SUMMARY
Secretion systems require high fidelity mechanisms to discriminate substrates amongst the vast cytoplasmic pool of proteins. Factors mediating substrate recognition by the type VI secretion system (T6SS) of Gram-negative bacteria, a widespread pathway that translocates effector proteins into target bacterial cells, have not been defined. We report that haemolysin co-regulated protein (Hcp), a ring-shaped hexamer secreted by all characterized T6SSs, binds specifically to cognate effector molecules. Electron microscopy analysis of an Hcp–effector complex from Pseudomonas aeruginosa revealed the effector bound to the inner surface of Hcp. Further studies demonstrated that interaction with the Hcp pore is a general requirement for secretion of diverse effectors encompassing several enzymatic classes. Though previous models depict Hcp as a static conduit, our data indicate it is a chaperone and receptor of substrates. These unique functions of a secreted protein highlight fundamental differences between the export mechanism of T6 and other characterized secretory pathways.
INTRODUCTION
Bacteria possess discrete pathways for delivering proteins to their surroundings. A diversity of secretion systems facilitates the necessary heterogeneity in size, structure and function within the proteins exported by these cells (Economou et al., 2006). For example, the type V secretion system can export exceptionally large proteins, such as filamentous adhesins, to the cell surface or the extracellular milieu (Dautin and Bernstein, 2007; Wagner et al., 2011), whereas types III, IV and VII systems exhibit the ability to directly translocate proteins into host cells (Abdallah et al., 2007; Fronzes et al., 2009; Galan, 2009; Izore et al., 2011). Of the thousands of proteins present in a cell, each secretion system acts with extraordinary fidelity to specifically recognize and release a distinct subset of substrates.
Substrate selection can be affected by several factors, including signal sequences, chaperones and receptors. For example, type III secretion system (T3SS) substrates are recruited to the secretory apparatus in complex with specialized chaperones (Akeda and Galan, 2005; Stebbins and Galan, 2001). A T3S-associated ATPase then engages the substrate by binding its N-terminal export signal, releasing it from the chaperone and driving substrate unfolding and translocation through the apparatus. Similar mechanisms appear to drive substrate recognition and export by the T4S pathway (Cambronne and Roy, 2007; Sutherland et al., 2012). While the factors involved in substrate selection for T3SSs and T4SSs are in part defined, comparatively little is known about this process in a related, but more recently described pathway, the type VI secretion system (T6SS).
The T6SS is a widely distributed protein translocation pathway that is able to directly target bacterial and eukaryotic cells (Silverman et al., 2012). The core T6 machinery consists of 13 essential subunits, encoded within large gene clusters on bacterial chromosomes. While the T6SS ultrastructure has not been determined, X-ray crystallographic and protein–protein interaction data suggest that an envelope-spanning complex and a bacteriophage tail-like structure are core components of the secretory machinery (Cascales and Cambillau, 2012). The phage-like complex consists of TssB (type six secretion B), TssC, Hcp (haemolysin co-regulated protein) and VgrG (valine-glycine repeat protein G). Visualization of Vibrio cholerae cells by fluorescence microscopy showed that TssB and TssC form a dynamic filamentous complex within the cytoplasm, switching between extended and contracted states while remaining anchored at the inner membrane (Basler et al., 2012). Hcp and VgrG are secreted components of the T6SS with significant structural homology to T4 bacteriophage tail tube (gp19) and tail spike proteins (gp5–gp27), respectively. By analogy to contractile bacteriophage, TssB and TssC are proposed to form a sheath enclosing a tube composed of Hcp, with VgrG localized at the tip to allow membrane breaching (Kanamaru, 2009; Kapitein and Mogk, 2013). Contraction of the TssB–TssC complex is postulated to drive Hcp and VgrG out of the cell. A recent report defining a complex composed of these proteins in Agrobacterium tumefaciens lends support to this model (Lin et al., 2013). Nevertheless, many aspects of the export mechanism for the T6SS remain undefined. In particular, how effectors are recognized for export and how they transit the T6S pathway is not known.
Hcp is a central component of the bacteriophage-like model of T6S-dependent intercellular effector transport. Studies have demonstrated that this universally and abundantly T6S-exported protein is essential for both the assembly of the T6S apparatus and the export of its effectors (Hood et al., 2010; Mougous et al., 2006; Pukatzki et al., 2007). Nevertheless, the precise function of Hcp in the T6SS has not been addressed. Crystal structures of diverse Hcp homologs from several species show the protein adopts a homohexameric ring conformation with a ~40 Å internal diameter; furthermore, Hcp rings stack to form tubular structures within the crystal lattices (Jobichen et al., 2010; Mougous et al., 2006; Osipiuk et al., 2011). It has been speculated based on these data that the protein functions as channel through which T6S effectors transit. However, Hcp tubes have only been observed under crystallographic conditions, or when stabilized in vitro with engineered disulfide bonds between ring subunits (Ballister et al., 2008). Also, since both stacking arrangements (head-to-head or head-to-tail) are found in Hcp X-ray crystal structures, the physiological significance of the Hcp tubule is uncertain.
The Hcp secretion island I (HSI-I)-encoded T6SS (H1-T6SS) of P. aeruginosa is a model system for studying the substrates, apparatus dynamics, and physiological significance of T6 (Basler and Mekalanos, 2012; Hood et al., 2010; Leroux et al., 2012; Russell et al., 2011). The H1-T6SS delivers at least three toxic effectors, Tse1-3 (type VI secretion exported 1–3), into target bacterial cells. Tse1 and Tse3 are translocated into the periplasm of target cells where they degrade peptidoglycan and cause cell lysis using amidase and muramidase activity, respectively (Russell et al., 2011). The precise molecular target of Tse2 is not yet known; however, this effector induces stasis and acts in the cytoplasm of recipient cells. Structure prediction algorithms indicate that Tse2 may function as a nuclease (Li et al., 2012). As the bacterial targeting activities of the H1-T6SS are also directed at kin, P. aeruginosa requires immunity proteins, Tsi1-3 (type VI secretion immunity 1–3), encoded alongside their cognate effector in bicistrons. P. aeruginosa harbors two additional T6SSs, the H2- and H3-T6SSs. Interestingly, these systems appear to export non-overlapping sets of effectors, though the factors mediating effector discrimination have not been elucidated (Russell et al., 2013).
In this study, we report that Hcp is a chaperone and receptor of T6S effectors. Focusing on Tse2, we show that a direct and highly specific interaction with the pore of its cognate Hcp, Hcp1 of the H1-T6SS, is required for the stability and export of the protein. We further demonstrate that effectors with unrelated sequences, Tse1 and Tse3, also require direct interactions with the pore of Hcp1 for secretion. Certain amino acid substitutions within the Hcp1 pore differentially affect the secretion of the three Tse proteins, indicating the capacity of Hcp1 to bind diverse proteins via distinct epitopes. Finally, we probe the generality of our results and find that specific interactions between T6S effectors and cognate Hcp proteins occur in diverse bacteria. Overall, our findings reveal a new paradigm for substrate recognition by a complex bacterial protein translocation machine.
RESULTS
Hcp1 Stabilizes Tse2 Posttranslationally
In the course of investigating factors that influence effector transport through the H1-T6SS of P. aeruginosa, we observed that strains lacking hcp1 displayed markedly reduced intracellular levels of Tse2 (Figure 1A). We conducted this, and subsequent studies in the ΔretS background, which is frequently employed for the study of the H1-T6SS (Basler and Mekalanos, 2012; Hachani et al., 2011; Mougous et al., 2006). The inactivation of retS overrides the requirement for cell–surface contact for T6S activation, and it results in elevated expression of both the H1-T6SS and its effectors, facilitating their detection via Western blot (Hood et al., 2010; Silverman et al., 2011). Deletions of other essential T6SS components including clpV1, tssM1, tssE1, and the functionally redundant vgrG1 and vgrG4 genes, did not lead to reduced Tse2 levels, indicating this phenotype is specific to the loss of hcp1 (Figure 1A). To measure more conclusively the effect of Hcp1 on overall Tse2 levels, we probed the consequences of hcp1 deletion in the ΔtssM1 background. The tssM1 gene encodes a core component of the secretory system; its deletion abrogates apparatus assembly and effector export (Felisberto-Rodrigues et al., 2011; Mougous et al., 2006). Tse2 levels were also strongly reduced in the Δhcp1 ΔtssM1 background, suggesting that Hcp1 is required for maintenance of intracellular Tse2 (Figure 1B). We also examined whether the levels of Tse1 and Tse3 are sensitive to the presence of Hcp1. Though not as dramatic as the effect on Tse2, we detected a significant reduction of intracellular Tse1 and Tse3 upon deletion of hcp1 from the ΔtssM1 background (Figure 1B). The effects of Δhcp1 on all effectors could be partially (Tse2) or fully (Tse1 and Tse3) genetically complemented.
Figure 1. Tse2 Requires Hcp1 for Intracellular Accumulation.

(A and B) Western blot analysis of intracellular levels of H1-T6S effectors (Tse1, Tse2 and Tse3) in the indicated P. aeruginosa backgrounds. RNA polymerase (RNAP) is included as a loading control. Unless otherwise indicated, the parental background in this and all subsequent figures is ΔretS. (C) Western blot analysis of intracellular Hcp1 and H1-T6S effector levels in a ClpXP-dependent Hcp1 depletion assay. Samples were processed 90 minutes after induced sspB expression in P. aeruginosa strains lacking the native sspB gene and containing wild-type hcp1 or hcp1–D4.
As an alternative approach for probing the influence of Hcp1 on effector levels, we used the ClpXP targeted degradation system to specifically deplete the protein from P. aeruginosa (Castang and Dove, 2012; McGinness et al., 2006). To accomplish this, we generated a P. aeruginosa strain wherein the native hcp1 open reading frame was modified to include a C-terminal fusion to the ssrA-like sequence, DAS+4 (Hcp1–D4). Consistent with our deletion studies, depletion of Hcp1–D4 resulted in a concomitant precipitous drop in cellular Tse2 levels (Figure 1C). Tse1 levels similarly decreased as observed in the Δhcp1 background, while Tse3 levels were not visibly affected by Hcp1 depletion. The cellular half-life of Tse3 may be too long to observe the relatively minor effect of Hcp1 during the 90 minute depletion assay (Figures 1B and 1C). Together, these data demonstrate that the Hcp1 protein is essential for the maintenance of intracellular Tse2 levels. Though Tse1 and Tse3 levels are measurably influenced by hcp1 deletion, significant intracellular accumulation of these effectors occurs independent of the protein. To investigate the mechanism underlying the influence of Hcp1 on H1-T6S effectors, we initially focused on Tse2, where a highly distinct Hcp1-dependent phenotype was observed.
Hcp1 Interacts Directly with Tse2
The dramatically lowered abundance of Tse2 in the absence of Hcp1 suggests that Hcp1 – acting directly or indirectly – plays a critical role either in regulating tse2 expression or stabilizing Tse2 posttranslationally. To test the former, we generated tse2 expression reporter strains containing a chromosomally-integrated construct consisting of the predicted tse2 promoter followed by an open reading frame consisting of the first 8 codons of tse2 fused to lacZ. Though the activity of the reporter increased as expected in the ΔretS background, a known posttranscriptional regulator of tse2, its activity was insensitive to the deletion of hcp1 (Figure 2A) (Brencic and Lory, 2009). Unable to implicate Hcp1 in a regulatory capacity, we probed for a physical interaction between the proteins using a co-immunoprecipitation (co-IP) assay. Interestingly, Tse2 specifically precipitated with Hcp1, suggesting a physical association between this effector and Hcp1 might underlie the Hcp1 requirement for Tse2 stability (Figure 2B). To test whether the observed interaction requires other T6S-associated factors, we next performed co-IP experiments from T6SS− E. coli strains co-expressing C-terminally hexahistidine-tagged hcp1 (hcp1-his6) and a vesicular stomatitis virus glycoprotein (VSV-G) epitope-tagged non-toxic allele of tse2 (tse2NT–V). In this heterologous host, the accumulation of Tse2 remained dependent upon the presence of Hcp1 (Figure 2C). Furthermore, co-expressed Tse2NT–V and Hcp1 associated tightly. To rule out nonspecific mechanisms that could explain our E. coli data, we tested the activity of another Hcp protein from P. aeruginosa, Hcp2, which does not participate in the H1-T6SS. Despite expression equivalent to or in excess of Hcp1, Hcp2 did not stabilize or interact with Tse2. From these data, we conclude that a direct interaction between Hcp1 and Tse2 likely promotes Tse2 stability.
Figure 2. Hcp1 Interacts Directly with Tse2.
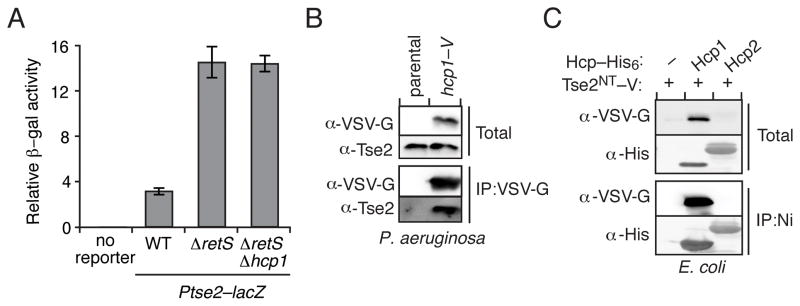
(A) Relative levels of β-galactosidase activity from the indicated P. aeruginosa strains containing a chromosomally-integrated lacZ reporter fused to the promoter region and first eight codons of tse2 (Ptse2–lacZ). Error bars represent standard deviation based on three independent replicates. (B) Western blot analysis of Tse2 from total and bead-associated fractions of an anti-VSV-G immunoprecipitation from P. aeruginosa strains encoding Hcp1 or ectopically expressing Hcp1. (C) Immunoblot detecting total and bead-associated fractions of a nickel-NTA precipitation assay from E. coli expressing a nontoxic, VSV-G epitope-tagged allele of tse2 (tse2NT–V (T79A S80A)) with empty vector (−) or a plasmid containing the indicated hcp (hcp1 or hcp2) homolog fused to a His6 tag.
Tse2 Interacts with the Inner Surface of Hcp1 Rings
X-ray crystallographic and electron microscopic (EM) studies of Hcp proteins, including Hcp1, have consistently observed the protein in a hexameric ring configuration (Jobichen et al., 2010; Leiman et al., 2009; Mougous et al., 2006; Osipiuk et al., 2011). Using sedimentation velocity analytical ultracentrifugation at low Hcp1 concentrations (10.3-3.2 μM), we further confirmed this quaternary state as the single detectable species in solution (Figure S1). These observations, taken together with the large hydrophobic interface observed between Hcp protomers, argue that the hexameric assembly is the relevant physiological state of Hcp proteins.
The Hcp1 hexamer reveals several potential protein–protein interaction surfaces. Sequence analysis of Hcp homologs shows that residues on the “top” and “bottom” faces of Hcp rings are highly conserved and may be important for ring–ring interactions (Figure S2) (Mougous et al., 2006). Thus, we reasoned that Tse2 likely interacts with either the inside or outside face of the Hcp1 ring. To define the regions of Hcp1 involved in Tse2 interaction, we pursued a site-directed mutagenesis approach, targeting all residues with more than 60% solvent accessibility located on the inner or outer surface of the Hcp1 ring. To this end, we mutated 34 positions; non-polar or small side chain amino acids were substituted with glutamine, and those with a polar or large side chain with alanine. The relative affect of each Hcp1 point mutation on Tse2 stability was determined by co-expression of the hcp1 alleles with tse2–V in E. coli. We found that approximately half of the mutations localized to the inside surface of Hcp1 exhibit 1% or less Tse2 stabilization relative to wild-type Hcp1 (Figures 3A, 3B and S3), whereas all Hcp1 variants with substitutions mappings to the outer surface of the ring displayed substantial Tse2-stabilization activity (1.0–100%). Interestingly, the majority of substitutions that destabilize Tse2 over 100-fold map together within a discrete patch inside the Hcp1 ring (Figure 3B).
Figure 3. Tse2 Binds to the Pore of the Hcp1 Ring.
(A) Representative Western blots showing the effect of the indicated Hcp1 amino acid substitutions on intracellular levels of Tse2NT in E. coli. The localization of the substitutions to the inside or outside surface of the Hcp1 ring is noted. (B) Surface representation of the Hcp1 ring colored to reflect Tse2 stabilization activity of each variant tested (white-red =100%-0.01%, gray = not tested). The lower image depicts a cutaway view of the Hcp1 hexamer. Levels of Tse2 were calculated based on the ratio of band intensity of Tse2NT and Hcp1 point mutants, normalized to Tse2 and wild-type Hcp1 (Figure S2). (C) Coomassie-stained SDS-PAGE gel of co-purified Tse2NT–His6 and Hcp1–V. (D) Class averages with applied six-fold symmetry from analysis of transmission electron micrographs of Tse2NT–His6 -Hcp1–V and a Hcp1–V-only control. The percentage of particles represented by each class average is indicated in the corresponding frame. See also Figure S1, S2, S3 and S4.
The ring shape of Hcp1 is readily discernable by negative stain transmission electron microscopy (TEM). Therefore, we reasoned that Hcp1 rings bound to Tse2 might be distinguished by TEM as “filled” particles. TEM analysis of approximately 3,000 randomly selected purified Hcp1–V–Tse2NT–His6 single particles showed that filled class averages constituted a significant fraction of total Hcp1 particles (Figures 3C, 3D and S4). In contrast, no class averages appeared filled in the control sample. The low fraction of filled rings observed in the Hcp1–Tse2 sample is likely explained by Tse2 degradation and precipitation during preparation (Li et al., 2012; Zou et al., 2012). Visualization of the Tse2–Hcp1 complex provides direct evidence that Tse2 interacts with the inner surface of Hcp1 rings.
Tse2 Secretion Requires Interaction with Hcp1
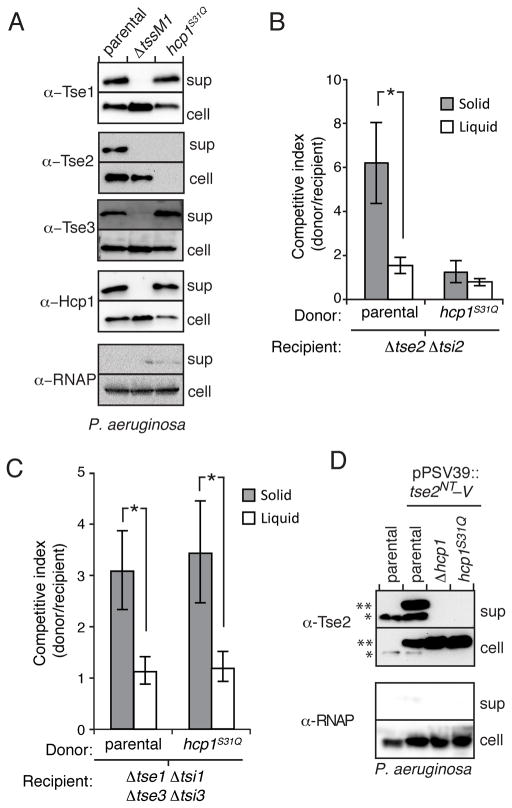
Motivated by our in vitro findings, we next sought to determine the influence of the interaction between Tse2 and the inner surface of Hcp1 on Tse2 stability and secretion in vivo. Using allelic exchange, we generated a P. aeruginosa strain encoding Hcp1S31Q at the native hcp1 locus. We selected this mutant from those identified by our in vitro studies, as this amino acid resides within the inner surface patch and strongly disrupts interaction with Tse2 via a relatively conservative substitution (small polar to large polar). Western blot analyses showed that although Hcp1S31Q is produced and secreted at levels comparable to the wild-type, strains bearing this mutation do not support Tse2 accumulation (Figure 4A). Furthermore, using interbacterial competition assays, we found that the hcp1S31Q background displays a marked defect in Tse2-dependent fitness (Figure 4B). The effects on H1-T6SS function appeared specific, as the levels of export and interbacterial delivery of Tse1 and Tse3 were not significantly impacted in the hcp1S31Q background (Figures 4A and 4C). Overall these data demonstrate that Tse2 stability requires interaction with the inner surface of the Hcp1 ring.
Figure 4. Tse2 Requires Interaction with Hcp1 for Secretion.
(A) Western blot analysis of cell and supernatant fractions of Hcp1 and H1-T6S effectors in P. aeruginosa strains harboring wild-type hcp1 or hcp1S31Q. Equally exposed α-RNA polymerase (RNAP) blots of equivalent fractions of total cell and supernatant (sup) samples are included as loading and cytoplasmic leakage controls in this and subsequent secretion assays. (B and C) Outcome of growth competition experiments between P. aeruginosa donor strains (parental or hcp1S31Q) and a Tse2-susceptible (Δtse2 Δtsi2) (B) or a Tse1- and Tse3-susceptible recipient strain (Δtse1 Δtsi1 Δtse3 Δtsi3) (C) on solid (gray) or liquid (white) media. The competitive index is calculated as the change (final/initial) in ratio of donor to recipient c.f.u. Error bars represent standard deviation based on four replicates. Asterisks denote statistical significance using ANOVA and Tukey’s post hoc test between the indicated conditions (P < 0.001). (D) Western blot analysis of cell and supernatant-associated fractions of chromosomal, endogenous Tse2 (one asterisk) and ectopically expressed Tse2NT–V (two asterisks) in the indicated P. aeruginosa strains.
Since Tse2 does not accumulate to detectable levels in hcp1S31Q, we could not explicitly measure the requirement of interaction with Hcp1 for Tse2 secretion. However, a previous report from our laboratory demonstrated that over-expressed Tse2 can be detected in P. aeruginosa backgrounds lacking hcp1 (Hood et al., 2010). Therefore, to overcome the Tse2 detection limit, we monitored the export of Tse2–V expressed with high induction from the lacUV5 promoter in the hcp1S31Q background. Western blot analysis confirmed that the Tse2–V protein is highly over-produced relative to endogenous Tse2 and that it is secreted in an Hcp1-dependent manner (Figure 4D). Interestingly, we found that despite intracellular accumulation, Tse2–V is not exported in the hcp1S31Q background. Thus, taken together with our observation that H1-T6SS function is generally preserved in the hcp1S31Q background, we conclude that interaction with Hcp1 is required both for the stabilization and export of Tse2.
Recognition of Tse2-like Effectors by Hcp is General
To determine the generality of our findings concerning P. aeruginosa Tse2 and Hcp1, we sought to identify and characterize functionally analogous proteins in other bacterial species. Protein BLAST analyses revealed Tse2 homologs within several T6SS+ Gram-negative bacteria, including Methylomonas methanica, Shewanella fridigimarina, Burkholderia ambifaria and Pseudoalteromonas sp. (Figure 5A). Sequence alignments of the homologs highlighted the conservation of several motifs, including a putative aspartic acid-containing nuclease-related catalytic site (Asp63 in Tse2) identified using protein structure prediction algorithms (Figure S5) (Li et al., 2012). As additional evidence of the functional relationship between Tse2 and its homologs, we found each homolog encoded in an apparent bicistron with a smaller gene encoding a protein with attributes similar to Tsi2, including length (Tsi2, 77; homologs, 75–79 amino acids) and an acidic pI (Tsi2, 3.9; homologs, 4.0–4.5) (Figure 5A).
Figure 5. Stabilization by Cognate Hcp is a General Feature of Tse2-like Effectors.
(A) Genomic organization of tse2 and tsi2 homologs from P. aeruginosa (PA) M. methanica (MM), S. frigidimarina (SF), B. ambifaria (BA) and Pseudoalteromonas sp. (Palt). Abbreviated locus tag numbers are indicated for tse2 (darker shade) and tsi2 (lighter shade) homologs from each organism. See also Figure S5. (B) Growth of E. coli containing plasmids with inducible expression of tse2 or tsi2 homologs from P. aeruginosa (upper panel) or M. methanica (lower panel). Serial ten-fold dilutions are indicated by numbers. (C and D) Western blot results of co-IP assays from E. coli co-expressing hcp–his6 homologs from P. aeruginosa (PA), M. methanica (MM) or P. protegens (PP) with tse2(D63N)–V (mutation denoted with asterisk) from M. methanica (C) or P. aeruginosa (D). (E) Analysis of cell and supernatant-associated fractions of the indicated P. aeruginosa strains expressing Tse2MM–V ectopically.
Confident we had identified bona fide Tse2 and Tsi2-related proteins, we selected M. methanica Tse2 (Tse2MM), which shares 71% identity with P. aeruginosa Tse2, for more detailed analyses. The genome of M. methanica contains a single T6S gene cluster and encodes a single clear homolog of P. aeruginosa Hcp1, Hcp1MM, making Tse2MM well suited for our study. To first establish that the M. methanica tse2 tsi2 pair is functionally orthologous to P. aeruginosa tse2 tsi2, we conducted toxicity studies in E. coli. Similar to tse2, expression of tse2MM in E. coli caused a dramatic drop in recovered c.f.u. (Figure 5B). Moreover, this toxicity was observed for Tse2MM(D63N), and it was inhibited by co-expression with tsi2MM.
We next tested whether Tse2MM, like Tse2, interacts with the Hcp1 homolog, Hcp1MM, encoded within the putative T6SS of M. methanica. Indeed, we found Tse2MM interacts directly with Hcp1MM and, additionally, requires Hcp1MM for intracellular accumulation (Figure 5C). Interestingly, we found that Tse2MM also interacts with and is stabilized by P. aeruginosa Hcp1, and likewise for the P. aeruginosa Tse2–Hcp1MM pair (Figures 5C and 5D). The specificity of these interactions is underscored by the observation that neither Tse2 nor Tse2MM interact with Hcp1 from Pseudomonas protegens (Hcp1PP), despite the fact that this Hcp1 homolog is nearly equally divergent from P. aeruginosa Hcp1 as Hcp1MM (Hcp1PP, 76%; Hcp1MM, 77%). Notably, Hcp1PP is encoded within an HSI-I-like gene cluster, yet P. protegens does not have a Tse2 homolog encoded within its genome. Finally, consistent with the hypothesis that interaction with Hcp is a key determinant for export via the T6S pathway, we observed efficient release of Tse2MM via the H1-T6SS (Figure 5E).
Amidase and Muramidase T6S Effector Classes Recognize Cognate Hcp Proteins
In the course of studying the behavior of Hcp1 point mutants in P. aeruginosa, we observed that while the effects of Hcp1S31Q are limited to Tse2, other Tse2-destabilizing Hcp1 pore substitutions also reduce the export of Tse1 and Tse3 (Figure 6A). These Hcp1 variants are themselves secreted, suggesting their effect on Tse1 and Tse3 export is not a consequence of misfolding or a failure to be recognized by the secretory apparatus. Moreover, certain substitutions exhibited differential effects on the secretion of Tse1 and Tse3. For example, Hcp1L28A inhibits Tse3 secretion, but not that of Tse1. On the contrary, Hcp1S115Q strongly affects both Tse1 and Tse3 secretion. Together with our observation that Tse1 and Tse3 display decreased abundance in strains lacking Hcp1 (Figure 1B), our findings led us to hypothesize that non-Tse2-type effectors also engage in specific interactions with the inner surface of the Hcp1 pore, and that these are required for H1-T6SS-dependent secretion.
Figure 6. Binding to the Pore of Cognate Hcp Proteins is Required for Export of Effectors with Amidase and Muramidase Activities.
(A) Western blot analysis of cell and supernatant fractions of Hcp1 and H1-T6S effectors in P. aeruginosa strains wherein the native hcp1 allele is substituted with the indicated mutant. (B and C) Western blot analysis of bead-associated fractions of a co-IP from E. coli co-expressing the indicated hcp–his6 alleles with Tse1–V (B) or Tse3–V (C). (D and E) Immunoblot detecting bead-associated fractions of a co-IP from E. coli co-expressing hcp–his6 homologs (TY, S. Typhi; BP, B. phytofirmans) and tae2TY–V (D) or tae1BP–V (E). See also Figure S6 and S7.
To determine whether Hcp1 directly interacts with Tse1 and Tse3, we performed co-IP assays using proteins co-expressed in E. coli. As a means of linking potential interactions to the secretion phenotypes observed, we utilized the secretion defective hcp1S115Q pore substitution mutant in addition to wild-type hcp1. Interbacterial competition assays confirmed this mutation functionally disrupts Tse1- and Tse3-dependent intoxication (Figure S6). While Tse1 and Tse3 did not efficiently co-IP with Hcp1S115Q, both proteins were detected in association with the wild-type protein (Figures 6B and 6C). From these data, we conclude that interaction with the inner surface of Hcp1 is a common requirement for the secretion of the three known H1-T6SS effectors. The observation that Hcp1 substitutions can have variable effects on the Tse proteins suggests that, consistent with their divergent sequences, the proteins each form a unique network of interactions with Hcp1.
Bioinformatic work conducted by our laboratory has found evidence of a non-random association between hcp genes and predicted T6S amidase effectors (Tae) (Figure S7) (Russell et al., 2012). Based on this observed genetic link, our finding that direct interactions between Tse1-3 and Hcp1 are required for secretion, and the results of a prior study demonstrating the interaction of a putative Edwardsiella tarda T6S effector with an Hcp homolog (Zheng and Leung, 2007), we posited that the recognition of effectors by cognate Hcp proteins might be a general determinant of substrate selectivity by the T6SS. As a first step toward validating this model, we selected two predicted cognate Tae–Hcp pairs based on genetic linkage (Salmonella enterica serovar Typhi Tae2) or homology to Tse1 (Burkholderia phytofirmans Tae1). As predicted, both effectors bound cognate, but not non-cognate Hcp proteins (Figures 6D and 6E). In total, our data suggest that the interaction between effectors and cognate Hcp proteins is a general phenomenon that is likely to – at least in part – define the particular effectors transported through a given T6SS.
DISCUSSION
We have identified Hcp as a critical gatekeeper protein of the T6SS. As a chaperone, a substrate receptor, and a secreted protein itself, the multipurpose nature of Hcp grants it a unique position among secretory system-associated proteins (Figure 7). This implies that the mechanism of substrate export by the T6SS is fundamentally different from other characterized secretion systems. Arguably, T3S and T4S, which both translocate macromolecules in a concerted fashion into target cells, are the most analogous secretion pathways to the T6SS (Cornelis, 2006; Fronzes et al., 2009). Indeed, T6S and T4S share two homologous proteins, TssL (DotU) and TssM (IcmF) (Cascales and Cambillau, 2012). In light of this relatedness, it is interesting that effector recognition by the T6SS occurs via a principally different process than these related pathways, which both utilize ATPases to directly engage substrates and provide the energy necessary for export (Akeda and Galan, 2005; Christie et al., 2005). The fact that the T6S pathway does not appear to directly link effector recognition to energizing secretion may be explained by its putative bacteriophage origins (Kanamaru, 2009). The energy driving unidirectional protein transport in the T6S pathway is thought to be stored in a filamentous structure composed of TssB and TssC, which contracts and is then recycled by a ClpB-like AAA+ family ATPase, ClpV (Basler et al., 2012; Bonemann et al., 2009).
Figure 7. Model depicting the role of Hcp in T6S effector recognition and export.
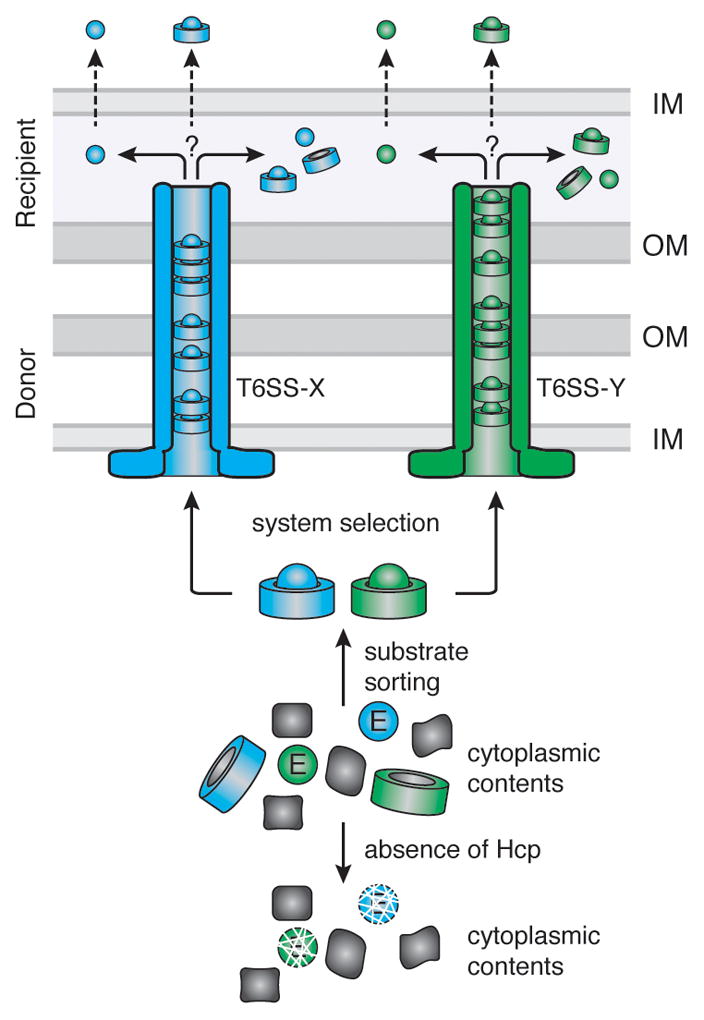
The schematic depicts the junction between two Gram-negative bacterial cells (OM, outer membrane; IM, inner membrane), a donor cell, harboring two T6SSs T6SS-X (blue) and T6SS-Y (green), and a recipient cell that is targeted by these systems. T6S effectors (Ex, Ey) are sorted from the cytoplasmic pool of proteins via interactions with cognate Hcp proteins (Hx, Hy). Interaction with Hcp prevents effector degradation. Hcp–effector complexes are recognized and transported through the appropriate T6SS via an unknown mechanism. Once the complex reaches the recipient cell, it may be either translocated into the periplasm intact, or the effector may dissociate from Hcp prior to periplasmic delivery. Likewise, effectors destined for the cytoplasm may be transported in complex or in isolation.
Prior models of the T6SS depict Hcp as a passive conduit for effectors (Basler et al., 2012; Bonemann et al., 2010; Cascales and Cambillau, 2012; Silverman et al., 2012). However, our data suggest that Hcp associates with effectors, protects them from proteolysis, and likely traffics with effectors during transport (Figure 7). Stoichiometric, or near stoichiometric, release of Hcp hexamers with effectors reconciles the massive accumulation of Hcp in the culture supernatants of T6S-activated strains (Mougous et al., 2006; Pieper et al., 2009; Pukatzki et al., 2006; Wu et al., 2008). Whether effectors in complex with Hcp are folded cannot be definitively established from our data. The structures of several amidase effectors indicate the proteins are of a size that could be accommodated within the Hcp pore (Chou et al., 2012; Dong et al., 2013a; Zhang et al., 2013). However, based on the instability of effectors in the absence of cognate Hcp proteins, we speculate they do not adopt their final folded configuration until, or accompanying, release from Hcp. It is of note that intra-molecular disulfide bonds have been observed in the structures of amidase effectors. The failure of these bonds to form in the cytoplasm could explain the instability of the proteins in this compartment, while their formation in the periplasm could drive release from Hcp. Interestingly, Tse2 lacks cysteine residues and ultimately accesses the cytoplasm of the recipient. In this case, interaction with the outer face of the inner membrane, or a receptor associated with the membrane, might drive effector release. It is also possible that the T6S apparatus remodels or modifies proteins during export to disrupt Hcp–substrate interactions.
Our data suggest that interactions with the pore of Hcp are, broadly, a critical determinant for the secretion of T6 effectors. However, we posit that other mechanisms may facilitate the recognition and export of disparate effector classes. Rather than a genetic linkage with hcp genes, Tle (type VI secretion lipase effector) and Rhs (recombination hot spot) effectors are encoded in association with vgrG genes (Koskiniemi et al., 2013; Russell et al., 2013). VgrG proteins often facilitate intercellular T6S-dependent delivery of effectors present as C-terminal fusion to their structural domain (Brooks et al., 2013; Pukatzki et al., 2007; Suarez et al., 2010). It is conceivable that Tle and Rhs effectors, which are considerably larger than Tse-type effectors, might gain access to recipient cells by non-covalently associating with VgrG proteins. A Tle protein from V. cholerae was found in the immunoprecipitate of a VgrG protein, however the genetically linked VgrG tested was not tested (Dong et al., 2013b).
While we have delineated a critical step in the process of effector secretion by the T6SS, many aspects of the overall structure-function model for the system remain untested. So far, the localization of Hcp inside the TssB–TssC sheath-like complex has not been demonstrated. Furthermore, there is some evidence suggesting that Hcp may not associate within of the TssB–TssC tubule. For instance, Hcp proteins bearing large C-terminal fusions have been identified, and it is difficult to envision how these much larger proteins would be accommodated inside the TssB–TssC structure (Blondel et al., 2009; Parret and De Mot, 2002). Irrespective of the association of Hcp–effector complexes with TssB–TssC, it also remains unresolved how Hcp–substrate complexes cross the inner and outer bacterial membranes. An envelope-spanning complex consisting of integral inner membrane proteins and an outer membrane lipoprotein has been identified, however the ultrastructure of this novel complex awaits visualization (Felisberto-Rodrigues et al., 2011).
Immunity to Tse2 is provided by the Tsi2 protein, a small acidic cytoplasmic protein that directly binds and inactivates the toxin. The current study adds previously unrecognized complexity to the subject of effector immunity, particularly cytoplasmic effectors like Tse2 that interact with cognate proteins prior to export. Based on the studies reported herein and the observation that Tsi2 is an essential gene, it is apparent that significant Tse2 immunity is not provided by Hcp1 (Hood et al., 2010; Jacobs et al., 2003). It is conceivable that Tsi2 interacts with Tse2 in the context of Tse2–Hcp1 complexes, however we did not detect this complex and favor the hypothesis that Tsi2 functions by scavenging free Tse2, including endogenous Tse2 and Tse2 delivered by other P. aeruginosa cells in trans.
Our discovery that effectors interact with the pore of Hcp may have implications for the identification of novel T6S substrates. The discovery of effectors by virtue of interaction with cognate Hcp proteins stands to overcome several difficulties encountered in such efforts. For instance, this method would not depend upon effector export, which is often repressed under in vitro culturing conditions (Silverman et al., 2012). Our findings may also guide efforts to engineer non-native substrate export into the T6S pathway. This approach could provide new ways to modulate the outcome of interbacterial interactions.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids and Growth Conditions
All P. aeruginosa strains were derived from the sequenced strain PAO1. Genomic DNA from M. methanica MC09, S. Typhi Ty2, P. protegens Pf-5 or P. aeruginosa PAO1 was used to amplify effector, immunity and hcp genes (Boden et al., 2011; Deng et al., 2003; Paulsen et al., 2005; Stover et al., 2000). E. coli strains DH5α, SM10 and BL21 were used for plasmid maintenance, conjugative transfer and gene expression, respectively. Plasmids used in this study are described and referenced in Table S1. See the Supplemental Experimental Procedures for specific growth conditions and cloning procedures.
Hcp1 Depletion Assays
We used a controllable protein degradation system originally developed for use in E. coli, but has also been optimized for use in P. aeruginosa (Castang et al., 2008; McGinness et al., 2006). Full details in Supplemental Experimental Procedures.
β-Galactosidase Assay
β-Galactosidase assays were performed as described previously with minor modifications. A chromosomal integration vector (mini-CTX-lacZ) carrying a fusion of lacZ to the putative promoter region and first eight codon of tse2 was used to generate strains for analysis. The plasmid was introduced into P. aeruginosa by conjugation (Hoang et al., 2000). Full details in Supplemental Experimental Procedures.
Co-Immunoprecipitation Assays
Full details for co-immunoprecipitation (co-IP) assays from P. aeruginosa or E. coli can be found in the Supplemental Experimental Procedures. Briefly, cell lysates were applied to anti-VSV-G agarose beads or Nickel-NTA sepharose beads and incubated for 1 hour at 4°C. Beads were washed 3 times with buffer before resuspending in SDS-PAGE buffer.
Tse2 Stability Assays with Hcp1 Point Mutants
E. coli BL21 plysS with plasmids containing each hcp1 allele (pET29b) and tse2NT (pSCRhaB2-CV) were co-transformed into E. coli. Overnight cultures were sub-inoculated (1:1000) into 3 mL of 2xYT and grown to an OD600 of 0.6. Cultures were induced with 0.1% rhamnose and 10 μM IPTG and harvested after 4 hours. 300 μl from each sample were centrifuged at 16,000 × g and pellets were resuspended in 30 μL Buffer 1 and 30 μL of SDS-PAGE loading dye, boiled and analyzed by Western blot. Densitometry was performed using AlphaView®Q software (ProteinSimple). The percentage of total accumulated Tse2 was determined by the following equation: (([Tse2*]/[Tse2])/([Hcp1*]/[Hcp1]))*100, where Tse2* = Tse2 accumulated in the presence of each Hcp1 point mutation (Hcp1*).
Secretion Assays
Overnight cultures of P. aeruginosa strains were used to inoculate 2 ml of LB (1:1000) supplemented with 0.5 mM IPTG. Cultures were grown at 37°C with shaking to mid-log phase and cell and supernatant fractions were processed as previously described (Hood et al., 2010).
Interbacterial Competitions Assays
Competition assays between P. aeruginosa strains were performed as previously described with minor variations that are described in Supplemental Experimental Procedures (Hood et al., 2010; Russell et al., 2011).
Purification of Tse2-Hcp1 Complex for Transmission Electron Microscopy
The Tse2-Hcp1 complex was purified from E. coli containing plysS, pET29b::tse2NT-His6 and pPSV35::hcp1-VSV-G using a two-step affinity chromatography method. Full details in Supplemental Experimental Procedures.
Single Particle Electron Microscopy of Hcp Rings and Hcp1-Tse2 Complex
Hcp1 rings and the Hcp1-Tse2 complex purified from E. coli were negatively stained by 0.75% uranyl formate as described previously (Ohi et al., 2004). Images were collected on a transmission electron microscope T12 (FEI) at room temperature under 120 kV, and recorded at a magnification of 67,000× on a 4k × 4k Gatan CCD rendering a final pixel size of 1.65 Å on the specimen level. Particles were selected and windowed into 120 × 120 pixel images using WEB (Frank et al., 1996). Full details in Supplemental Experimental Procedures.
E. coli Toxicity Measurements
E. coli toxicity assays were performed as previously described with some variations described in Supplemental Experimental Procedures (Russell et al., 2011).
Supplementary Material
HIGHLIGHTS.
Hcp is a chaperone required for the intracellular accumulation of Tse2
Export of type VI secretion effectors requires direct interaction with the pore of Hcp.
Cognate Hcp–effector interactions can define substrate specificity of the T6SS.
Substrate recognition and energization of secretion are uncoupled in the T6SS.
Acknowledgments
The authors wish to thank Colin Murrell for providing M. methanica genomic DNA, Simon Dove for sharing the sspB knockout and expression constructs, Michele LeRoux and Brook Peterson for critical review of the manuscript, Alistair Russell for assistance with bioinformatics, and Spencer Anthony-Cahill, Peter Brzovic, Rachel Klevit, and members of the Mougous laboratory for helpful discussions. The Catalano laboratory is supported by National Institutes of Health Grant GM088186 and National Science Foundation Grant MCB-1158107. The Gonen laboratory is supported by the Howard Hughes Medical Institute. Work in the Mougous laboratory was funded by a grant from the NIH (AI080609). J.M.S. was supported by Public Health Service National Research Service Award T32 GM07270 from NIGMS and by a Helen Riaboff Whiteley Fellowship from the Department of Microbiology. J.D.M. holds an Investigator in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdallah AM, Gey van Pittius NC, Champion PA, Cox J, Luirink J, Vandenbroucke-Grauls CM, Appelmelk BJ, Bitter W. Type VII secretion--mycobacteria show the way. Nat Rev Microbiol. 2007;5:883–891. doi: 10.1038/nrmicro1773. [DOI] [PubMed] [Google Scholar]
- Akeda Y, Galan JE. Chaperone release and unfolding of substrates in type III secretion. Nature. 2005;437:911–915. doi: 10.1038/nature03992. [DOI] [PubMed] [Google Scholar]
- Ballister ER, Lai AH, Zuckermann RN, Cheng Y, Mougous JD. In Vitro Self-Assembly of Tailorable Nanotubes from a Simple Protein Building Block. Proc Natl Acad Sci USA. 2008;105:3733–3738. doi: 10.1073/pnas.0712247105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler M, Mekalanos JJ. Type 6 Secretion Dynamics Within and Between Bacterial Cells. Science. 2012;337:815. doi: 10.1126/science.1222901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature. 2012;483:182–186. doi: 10.1038/nature10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel CJ, Jimenez JC, Contreras I, Santiviago CA. Comparative genomic analysis uncovers 3 novel loci encoding type six secretion systems differentially distributed in Salmonella serotypes. BMC genomics. 2009;10:354. doi: 10.1186/1471-2164-10-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boden R, Cunliffe M, Scanlan J, Moussard H, Kits KD, Klotz MG, Jetten MS, Vuilleumier S, Han J, Peters L, et al. Complete genome sequence of the aerobic marine methanotroph Methylomonas methanica MC09. Journal of bacteriology. 2011;193:7001–7002. doi: 10.1128/JB.06267-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonemann G, Pietrosiuk A, Diemand A, Zentgraf H, Mogk A. Remodelling of VipA/VipB tubules by ClpV-mediated threading is crucial for type VI protein secretion. The EMBO journal. 2009;28:315–325. doi: 10.1038/emboj.2008.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonemann G, Pietrosiuk A, Mogk A. Tubules and donuts: a type VI secretion story. Mol Microbiol. 2010;76:815–821. doi: 10.1111/j.1365-2958.2010.07171.x. [DOI] [PubMed] [Google Scholar]
- Brencic A, Lory S. Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol Microbiol. 2009;72:612–632. doi: 10.1111/j.1365-2958.2009.06670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks TM, Unterweger D, Bachmann V, Kostiuk B, Pukatzki S. Lytic activity of the Vibrio cholerae type VI secretion toxin VgrG-3 is inhibited by the antitoxin TsaB. The Journal of biological chemistry. 2013;288:7618–7625. doi: 10.1074/jbc.M112.436725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambronne ED, Roy CR. The Legionella pneumophila IcmSW complex interacts with multiple Dot/Icm effectors to facilitate type IV translocation. PLoS pathogens. 2007;3:e188. doi: 10.1371/journal.ppat.0030188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales E, Cambillau C. Structural biology of type VI secretion systems. Philos Trans R Soc Lond B Biol Sci. 2012;367:1102–1111. doi: 10.1098/rstb.2011.0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castang S, Dove SL. Basis for the essentiality of H-NS family members in Pseudomonas aeruginosa. J Bacteriol. 2012;194:5101–5109. doi: 10.1128/JB.00932-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castang S, McManus HR, Turner KH, Dove SL. H-NS family members function coordinately in an opportunistic pathogen. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:18947–18952. doi: 10.1073/pnas.0808215105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou S, Bui NK, Russell AB, Lexa KW, Gardiner TE, Leroux M, Vollmer W, Mougous JD. Structure of a Peptidoglycan Amidase Effector Targeted to Gram-Negative Bacteria by the Type VI Secretion System. Cell Rep. 2012;1:656–664. doi: 10.1016/j.celrep.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie PJ, Atmakuri K, Krishnamoorthy V, Jakubowski S, Cascales E. Biogenesis, architecture, and function of bacterial type iv secretion systems. Annual review of microbiology. 2005;59:451–485. doi: 10.1146/annurev.micro.58.030603.123630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis GR. The type III secretion injectisome. Nat Rev Microbiol. 2006;4:811–825. doi: 10.1038/nrmicro1526. [DOI] [PubMed] [Google Scholar]
- Dautin N, Bernstein HD. Protein secretion in gram-negative bacteria via the autotransporter pathway. Annual review of microbiology. 2007;61:89–112. doi: 10.1146/annurev.micro.61.080706.093233. [DOI] [PubMed] [Google Scholar]
- Deng W, Liou SR, Plunkett G, 3rd, Mayhew GF, Rose DJ, Burland V, Kodoyianni V, Schwartz DC, Blattner FR. Comparative genomics of Salmonella enterica serovar Typhi strains Ty2 and CT18. Journal of bacteriology. 2003;185:2330–2337. doi: 10.1128/JB.185.7.2330-2337.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Zhang H, Gao ZQ, Wang WJ, She Z, Liu GF, Shen YQ, Su XD, Dong YH. Structural insights into the inhibition of type VI effector Tae3 by its immunity protein Tai3. The Biochemical journal. 2013a doi: 10.1042/BJ20130193. [DOI] [PubMed] [Google Scholar]
- Dong TG, Ho BT, Yoder-Himes DR, Mekalanos JJ. Identification of T6SS-dependent effector and immunity proteins by Tn-seq in Vibrio cholerae. Proceedings of the National Academy of Sciences of the United States of America. 2013b;110:2623–2628. doi: 10.1073/pnas.1222783110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Economou A, Christie PJ, Fernandez RC, Palmer T, Plano GV, Pugsley AP. Secretion by numbers: protein traffic in prokaryotes. Mol Microbiol. 2006;62:308–319. doi: 10.1111/j.1365-2958.2006.05377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felisberto-Rodrigues C, Durand E, Aschtgen MS, Blangy S, Ortiz-Lombardia M, Douzi B, Cambillau C, Cascales E. Towards a structural comprehension of bacterial type VI secretion systems: characterization of the TssJ-TssM complex of an Escherichia coli pathovar. PLoS Pathog. 2011;7:e1002386. doi: 10.1371/journal.ppat.1002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Fronzes R, Christie PJ, Waksman G. The structural biology of type IV secretion systems. Nat Rev Microbiol. 2009;7:703–714. doi: 10.1038/nrmicro2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan JE. Common themes in the design and function of bacterial effectors. Cell host & microbe. 2009;5:571–579. doi: 10.1016/j.chom.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachani A, Lossi NS, Hamilton A, Jones C, Bleves S, Albesa-Jove D, Filloux A. Type VI secretion system in Pseudomonas aeruginosa: secretion and multimerization of VgrG proteins. The Journal of biological chemistry. 2011;286:12317–12327. doi: 10.1074/jbc.M110.193045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang TT, Kutchma AJ, Becher A, Schweizer HP. Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid. 2000;43:59–72. doi: 10.1006/plas.1999.1441. [DOI] [PubMed] [Google Scholar]
- Hood RD, Singh P, Hsu F, Guvener T, Carl MA, Trinidad RR, Silverman JM, Ohlson BB, Hicks KG, Plemel RL, et al. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell host & microbe. 2010;7:25–37. doi: 10.1016/j.chom.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izore T, Job V, Dessen A. Biogenesis, regulation, and targeting of the type III secretion system. Structure. 2011;19:603–612. doi: 10.1016/j.str.2011.03.015. [DOI] [PubMed] [Google Scholar]
- Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, Will O, Kaul R, Raymond C, Levy R, et al. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2003;100:14339–14344. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobichen C, Chakraborty S, Li M, Zheng J, Joseph L, Mok YK, Leung KY, Sivaraman J. Structural basis for the secretion of EvpC: a key type VI secretion system protein from Edwardsiella tarda. PLoS One. 2010;5:e12910. doi: 10.1371/journal.pone.0012910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamaru S. Structural similarity of tailed phages and pathogenic bacterial secretion systems. Proc Natl Acad Sci U S A. 2009;106:4067–4068. doi: 10.1073/pnas.0901205106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitein N, Mogk A. Deadly syringes: type VI secretion system activities in pathogenicity and interbacterial competition. Current opinion in microbiology. 2013 doi: 10.1016/j.mib.2012.11.009. [DOI] [PubMed] [Google Scholar]
- Koskiniemi S, Lamoureux JG, Nikolakakis KC, t’Kint de Roodenbeke C, Kaplan MD, Low DA, Hayes CS. Rhs proteins from diverse bacteria mediate intercellular competition. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:7032–7037. doi: 10.1073/pnas.1300627110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiman PG, Basler M, Ramagopal UA, Bonanno JB, Sauder JM, Pukatzki S, Burley SK, Almo SC, Mekalanos JJ. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc Natl Acad Sci U S A. 2009;106:4154–4159. doi: 10.1073/pnas.0813360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroux M, De Leon JA, Kuwada NJ, Russell AB, Pinto-Santini D, Hood RD, Agnello DM, Robertson SM, Wiggins PA, Mougous JD. Quantitative single-cell characterization of bacterial interactions reveals type VI secretion is a double-edged sword. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:19804–19809. doi: 10.1073/pnas.1213963109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Le Trong I, Carl MA, Larson ET, Chou S, De Leon JA, Dove SL, Stenkamp RE, Mougous JD. Structural basis for type VI secretion effector recognition by a cognate immunity protein. PLoS Pathog. 2012;8:e1002613. doi: 10.1371/journal.ppat.1002613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JS, Ma LS, Lai EM. Systematic dissection of the agrobacterium type VI secretion system reveals machinery and secreted components for subcomplex formation. PLoS One. 2013;8:e67647. doi: 10.1371/journal.pone.0067647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinness KE, Baker TA, Sauer RT. Engineering controllable protein degradation. Molecular cell. 2006;22:701–707. doi: 10.1016/j.molcel.2006.04.027. [DOI] [PubMed] [Google Scholar]
- Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, Goodman AL, Joachimiak G, Ordonez CL, Lory S, et al. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science. 2006;312:1526–1530. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi M, Li Y, Cheng Y, Walz T. Negative Staining and Image Classification - Powerful Tools in Modern Electron Microscopy. Biol Proced Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osipiuk J, Xu X, Cui H, Savchenko A, Edwards A, Joachimiak A. Crystal structure of secretory protein Hcp3 from Pseudomonas aeruginosa. J Struct Funct Genomics. 2011;12:21–26. doi: 10.1007/s10969-011-9107-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parret AH, De Mot R. Escherichia coli’s uropathogenic-specific protein: a bacteriocin promoting infectivity? Microbiology (Reading, England) 2002;148:1604–1606. doi: 10.1099/00221287-148-6-1604. [DOI] [PubMed] [Google Scholar]
- Paulsen IT, Press CM, Ravel J, Kobayashi DY, Myers GS, Mavrodi DV, DeBoy RT, Seshadri R, Ren Q, Madupu R, et al. Complete genome sequence of the plant commensal Pseudomonas fluorescens Pf-5. Nat Biotechnol. 2005;23:873–878. doi: 10.1038/nbt1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieper R, Huang ST, Robinson JM, Clark DJ, Alami H, Parmar PP, Perry RD, Fleischmann RD, Peterson SN. Temperature and growth phase influence the outer-membrane proteome and the expression of a type VI secretion system in Yersinia pestis. Microbiology (Reading, England) 2009;155:498–512. doi: 10.1099/mic.0.022160-0. [DOI] [PubMed] [Google Scholar]
- Pukatzki S, Ma AT, Revel AT, Sturtevant D, Mekalanos JJ. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc Natl Acad Sci U S A. 2007;104:15508–15513. doi: 10.1073/pnas.0706532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC, Heidelberg JF, Mekalanos JJ. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci U S A. 2006;103:1528–1533. doi: 10.1073/pnas.0510322103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell AB, Hood RD, Bui NK, LeRoux M, Vollmer W, Mougous JD. Type VI secretion delivers bacteriolytic effectors to target cells. Nature. 2011;475:343–347. doi: 10.1038/nature10244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell AB, LeRoux M, Hathazi K, Agnello DM, Ishikawa T, Wiggins PA, Wai SN, Mougous JD. Diverse type VI secretion phospholipases are functionally plastic antibacterial effectors. Nature. 2013;496:508–512. doi: 10.1038/nature12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell AB, Singh P, Brittnacher M, Bui NK, Hood RD, Carl MA, Agnello DM, Schwarz S, Goodlett DR, Vollmer W, et al. A widespread bacterial type VI secretion effector superfamily identified using a heuristic approach. Cell host & microbe. 2012;11:538–549. doi: 10.1016/j.chom.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JM, Austin LS, Hsu F, Hicks KG, Hood RD, Mougous JD. Separate inputs modulate phosphorylation-dependent and -independent type VI secretion activation. Mol Microbiol. 2011;82:1277–1290. doi: 10.1111/j.1365-2958.2011.07889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JM, Brunet YR, Cascales E, Mougous JD. Structure and Regulation of the Type VI Secretion System. Annual review of microbiology. 2012;66:453–473. doi: 10.1146/annurev-micro-121809-151619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins CE, Galan JE. Maintenance of an unfolded polypeptide by a cognate chaperone in bacterial type III secretion. Nature. 2001;414:77–81. doi: 10.1038/35102073. [DOI] [PubMed] [Google Scholar]
- Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M, et al. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- Suarez G, Sierra JC, Erova TE, Sha J, Horneman AJ, Chopra AK. A type VI secretion system effector protein VgrG1 from Aeromonas hydrophila that induces host cell toxicity by ADP-ribosylation of actin. J Bacteriol. 2010;192:155–168. doi: 10.1128/JB.01260-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland MC, Nguyen TL, Tseng V, Vogel JP. The Legionella IcmSW complex directly interacts with DotL to mediate translocation of adaptor-dependent substrates. PLoS pathogens. 2012;8:e1002910. doi: 10.1371/journal.ppat.1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner C, Polke M, Gerlach RG, Linke D, Stierhof YD, Schwarz H, Hensel M. Functional dissection of SiiE, a giant non-fimbrial adhesin of Salmonella enterica. Cellular microbiology. 2011;13:1286–1301. doi: 10.1111/j.1462-5822.2011.01621.x. [DOI] [PubMed] [Google Scholar]
- Wu HY, Chung PC, Shih HW, Wen SR, Lai EM. Secretome analysis uncovers an Hcp-family protein secreted via a type VI secretion system in Agrobacterium tumefaciens. J Bacteriol. 2008;190:2841–2850. doi: 10.1128/JB.01775-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Gao ZQ, Wang WJ, Liu GF, Xu JH, Su XD, Dong YH. Structure of the type VI effector-immunity complex (Tae4-Tai4) provides novel insights into the inhibition mechanism of the effector by its immunity protein. The Journal of biological chemistry. 2013;288:5928–5939. doi: 10.1074/jbc.M112.434357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Leung KY. Dissection of a type VI secretion system in Edwardsiella tarda. Mol Microbiol. 2007;66:1192–1206. doi: 10.1111/j.1365-2958.2007.05993.x. [DOI] [PubMed] [Google Scholar]
- Zou T, Yao X, Qin B, Zhang M, Cai L, Shang W, Svergun DI, Wang M, Cui S, Jin Q. Crystal structure of Pseudomonas aeruginosa Tsi2 reveals a stably folded superhelical antitoxin. Journal of molecular biology. 2012;417:351–361. doi: 10.1016/j.jmb.2012.01.040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.