Summary
Polo-like Kinase 4 (Plk4) is a conserved master-regulator of centriole assembly [1]. Previously, we found that Drosophila Plk4 protein levels are actively suppressed during interphase [2]. Degradation of interphase Plk4 prevents centriole overduplication and is mediated by the ubiquitin-ligase complex, SCFSlimb/βTrCP [3, 4]. Since Plk4 stability depends on its activity [5, 6], we studied the consequences of inactivating Plk4 or perturbing its phosphorylation-state within its Slimb-recognition motif (SRM). Mass spectrometry of in vitro phosphorylated Plk4 and Plk4 purified from cells reveals that it is directly responsible for extensively autophosphorylating and generating its Slimb-binding phosphodegron. Phosphorylatable residues within this regulatory region were systematically mutated to determine their impact on Plk4 protein levels and centriole duplication when expressed in S2 cells. Notably, autophosphorylation of a single residue (Ser293) within the SRM is critical for Slimb binding and ubiquitination. Our data also demonstrate that autophosphorylation of numerous residues flanking S293 collectively contribute to establishing a high-affinity binding site for SCFSlimb. Taken together, our findings suggest that Plk4 directly generates its own phosphodegron and can do so without the assistance of an additional kinase(s).
Results and Discussion
Centriole over-duplication is prevented by limiting centriole duplication to one occurrence per cell cycle, and while numerous proteins affect centriole number, much remains to be discovered about their coordination and regulation [7]. A key component is Polo-like kinase 4 (Plk4)/Sak, a conserved master-regulator of centriole duplication [8, 9]. During interphase, Plk4 is degraded which prevents centriole over-duplication [3, 4]. Features of this degradation mechanism support the following model: Plk4 homodimerizes and phosphorylates several residues within a hydroxyl-rich region called the Downstream Regulatory Element (DRE) (Figure 1A), although precise sites of phosphorylation are unknown [5, 6, 10, 11]. Embedded within the DRE is a conserved Slimb-Recognition Motif (SRM: DpSGXXpT) whose serine/threonine residues are phosphorylated, generating a phosphodegron recognized by the F-box protein Slimb in Drosophila or β-TrCP in humans [3–6, 12]. F-box proteins are substrate-targeting subunits of the SCF ubiquitin-ligase which ubiquitinates Plk4, marking the kinase for proteasomal degradation [3–6, 12].
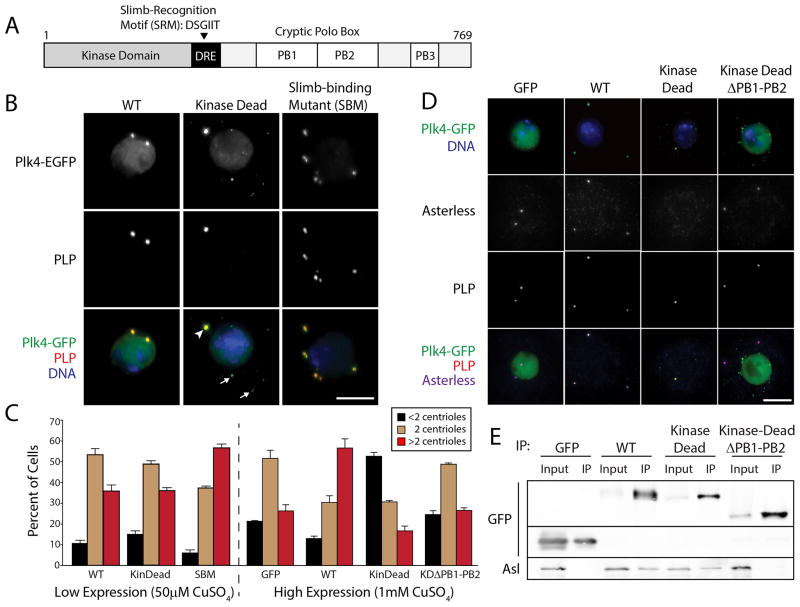
Figure 1. High Expression of Kinase-dead Plk4 Blocks Centriole Duplication by Preventing Asterless Targeting to Centrioles.
(A) Linear map of Drosophila Plk4 showing functional and structural domains. The Downstream Regulatory Element (DRE) is a span of approximately 50 amino acids containing the phylogenetically conserved Slimb-recognition motif (SRM). Plk4 contains three Polo-boxes (PB) [10]. PB1 and PB2 comprise the Asl-binding region [10, 20–22].
(B) S2 cells co-expressing the indicated Plk4-EGFP construct (green) and Nlp-EGFP (a nuclear protein used as a transfection marker; green nuclei) were immunostained for PLP (red) to mark centrioles. DNA (blue). Expression of Plk4 constructs was induced with 50μM CuSO4. KD-Plk4 targets centrioles (arrowhead) but also forms cytoplasmic punctate aggregates (arrows). Scale, 5μm.
(C) Transfected S2 cells were induced to express Plk4-EGFP constructs at low (50μM CuSO4) or high (1mM CuSO4) levels for three days, immunostained for PLP, and their centrioles counted. Centriole amplification (a significant, increased frequency of >2 centrioles/cell) occurs in cells expressing SBM-Plk4 (P=0.0001) or a high level of WT-Plk4 (P=0.0002). In contrast, centriole duplication is inhibited (a significant, increased frequency of <2 centrioles/cell) in cells expressing a high level of KD-Plk4 (P<0.0001) but not KD-Plk4-ΔPB1-PB2 (P=0.29). At least three experiments were performed per construct (n = 600 cells/construct). Error bars in all figures, S.E.M.
(D) S2 cells co-expressing the indicated Plk4-EGFP construct (green) and the transfection marker, Nlp-EGFP (green nuclei), were immunostained for PLP (red) and Asterless (blue; bottom row). DNA (blue; top row). Expression of Plk4 constructs was induced with 1mM CuSO4. Scale, 5μm.
(E) Anti-GFP immunoprecipitates from S2 cell lysates transiently-expressing the indicated Plk4-EGFP construct (or control GFP) were probed for GFP and endogenous Asterless. Note that co-precipitating Asl is absent in the control and KDΔPB1-PB2 samples.
Several important questions concerning Plk4 regulation remain to be answered. First, although purified Plk4 extensively phosphorylates its DRE in vitro [5], the specific phosphorylation sites required for Slimb recognition have not been defined. Second, it is not known which specific residues are autophosphorylated. Third, it is not clear whether the Plk4 phosphodegron is generated directly by autophosphorylation or whether another unidentified kinase is required [6, 13, 14]. Thus, important features of Plk4 down-regulation remain to be discovered. To address these mechanistic unknowns, we examined which DRE residues are autophosphorylated, explored the consequences of inactivating Plk4, and determined what happens to Plk4 stability when the normal DRE phosphorylation pattern is perturbed.
Previous work has shown that kinase-dead (KD)-Plk4 is stable when expressed in cells, suggesting that kinase activity precedes its degradation [2, 5, 6, 15, 16]. KD-Plk4 expression also induces centriole over-duplication in tumor-derived cell lines when endogenous Plk4 is present [5, 6, 9]. The latter result is counterintuitive, but it was proposed that the SRM within KD-Plk4 is phosphorylated in trans after heterodimerizing with endogenous Plk4, and that phosphorylated KD-Plk4 then sequesters β-TrCP/Slimb, decreasing the available β-TrCP. Thus, endogenous Plk4 levels would increase and stimulate centriole amplification [6].
We examined whether centriole amplification is a universal consequence of KD-Plk4 expression or unique to transformed cells. We expressed metallothionein-inducible KD-Plk4 and other Plk4-EGFP constructs in S2 cells, an immortalized Drosophila cell line. After 3 days of expression, centrioles were visualized by immunostaining for PLP [17], a centriole protein that coats the surface of mature centrioles [18], and manually counted. Due to their small size, mother and daughter centrioles cannot be distinguished within an engaged pair using standard light microscopy in most fly cells. Nevertheless, the number of PLP spots is an accurate readout of centriole loss (<2 spots) and amplification (>2 spots) in these cells [2–4, 8, 10]. As expected, all Plk4 constructs localized to centrioles, though KD-Plk4 also formed small puncta in the cytoplasm that lacked PLP (Figure 1B). Under low expression conditions, only non-degradable Plk4 Slimb-binding mutant (SBM; a S293A/T297A double mutation within the SRM that blocks Slimb binding [3, 4]) significantly increased the percentage of cells with >2 centrioles (Figure 1C), as shown previously [3, 4]. However, under high expression conditions, wild-type (WT)-Plk4 induced significant centriole amplification, consistent with the model that increased Plk4 activity drives centriole over-duplication (Figure 1C). Surprisingly, KD-Plk4 high expression had the opposite effect: the percentage of cells with <2 centrioles significantly increased (Figure 1C). Therefore, our results suggest that KD-Plk4 behaves as a dominant/negative in S2 cells, inhibiting centriole duplication. A similar result was also observed in KD-Plk4-overexpressing early Drosophila embryos [19].
Because KD-Plk4 formed puncta within the cytoplasm, we reasoned that KD-Plk4 overexpression might perturb localization of essential centriole components, such as the Plk4-binding protein Asterless (Asl) [20–22]. To test this, anti-Asl antibodies (Figure S1A) were used to examine the association of Asl with KD-Plk4 and centrioles. Endogenous Asl co-immunoprecipitated with WT- and KD-Plk4-EGFP but not control EGFP (Figure 1E). Unlike WT-Plk4, high expression of KD-Plk4 completely blocked Asl localization to centrioles (Figure 1D). This effect was directly due to KD-Plk4 binding because expression of a mutant form of KD-Plk4 lacking the Asl-binding domain (KD-Plk4-ΔPB1-PB2-EGFP) (Figure 1E) [10, 20–22] displayed normal centriole numbers and Asl localization (Figure 1C, D). These results suggest that KD-Plk4 blocks centriole duplication by sequestering Asl, thus preventing Asl from localizing endogenous Plk4 to centrioles (Figure S1B, C). The disparity in KD-Plk4 overexpressed in human cancer and Drosophila cells might reflect a fundamental difference in Plk4 regulation in these systems.
Previous work in transformed cells suggests that Plk4 homodimerizes and trans-phosphorylates to induce its degradation [5, 6]. To test whether trans-phosphorylation is a prerequisite for SCFSlimb recognition, we co-expressed EGFP- and myc-tagged KD- and WT-Plk4 in cells and evaluated their relative stabilities using quantitative immunoblotting. As expected, Plk4 was almost undetectable in cells co-expressing two WT constructs (Figure 2A, lane 1). (We verified WT-Plk4-Myc expression by depleting Slimb to stabilize this protein; Figure S2A.) In contrast, KD-Plk4 is strongly stabilized when co-expressed with another KD protein (Figure 2A, lane 3). However, KD-Plk4 is clearly less stable when co-expressed with WT-Plk4 and migrates as a smear on SDS-PAGE (Figure 2A, lane 2), suggesting that KD-Plk4 has been phosphorylated. Therefore, the ability to trans-phosphorylate controls Plk4 stability.
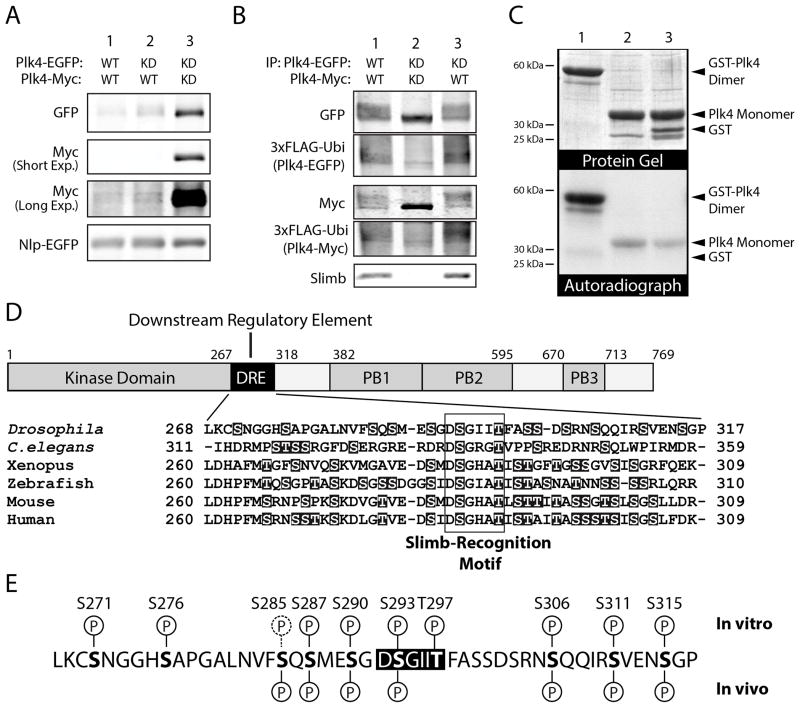
Figure 2. Plk4 is Destabilized by Trans-Autophosphorylation and Directly Autophosphorylates its SRM In Vitro.
(A) The relative protein stabilities of different combinations of Plk4 constructs were analyzed by immunoblotting lysates of S2 cells transiently co-expressing the indicated EGFP- and Myc-tagged Plk4 constructs and Nlp-EGFP (used as a loading control and expressed under its endogenous promoter). Anti-Myc immunoblots are shown at short and long exposures.
(B) Anti-GFP immunoprecipitates (IPs) were prepared from lysates of S2 cells transiently expressing 3xFLAG-ubiquitin and the indicated combinations of EGFP- and Myc-tagged Plk4 constructs. Blots of the IPs were probed with anti-GFP, FLAG, and Slimb antibodies. Note that robust ubiquitination of Plk4 corresponds to the presence of endogenous Slimb, and that co-expression of KD-Plk4 (lane 2) prevents phosphorylation (as indicated by the lack of gel-shift), Slimb binding, and ubiquitination.
(C) Autophosphorylation of Plk4 kinase domain is more efficient as a dimer compared to a monomeric species. Lanes 1–3, both purified recombinant GST-tagged (dimeric) human Plk4 kinase domain + SRM and monomeric kinase (cleaved of its GST-tag) autophosphorylates in vitro. Plk4 does not phosphorylate purified GST (lane 3). Top panel, Coomassie-stained SDS-PAGE gel; bottom panel, corresponding autoradiograph. Equimolar amounts of dimeric and monomeric kinase were assayed.
(D) The DRE contains a conserved, high percentage (~20–40%) of hydroxyl amino acids (highlighted) that are potential sites of phosphorylation. These include the conserved Ser and Thr residues within the SRM (boxed).
(E) In vitro phosphorylation sites within the DRE were identified by tandem mass spectrometry (MS) analysis of purified fly His6-Plk4 (amino acid residues 1–317, comprising the kinase and DRE domains) incubated with MgATP. Above the DRE sequence, in vitro phosphorylation sites identified with high confidence are indicated with a ‘P’ encircled with a solid line; a low confidence site (S285) is indicated with a ‘P’ encircled with a dashed line. In vivo phosphorylation sites within the DRE were identified by MS analysis of full-length Plk4-EGFP immunoprecipitated from S2 cell lysates. Identified in vivo sites are marked below the DRE sequence. (A very low confidence site, S271, is not marked.) At least nine of the hydroxyl residues within the DRE are phosphorylated in vitro, and seven of these residues are also phosphorylated in vivo (bottom row).
We tested this further by assaying Slimb association with Plk4 and its ubiquitination state. To assess Plk4 ubiquitination, S2 cells were co-transfected with 3xFLAG-Ubiquitin (Ubi). Immunoprecipitation of EGFP-tagged WT-Plk4 retrieved the second Myc-tagged WT-Plk4 as well as endogenous Slimb, and both Plk4 proteins were robustly ubiquitinated (Figures 2B, lane 1; S2B) while negative control GFP was not (Figure S2C). In contrast, ubiquitination diminished when two KD-Plk4 constructs were co-expressed (Figures 2B, lane 2; S2B), and, as predicted from their increased stabilities, Slimb failed to associate (Figure 2B, lane 2).
If trans-phosphorylation stimulates Slimb binding, then dimerization of KD-Plk4 with WT-Plk4 should result in KD-Plk4’s trans-phosphorylation, ubiquitination, and destabilization (Figure S2B). In this case, Slimb associated with KD-Plk4-EGFP, displayed the smeary appearance of a phosphorylated species, and was robustly ubiquitinated (Figure 2B, lane 3).
The above observations support a conserved mechanism of degradation whereby trans-phosphorylation within a Plk4 homodimer induces its ubiquitination. We next used an in vitro system to ask whether dimerization affects Plk4 autophosphorylation. A construct of human Plk4 encoding the kinase domain and its SRM (amino acids 1–289) was tagged with GST to induce dimerization. Autophosphorylation of the dimeric Plk4 and the monomeric kinase (generated by cleaving GST) was compared. Whereas monomeric Plk4 displayed some capacity to trans-phosphorylate, dimeric Plk4 kinase strongly autophosphorylated under the same conditions (Figure 2C, lanes 1 and 2). Phosphorylation was specific as monomeric Plk4 did not phosphorylate GST added to the reaction (Figure 2C, lane 3). Thus, Plk4 dimerization is an efficient mechanism for trans-phosphorylation and self-destruction.
Previous studies revealed that Slimb interacts with Plk4 through a phosphorylated motif (the SRM) near the kinase domain (Figure 2D) [3, 4, 23]. This region of 50 amino acids, the Downstream Regulatory Element (DRE), contains the SRM, is serine/threonine rich, and is extensively autophosphorylated in vitro [2, 5]. However, specific autophosphorylated DRE residues are unknown, and it is not clear whether Plk4 directly generates its own phosphodegron to recruit Slimb or whether another kinase is responsible [6, 13, 14]. First, we mapped autophosphorylated residues in vitro using tandem mass spectrometry (MS) of Drosophila Plk4 containing the kinase domain and DRE (amino acids 1–317) and identified 10 (of 13 possible) autophosphorylated DRE residues (Figures 2E and S2D; Table S1). Significantly, both SRM residues S293 and T297 were autophosphorylated as were 8 phosphorylation sites flanking the SRM. Tandem MS was also used to examine the in vivo phosphorylation state of the DRE of Plk4-EGFP immunoprecipitated from S2 cell lysates. Of the 10 residues identified in vitro, 7 were phosphorylated in vivo, including S293 (Figures 2E and S2E; Table S2). Although our results do not exclude the possibility that additional DRE residues are autophosphorylated in cells, our findings demonstrate that Plk4 can directly autophosphorylate its DRE, including the key SRM residues, S293 and T297.
To investigate what impact the phosphorylation states of DRE residues have on Plk4 stability, we mutated each hydroxyl residue to alanine within full-length Plk4. To avoid neglecting a phosphorylated residue not identified in our MS analysis, we individually mutated all 13 serine/threonine DRE residues (Figure 3A). Each Plk4-EGFP mutant was expressed in S2 cells and quantitative immunoblots were used to measure protein levels. Unexpectedly, mutation of only one residue, S293, extensively stabilized Plk4 (Figure 3B). Interestingly, the second SRM residue, T297, had only a small impact on Plk4 stability (Figure 3B). Thus, S293 is the key residue for Slimb recognition whereas T297 makes a minor contribution. Mutation of other residues downstream of T297A also slightly increased Plk4 stability compared to WT-Plk4, suggesting that phosphorylation of these residues may also contribute to Slimb binding (Figure 3B).
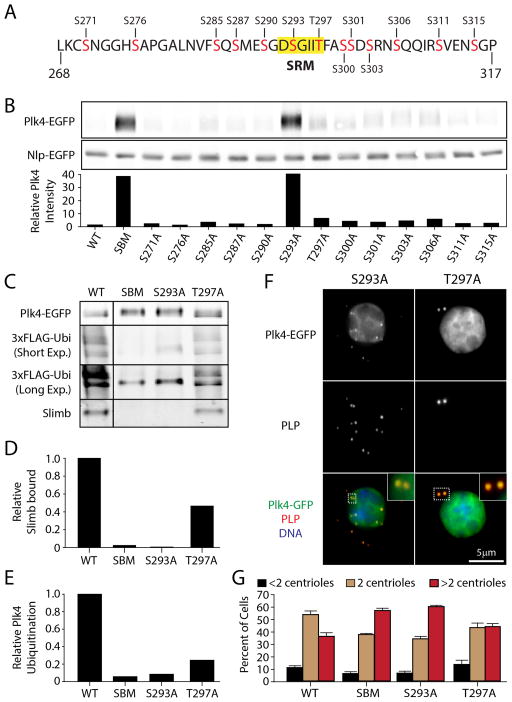
Figure 3. S293 of the SRM is the Critical DRE Residue for Slimb Recognition.
(A) 13 hydroxyl amino acids (red) within the DRE of full-length fly Plk4 were individually mutated to alanines and used to evaluate the impact of each residue on Plk4 stability and centriole duplication. S293 and T297 reside within the SRM (yellow highlight).
(B) (Top) Anti-GFP immunoblot of lysates prepared from S2 cells transiently co-expressing the indicated Plk4-EGFP construct and Nlp-EGFP (loading control). All Plk4 mutants are single alanine mutants except SBM (a double mutant of S293A/T297A). (Bottom) Plk4-EGFP intensities were measured by densitometry of the anti-GFP immunoblot and normalized with their respective Nlp-EGFP loading controls. The plotted values are the normalized Plk4 intensities relative to the WT-Plk4 treatment.
(C) Anti-GFP immunoprecipitates of lysates prepared from S2 cells transiently expressing 3xFLAG-ubiquitin and the indicated Plk4-EGFP construct were probed with anti-GFP, FLAG, and Slimb antibodies. Short and long exposures of the anti-FLAG immunoblot are shown.
(D) Amounts of associated endogenous Slimb were determined by densitometry of the anti-Slimb immunoblot and then normalizing the measurements with the amounts of Plk4-EGFP present in the IPs. The plotted values are relative to the WT-Plk4 treatment.
(E) The relative amounts of total Plk4 FLAG-Ubi were calculated using the densitometry method described in (D).
(F) S2 cells co-expressing the indicated Plk4-EGFP construct (green puncta) and Nlp-EGFP (green nuclei) were immunostained for PLP (red) to mark centrioles. DNA (blue). Insets show higher magnifications of the boxed regions.
(G) The centrioles of S2 cells treated and immunostained as in (F) were counted. Centrioles are amplified in cells expressing SBM (P<0.0001) or S293A (P<0.0001) but not T297A (P=0.06). There is no significant difference in centriole loss (black bars) in these treatments. Four experiments were performed per construct (n = 600 cells/construct).
To determine if mutation of S293 alone blocks Slimb binding, immunoprecipitations were performed from cells co-expressing 3xFLAG-Ubi. S293A failed to bind Slimb and ubiquitination decreased ~10-fold, whereas T297A had a moderate impact (Figure 3C–E), consistent with the observation that T297A is slightly more stable than WT-Plk4 (Figure 3B). Overexposure of the FLAG immunoblot revealed one persistent ubiquitinated species for S293A and SBM-Plk4 (Figure 3C), suggesting that an unidentified Slimb-independent ubiquitin-ligase may also regulate Plk4.
Mutations that increase Plk4 stability should induce centriole amplification [3, 4]. To test this, Plk4-EGFP constructs were expressed for 3 days and centriole numbers were measured. As expected, both S293A and T297A mutants localized to centrioles (Figure 3F). Whereas S293A significantly increased centriole number (>2 centrioles per cell) to an extent nearly identical to SBM-Plk4, T297A did not produce a significant increase (Figure 3G). Thus, S293 is the key residue for Slimb recognition, whereas T297 modestly increases the efficiency of Slimb binding but is nonessential.
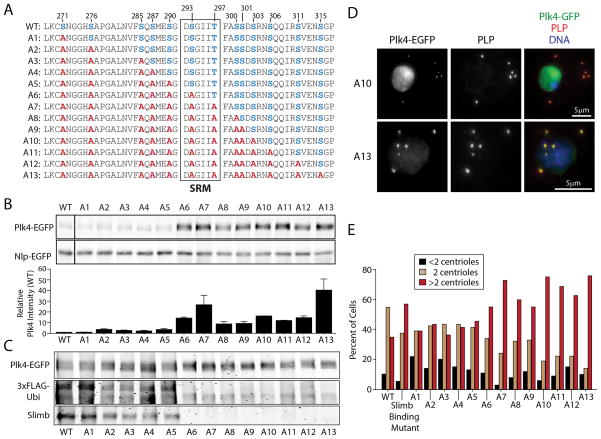
We next examined whether phosphorylated DRE residues collectively regulate Plk4 by systematically mutating every hydroxyl residue, such that each new Plk4 construct contained a steadily accumulating number of alanine mutations (Figure 4A). Mutation of the 5 residues upstream of S293 had little effect on Plk4 stability, even when mutated together (Figure 4B, A1–A5). However, once S293A was included, Plk4 stability increased significantly (Figure 4B, A6). Plk4 mutants with additional alanine substitutions downstream of S293A did not significantly elevate levels greater than A6 (Figure 4B, A7–A12) with the exception of Plk4 harboring all 13 alanine mutations (A13) which displayed a ~3-fold average increase in protein level (compared to A6; P=0.0001). These results suggest that numerous hydroxyl DRE residues, in combination with S293, function collectively to recruit Slimb.
Figure 4. Mutation of All 13 Hydroxyl DRE Residues Display Only a Subtle Difference in Plk4 Stability Compared to the S293A Mutant.
(A) Hydroxyl residues (blue) within the DRE were systematically mutated to non-phosphorylatable alanines (red) to generate a series of Plk4 constructs containing an increasing number of mutations.
(B) Plk4 is stabilized by mutation of residue S293, but this stability is slightly modulated by neighboring phosphorylatable residues within the DRE. (Top) Anti-GFP immunoblot of lysates from S2 cells transiently co-expressing the indicated Plk4-EGFP construct and Nlp-EGFP (loading control). (Bottom) Relative amounts of Plk4 protein were determined by measurement of the integrated intensities of the Plk4 bands, normalized to their respective loading controls, and then plotted relative to the normalized intensity of WT-Plk4. Measurements were obtained from three experiments.
(C) Blots of anti-GFP immunoprecipitates from lysates of S2 cells transiently expressing 3xFLAG-Ubi and the indicated Plk4-EGFP construct were probed with anti-GFP, FLAG, and Slimb antibodies. Slimb binding is reduced by mutation of upstream DRE serines (A2–A5). As expected, Slimb binding is eliminated in mutants containing the S293A mutation (A6–A13).
(D) S2 cells co-expressing the indicated Plk4-EGFP plasmid (green puncta) and Nlp-EGFP (green nuclei) were immunostained for PLP to mark centrioles (red). DNA (blue).
(E) Expression of stabilized Plk4 mutants increases the percent of cells with excess centrioles (>2). Centrioles in 100 cells were measured per construct.
Since S293A alone prevents Slimb binding, we predicted that Plk4 mutants harboring the S293A mutation (A6–A13) would not bind Slimb and exhibit diminished ubiquitination. Surprisingly, mutants that include the S276A mutation (A2–A5) displayed reduced Slimb binding and ubiquitination compared to WT-Plk4. As expected, mutants of Plk4 lacking S293 (A6–A13) fail to bind Slimb and are not extensively ubiquitinated (Figure 4C). Therefore, the phosphorylation state of hydroxyl DRE residues preceding the SRM can influence Slimb binding. All A1–A13 mutants also localized properly to centrioles and those constructs with increased stability (A6–A13) increased the percent of cells with >2 centrioles (Figure 4D, E, and data not shown). Thus, the trend in centriole amplification duplicates the trend in Plk4 stability.
The previous mutant series revealed that some hydroxyl residues flanking the SRM contribute to Slimb recognition and ubiquitination. To examine how these residues affect regulate Plk4, we generated a new mutant series which progressively accumulated alanine substitutions without mutating S293 (Figure S3A). Strikingly, though Plk4 stability is minimally affected by either the single T297A mutant or the aggregate mutation of the 5 hydroxyl residues upstream of the SRM (A5), the combination of these mutations (A14) stabilized Plk4 to a level almost 4-fold greater than T297A, and constructs containing additional mutations downstream of T297A are just as stable (Figure S3B). Elimination of upstream hydroxyl residues and T297 (A14) reduced Slimb binding and the extent of ubiquitination by ~50% (Figure S3C), confirming that upstream hydroxyl DRE residues contribute to Slimb binding. Additional mutations downstream of the SRM decreased Slimb binding by another 2-fold (Figure S3C, A15–A20), but did not further increase Plk4 stability consistently (Figure S3B, A15–A20). Thus, DRE residues besides S293 collectively contribute to Slimb binding and ubiquitination.
To completely isolate the contribution of non-SRM residues to Plk4 stability, we generated constructs possessing an unaltered SRM but with all hydroxyl residues flanking S293/T297 mutated to alanine (Figure S4A). This series showed a clear pattern: progressively accumulating mutations had little effect on Plk4 stability until residues downstream of the SRM were mutated (compare A1–A5 of Figure 4B with Figure S4B). Therefore, mutation of upstream hydroxyl residues has limited impact on Plk4 stability and only becomes appreciable when T297 (Figure S3B) and/or downstream residues are also mutated. As further downstream residues were mutated, protein levels displayed a near steady increase; this was accompanied by a ~5-fold reduction in the amount of associated Slimb (Figure S4B, C). All mutants localized to centrioles (Figures S3D, S4D and data not shown), and all stabilizing mutations increased centriole numbers (Figures S3E, S4E). Thus, our findings suggest that the SRM is flanked by phosphorylatable residues that positively regulate Slimb recognition. Their cumulative effect is substantial as Slimb binding is inhibited to almost as great an extent by loss of flanking hydroxyl residues (e.g., A26) as by KD or SBM (Figure S4C). Even though S293 is required for Slimb binding (Figure 3C), loss of the other hydroxyl residues can diminish ubiquitination as strongly as S293A (Figure S3C). Our data suggest that hydroxyl residues downstream of the SRM (Figure S4, A21–A26) exert a greater influence on Slimb binding, Plk4 stability, and centriole duplication than those upstream of the SRM (Figure S3, A1–A5), although the mechanistic basis for this is unknown.
Our findings demonstrate that the Plk4 DRE influences Slimb binding, ubiquitination, and centriole duplication. Previous studies demonstrated that S293 and T297 are critical in regulating Plk4 stability [3–6], and our results support functional roles for other phosphorylatable DRE residues [5]. Furthermore, self-regulation by trans-autophosphorylation is conserved [5, 6], because co-expression of WT-Plk4 converts KD-Plk4 into a Slimb-binding species and promotes its ubiquitination and degradation in Drosophila. Moreover, dimerized Plk4 autophosphorylates in vitro more readily than monomeric Plk4.
Although autophosphorylation is necessary for Plk4 degradation, it was not previously known whether Plk4 directly generates its own phosphodegron or whether autophosphorylation serves to prime and recruit an additional kinase(s) to phosphorylate the SRM. The latter is true for the Slimb/β-TrCP substrates β-catenin, Ci, and Wee1; multiple kinases sequentially phosphorylate and generate the phosphodegron in these Slimb/β-TrCP substrates [24–26]. Significantly, a majority of the Plk4 hydroxyl DRE residues are autophosphorylated. Therefore, our data supports a model whereby Plk4 directly generates its phosphodegron unassisted. This is consistent with our previous RNAi screen of the Drosophila kinome which did not reveal the involvement of an additional kinase in regulating Plk4 levels [2]. Our systematic mutagenesis of the DRE indicates that S293 is the critical phosphoresidue for Slimb binding, and that multiple residues flanking S293, particularly those downstream, collectively generate a higher-affinity Slimb-binding region. However, mutation of all hydroxyl DRE residues except for S293 significantly increases Plk4 stability, suggesting that phosphorylation of S293 alone does not promote efficient degradation. Notably, the Plk4 DRE is not the only autophosphorylated region [13]. The importance of these additional phospho-residues in regulating Plk4 activity and/or its association with binding partners remains to be determined.
Supplementary Material
Highlights.
Expression of kinase-dead Plk4 inhibits centriole duplication.
Residues within and flanking the Slimb Recognition Motif (SRM) are phosphorylated.
Phosphorylation of only S293 of the SRM is required for Slimb binding to Plk4.
Collective phosphorylation of flanking residues increases Slimb binding.
Acknowledgments
We thank P. Krieg for comments on the manuscript as well as Monica Bettencourt-Dias and her lab for sharing results prior to publication. G.C.R. is grateful for support from the NCI P30 CA23074 and GI SPORE (NCI/NIH P50 CA95060), the National Science Foundation (1158151), and the Arizona Biomedical Research Commission (1210) as well as the TRIF Imaging Fellowship Program to J.E.K. S.S., M.G., and N.M.R. are supported by the Division of Intramural Research at the NIH/NHLBI (1ZIAHL006104).
Footnotes
Supplemental Data include Supplemental Experimental Procedures, four figures, and two tables.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brito DA, Gouveia SM, Bettencourt-Dias M. Deconstructing the centriole: structure and number control. Curr Opin Cell Biol. 2012;24:4–13. doi: 10.1016/j.ceb.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 2.Brownlee CW, Klebba JE, Buster DW, Rogers GC. The protein phosphatase 2A regulatory subunit Twins stabilizes Plk4 to induce centriole amplification. J Cell Biol. 2011;195:231–243.3. doi: 10.1083/jcb.201107086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rogers GC, Rusan NM, Roberts DM, Peifer M, Rogers SL. The SCFSlimb ubiquitin ligase regulates Plk4/Sak levels to block centriole reduplication. J Cell Biol. 2009;184:225–239. doi: 10.1083/jcb.200808049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cunha-Ferreira I, Rodrigues-Martins A, Bento I, Riparbeli M, Zhang W, Laue E, Callaini G, Glover DM, Bettencourt-Dias M. The SCF/Slimb ubiquitin ligase limits centrosome amplification through degradation of SAK/PLK4. Curr Biol. 2009;19:43–49. doi: 10.1016/j.cub.2008.11.037. [DOI] [PubMed] [Google Scholar]
- 5.Holland AJ, Lan W, Niessen S, Hoover H, Cleveland DW. Polo-like kinase 4 kinase activity limits centrosome overduplication by autoregulating its own stability. J Cell Biol. 2010;188:191–198. doi: 10.1083/jcb.200911102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guderian G, Westendorf J, Uldschmid A, Nigg EA. Plk4 trans-autophosphorylation regulates centriole number by controlling beta TrCP-mediated degradation. J Cell Sci. 2010;123:2163–2169. doi: 10.1242/jcs.068502. [DOI] [PubMed] [Google Scholar]
- 7.Gonczy P. Towards a molecular architecture of centriole assembly. Nat Rev Mol Cell Biol. 2012;13:425–435. doi: 10.1038/nrm3373. [DOI] [PubMed] [Google Scholar]
- 8.Bettencourt-Dias M, Rodrigues-Martins A, Carpenter L, Riparbelli M, Lehmann L, Gatt MK, Carmo N, Balloux F, Callaini G, Glover DM. SAK/PLK4 is required for centriole duplication and flagella development. Curr Biol. 2005;15:2199–2207. doi: 10.1016/j.cub.2005.11.042. [DOI] [PubMed] [Google Scholar]
- 9.Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA. The Polo kinase Plk4 functions in centriole duplication. Nat Cell Biol. 2005;7:1140–1146. doi: 10.1038/ncb1320. [DOI] [PubMed] [Google Scholar]
- 10.Slevin LK, Nye J, Pinkerton DC, Buster DW, Rogers GC, Slep KC. The structure of the plk4 cryptic polo box reveals two tandem polo boxes required for centriole duplication. Structure. 2012;20:1905–1917. doi: 10.1016/j.str.2012.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leung GC, Hudson JW, Kozarova A, Davidson A, Dennis JW, Sicheri F. The Sak polo-box comprises a structural domain sufficient for mitotic subcellular localization. Nat Struct Biol. 2002;9:719–724. doi: 10.1038/nsb848. [DOI] [PubMed] [Google Scholar]
- 12.Korzeniewski N, Zheng L, Cuevas R, Parry J, Chatterjee P, Anderton B, Duensing A, Munger K, Duensing S. Cullin 1 functions as a centrosomal suppressor of centriole multiplication by regulating polo-like kinase 4 protein levels. Cancer Res. 2009;69:6668–6675. doi: 10.1158/0008-5472.CAN-09-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holland AJ, Lan W, Cleveland DW. Centriole duplication: a lesson in self-control. Cell Cycle. 2010;9:2731–2736. doi: 10.4161/cc.9.14.12184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sillibourne JE, Bornens M. Polo-like kinase 4: the odd one out of the family. Cell Div. 2010;5 doi: 10.1186/1747-1028-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sillibourne JE, Tack F, Vloemans N, Boeckx A, Thambirajah S, Bonnet P, Ramaekers FC, Bornens M, Grand-Perret T. Autophosphorylation of polo-like kinase 4 and its role in centriole duplication. Mol Biol Cell. 2010;21:547–561. doi: 10.1091/mbc.E09-06-0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leung GC, Ho CS, Blasutig IM, Murphy JM, Sicheri F. Determination of the Plk4/Sak consensus phosphorylation motif using peptide spots arrays. FEBS Lett. 2007;581:77–83. doi: 10.1016/j.febslet.2006.11.080. [DOI] [PubMed] [Google Scholar]
- 17.Martinez-Campos M, Basto R, Baker J, Kernan M, Raff JW. The Drosophila pericentrin-like protein is essential for cilia/flagella function, but appears dispensable for mitosis. J Cell Biol. 2004;165:673–683. doi: 10.1083/jcb.200402130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mennella V, Keszthelyi B, McDonald KL, Chhun B, Kan F, Rogers GC, Huang B, Agard DA. Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization. Nat Cell Biol. 2012;14:1159–1168. doi: 10.1038/ncb2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodrigues-Martins A, Riparbelli M, Callaini G, Glover DM, Bettencourt-Dias M. From centirole biogenesis to cellular function: centrioles are essential for cell division at critical developmental stages. Cell Cycle. 2008;7:11–16. doi: 10.4161/cc.7.1.5226. [DOI] [PubMed] [Google Scholar]
- 20.Dzhindzhev NS, Yu QD, Weiskopf K, Tzolovsky G, Cunha-Ferreira I, Riparbelli M, Rodrigues-Martins A, Bettencourt-Dias M, Callaini G, Glover DM. Asterless is a scaffold for the onset of centriole assembly. Nature. 2010;467:714–718. doi: 10.1038/nature09445. [DOI] [PubMed] [Google Scholar]
- 21.Hatch EM, Kulukian A, Holland AJ, Cleveland DW, Stearns T. Cep152 interacts with Plk4 and is required for centriole duplication. J Cell Biol. 2010;191:721–729. doi: 10.1083/jcb.201006049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cizmecioglu O, Arnold M, Bahtz R, Settele F, Ehret L, Haselmann-Weiss U, Antony C, Hoffmann I. Cep152 acts as a scaffold for recruitment of Plk4 and CPAP to the centrosome. J Cell Biol. 2010;191:731–739. doi: 10.1083/jcb.201007107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat Rev Cancer. 2008;8:438–49. doi: 10.1038/nrc2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu C, Li Y, Semenov M, Han C, Baeg GH, Tan Y, Zhang Z, Lin X, He X. Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell. 2002;108:837–847. doi: 10.1016/s0092-8674(02)00685-2. [DOI] [PubMed] [Google Scholar]
- 25.Smelkinson M, Kalderon D. Processing of the Drosophila hedgehog signaling effector Ci-155 to the repressor Ci-75 is mediated by direct binding to the SCF component Slimb. Curr Biol. 2006;16:110–116. doi: 10.1016/j.cub.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 26.Watanabe N, Arai H, Nishihara Y, Taniguchi M, Hunter T, Osada H. M- phase kinases induce phospho-dependent ubiquitination of somatic Wee1 by SCFbeta-TrCP. Proc Natl Acad Sci USA. 2004;101:4419–4424. doi: 10.1073/pnas.0307700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rogers SL, Rogers GC. Culture of Drosophila S2 cells and their use for RNAi-mediated loss-of-function studies and immunofluorescence microscopy. Nat Protoc. 2008;3:606–611. doi: 10.1038/nprot.2008.18. [DOI] [PubMed] [Google Scholar]
- 28.Rothbauer U, Zolghadr K, Muyldermans S, Schepers A, Cardoso MC, Leonhardt H. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol Cell Proteomics. 2008;7:282–289. doi: 10.1074/mcp.M700342-MCP200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.