (80.42%)f |
24 (60%)g |
0.032* |
| PR and Refractory |
|
32 |
20 |
|
| Induction death |
|
1 |
0 |
|
| Reach CR timee |
|
|
|
|
| 38 ± 38.0 | 40.5 ± 44.3 | 36 ± 29.7 | 0.526 |
aThe median value of BDH2 expression in the total population was used as the cutoff level of 9.006 to define low- and high-expression groups.
bNumber of patients (%).
cMedian (range).
dOnly the 86 patients who received conventional intensive induction chemotherapy, and then consolidation chemotherapy if CR was achieved, were included in the analyses.
ePatients received conventional intensive induction chemotherapy and achieved CR, median, days ± S.D.
fOnly 46 patients received I3A7.
gOnly 40 patients received I3A7 (Idarubicin and Ara-C).
*Statistically significant (P < 0.05).
The incidences of common genetic alterations in the BDH2high and BDH2low groups are shown in Table 2. On the whole cohort analysis, our patients showed similar incidences of FLT3-ITD and FLT3-TKD mutations when compared with data from Taiwan National University [28]; however, the incidences of NPM1, MLL and CEBPA mutations were higher and the incidence of IDH1 mutation was lower. FLT3-ITD showed a higher mutation rate in the BDH2high group (P = 0.030, Table 2) and DNMT3A showed a higher mutation rate in the BDH2low group (P = 0.009, Table 2). We did not observe differences in NPM1, FLT3-TD, CEBPA, and IDH1/2 mutations between the 2 groups (Table 2).
Table 2.
Comparison of other genetic alterations between AML patients in low and high BDH2 expression groups
| Variant |
Number of patients with the gene alternation (percentage) |
P | ||
|---|---|---|---|---|
| Whole cohort | Low BDH2, expression | High BDH2, expression | ||
|
NPM1mut |
36 (31.85%) |
16 (28.07%) |
20 (35.71%) |
0.271 |
|
FLT3-ITD |
23 (20.35%) |
7 (12.28%) |
16 (28.57%) |
0.030* |
|
FLT3-TKD |
8 (7.08%) |
5 (8.77%) |
3 (5.36%) |
0.338 |
|
NPM1mut/FLT3-ITDneg |
20 (17.70%) |
11(19.30%) |
9 (16.07%) |
0.368 |
|
CEBPAa |
34 (30.09%) |
16 (28.07%) |
18 (32.14%) |
0.444 |
|
CEBPADouble mutation |
9 (7.96%) |
3 (5.26%) |
6 (10.71%) |
0.593 |
|
IDH1b |
3 (3.75 %) |
1 (2.56 %) |
2 (4.88%) |
0.592 |
|
IDH2b |
8 (10 %) |
3 (7.69%) |
5 (12.20%) |
0.508 |
|
DNMT3Ab |
12 (15 %) |
10 (25.641%) |
2 (4.88%) |
0.009* |
|
MLLb |
7 (8.75%) |
3 (7.69%) |
4 (9.76%) |
0.426 |
|
ERGc |
11.17 (10.26-12.08) |
11.60 (10.48-12.72) |
10.68 (9.17-12.20) |
0.320 |
|
MN1c |
12.98 (12.28-13.68) |
13.35 (12.42-14.28) |
12.55 (11.47-13.64) |
0.257 |
|
miR-181ac |
3.12 (2.57-3.67) |
3.17 (2.35-3.98) |
3.07 (2.30-3.84) |
0.864 |
| miR-3151c | 12.35 (11.90-12.80) | 12.18 (11.53-12.84) | 12.53 (11.89-13.16) | 0.448 |
Values are number (%) of patients with alteration.
aCEBPAsingle and double mutations.
bOnly 80 patients with high quality of DNA to sequence; 39 patients are low BDH2 expression and 41 patients are high expression.
cMean (95% CI).
*Statistically significant (P < 0.05).
Gene alternations frequencies between younger and elder patients
As shown in Table 3, the frequency of FLT3-TKD mutation is higher in patients more than 60 years-old. And the CEBPA double mutation rate is higher in younger patients group. There are no different of NPM1, FLT3-ITD, IDH1/2, DNMT3A and MLL gene mutations, and no difference in BDH2, ERG, MN1, miR-181a and miR-3151 expression levels, between different age group.
Table 3.
Comparison of other genetic alterations between older and younger AML patientsa
| Variant |
Number of patients with the gene mutation (percentage); Median of RNA expression (delta CT) |
P | ||
|---|---|---|---|---|
| Whole cohort | 60 years or younger, (n=69) | Older than 60 y/o, (n=44) | ||
|
NPM1mut |
38 (33.33%) |
21 (30%) |
17 (38.64%) |
0.227 |
|
FLT3-ITD |
23 (20.18%) |
15 (21/43%) |
8 (18.18%) |
0.433 |
|
FLT3-TKD |
8 (7.02%) |
2 (2.86%) |
6 (13.64%) |
0.036* |
|
NPM1mut/FLT3-ITDneg |
22 (19.30%) |
11(15.71%) |
11 (25%) |
0.164 |
|
CEBPAa |
34 (30.36%) |
18 (26.09%) |
16 (37.20%) |
0.151 |
|
CEBPADouble mutation,b |
9 (78.04%) |
8 (11.59%) |
1 (2.23%) |
0.016* |
|
IDH1c |
3 (3.75 %) |
2 (4%) |
1 (3.3%) |
0.686 |
|
IDH2c |
8 (10 %) |
4 (8%) |
4 (13.2%) |
0.454 |
|
DNMT3Ac |
12 (15 %) |
10 (25.641%) |
2 (4.88%) |
0.095 |
|
MLL |
7 (8.75%) |
4 (8%) |
3 (10%) |
0.528 |
|
Delta BDH2d |
9.006 (4.53-12.36) |
9.051 (4.53-12.36) |
8.915 (6.4-12.15) |
0.102 |
|
ERGd |
10.595 (4.52-19.92) |
10.88 (5.17-19.92) |
9.74 (4.52-19.85) |
0.435 |
|
MN1d |
13.25 (7.75-19.87) |
13.06 (7.76-19.87) |
13.25 (7.75-18.38) |
0.992 |
|
miR-181ad |
3.21 (−3.8-8.44) |
3.3 (−0.03-8.44) |
2.49 (−3.8-8.2) |
0.140 |
| miR-3151d | 12.685 (4.97-15.29) | 12.685 (9.16-15.07) | 12.6 (4.97-15.29) | 0.247 |
Values are number (%) of patients with alteration.
aCEBPAsingle and double mutations.
bCEBPAdouble mutationvs CEBPAsingle mutationvs no mutation.
cOnly 80 patients with high quality of DNA to sequence; 50 patients are 60 y/o or younger patients and 30 patients are high expression.
dMedian delta CT(range).
*Statistically significant (P < 0.05).
BDH2 expression as a prognostic marker
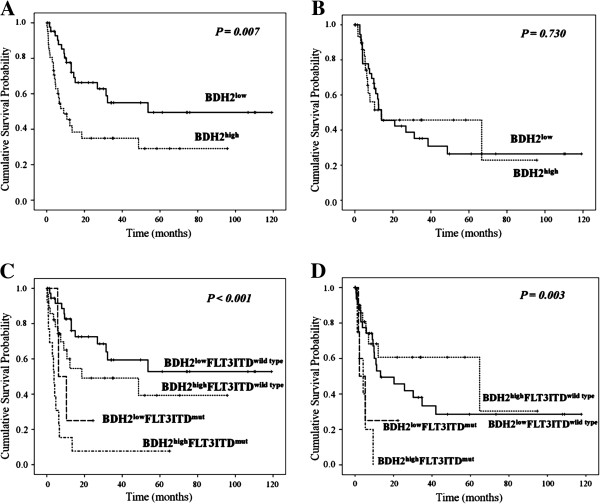
We analyzed 86 patients who received a standard intensive chemotherapy. In response rate analysis, patients in the BDH2high group showed a lower complete response rate (60%) than those in the BDH2low group (80.42%). However, no difference was observed between the 2 groups with respect to the time required to reach a complete response (Table 1). We also analyzed complete response (CR) rate based on genetic alterations and noticed that patients with DNMT3A mutations had significant higher CR rate than patients without DNMT3A mutation (P = 0.012). We did not find significant difference in CR rate between FLT3-ITD, NPM1, CEBPA and IDH1/2 mutations (data not shown). Results of the survival analysis showed that patients in the BDH2high group had a lower overall survival (OS) with a medium survival of 9 months than those in the BDH2low group with a median survival of 53.667 months (P = 0.007, Figure 2A). However, we did not note any difference in the LFS rates between the BDH2high and BDH2low groups, with median survivals of 12.033 months and 13.2 months, respectively (P = 0.730, Figure 2B).
Figure 2.
Outcome of patients with CN-AML according to BDH2 expression levels and FLT3-ITD mutation status. In 85 patients receiving intensive induction chemotherapy, we analyzed A) overall survival; B) leukemia-free survival between 2 different BDH2 expression groups; and C) overall survival; D) leukemia-free survival between groups differentiated by BDH2 expression level, combined with the FLT3-ITD mutation status.
In univariate analysis of the impact factors on OS, old age, high BDH2 expression, and FLT3-ITD mutation adversely affected OS with statistical significance (P = 0.001, 0.007, and < 0.001, respectively; Table 4). NPM1, FLT2-TKD, CEBPA, IDH1/2, DNMT3A and MLL mutations did not show impact on OS. Multivariate analysis showed that FLT3-ITD, FLT3-TKD and MN1 mutations adversely affected RFS. While NPM1 mutation showed positive impact on RFS. High BDH2 expression had a mild adverse effect on RFS without statistical significance. However, a significant, independent adverse impact of high BDH2 expression, FLT3-ITD mutation, MN1 mutation, and old age were observed in the OS multivariate analysis (Table 5). In patients with 60 year-old or younger, FLT3-ITD, FLT3-TKD, MN1 mutations, and ERG and BDH2 higher expression level showed adverse impact on survival. In elder patients, we saw ERG and BDH2 higher expression level had poor survival (Table 6). No patients in elderly group had DNMT3A mutation.
Table 4.
Univariate analyses of the impact on overall survival in patients with CN-AML who received intensive induction chemotherapy
| Variable |
No. of patients |
Overall survival |
|
|---|---|---|---|
| Mediana | P | ||
| Age, years |
|
|
0.001* |
| 60 or younger |
66 |
32.2 |
|
| Older than 60 |
22 |
4.5 |
|
| WBC |
|
|
0.469 |
| 50000/uL or less |
49 |
31.433 |
|
| Greater than 50000/uL |
37 |
13 |
|
|
BDH2 |
|
|
0.007* |
| Lower expression |
46 |
53.667 |
|
| Higher expression |
40 |
9 |
|
|
CEBPA |
|
|
0.755 |
| Double mutation |
9 |
31.433 |
|
| Single mutation |
16 |
53.667 |
|
| No mutation |
61 |
16.267 |
|
|
NPM1 |
|
|
0.179 |
| Mutated |
28 |
10.4 |
|
| Wild |
58 |
32.2 |
|
|
FLT3-ITD- |
|
|
<0.001* |
| Mutated |
17 |
4.767 |
|
| Wild |
70 |
48.633 |
|
|
FLT3-TKD |
|
|
0.596 |
| Mutated |
7 |
13.00 |
|
| Wild |
79 |
18.567 |
|
|
NPM1/FLT3-ITD |
|
|
0.314 |
| NPM1+/FLT3-ITD- |
15 |
NR |
|
| Others |
71 |
15.067 |
|
|
IDH1b |
|
|
0.796 |
| Mutated |
3 |
22.21 |
|
| Wild |
61 |
27.055 |
|
|
IDH2b |
|
|
0.749 |
| Mutated |
4 |
21.91 |
|
| Wild |
60 |
27.16 |
|
|
DNMT3Ab |
|
|
0.508 |
| Mutated |
11 |
32.58 |
|
| Wild |
53 |
25.63 |
|
|
MLLb |
|
|
0.178 |
| Mutated |
5 |
12.63 |
|
| Wild |
59 |
13.12 |
|
|
BDH2low/FLT3-ITD |
|
|
<0.001* |
| BDH2low/FLT3-ITDwild type |
40 |
722.16 |
|
| BDH2low/FLT3-ITDmutation |
5 |
519.73 |
|
| BDH2high/FLT3-ITDwild type |
27 |
371.56 |
|
| BDH2high/FLT3-ITDmutation | 13 | 182.27 | |
Abbreviation: NR indicates not reached.
aMedian, months.
bOnly 64 patients who had high quality DNA preserved for sequence received standard intensive induction chemotherapy.
*Statistically significant (P < 0.05).
Table 5.
Multivariate analyses (cox regression) of relapse-free survival and overall survivala
| Variables |
Relapse-free survival |
Overall survival |
||||||
|---|---|---|---|---|---|---|---|---|
| |
95% CI |
|
|
95% CI |
|
|||
| HR | Lower | Upper | P | HR | Lower | Upper | P | |
| Whole cohort (n = 86) |
|
|
|
|
|
|
|
|
| Ageb |
3.083 |
0.184 |
51.762 |
0.434 |
3.252 |
1.322 |
8.002 |
0.010* |
|
NPM1c |
0.087 |
0.012 |
0.604 |
0.014* |
0.392 |
0.128 |
1.195 |
0.100 |
|
FLT3-ITDd |
21.079 |
1.427 |
311.302 |
0.026* |
4.532 |
1.395 |
14.723 |
0.012* |
|
FLT3-TKD |
19.630 |
1.539 |
250.445 |
0.022* |
1.801 |
0.184 |
17.603 |
0.613 |
|
CEBPAe |
0.408 |
0.144 |
1.154 |
0.091 |
0.398 |
0.147 |
1.080 |
0.070 |
|
IDH1c |
0.448 |
0.039 |
5.167 |
0.520 |
0.648 |
0.128 |
3.268 |
0.599 |
|
IDH2c |
0.583 |
0.025 |
13.640 |
0.737 |
0.254 |
0.036 |
1.781 |
0.168 |
|
DNMT3Ac |
0.292 |
0.041 |
2.098 |
0.221 |
0.731 |
0.158 |
3.380 |
0.689 |
|
MLLc |
31.310 |
0.729 |
1344.29 |
0.073 |
1.040 |
0.215 |
5.020 |
0.961 |
|
MN1 |
1.391 |
1.027 |
1.883 |
0.033* |
1.195 |
1.025 |
1.394 |
0.023* |
|
ERG |
5.799 |
0.548 |
61.325 |
0.144 |
2.545 |
0.993 |
6.520 |
0.052 |
|
miR-181a |
0.824 |
0.598 |
1.135 |
0.236 |
0.860 |
0.692 |
1.069 |
0.174 |
|
miR-3151 |
1.183 |
0.750 |
1.867 |
0.469 |
1.285 |
0.884 |
1.866 |
0.189 |
| BDH2f | 2.035 | 0.472 | 8.782 | 0.341 | 2.547 | 1.094 | 6.685 | 0.050* |
Abbreviations: HR indicates Hazard ratio; CI, confidence interval.
aOnly patients with intensive induction chemotherapy enrolled.
bAge > 60 relative to Age ≤ 60 (the reference).
cNPM1mut versus NPM1wild type; IDH1mut versus IDH1wild type; IDH2mut versus IDH2wild type; DNMT3Amut versus DNMT3Awild type ; MLLmut versus MLLwild.
dFLT3-ITDmut versus FLT3-ITDneg.
eCEBPAdouble-mutation versus CEBPAsingle mutation versus CEBPAno mutation.
fBDH2high relative to BDH2low group.
*Statistically significant (P < 0.05).
Table 6.
Multivariate analyses (cox regression) of overall survivala in younger and older patients
| Variables |
Patients ≦ 60 y/o |
Patients > 60 y/o |
||||||
|---|---|---|---|---|---|---|---|---|
| |
95% CI |
|
|
95% CI |
|
|||
| HR | Lower | Upper | P | HR | Lower | Upper | P | |
|
NPM1b |
0.000 |
0.000 |
1.490E89 |
0.898 |
0.263 |
0.021 |
3.377 |
0.305 |
|
FLT3-ITDc |
7.505 |
1.538 |
36.614 |
0.013* |
3.235 |
0.424 |
24.699 |
0.258 |
|
FLT3-TKD |
9765.104 |
32.249 |
2.95E6 |
0.002* |
144.617 |
0.000 |
3.75E169 |
0.980 |
|
CEBPAd |
0.357 |
0.125 |
1.020 |
0.054 |
1.090 |
0.292 |
4.066 |
0.898 |
|
IDH1b |
14220.871 |
0.000 |
3.9E99 |
0.932 |
174.990 |
0.000 |
4.56E169 |
0.979 |
|
IDH2b |
0.000 |
0.000 |
2.695E210 |
0.958 |
0.459 |
0.050 |
4.242 |
0.492 |
|
DNMT3Ab |
146.313 |
0.000 |
4.16E97 |
0.965 |
|
|
|
|
|
MLLb |
0.059 |
0.000 |
1.78E94 |
0.980 |
2.245 |
0.173 |
29.122 |
0.536 |
|
MN1e |
1.923 |
1.130 |
3.273 |
0.016* |
1.198 |
0.916 |
1.567 |
0.187 |
|
ERG |
53.777 |
1.820 |
1589.377 |
0.021* |
32.349 |
2.889 |
362.237 |
0.005* |
|
miR-181a |
0.514 |
0.260 |
1.014 |
0.055 |
1.361 |
0.933 |
1.986 |
0.110 |
|
miR-3151 |
1.164 |
0.580 |
2.337 |
0.669 |
0.423 |
0.191 |
0.935 |
0.033* |
| BDH2e | 4.829 | 1.002 | 23.274 | 0.050* | 18.937 | 1.571 | 228.308 | 0.021* |
Abbreviations: HR indicates Hazard ratio; CI, confidence interval.
aOnly patients with intensive induction chemotherapy enrolled.
bNPM1mut versus NPM1wild type; IDH1mut versus IDH1wild type; IDH2mut versus IDH2wild type; DNMT3Amut versus DNMT3Awild type.
cFLT3-ITDmut versus FLT3-ITDneg.
dCEBPAdouble-mutation versus CEBPAsingle mutation versus CEBPAno mutation.
eBDH2high relative to BDH2low group.
*Statistically significant (P < 0.05).
By combining two independent prognostic factors, BDH2 expression and FLT3-ITD mutation, we found that patients with BDH2lowFLT3-ITD-wild type had the highest OS, with a median survival surpassing 10 years. On the other hand, patients with BDH2highFLT3-ITD-mutation had the worst overall survival, with a median survival of 3.833 months. We observed significant differences in the overall survival between the BDH2 expression groups with and without the FLT3-ITD mutation (median survival: BDH2lowFLT3-ITD-wild type vs. BDH2highFLT3-ITD-wild type vs. BDH2lowFLT3-ITD-mutation vs. BDH2highFLT3-ITD-mutation = not reached vs. 18.567 months vs. 6.2 months vs. 3.833 months, P < 0.001, Figure 2C and 2D).
We analyzed 22 patients who received allogeneic hematopoietic stem cell transplantation, including 14 patients in the BDH2 lower expression group and 8 in the BDH2 higher expression group. Though there was no statistically significant difference, we saw a trend of longer survival from hematopoietic stem cell infusion in the BDH2low group (P = 0.520, Additional file 3: Figure S3).
Survivin reduction was responsible for inducing apoptosis in BDH2-KD cells under hyperoxidative stress via a caspase-3-independent pathway
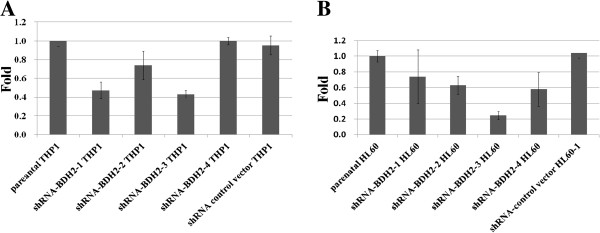
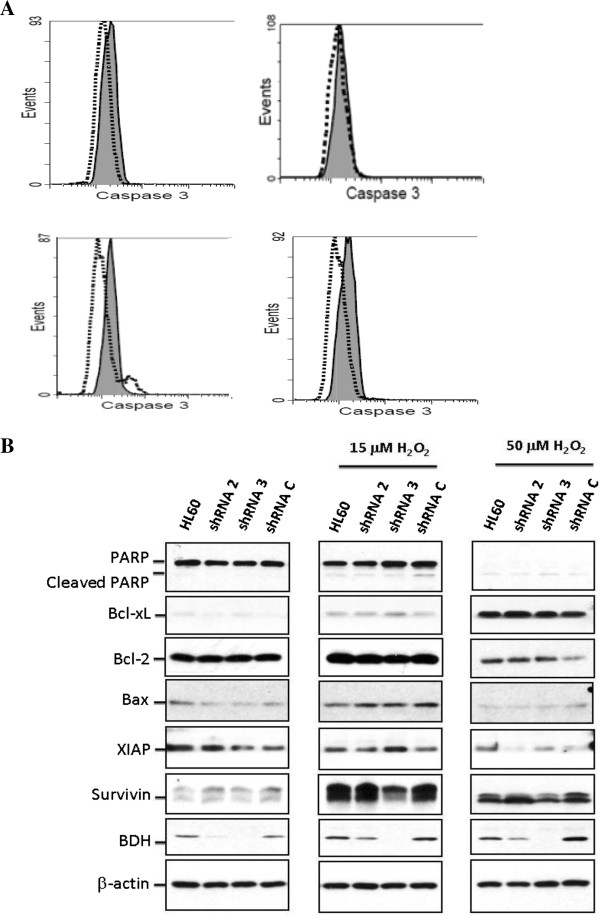
Reactive oxygen species (ROS) can induce apoptosis [29]. Devireddy et al. showed that the apoptosis rate increased in BDH2-KD FL5.12 cells upon H2O2 treatment [22]. The impact of BDH2 was evaluated by using RNA interference-mediated BDH2-KD in THP1 and HL60 leukemia cell lines. The efficacy of BDH2-KD was confirmed at both RNA (Figure 3A and 3B) and protein levels (Figure 4B). Microscopic analysis under hyperoxidative stress showed that shRNA-BDH2-3 HL60 had more apoptotic cells (Additional file 4: Figure S4). This result was confirmed by Annexin V/PI staining analysis. Figure 5 shows the apoptotic population in different levels of BDH2-KD HL60 cells. After H2O2 treatment, BDH2-KD HL60 cells showed a more prominent increase in the secondary and forth quadrant, indicating that BDH2 exerted an anti-apoptotic effect (Figure 5). The same effect was also observed in BDH2-KD THP1 cells (data not shown). Nevertheless, the conventional apoptosis-related proteins, including PARP, caspase-3, Bcl-xL, Bcl-2 and Bax, did not mediate H2O2-induced apoptosis in BDH2-KD HL60 cells (Figure 4B). Western blotting data also failed to show caspase-3 activation using flow cytometry analysis (Figure 4A), regardless of whether the expression level of BDH2 was high or low. Taken together, the results did not show a correlation between BDH2-KD and the activation of the Bcl-2/Bcl-xL-mediated intrinsic pathway via the caspase cascade to trigger apoptosis. This phenomenon correlated with mitochondrial membrane potential analysis (Additional file 5: Figure S5).
Figure 3.
shRNA interference-mediated knockdown efficiency in leukemia cell lines. BDH2 mRNA expression level in BDH2-knockdown, control vector infected, and parental cells. A) THP1 cells and B) HL60 cells, as determined by qRT-PCR analyses.
Figure 4.
Caspase-3 activity after treatment with 50 μM H2O2 for 2 h in the HL60 cell line. A) Cleaved caspase-3 detected by flow cytometry. The gray area represents cells that were not subjected to the H2O2 treatment, and the dot- lines denote cells treated with H2O2. Top left: parental HL60; top right: shRNA-BDH2-2 HL60; bottom left: shRNA-BDH2-3 HL60; bottom right: shRNA control vector HL60. B) Bcl-2, Bcl-xL, BAX, PARP, survivin, XIAP, and BDH2 protein expression on a western blot analyses.
Figure 5.
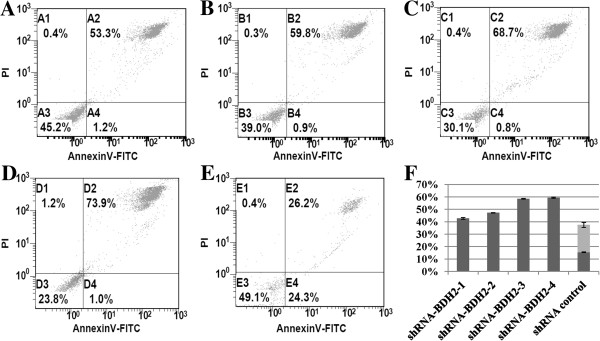
Apoptosis rate in HL60 cells after treatment with 50 μM H2O2 for 2 h. The apoptosis rate was dependent on the BDH2-knockdown efficiency: A) shRNA-BDH2-1 HL60; B) shRNA-BDH2-2 HL60; C) shRNA-BDH2-3 HL60; D) shRNA-BDH2-4 HL60; and E) shRNA control vector HL60; F) the apoptosis rate of each cell lines. Dark gray: late apoptosis bar; light gray bar: early apoptosis.
There is a family of functionally and structurally related proteins that serve as endogenous inhibitors of apoptosis (IAP) [30]. Survivin, the smallest member of the IAP family, increases during ROS stimulation to protect cells from apoptosis [30,31]. A marked induction of survivin upon H2O2 treatment was observed in our study, whereas XIAP was not altered in cells with different BDH2 expression levels. Of note, the induction of survivin was abrogated in shRNA-BDH2-3 HL60 cells. These data suggest that survivin is responsible for elevating the apoptosis rate upon ROS treatment in BDH2-KD cells.
Discussion
The prognosis of CN-AML depends on the molecular status (e.g., FLT3, NPM1, CEBPA, IDH1/2, DNMT3A, MLL, ERG, MN1 and micro-RNA-181a and 3151); however, not all candidate markers have been detected so far. Energy metabolism differs in normal cells and cancer cells. Inborn errors of metabolism can induce cancer development [32]. Cancer can also result from deficiency or overactivity of enzymes, deficiency of a cofactor required for enzymatic activity, an abnormality in degradation or transport processes that lead to the accumulation of upstream metabolites, deficiency of a downstream metabolite, or diversion of the affected metabolic flux to secondary pathways [33].
There are 3 different types of insults related to the inborn errors of metabolism: (1) toxic accumulation of metabolites, (2) metabolite channeling, and (3) mitochondrial dysfunction. Toxin accumulation leads to an increase in oxidative stress and ROS, as seen with iron accumulation in hemochromatosis [34]. In addition, accumulation of toxic metabolites could affect gene expression or cause a shift to alternative metabolic pathways, which could lead to tumorigenesis [29].
Human BDH2 (DHRS6) is an enzyme that participates in the citric acid cycle metabolism and ketogenesis [32], which may play a crucial role in promoting tumorigenesis [33,34]. In our results, BDH2 mRNA expression was higher in the BM of patients with CN-AML and AML with multiple chromosome abnormalities, compared with normal BM samples. We focused on CN-AML patients. No differences were observed in the incidence of clinical pictures, including blast amount in BM, FAB classification, and alterations in genes, except FLT3-ITD and DNMT3A, between BDH2high and BDH2low groups. High BDH2 expression is an independent indicator of poor prognosis of CN-AML, which may be related to a poor response to conventional intensive chemotherapy and a low CR rate. Given the independent adverse impact on survival and the low response rate in patients in the BDH2high group in both younger and elder patients, we suggest that higher BDH2 expression makes patients resistant to intensive induction chemotherapy. Although high BDH2 expression does not shorten the duration of relapse once patients obtain CR, patients with low BDH2 expression may benefit from further treatment, including chemotherapy and allogenetic hematopoietic stem cell transplantation, which may prolong survival time.
In outcome analysis by gene alternations, FLT3-ITD and MN1 mutations, and BDH2 were independent adverse prognostic factor for survival, with statistics significant. ERG and MLL mutations, and higher miR-3151 expression level showed a trend of adverse impact on survival. NPM1, CEBPA mutations and miR-181a showed positive outcome in CN-AML patients. However, DNMT3A mutations showed a good impact on survival, that could not compatible with previous reports. The mutations in DNMT3A in eleven of twelve of our patients are R882 (4279129C->T). One patient with DNMT3A mutation is R882 (4279073G->T). Only 12 CN-AML patients have DNMT3A mutation. Because of small number of patients, we cannot say that DNMT3A mutations had positive impact in overall survival (OS) in our patients. And that is the reasons of non-significant impact of other well-known genes alternations in CN-AML in our study.
To test if BDH2 can induce chemoresistance, we generated BDH2-KD leukemia cell lines. Oxidative damage is thought to be an important mechanism by which agents such as alkylators can damage DNA [35]. The intensive chemotherapy agents we used were cytarabine and idarubicin, which is an anthracycline. Cytarabine is an antimetabolic agent that causes DNA damage when the cell cycle holds in the S-phase [35]. Anthracycline kills leukemia cells via 3 mechanisms: inhibiting DNA and RNA synthesis by intercalating between base pairs of the DNA/RNA strand; inhibiting topoisomerase II enzyme, preventing the relaxation of supercoiled DNA, and thus blocking DNA transcription and replication; and creating iron-mediated free oxygen radicals that damage DNA and cell membranes [36]. In our study, BDH2-KD cells were more sensitive to ROS stimulation and more susceptible to apoptosis than parental and shRNA control vector transfected cells. Among the regulators of programmed cell death, or apoptosis, members of the Bcl-2 family control the release of apoptogenic proteins from mitochondria [37], whereas members of the IAP gene family act as endogenous inhibitors of caspases [38]. No difference was observed between Bcl-2 and Bcl-XL expression in parental, control vector, and BDH2-KD leukemia cell lines, before or after H2O2 treatment. The data showed that survivin was less in BDH2-KD cells than control cells, after ROS stimulation, but did not change before ROS stimulation. The other IAP, XIAP, did not differ significantly between BDH2-KD and control cells, before or after ROS stimulation. We also observed a limited increase in the cleaved form of caspase-3 in BDH2-KD cells by flow cytometry analysis. Survivin, the smallest member of the IAP family, has a synergic effect with XIAP in cytoprotection [39]. Survivin inhibits active caspase-9, but not active caspase-3 [40]. Survivin is selectively expressed in most human cancers, including lung, breast, pancreatic, and colon carcinomas; soft tissue sarcomas; brain tumors; melanoma; neuroblastoma; and hematologic malignancies [40]. We observed that BDH2-KD cells had a higher apoptotic rate under ROS stimulation, mediated by suppression of survivin release after ROS exposure.
Here, we showed that BDH2 overexpression could shorten overall survival and decrease the response rate from intensive induction chemotherapy. The mechanism by which BDH2 works as an anti-apoptotic factor is mediated by survivin through a caspase-3 independent pathway. To the best of our knowledge, we are the first to report that BDH2 is newly identified as a poor independent prognostic factor for CN-AML. Lower responsiveness to chemotherapy is predicted and hematological stem cell transplantation should be considered for patients with high expression of BDH2. The limitations of our work are related small patients’ number and not purify leukemia or CD34 (+) hematopoietic cells while collected bone marrow samples. We did not find difference of CD34 content in bone marrow samples between BDH2 higher and lower expression groups. On the other hand, we performed a study to induce THP1 to mature monocytes by using 1,25 Vitamine D3, and detect the BDH2 mRNA expression level. We did not find difference in native THP1 cells and mature treated THP1 cells (data not shown) [23].
In our future work, we will enroll more AML patients, especially patients receiving hematological stem cell transplantation to analysis the impact of BDH2. We will prospectively collect bone marrow from AML patients and sort CD34+ hematopoietic cells and perform those experiments for analysis prognosis. On the other hand, since BDH2 is an anti-apoptosis factor, we will analyze its impact with other factors which are related to apoptosis, like Wilms tumor 1[41], and DNA methyltransferase 3[42].
Conclusion
We reported that BDH2 overexpression could shorten overall survival and decrease the response rate from intensive induction chemotherapy. The mechanism by which BDH2 works as an anti-apoptotic factor is mediated by survivin through a caspase-3 independent pathway.
Abbreviations
BDH2: Type 2-hydroxybutyrate dehydrogenase; LCN2: Lipocalin-2; CEBPA: CCAAT/enhancer binding protein (C/EBP), alpha; FLT3-ITD: Fms-related tyrosine kinase 3 Internal tandem duplications; NPM1: Nucleophosmin 1; IDH1/2: Isocitrate dehydrogenase 1/2; DNMT3A: DNA (cytosine-5)-methyltransferase 3A; MLL: Mixed lineage leukemia gene; MN1: Meningionma1; ERG: ETS-related gene; miR-181a: Micro-RNA-181a; miR-3151: Micro-RNA-3151; CR: Complete remission; OS: Overall survival; RFS: Relapse free survival; LFS: Leukemia free survival.
Competing interests
The authors declare no competing financial interests.
Authors’ contributions
WC designed and performed research experiments, analyzed data, and wrote the manuscript, WT and PL performed some research experiments, MY provided cells and reagents, and reviewed and edited the manuscript, YL collected patient data and samples, CC provided suggestions to the research and data analysis, WY performed research experiments, SL reviewed and edited the manuscript, provided suggestions to the research. All authors read and approved the final manuscript.
Supplementary Material
mRNA expression levels of BDH2 in bone marrow. BDH2mRNA expression levels in bone marrow from patients with normal bone marrow, cytogenetically normal AML (CN-AML), good risk patients with AML-ETO fusion gene and chromosome inv (16), and poor risk patients with multiple chromosome abnormalities. P<0.001. Higher ΔCT-BDH2 means lower mRNA expression levels. BM: bone marrow. Sample number of bone marrows from normal bone marrow, CN-AML, AML with AML-ETO (+), AML with inv (16), and AML with multiple chromosome abnormalities are 43, 113, 10, 3 and 25.
Correlation between BDH2 and LCN2 mRNA expression in bone marrow. There is no correlation between the mRNA expression of BDH2 and LCN2.
The Kaplan-Meier overall survival curves in different BDH2 expression groups. HSCT indicates hematopoietic stem cell transplantation.
Apoptosis after 50 μM H2O2 treatment for 2 h in each cell line, assessed under a light microscope at 200×. Blue arrows indicate apoptotic cells. shRNA-BDH2-3 HL60 cells had more apoptotic cells.
The mitochondrial membrane potential change. That was analyzed using a MitoProbe™ JC-1 Assay Kit and no difference between BDH2-knockdown THP1 cells and control cells. shRNA1, shRNA-BDH2-1 THP1; shRNA3, shRNA-BDH2-3 THP1; shRNAc, shRNA empty vector infected THP1; CCCP, carbonyl cyanide 3-chlorophenylhydrazone, positive control.
Contributor Information
Wen-Chi Yang, Email: wenchi9251103@yahoo.com.tw.
Wan-Chi Tsai, Email: wanchi@cc.kmu.edu.tw.
Pai-Mei Lin, Email: paimei@isu.edu.tw.
Ming-Yu Yang, Email: yangmy@gmail.com.
Yi-Chang Liu, Email: ycliu@cc.kmu.edu.tw.
Chao-Sung Chang, Email: ccschang@gmail.com.
Wen-Hui Yu, Email: s0933188@mail.ncyu.edu.tw.
Sheng-Fung Lin, Email: shlin@kmu.edu.tw.
Acknowledgments
The authors thank the Statistical Analysis Laboratory, Department of Internal Medicine, Kaohsiung Medical University Hospital, Taiwan.
References
- Byrd JC, Mrozek K, Dodge RK. et al. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: results from Cancer and Leukemia Group B (CALGB 8461) Blood. 2002;100:4325–4336. doi: 10.1182/blood-2002-03-0772. [DOI] [PubMed] [Google Scholar]
- Dohner H, Estey EH, Amadori S, Appelbaum FR. et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international report panel, on behalf of the European Leukemia Net. Blood. 2010;115:453–474. doi: 10.1182/blood-2009-07-235358. [DOI] [PubMed] [Google Scholar]
- Campo E, Swerdlow SH, Harris NL, Pileri S, Stein H, Jaffe ES. The 2008 WHO classification of lymphoid neoplasms and beyond: evolving concepts and practical applications. Blood. 2011;117(19):5019–5032. doi: 10.1182/blood-2011-01-293050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang L, Jin S, Su SM. KDH mutations in glioma and acute myeloid leukemia. Trends Mol Med. 2010;16:387–397. doi: 10.1016/j.molmed.2010.07.002. [DOI] [PubMed] [Google Scholar]
- Delhommeau F, Dupont S, Della Valle V. et al. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360:2289–2301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- Gaidzik VI, Bullinger L, Schlenk RF. et al. RUNX1 mutations in acute myeloid leukemia: results from a comprehensive genetic and clinical analysis from the AML Study Group. J Clin Oncol. 2011;29:1364–1372. doi: 10.1200/JCO.2010.30.7926. [DOI] [PubMed] [Google Scholar]
- Langer C, Marcucci G, Holland KB. et al. Prognostic importance of MN1 transcript levels, and biologic insights from NM1-associated gene and microRNA expression signatures in cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol. 2009;27:3198–3204. doi: 10.1200/JCO.2008.20.6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer C, Radmacher MD, Ruppert AS. et al. High BAALC expression associates with other molecular prognostic markers, poor outcome, and a distinct gene-expression signature in cytogenetically normal patients younger than 60 years with acute myeloid leukemia: a Cancer and Leukemia Group B (CALGB) study. Blood. 2008;111:5371–5379. doi: 10.1182/blood-2007-11-124958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcucci G, Maharry K, Whitman SP. et al. High expression levels of the ETS-related gene, ERG, predict adverse outcome and improve molecular risk-based classification of cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol. 2007;25:3337–3343. doi: 10.1200/JCO.2007.10.8720. [DOI] [PubMed] [Google Scholar]
- Marcucci G, Maharry K, Wu YZ. et al. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol. 2010;28:2348–2355. doi: 10.1200/JCO.2009.27.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzeler KH, Dufour A, Benthaus T. et al. ERG expression is an independent prognostic factor and allows refined risk stratification in cytogenetically normal acute myeloid leukemia: a comprehensive analysis of ERG, NM1 and BAALC transcript levels using oligonucleotide microarrays. J Clin Oncol. 2009;27:5031–5038. doi: 10.1200/JCO.2008.20.5328. [DOI] [PubMed] [Google Scholar]
- Metzeler KH, Maharry K, Radmacher MD. et al. TET2 mutations improve the new European Leukemia Net risk classification of acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol. 2011;29(10):1378–1381. doi: 10.1200/JCO.2010.32.7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubauer A, Maharry K, Mrozek K. et al. Patients with acute myeloid leukemia and RAS mutations benefit most from postremission high-dose cytarabine: a Cancer and Leukemia Group B study. J Clin Oncol. 2008;26:4603–4609. doi: 10.1200/JCO.2007.14.0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschka P, Marcucci G, Ruppert AS. et al. Wilms tumor 1 gene mutations independently predict poor outcome in adults with cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol. 2008;26:4595–4602. doi: 10.1200/JCO.2007.15.2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschka P, Schlenk RF, Gaidzik VI. et al. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J Clin Oncol. 2010;28:3636–3643. doi: 10.1200/JCO.2010.28.3762. [DOI] [PubMed] [Google Scholar]
- Tang JL, Hou HA, Chen CY. et al. AML1/RUNX1 mutations in 470 adults patients with de novo acute myeloid leukemia: prognostic implication and interaction with other gene alterations. Blood. 2009;114:5352–5361. doi: 10.1182/blood-2009-05-223784. [DOI] [PubMed] [Google Scholar]
- Mithat Gönen JP, Figueroa ME, Fernandez H. et al. Prognostic Relevance of Integrated Genetic Profiling in Acute Myeloid Leukemia. New Eng J Med. 2012;366:1079–1089. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warbur effect: The metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Manning BD, Cantley LC. Targeting the PI3K-Akt pathway in human cancer: Rationale and promise. Cancer Cell. 2003;4:257–262. doi: 10.1016/S1535-6108(03)00248-4. [DOI] [PubMed] [Google Scholar]
- Guo K, Lukacik P, Papagrigoriou E. et al. Characterization of human DHRS6, an orphan short chain dehydrogenase/reductase enzyme: a novel, cytosolic type 2 R-beta-hydroxybutyrate dehydrogenase. J Biol Chem. 2006;281:10291–10297. doi: 10.1074/jbc.M511346200. [DOI] [PubMed] [Google Scholar]
- Devireddy LR, Hart DO, Goetz DH, Green MR. A mammalian siderophore synthesized by an enzyme with a bacterial homolog involved in enterobactin production. Cell. 2010;141:1006–1017. doi: 10.1016/j.cell.2010.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WC, Lin PM, Yang MY, Higher lipocalin 2 expression may represent an independent favorable prognostic factor in cytogenetically normal acute myeloid leukemia. Leukemia Lymphoma. 2012. 10.3109/10428194.2012.749402. [DOI] [PubMed]
- Schnittger S, Kinkelin U, Schoch C. et al. Screening for MLL tandem duplication in 387 unselected patients with AML identify a prognostically unfavorable subset of AML. Leukemia. 2000;14:796–804. doi: 10.1038/sj.leu.2401773. [DOI] [PubMed] [Google Scholar]
- Falini B, Mecucci C, Tiacci E. et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med. 2005;352:254–266. doi: 10.1056/NEJMoa041974. [DOI] [PubMed] [Google Scholar]
- Kiyoi H, Naoe T, Nakano Y. et al. Prognostic implication of FLT3 and N-RAS gene mutations in acute myeloid leukemia. Blood. 1999;93:3074–3080. [PubMed] [Google Scholar]
- Pabst T, Mueller BU, Zhang P. et al. Dominant-negative mutations of CEBPA, encoding CCAAT/enhancer binding protein-alpha (C/EBPalpha), in acute myeloid leukemia. Nat Genet. 2001;27:263–270. doi: 10.1038/85820. [DOI] [PubMed] [Google Scholar]
- Chou WC, Hou HA, Chen CY. et al. Distinct clinical and biologic characteristics in adult acute myeloid leukemia bearing the the isocitrate dehydrogenase 1 mutation. Blood. 2010;115:2749–2754. doi: 10.1182/blood-2009-11-253070. [DOI] [PubMed] [Google Scholar]
- Simon HU, Haj-Yehia A, Levi-Schaffer F. Role of reactive oxygen species (ROS) in apoptosis induction. Apoptosis. 2000;5:415–418. doi: 10.1023/A:1009616228304. [DOI] [PubMed] [Google Scholar]
- Chu J, Wu S, Xing D. Survivin mediates self-protection through ROS/cdc25c/CDK1 signaling pathway during tumor cell apoptosis induced by high fluence low-power laser irradiation. Cancer Lett. 2010;297:207–219. doi: 10.1016/j.canlet.2010.05.013. [DOI] [PubMed] [Google Scholar]
- Yao LL, Wang YG, Cai WJ, Yao T, Zhu YC. Survivin mediates the anti-apoptotic effect of delta-opioid receptor stimulation in cardiomyocytes. J Cell Sci. 2007;120:895–907. doi: 10.1242/jcs.03393. [DOI] [PubMed] [Google Scholar]
- Laffel L. Ketone Bodies: a review of physiology, pathophysiology and application of monitoring to diabetes. Diabetes Metab Res Rev. 1999;15:412–426. doi: 10.1002/(SICI)1520-7560(199911/12)15:6<412::AID-DMRR72>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Pavlides S, Tsirigos A, Mibneco G. et al. The autophagic tumor stroma model of cancer: role of oxidative stress and ketone production in fueling tumor cell metabolism. Cell Cycle. 2010;9:3485–3505. doi: 10.4161/cc.9.17.12721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer GD, Brucker DP, Bahr O. et al. Differential utilization of ketone bodies by neurons and glioma cell lines: a rationale for ketogenic diet as experimental glioma therapy. BMC Cancer. 2011;11:315–321. doi: 10.1186/1471-2407-11-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael E. Trigg, MD, Anne Flanigan-Minnick: Mechanisms of action of commonly used drugs to treat cancer (Review) Comm Oncol. 2011;8:357–369. doi: 10.1016/S1548-5315(12)70051-7. [DOI] [Google Scholar]
- Müller I, Niethammer D, Bruchelt G. Anthracycline-derived chemotherapeutics in apoptosis and free radical cytotoxicity (Review) Int J Mol Med. 1998;1:491–494. doi: 10.3892/ijmm.1.2.491. [DOI] [PubMed] [Google Scholar]
- Cory S, Adams JM. The Bcl2 family: regulators of the cellular life-or-death switch. Nat Rev Cancer. 2002;2:647–656. doi: 10.1038/nrc883. [DOI] [PubMed] [Google Scholar]
- Shi Y. Mechanisms of caspase activation and inhibition during apoptosis. Mol Cell. 2002;9:459–470. doi: 10.1016/S1097-2765(02)00482-3. [DOI] [PubMed] [Google Scholar]
- Dohi T, Okada K, Xia F. et al. An IAP-IAP complex inhibits apoptosis. J Biol Chem. 2004;279:34087–34090. doi: 10.1074/jbc.C400236200. [DOI] [PubMed] [Google Scholar]
- Mita AC, Mita MM, Nawrocki ST, Giles FJ. Survivin: key regulator of mitosis and apoptosis and novel target for cancer therapeutics. Clin Cancer Res. 2008;14:5000–5005. doi: 10.1158/1078-0432.CCR-08-0746. [DOI] [PubMed] [Google Scholar]
- Bansal H, Seifert T, Bachier C. et al. The transcription factor Wilms tumor 1 confers resistance in myeloid leukemia cells against the proapoptotic therapeutic agent TRAIL by regulating the antiapoptotic protein Bcl-xL. J Biol Chem. 2012;287(39):32875–32880. doi: 10.1074/jbc.C112.366559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro AF, Pratcorona M, Erpelinck-Verschueren C. et al. Mutant DNMT3A: a marker of poor prognosis in acute myeloid leukemia. Blood. 2012;119:5824–5831. doi: 10.1182/blood-2011-07-367961. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
mRNA expression levels of BDH2 in bone marrow. BDH2mRNA expression levels in bone marrow from patients with normal bone marrow, cytogenetically normal AML (CN-AML), good risk patients with AML-ETO fusion gene and chromosome inv (16), and poor risk patients with multiple chromosome abnormalities. P<0.001. Higher ΔCT-BDH2 means lower mRNA expression levels. BM: bone marrow. Sample number of bone marrows from normal bone marrow, CN-AML, AML with AML-ETO (+), AML with inv (16), and AML with multiple chromosome abnormalities are 43, 113, 10, 3 and 25.
Correlation between BDH2 and LCN2 mRNA expression in bone marrow. There is no correlation between the mRNA expression of BDH2 and LCN2.
The Kaplan-Meier overall survival curves in different BDH2 expression groups. HSCT indicates hematopoietic stem cell transplantation.
Apoptosis after 50 μM H2O2 treatment for 2 h in each cell line, assessed under a light microscope at 200×. Blue arrows indicate apoptotic cells. shRNA-BDH2-3 HL60 cells had more apoptotic cells.
The mitochondrial membrane potential change. That was analyzed using a MitoProbe™ JC-1 Assay Kit and no difference between BDH2-knockdown THP1 cells and control cells. shRNA1, shRNA-BDH2-1 THP1; shRNA3, shRNA-BDH2-3 THP1; shRNAc, shRNA empty vector infected THP1; CCCP, carbonyl cyanide 3-chlorophenylhydrazone, positive control.