Abstract
Cell-mediated immunity critically depends on lymphocyte localization at sites of infection. While some memory T cells recirculate, a distinct lineage (resident memory T cells; TRM) are embedded in non-lymphoid tissues (NLTs) and mediate potent protective immunity. However, the defining transcriptional basis for TRM establishment is unknown. We report that CD8+ TRM cells lacked expression of the transcription factor KLF2 and its target gene S1pr1 (encoding sphingosine 1-phosphate receptor 1). Forced S1PR1 expression prevented establishment of TRM. Cytokines inducing TRM phenotype (including TGF-β, IL-33 and TNF) provoked KLF2 downregulation in a phosphatidylinositol-3-OH kinase (PI(3)K)–Akt-dependent pathway, suggesting environmental regulation. Hence KLF2 and S1PR1 regulation provides a switch, dictating whether CD8+ T cells commit to the recirculating or tissue resident memory populations.
Introduction
During an immune response, antigen-specific T cells undergo massive clonal expansion, contribute to antigen clearance and then generate a memory population capable of more rapid and efficient recall responses. An important feature of memory T cells is their altered trafficking capacity which allows them (but not naïve T cells) to survey non-lymphoid tissues (NLTs)1, 2. It has become clear that a subset of memory CD8+ T cells, TRM, do not recirculate through the body, but are instead maintained in diverse NLTs (including the small intestine, brain, salivary glands, skin and female reproductive tract)3–9. TRM cells have been shown to provide superior protection (compared to circulating memory cells) against local secondary infections5–10, and TRM cells are now recognized as critical sentinels for protective immunity11–15.
However, an essential and unresolved question is the mechanism through which TRM residency becomes established11–14. For some NLTs, TRM cell expression of integrin CD103 (or its ligand, E-cadherin) contributes to TRM maintenance5, 16. However, these molecules are not expressed by TRM cells in all NLTs5, 16, suggesting such interactions do not constitute a universal mechanism for TRM retention. Indeed, while CD103 was required for maintenance of TRM cells in the small intestinal intraepithelial lymphocyte (IEL) population, it was found to be dispensable for memory cell establishment in the lamina propria lymphocyte (LPL) population of the same organ16.
A more consistent marker for TRM populations from multiple NLTs is expression of CD69 (refs. 13, 16). CD69 upregulation is often correlated with T cell receptor (TCR) stimulation – yet foreign antigen persistence is dispensable for establishment and/or maintenance of TRM in various NLTs8, 16. Hence the factors that promote residency of TRM remain ill-defined, and nothing is known about the transcriptional regulation that distinguishes cells committing to the recirculating versus resident populations.
Kruppel-like factor 2 (KLF2) is a zinc-finger transcription factor that directly promotes expression of the genes encoding sphingosine-1 phosphate receptor 1 (S1PR1) and L-selectin (CD62L), two molecules that are critical for naïve T cell recirculation17, 18. S1PR1, through detection of its ligand S1P in the blood and lymph, is essential for naïve lymphocytes to access the circulatory system from the thymus and lymph nodes19. Consequently, deficiency in KLF2 (ref. 17) or S1PR1 (ref. 19) causes retention of naïve T cells in lymphoid tissues. TCR stimulation induces rapid loss of KLF2 (and S1PR1), providing a mechanism for initial retention of activated T cells in lymphoid tissues, while these molecules are re-expressed in memory CD8+ T cells isolated from lymphoid tissues19–22. However, potential heterogeneity in KLF2 and S1PR1 expression by distinct memory T cell subsets (including TRM cells) has not been investigated. In this study, we show that CD8+ TRM cells in NLTs were characterized by low expression of KLF2 and S1PR1, and that transcriptional downregulation of S1PR1 was critical for the establishment of this resident memory pool.
Results
KLF2 is downregulated in CD8+ T cells found in NLTs
While KLF2 is expressed in bulk naïve and memory CD8+ T cell populations20, 21, it was unclear whether distinct memory subsets differed in KLF2 expression. To test this, we utilized mice in which gfp (encoding green fluorescent protein, or GFP) was knocked into the endogenous Klf2 gene, creating a functional GFP-KLF2 fusion protein (KLF2GFP) as a reporter for KLF2 expression23. Similarly abundant KLF2GFP expression was observed in bulk splenic CD62L+ (central memory) and CD62L− (effector memory) memory-phenotype CD8+ T cells (Fig. 1a). Thus, despite the fact that KLF2 promotes transcription of Sell (the gene encoding CD62L)17, 18, KLF2 expression alone does not accurately predict active Sell transcription. The KLF2GFP gene was also crossed with P14 TCR-transgenic cells (which recognize the Db restricted epitope gp33–41 epitope [sequence KAVYNFATC], derived from LCMV GP). KLF2GFP P14 CD8+ T cells were adoptively transferred and primed by infection with lymphocytic choriomeningitis virus (LCMV) Armstrong strain, and normal KLF2GFP transgenic animals were infected with LCMV in parallel. At memory-stage, both KLF2GFP P14 CD8+ T cells and polyclonal gp33–41/Db specific KLF2GFP CD8+ T cells showed uniformly high KLF2 expression in spleen and lymph nodes (Fig. 1b). In contrast, however, memory cells in the small intestinal LPL and IEL populations showed KLF2 downregulation (Fig. 1b). These findings complement our earlier findings on gut mucosal T cells of undefined specificity and activation status in unimmunized mice24.
Figure 1. Differential KLF2 and S1PR1 expression by memory CD8+ T cells in lymphoid and non-lymphoid tissues.
(a) KLF2GFP expression in splenic memory-phenotype CD8+ T cells (representative of n=9 from 3 independent experiments). (b) Expression of KLF2GFP in antigen-specific CD8+ T cells (endogenous Db/gp33 tetramer+ or adoptively transferred P14 CD8+ T cells) from indicated tissues, >28 days after LCMV infection. Data show fluorescence of KLF2GFP CD8+ T cells (black line) overlaid on CD8+ T cells from matched WT controls (grey filled). Data are representative of 6–12 animals from at least 3 independent experiments. (c, d) Analysis of KLF2GFP expression in adoptively transferred P14 CD8+ T cells, >28 days post LCMV infection. (c) IV anti-CD8 antibody was used to distinguish P14 T cells in tissue parenchyma (solid line) versus vascular-associated cells (dashed line). Data are overlaid with WT P14 CD8+ T cells (grey filled) as controls (representative of n=9 from 3 independent experiments). (d) CD69 versus KLF2GFP expression for parenchymal P14 T cells within indicated tissues (representative of n=9 from 3 independent experiments). (e) RNA was isolated from sorted P14 CD8+ T cells, isolated from spleen, salivary glands and LPL 30 days post LCMV infection, and subjected to RT-PCR for the indicated genes. Gene expression (relative to HPRT control) was normalized to the spleen for comparison between experiments. Compiled from four independent experiments (9–12 pooled mice each experiment), graphs show mean +/− SD. (f) Activated P14 CD8+ T cells were transduced with retroviral vectors encoding KLF2 (or a control, “Empty” vector). Two days later, cells expressing the transduction marker (Thy-1.1) were enriched and subjected to RT-PCR for the indicated genes. Gene expression, relative to HPRT controls, was normalized on empty vector controls. Data are compiled from 3 separate transduction experiments. At the same time point, cell surface expression of CD69 was determined on Thy-1.1+ve transduced cells (inset) - data shown are representative of at least 3 transduction experiments. Statistical analysis utilized one-way ANOVA (with Dunnett’s Multiple Comparison Test in (e)) and significance is indicated as follows: ***, p<0.001; **, p<0.01; *, p<0.05; NS, p>0.05.
We extended this analysis to other NLTs, assaying KLF2GFP P14 memory CD8+ T cells from salivary gland, kidney and brain. Since blood lymphocyte contamination can complicate identification of cells in the parenchyma of certain tissues25, we administered intravenous CD8-specific antibody to label (and exclude) vascular-associated cells prior to tissue isolation. Parenchymal KLF2GFP P14 T cells in salivary gland, kidney and brain of mice infected with LCMV 8 or 30 days earlier showed KLF2 downregulation, while cells in the LN and blood, as well as NLT vascular-associated cells, displayed abundant KLF2GFP expression (Fig. 1c, Supplementary Fig. 1a). CD69 expression was evident for parenchymal T cells in NLTs (Supplementary Fig. 1b), consistent with typical TRM cell phenotype3–5, 26. Accordingly, CD69 and KLF2 expression showed a tight inverse relationship (Fig. 1d).
Low KLF2GFP expression might arise from induced KLF2 protein degradation21, 27, yet real-time RT-PCR analysis showed significantly less Klf2 mRNA expression in antigen-primed memory P14 CD8+ T cells sorted from NLTs (LPLs and salivary gland) compared to the spleen (Fig. 1e). Furthermore, expression of the KLF2 targets S1pr1 and Sell (encoding CD62L) was also severely reduced in the NLT populations (Fig. 1e). Detecting cell surface S1PR1 expression on CD8+ T cells is difficult with current reagents, precluding accurate comparison between tissues (data not shown). However, previous studies showed that S1PR1 deficiency leads to cell surface expression of CD69 (due to the loss of competitive protein-protein interactions between CD69 and S1PR1; refs. 28, 29): hence the finding that TRM cells are uniformly CD69+ might correlate with reduced S1pr1 expression. Indeed, despite substantial differences in cell surface CD69 (Supplementary Fig. 1b), Cd69 transcript abundance was not significantly different among memory P14 cells from spleen, salivary glands and LPLs (Fig. 1e), although there was a trend of moderately increased Cd69 mRNA expression in cells from the LPLs.
To further explore how KLF2 influences CD69 expression, we evaluated the impact of forced KLF2 expression using transduction of activated P14 CD8+ T cells with a retroviral vector encoding mouse KLF2 (or a control, “empty” vector). As anticipated17, 18, ectopic KLF2 expression increased mRNA abundance for S1pr1 and Sell, but not Cd8a (Fig. 1f). Forced KLF2 expression also caused a modest increase in Cd69 mRNA abundance – a result that strikingly contrasts with the reduction in cell surface expression of CD69 protein (Fig. 1f). These findings show that KLF2 does not repress CD69 transcription, and support the model that elevated CD69 protein surface expression in KLF2lo cells is a consequence of reduced S1PR1 expression. Taken together, our data suggest that TRM cells in NLTs are distinguished from their recirculating counterparts by low expression of KLF2 and its transcriptional targets, including S1PR1.
KLF2 downregulation occurs during TRM establishment
KLF2 is downregulated following TCR activation, but is re-expressed in the effector phase of the T cell response20, 21, which is the same stage at which antigen-specific CD8+ T cells become established in NLTs11–14, 30. Hence it was important to define the kinetics of changes in T cell KLF2 expression during the immune response. Congenically distinct KLF2GFP transgenic and wild-type P14 CD8+ T cells were co-transferred into recipient mice and analyzed throughout the effector and memory response provoked by LCMV infection. KLF2 expression dropped precipitously in responding P14 cells at 48h post-infection (PI) (Fig. 2a), but in lymphoid tissues and blood, KLF2 expression was fully recovered by 5d PI and maintained into the memory phase (Fig. 2b, c). In stark contrast, P14 cells from the parenchyma of NLTs showed variable KLF2 expression at d5 PI, but were KLF2lo in all NLTs from 8d PI (Fig. 2b, c, Supplementary Fig. 2). This timeframe is synchronous with the establishment of TRM cells, as shown by the peak of P14 cell numbers accumulating in lymphoid and NLTs (Fig. 2d). Such data are consistent with a model in which CD8+ T cells enter the NLTs (from blood) as a KLF2hi population, and downregulate KLF2 while establishing the TRM pool. KLF2 expression declined with similar kinetics in CD8+ T cells from all NLTs studied in contrast to much more variability in CD103 upregulation, which was rapid and prominent in some NLTs but barely detected in others during this timecourse (Fig. 2c)16. The rapid termination of KLF2 expression in CD8+ T cells while settling in NLTs, together with the loss of trafficking molecules regulated by KLF2 (Fig. 1e), suggests that KLF2 downregulation may be important in establishment of TRM at various sites.
Figure 2. KLF2 is downregulated during CD8+ T cell seeding of non-lymphoid tissues.
Congenically distinct WT P14 and KLF2GFP P14 CD8+ T cells were adoptively co-transferred, and host mice infected with LCMV. (a, b) Histograms show fluorescence of KLF2GFP P14 CD8+ T cells (black line) and P14 CD8+ T cells (grey filled) for (a) cells analyzed before transfer (“naïve”) and from spleen/LN at 2 days following LCMV infection, and (b) cells from indicated tissues at listed time points following LCMV infection. Data are representative of n=9 animals from 3 independent experiments. (c) Percentage of CD103+ and KLF2GFP+ P14 CD8+ T cells at the indicated time points in lymphoid tissues and blood (left graphs, open symbols) and in NLTs (right graphs, closed symbols). Data are compiled from 3 independent experiments (n=9 animals) and graphs show mean +/− SD. Dotted line indicates threshold for detecting KLF2GFP+ cells, based on analysis of WT P14 T cells. (d) Absolute number of P14 CD8+ T cells isolated from indicated tissues at specified timepoints following LCMV infection. Data are compiled from a minimum of 15 animals (derived from a minimum of 4 experiments) with the exception of the brain (day 5; 12 animals from 4 experiments and day 30; 9 animals from 3 experiments). Data included cell numbers from empty-vector transduced P14 CD8+ T cells, discussed in Figure 4. Graphs show mean +/− SEM.
KLF2 expression in TRM and recirculating CD8+ T cells
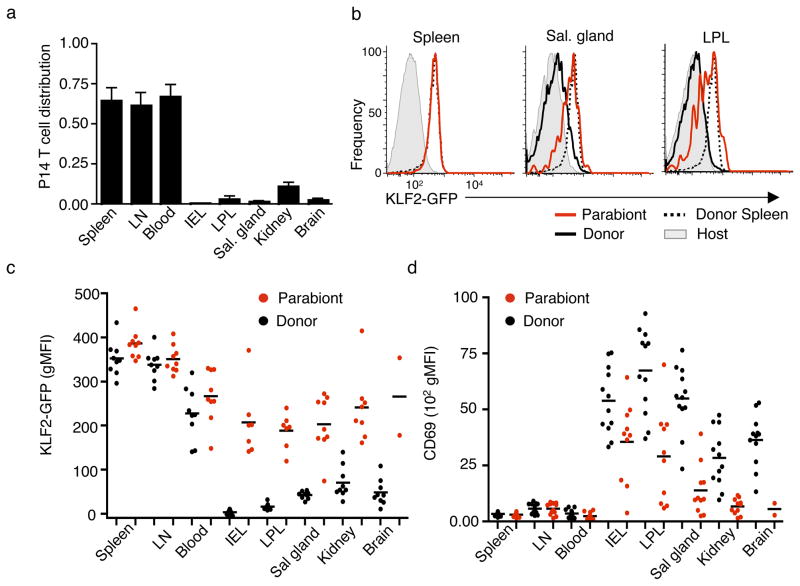
Some recirculating memory CD8+ T cells – the classically defined effector-memory subset31 – would be expected to traffic through NLTs, and we sought to analyze KLF2 expression by this population compared to the non-recirculating TRM cells. Hence, we conducted parabiosis experiments in which mice containing memory KLF2GFP P14 T cells were surgically joined to infection-matched partners, leading to shared circulation and permitting recirculating memory cells to access NLTs (Supplementary Fig. 3)2.
As expected (based on previous studies2, 7, 9), memory P14 CD8+ T cells were well represented in lymphoid tissues of the parabiont, but were scarce in multiple NLTs (Fig. 3a). This finding reinforces the concept that the majority of antigen-primed memory CD8+ T cells in non-lymphoid sites are TRM cells, but also allowed us to interrogate KLF2 and CD69 expression on the rare population of recirculating effector memory cells that reach the NLT parenchyma. Interestingly, such cells exhibited intermediate expression of KLF2 and CD69, differing from the KLF2lo, CD69hi phenotype of TRM cells (which are prominent in the “donor” animal in the parabiotic pair) (Fig. 3b–d). Hence, the small fraction of CD8+ T cells that enter the NLTs from the blood at memory time points (chiefly recirculating effector memory cells) display elevated KLF2 expression compared to the established TRM population. Such recirculating cells may correspond to the small population of KLF2+ memory CD8+ T cells observed within NLT in the steady state.
Figure 3. Distinct KLF2GFP expression by recirculating memory CD8+ T cells in NLT.
(a–d) Parabiotic mice were generated (Supplementary Figure 3), in which animals bearing memory KLF2GFP P14 cells (induced by LCMV infection) were conjoined with infection-matched animals. At 13–17 days after surgery, paired animals were sacrificed and tissues harvested. The mice originally carrying the KLF2GFP P14 CD8+ T cell population were termed the “Donor” while the other animal in each parabiotic pair was termed the “Parabiont”. Data were compiled from 5 independent experiments (n=12 pairs), and gating was on live, non-vascular-associated P14 CD8+ T cells. (a) Relative abundance of P14 T cells in indicated tissues of donor and parabiont. This is shown as the percent of P14 T cells among total non-vascular-associated CD8+ cells in the parabiont, divided by the percent P14 T cells among total non-vascular-associated CD8+ cells in the donor. (b) KLF2GFP expression in spleen, salivary gland and LPL from parabiotic pairs. Black lines show GFP fluorescence of P14 KLF2GFP CD8+ T cells isolated from the donor (black line) or parabiont (red line). Donor spleen P14 KLF2GFP CD8+ T cells (dashed line) is included in all graphs for reference. Grey histograms show background fluorescence (host CD8+ T cells). (c and d) Geometric MFI (gMFI) for KLF2GFP (c) or CD69 (d) on P14 CD8+ T cells isolated from parabiont (red symbols) or donor (black symbols) animals. In (c), the background gMFI (of host CD8+ T cells) was subtracted. KLF2GFP and CD69 levels are not shown when <25 P14 T cells could be detected in a tissue.
Forced expression of S1PR1 inhibits TRM establishment
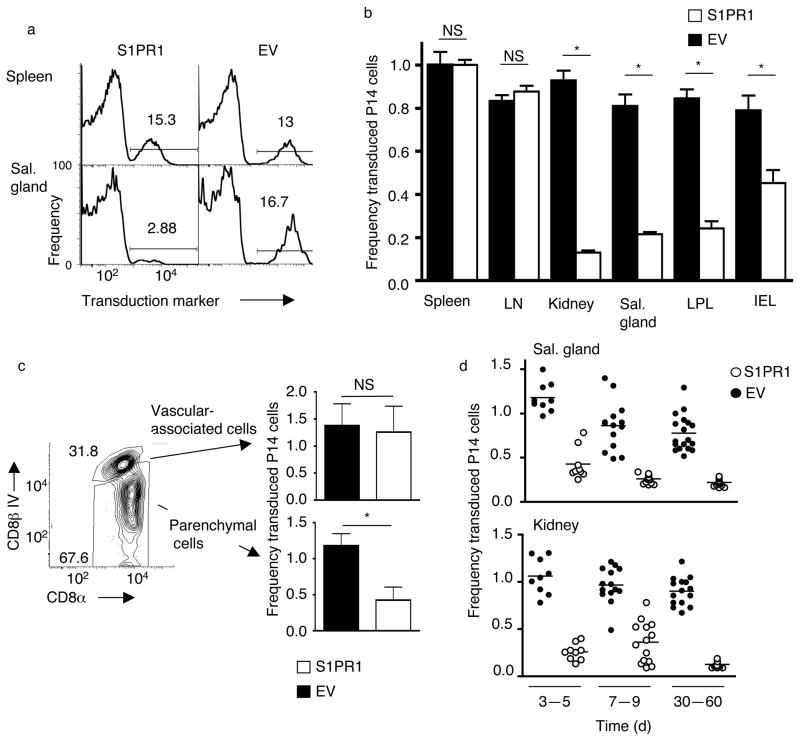
Although reduced expression of KLF2 (and its targets) correlates with formation of TRM cells, the mechanistic relevance of these changes was unclear. Of KLF2 transcriptional targets, S1pr1 was a strong candidate for regulating TRM retention, given the well-documented role of S1PR1 in controlling T cell egress19,22. Hence we explored how forced S1PR1 expression impacted NLT residency. Activated P14 CD8+ T cells were transduced with retroviral vectors28, 32 encoding S1PR1 or the transduction marker (Thy-1.1) alone (“empty-vector”) (Supplementary Fig. 4a), and co-transferred into recipients that were subsequently infected with LCMV. Forced S1PR1 expression was verified in transduced cells maintained in vitro (Supplementary Fig. 4b). A potential caveat with this approach arises from studies on S1PR1-transgenic mice, which indicated that forced S1PR1 impairs T cell expansion following TCR activation33. However, this effect was not observed in cultures of retrovirally transduced P14 cells (Supplementary Fig. 4c, d).
While the frequency of S1PR1 and empty-vector transduced P14 cells was similar in lymphoid tissues, the frequency of S1PR1-transduced P14 cells was considerably lower in NLTs (Fig. 4a). Percentages of S1PR1 and control-transduced P14 cells for each tissue were plotted relative to those present in the spleen in order to normalize between experiments (Supplementary Fig. 5a). S1PR1 transduction had little impact in lymphoid tissues, but profoundly reduced the representation of P14 T cells in NLTs (Fig. 4b). This result was observed in NLT parenchyma but not the corresponding vascular-associated population (Fig. 4c), ruling out effects of tissue processing on this result. The reduction in NLT S1PR1-transduced P14 T cells was observed from early timepoints (3–5 days: Fig. 4d), suggesting that forced S1PR1 prevents initial formation, or “settling” of the NLT resident pool. Analysis of absolute numbers of transduced P14 cells validated this conclusion, with S1PR1 transduction having minimal impact on recovery from lymphoid sites, while leading to a substantial reduction in the population isolated from multiple NLTs (Supplementary Fig. 5b, c).
Figure 4. Forced S1PR1 prevents establishment of TRM.
Activated P14 CD8+T cells were transduced with retroviral vectors encoding S1PR1 and the transduction marker Thy-1.1 (“S1PR1”) or Thy-1.1 alone (Empty vector; “EV”) (see Supplementary Fig. 4a). Congenically distinct S1PR1 and empty-vector transduced cells were co-transferred into recipients subsequently infected with LCMV. (a) Histograms (representative of n=12 from 4 independent experiments) showing the frequency of cells expressing the transduction marker from the indicated retroviral vector, for P14 CD8+ T cells cells isolated from spleen and salivary gland >30 days post LCMV infection. (b) Transduction frequency of P14 CD8+ T cells, transduced with empty vector (black bars) or S1PR1 vector (white bars) in the parenchyma of the indicated tissues, 28–60 days post LCMV. Data are normalized to the percent transduction for the spleen from the same animal. Graphs show mean +/− SEM for 11–18 samples per group, compiled from at least 4 independent experiments. (c) Relative transduction for S1PR1 (white bars) or empty vector (black bars) transduced P14 CD8+ T cells present in vascular-associated versus tissue parenchyma of the salivary gland, isolated 5 days after LCMV infection. Bar graphs are compiled from 3 independent experiments (n=9). (d) Frequency of transduction (relative to spleen) of empty vector (black) versus S1PR1 (white) transduced P14 cells from kidney and salivary gland, at indicated time points following LCMV infection. N=9–18 from at least 3 independent experiments. Similar time-course trends were observed for other NLTs (data not shown). In all panels, analysis gated on live P14 CD8+ T cells, and (except panel c) with exclusion of vascular-associated cells. Statistical significance in this figure is indicated as follows: *, p<0.001; NS, p>0.05.
The impact of S1PR1 transduction on IEL cells was more variable (albeit still significant) compared to other tissues (Supplementary Fig. 6a) and since CD103 is important for TRM cell maintenance in this site16, we considered whether CD103 upregulation might override S1PR1 in promoting residency in other NLTs. If this occurred, we might expect to find that the small S1PR1-transduced population present in other NLTs exhibited faster and/or more efficient induction of CD103, compared to the empty-vector control. However, CD103 induction profiles were similar in both groups (Supplementary Fig. 6b), arguing against this model.
Our conclusions contrast with studies suggesting S1PR1 induces T cell tissue residency34. However, that report examined migration of naïve T cells (and chiefly focused on CD4+ T cells) after footpad injection, while our studies track physiological trafficking of activated CD8+ T cells during an immune response. Such differences might account for the discrepancy in these findings.
S1PR1 is well defined at promoting lymphocyte egress19, 22, but it was possible that it served to limit entry into NLTs in our studies. To explore this issue we used a distinct model system, permitting us to more tightly synchronize CD8+ T cell entry into NLTs. Skin inflammation induced by dinitrofluorobenzene (DNFB) leads to acute recruitment of in vitro-activated effector CD8+ T cells, and local generation of TRM cells8. Consistent with our data in other NLTs, activated P14 cells present in DNFB-treated skin downregulated KLF2GFP (Fig. 5a). We next used in vitro-activated P14 cells transduced with S1PR1 or empty vectors and cultured to allow time for S1PR1 expression, prior to adoptive transfer into animals subsequently treated with DNFB. S1PR1 and empty-vector transduced cells entered DNFB-treated skin in similar numbers at day 2 after transfer, suggesting no effect of S1PR1 expression on entry to the inflamed site (Fig. 5b). However, during successive days the numbers of S1PR1-transduced cells declined significantly in the skin but not the spleen (Fig. 5b). This effect was also observed by monitoring the frequency of S1PR1 versus EV transduced cells in spleen and skin (Fig. 5c). These results are consistent with forced S1PR1 expression driving egress from the inflamed skin. Taken together, these data suggest that sustained S1PR1 expression is incompatible with formation of the TRM population.
Figure 5. Accumulation of activated CD8+ T cells in inflamed skin is dependent on S1PR1 downregulation.
(a) In vitro activated effector P14 and KLF2GFP P14 were co-transferred into recipients, which were subsequently treated with DNFB on the flank skin. After 5 days, the KLF2GFP gMFI was measured in indicated tissues (and corrected by subtracting the background for WT P14). Data are compiled from 3 independent experiments (n=10) and show mean +/− SEM. (b–c) Activated P14 CD8+ T cells transduced with S1PR1 (white symbols) and empty (black symbols) vectors were co-transferred into recipients that were subsequently treated with DNFB on the flank skin. At the indicated days post DNFB treatment, the number of transduced P14 CD8+ T cells in skin and spleen (b), and the percentage of transduced cells (normalized to the percent transduction in the spleen from the same animal) (c) was calculated. In (b), P14 T cell numbers are shown for both DNFB treated and untreated contralateral (“ctrl”) flank skin. Data are compiled from 3 independent experiments, with n=9. (d) Congenically distinct P14 CD8+ T cells were activated in vitro and transduced with retroviruses encoding KLF2 or an empty vector. Transduced cells were co-transferred into congenic recipients treated with DNFB as in (b, c). At the indicated time points, the frequency of each transduced donor population was determined in the treated skin. Data are compiled from 3 independent experiments (n=9). Statistical significance is indicated (**, p<0.001; *, p<0.05; NS, p>0.05).
We extended these studies to test the impact of forced KLF2 expression on generation of tissue-resident cells. Activated P14 cells were transduced with retroviral vectors encoding KLF2 (or a control, “empty” vector) and adoptively transferred into animals that were subsequently treated with DNFB. Transduction with the retrovirus encoding KLF2 led to an increase in mRNA for S1PR1 (and CD62L) (Fig. 1f), suggesting we had achieved adequate expression of KLF2 to impact target gene transcription. Similar to forced S1PR1, ectopic KLF2 expression had no effect on early accumulation of donor T cells into the inflamed skin (Fig. 5d: day 2), but did lead to a significant decline in the maintenance of this population at later time points (Fig. 5d: day 8). While the pleotropic effects of overexpressed KLF2 (ref. 35) could complicate interpretation of these experiments, the similar effects of forced S1PR1 and KLF2 supports the model that reduced expression of KLF2, and a subsequent decrease in S1PR1, are important steps in establishment of the tissue-resident T cell pool.
KLF2 downregulation in NLT is not due to TCR stimulation
The loss of KLF2 during TRM generation indicates a basis for impaired S1PR1 expression – but what signals induce downregulation of KLF2 in NLT? TCR stimulation is well known to terminate KLF2 expression21, 27 and, even though our LCMV model system involves acute infection (with the Armstrong strain), it was possible TCR engagement persists in NLTs.
First, we tested whether KLF2 downregulation was induced in a situation where no cognate foreign antigen was present in vivo. KLF2GFP P14 CD8+ T cells were stimulated with LCMV gp33–41 peptide in vitro, cultured for 48 h and then transferred into uninfected recipient mice. KLF2GFP expression was assessed 12–15 days later. Although P14 CD8+ T cell migration to NLTs was not as robust as observed in LCMV-infected hosts (data not shown), similar patterns of KLF2GFP expression were detected – with cells in lymphoid sites retaining high expression of KLF2, while donor T cells in NLT were KLF2lo (Fig. 6a). Together with the finding that KLF2 was downregulated on P14 CD8+ T cells migrating into DNFB-treated skin, these data suggest that engagement with specific foreign peptide-MHC ligands is not required for KLF2 downregulation in the developing TRM population.
Figure 6. The KLF2low phenotype of TRM does not correlate with sustained TCR engagement.
(a) In vitro activated effector KLF2GFP P14 splenocytes were transferred into uninfected hosts, and 12–15 days later the KLF2GFP status of live non-vascular-associated P14 cells in spleen, blood and indicated NLTs were analyzed. N=8 from 3 independent experiments. The dotted line indicates threshold detection of KLF2GFP expressing cells (as in Fig. 2c). (b) To evaluate TCR activation status of CD8+ T cells in NLT, Nur77GFP P14 CD8+ T cells were transferred into C57BL/6 and either infected with LCMV or left uninfected (grey-filled). Thirty days after infection, Nur77GFP expression in P14 CD8+ T cells isolated from tissue parenchyma of indicated NLTs (black line) was compared to cells from the spleen (dotted line). Black filled histograms indicate Nur77GFP expression in P14 CD8+ T cells from animals injected with gp33–44 peptide 8 hours before harvest. Bar graph shows Nur77GFP expression as mean +/− SD, normalized to spleen for each group. Data are compiled from 4 independent experiments (n=11–12). Statistical analysis is relative to spleen. (**, p<0.001; *, p<0.05; NS, p>0.05).
It was possible, however, that TCR engagement with other cross-reactive foreign antigens (for example, from commensal microbes) or even self-peptide-MHC ligands might drive the KLF2lo phenotype of the TRM population. To investigate this hypothesis, we utilized Nur77GFP transgenic mice, which sensitively report TCR stimulation36. Nur77GFP expression on P14 memory cells in spleen and most NLTs (IEL, salivary gland and kidney) did not differ from that in naïve cells (Fig. 6b). Systemic peptide immunization provoked strong Nur77GFP induction in NLTs (Fig. 6b), suggesting the reporter was functional. We did observe modest (yet reproducible) increases in Nur77GFP expression in cells isolated from the LPL (echoing the mildly increased Cd69 transcripts in that population: Fig. 1e). Hence, with the possible exception of the small intestine-LPL following LCMV infection, our data suggest basal stimulation of TCRs are comparable in memory CD8+ T cells from both lymphoid and non-lymphoid sites, arguing against peptide-MHC engagement as a mechanism of KLF2 downregulation.
Cytokines induce KLF2 downregulation through P13K/AKT
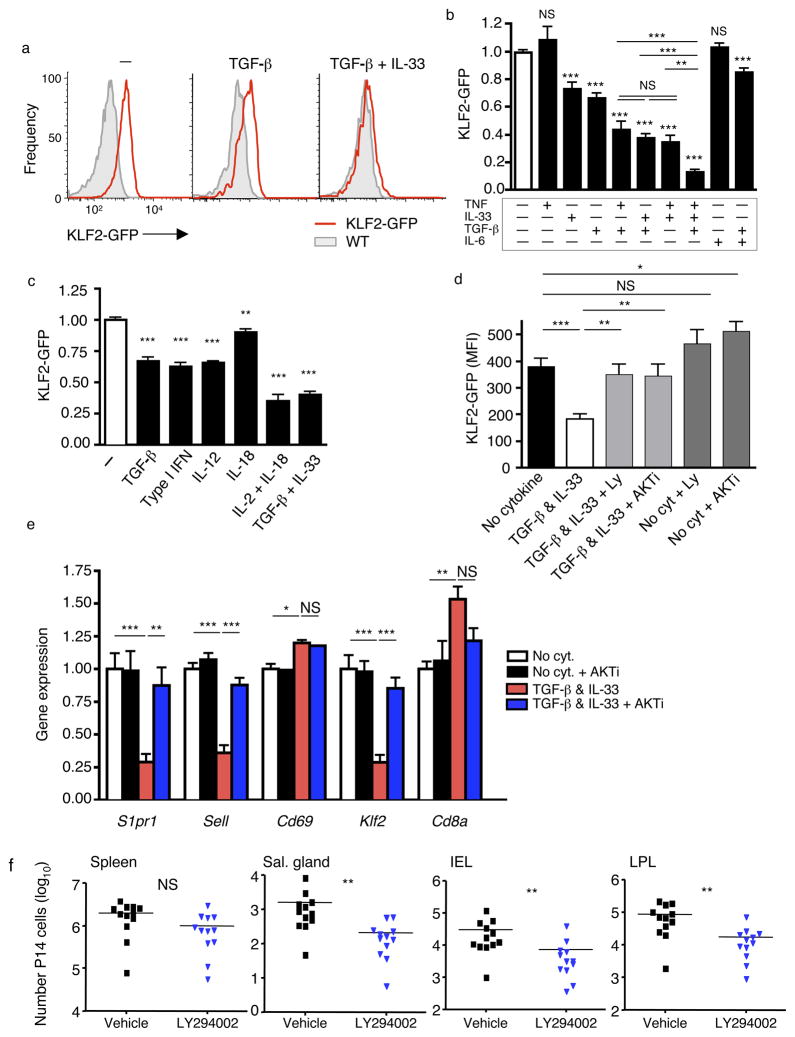
Cytokines can also induce KLF2 downregulation in activated CD8+ T cells18, 21, 37. Recent studies demonstrated that tumor growth factor-β (TGF-β) paired with various cytokines induce activated CD8+ T cells to acquire the CD69hi, CD103+ phenotype of IEL TRM cells16. Hence, we tested whether these cytokines also regulate KLF2GFP expression. Using in vivo-activated KLF2GFP P14 CD8+ T cells, we observed that culture with pairwise combinations of tumor necrosis factor (TNF), interleukin 33 (IL-33), and TGF-β cooperate to reduce KLF2GFP expression, while addition of all 3 cytokines leads to even more robust KLF2 loss (Fig. 7a, b). Kinetically, KLF2GFP downregulation was detectable within the first day of TGF-β and IL-33 exposure but was more substantial at 40 hours (Supplementary Fig. 7a). TGF-β and IL-6 (a cytokine combination that failed to drive a complete IEL TRM phenotype16) more modestly reduced KLF2GFP expression (Fig. 7b). IL-15 is important for memory CD8+ T cell survival38, and this cytokine induced KLF2 expression in activated CD8+ T cells, as expected37, 39 (Supplementary Fig. 7b). Yet exposure to IL-15 together with TGF-β and IL-33 led to KLF2 loss (Supplementary Fig. 7b), indicating that the downregulation pathway dominates.
Figure 7. Diverse cytokines induce KLF2 downregulation in activated CD8+ T cells, through a PI3K/Akt sensitive pathway.
(a–c) WT and KLF2GFP P14 CD8+ T cells were co-adoptively transferred and recipients infected with LCMV for 4.5 days. Cells were then cultured with indicated cytokines for 40 hrs. Representative histograms (a) show GFP fluorescence of KLF2GFP P14 cells (red) and WT P14 cells (grey filled), while the bar graphs (b, c) indicate compiled data (from at least 4 experiments; n=11–18 in (b) and 3 experiments; n=9 in (c)), normalized on cells cultured with no added cytokines. Data show mean +/− SEM, with statistical analysis relative to the no cytokine group (except where indicated by horizontal bars). (d) P14 T cells were activated and cultured as in (a–c) with or without addition of LY294002 (10uM) or AKTi (1uM), to inhibit PI3K and AKT, respectively (n=11–12 from 4 independent experiments). (e) WT and KLF2GFP P14 CD8+ T cells were activated in vitro for 48 hours and then cultured with cytokines and/or inhibitors as in (d). Bar graph shows indicated gene expression determined by RT-PCR of sorted P14 CD8+ T cells. Data are compiled from 4 independent experiments for all genes except CD69 (3 experiments for each group with the exception of “no cytokine + Akti” which shows data from 2 experiments for comparison). (f) WT and KLF2GFP P14 CD8+ T cells were co-transferred, and recipients were then infected with LCMV for 4 days. Animals were treated twice (12 hours apart) with 50mg/kg LY294002 (blue symbols) or vehicle only (black symbols), and the animals sacrificed 12 hours later. Total live non-vascular-associated P14 cells were determined for tissues indicated. Data are compiled from 5 independent experiments (n=12 animals). Statistical significance for all panels is indicated as follows: ***, p<0.001; **, p<0.01; *, p<0.05; NS, p>0.05.
We had shown previously that IL-12 could also provoke KLF2 loss21, and various combinations of cytokines have been shown to promote cell-surface CD69 expression40, so we extended these studies to other cytokines. Both Type-I IFN and IL-12 induced a substantial reduction in KLF2GFP expression, while IL-18 had a more modest (although still significant) effect (Fig. 7c). Interestingly, the combination of IL-12 with IL-18 (a signal that can induce effector functions in NK and CD8+ memory cells) led to substantial loss of KLF2GFP, comparable to the effects of TGF-β and IL-33 (Fig. 7c). Hence, these data reinforce the concept that multiple combinations of distinct cytokines can lead to the KLF2lo phenotype.
Klf2 transcription is induced by Foxo1, and the latter is negatively regulated by PI(3)K–Akt signaling41. Furthermore, IL-2 induced downregulation of KLF2 expression is dependent on PI(3)K–Akt activation37. Although TGF-β primarily signals through the Smad pathway, it also activates PI(3)K in some situations42. Indeed, we found that the capacity of TGF-β and IL-33 to induce loss of KLF2GFP expression was substantially reversed by blockade of either PI(3)K or Akt (Fig. 7d, Supplementary Fig. 7c). Similar effects were observed for in vitro-activated P14 CD8+ T cells (Supplementary Fig. 7d), minimizing concerns that the cytokines (or inhibitors) mediate their effects through other cell types.
To test the mechanism of cytokine-induced KLF2 downregulation, we performed RT-PCR analysis on in vitro-activated cells. These studies revealed TGF-β and IL-33 induced transcriptional downregulation of Klf2 and its targets S1pr1 and Sell, but did not reduce Cd69 or Cd8a transcription (Fig. 7e). Furthermore, this pattern was reversed by Akt inhibition (Fig. 7e). Hence, TGF-β and IL-33, in a PI(3)K-Akt dependent fashion, induce changes in gene expression similar to those observed in TRM cells. Whether these cytokines directly activate PI(3)K-Akt or act through intermediate factors is, however, not clear. Interestingly, CD103 induction by TGF-β–IL-33 treatment was unaffected by PI(3)K or Akt inhibitors (Supplementary Fig. 7e), suggesting that the signaling pathways controlling CD103 and KLF2 expression are distinct.
We next sought to explore the pathways regulating KLF2 expression in vivo. Our data showed that multiple cytokines were capable of inducing KLF2 loss in activated CD8+ T cells, making it unlikely that individual cytokines (or pairs of cytokines) would play a non-redundant role in KLF2 regulation and thus limiting the utility of cytokine blockade or gene knockout approaches. However, the finding that PI(3)K signaling appeared crucial for cytokine induced KLF2 downregulation prompted us to test whether transient PI(3)K inhibition in vivo would affect settling of cells in the NLTs. P14 CD8+ T cells were adoptively transferred into normal hosts that were infected with LCMV. Some of these animals were subsequently treated with the PI(3)K inhibitor LY294002 in vivo for 24 h, and both KLF2GFP expression and numbers of donor T cells were determined in lymphoid and non-lymphoid sites. Interestingly, PI(3)K blockade led to a significant decrease in P14 numbers in NLT but not the spleen (Fig. 7f). Furthermore, elevated KLF2GFP expression was observed in P14 cells in several NLTs (Supplementary Fig. 8a). Such data are consistent with the PI(3)K–Akt pathway in T cells being important for KLF2 loss and TRM generation (although interpretation of these studies is complicated by the fact that systemic PI(3)K blockade will affect diverse cell populations). Lastly, PI(3)K–Akt signaling can elicit Foxo1 protein degradation43, and we observed less Foxo1 in memory P14 cells from salivary gland versus spleen (Supplementary Fig. 8b), consistent with induction of this pathway in NLTs in vivo.
Discussion
Overall our data indicate that TRM in NLT are characterized by low expression of KLF2, and that this is functionally relevant for establishment and/or retention of the TRM pool through transcriptional downregulation of the key KLF2 target S1pr1.
In various systems, CD8+ T cell migration into NLTs peaks during the effector phase of the immune response, and parabiosis experiments (including those in the current study) suggest that there is minimal replacement of TRM cells by T cells from the circulation at memory timepoints11–14, 30. Hence the TRM pool is typically seeded early in the immune response. This pattern is consistent with transient expression of key trafficking molecules on effector CD8+ T cells (such as the brief induction of α4β7 required for generation of CD8+ TRM cells in the small intestine-IEL pool3). However, effector cells that enter NLTs are faced with the decision of whether to rejoin the circulation or commit to long-term residency. Our studies suggest that commitment to the TRM subset involves loss of KLF2 (and consequently S1PR1) expression. These findings are consistent with the new study from Mackay et al.44, that showed decreased S1pr1 transcripts in mouse CD8+ TRM settling in the skin epidermis during the response to HSV infection. Similar loss of S1PR1 expression was reported by Zhu et al., studying gene expression of human CD8+ T cells at sites of HSV-2 reactivation in the skin45. Such data support the hypothesis that transcriptional downregulation of S1pr1 is a defining feature of TRM cells in humans as well as mice.
In the acute infection-inflammation systems we explored, the decline in KLF2 did not correlate with sustained TCR stimulation by the TRM population. The fact that TRM uniformly express CD69 – a marker often taken as a surrogate for T cell stimulation – is best explained by a decline in S1PR1 through the well documented mutual antagonism between CD69 and S1PR1 for cell surface expression28, 29. However, in situations of persistent local infections – such as VSV infection in the brain4 or HSV-1 infection in mouse skin6 – the TRM population may also undergo recurrent TCR activation. Such stimulation would also drive loss of KLF2 expression, as well as upregulate CD69 transcription (further compromising S1PR1 expression). Thus persistence of foreign antigen may reinforce the mechanisms for dampening effective S1PR1 expression by the TRM subset.
Our studies focus on establishment of the TRM pool, and demonstrate that sustained S1PR1 expression is incompatible with generation of this population. Other molecules may further “anchor” TRM in NLTs. A good candidate is CD103, which is upregulated in many (but not all) non-lymphoid sites. Interestingly, CD103 deficiency does not compromise initial settling of activated CD8+ T cells into the small intestine -IEL (in contrast to the effect of forced S1PR1 reported here), but does lead to a decline in the small intestine-IEL (but not small intestine-LPL) population at later timepoints16. Hence, in at least some tissues, CD103 expression may provide an important long-term retention signal. However, our studies suggest that control of S1PR1 expression is a necessary prelude to this step.
NLTs are not exclusively populated by TRM cells – our parabiosis studies confirm that a small subset of recirculating TEM enter the non-lymphoid sites from the blood. Interestingly, these cells show intermediate expression of KLF2 and CD69. Whether this phenotype indicates that such cells are in the process of joining the TRM pool or, instead, indicates a transient decline in KLF2 expression by cells destined to rejoin the circulation (as part of the classic recirculating TEM pool) is not clear. Studies that examined extended parabiosis periods observed that recirculating cells continue to constitute a small fraction of the NLT parenchymal population2, 7, indicating minimal replacement of TRM over time. Regardless, our data strongly indicate that the KLF2lo phenotype marks cells that do not themselves recirculate, and hence correspond to the TRM population.
Other factors beyond KLF2 and S1PR1 expression may support recirculation. For example, CCR7 expression has been shown to promote lymphocyte egress from NLTs46, 47. Nevertheless, the fact that there is an abundant population of CCR7lo TEM in the blood and spleen indicates that this expression pattern alone does not lead cells to be retained in non-lymphoid sites. In contrast, we found that most TEM in lymphoid tissues are KLF2hi, while the vast majority of TRM are KLF2lo – hence the KLF2lo phenotype is more reliable than the CCR7lo phenotype at predicting resident versus recirculating properties of memory CD8+ T cells.
We found that multiple cytokines were capable of promoting downregulation of KLF2. We especially focused on TNF, IL-33 and TGF-β since they had previously been shown to promote other TRM phenotypic traits16. However, it is important to note that cytokines may control multiple pathways involved in TRM establishment: For example, TGF-β promotes not only KLF2 downregulation but also CD103 upregulation. TGF-β reactivity is critical for establishment of TRM in the small intestine-IEL16, but defining the relative contributions of KLF2 versus CD103 regulation is difficult. At a more general level, our data suggest induction of the PI(3)K pathway by these cytokines may be a core element in control of KLF2 expression. These findings resonate with recent studies suggesting that the PI(3)K signaling cascade, while typically considered from the perspective of cell survival and metabolism, has a crucial role in T cell migration48. Hence, we propose that diverse cytokines (potentially distinct combinations in distinct NLTs) converge on the PI(3)K-Akt pathway as a mechanism of enforcing KLF2 downregulation.
It is already clear that CD8+ TRM play a critical role as a first line of defense against local infection5–8, 11–14. Our data suggest that induced downregulation of KLF2 or S1PR1 could be used as a method to efficiently populate or enhance the formation of TRM during vaccination. Treatments which cause local induction of PI(3)K-Akt signals might be used to enhance establishment of TRM in desired sites, potentially in combination with approaches that increase local recruitment of circulating T cells into NLTs8, 15. At the same time, TRM may also contribute to pathological conditions, such as fixed drug eruptions in the skin11, 13. Hence, our studies indicate potential therapeutic targets through which generation of resident memory CD8+ T cells could be promoted or reversed.
S1PR1 expression is firmly established as being critical for lymphocyte recirculation – our data indicate that the immune system co-opts regulation of this pathway to dictate residency of TRM cells. Furthermore, these studies argue that expression of KLF2 constitutes a defining transcription factor of recirculating versus resident memory CD8+ T cells. There has been considerable advancement in relating functional characteristics of T cell subsets to key transcriptional regulators49–51: Our work suggests another dimension in this system, whereby control of T cell recirculation through regulated KLF2 expression determines tissue distribution characteristics of T cell subsets – and hence dictates how effector functions are deployed during recall responses.
Methods
Mice
C57BL/6 and CD45.1 congenic B6 mice were purchased from the National Cancer Institute and used at 6–8 weeks of age. KLF2GFP and Nur77GFP reporter mice were described earlier22,34 and were crossed to P14 TCR transgenic mice (specific for the H-2Db restricted LCMV gp33–41 epitope [KAVYNFATC]). For adoptive co-transfer studies, combinations of CD45.2, CD45.1 and CD45.1/CD45.2 mice were used for donor and host strains, to allow for discrimination of each donor population versus host cells. Animals were maintained under specific pathogen-free conditions at the University of Minnesota. All experimental procedures were approved by the Institutional Animal Care and Use Committee at the University of Minnesota.
Adoptive transfer and infections
Splenocytes were prepared from KLF2GFP (or Nur77GFP) P14 and congenic wild-type P14 mice. The cell populations were mixed 1:1 and 25,000–45,000 CD8+ T cells were transferred into recipient B6 mice, which were infected the next day with 2 × 105 PFU LCMV (Armstrong strain), via ip injection. Mice were maintained under BSL-2 housing and, at indicated times following infection, were sacrificed for analysis.
In vivo anti-CD8 antibody IV and lymphocyte isolation
To label cells in the circulation, mice were injected with fluorochrome conjugated anti-CD8 antibodies (via the tail vein), as previously described25. At 2.5 min following antibody injection, mice were bled and 30 s later mice were sacrificed. Heart perfusion, using 10 ml of cold PBS was performed (in some experiments this step was eliminated) and tissues were harvested and lymphocytes isolated as described16,25. Briefly, lymph nodes were mashed to achieve a single cell suspension, while the salivary glands, kidney, brain, and spleen were mashed and incubated at 37 °C with 100 U/ml collagenase type I (Worthington Biochemical Corporation), 10% FBS, MgCl2/CaCl2, HEPES (pH 7.2), L-glutamine RPMI solution and subjected to mixing (45 min shaking in spinner plates, with magnetic stirrers rotating at 450 rpm). Skin tissues were minced and incubated for 90 min at 37 °C in 2% FCS RPMI medium including Collagenase D (1 mg/ml, Roche applied science) and DNase I (5 μg/ml, SIGMA). For the small intestine, Peyer’s patches were dissected out and the remaining tissue cut longitudinally and then into 1cm pieces. These were rinsed and incubated with 15.4 mg/ml dithiothreitol in HBSS/HEPES (pH 7.2) bicarbonate buffer for 30 min (at 37°C, in spinner plates, as described above) to extract IELs. The remaining pieces were additionally incubated with the collagenase solution and conditions as listed above, to isolate LPL. All tissues were then subjected to a 44/67% percoll gradient (858g at 21°C for 20 min) to isolate lymphocytes.
Flow cytometry and antibodies
Single-cell suspensions were prepared and cells were resuspended at 3–6 × 107 cells/ml in flow cytometry buffer (1% FBS, 0.1% sodium azide) and stained in 100 μl. Isolated lymphocytes in all experiments were incubated with LIVE/DEAD Fixable Aqua Dead Cell Stain Kit (Invitrogen) for 15 min at 4 °C to detect dying cells before staining with a cell surface antibody staining cocktail. For detection of Foxo1 protein, lymphocytes were isolated from salivary glands and spleens of adoptively transferred P14 mice infected 30–60 days previously with LCMV. After surface staining, cells were fixed with a fix/perm solution (eBioscience) for 45 min at 4 °C. Cells were then stained with anti-Foxo1 (Cell signaling clone c29H4) or rabbit IgG isotype control (Cell signaling) at 75–150 ng/test for 45 min at 4 °C. This was followed by a secondary donkey anti-rabbit PE antibody (Immunoresearch) for 30 min on ice. All additional fluorochome-conjugated antibodies (clones listed in the Supplementary Table 1) were purchased from eBioscience, BD BioScience, R&D Systems, or BioLegend. Cells were analyzed using a BD Pharmingen LSR II flow cytometer and data analyzed using FlowJo (TreeStar) software.
For analysis of P14 CD8+ T cells in all tissues, the gating strategy included: live, singlet CD8+ lymphocytes, non-vascular-associated (stained negative for IV administered anti-CD8b)25, with respective congenic markers.
Quantitative RT PCR
Congenically marked WT and KLF2GFP P14 were co-transferred into C57BL/6 mice and infected with LCMV the following day. At 28–36 days post LCMV infection, spleen, salivary glands and small intestine LPL were isolated and P14 CD8+ T cells were sorted on a FACS Aria (Becton Dickinson) based on congenic markers. In other studies, cells were activated in vitro and cultured for 40 h in indicated cytokines, or were transduced with KLF2 or control retrovirus (as indicated), cultured for 2 d and magnetically enriched for cells bearing the transduction marker (Thy-1.1) before analysis. In all cases, RNA was isolated using the RNeasy microkit (Qiagen) and reverse transcription was performed (qScript cDNA synthesis kit, Quanta Biosciences). Gene expression was determined (in triplicate for cultured cells, duplicate for ex vivo samples) using the ABI 7700 sequence detection system and amplification was detected using the SYBR Green PCR Master Mix (Applied Biosystems). Primer sequences are shown in Supplementary Table 2.
Cycle threshold values for the control target (Hprt or Gapdh, as indicated) were subtracted from cycle threshold values for the gene of interest. Next, the mean values (averaged across all repeated experiments) for gene expression of P14 in the spleen (Fig. 1) or P14 cultured without additional cytokines (Fig. 7) was used to normalize between experiments.
Parabiosis
Congenically marked KLF2GFP P14 T cells were transferred into B6 mice. The following day, the animals were infected ip with LCMV and 30–65 days later, mice underwent parabiotic surgery following the schematic described in Supplementary Fig. 3. Procedures were as described previously2. Briefly, mice were anesthetized with ketamine, flank hair removed using Nair® and the skin cleaned using betadine. A lateral incision was made on each mouse from knee to elbow. Mice were joined with a continuous sub-cuticular suture on both the dorsal and ventral sides, with mattress and cruciate sutures joining the skin layer. Mattress sutures just under the armpit and knee were made to secure the parabiosed mice together. At 7–9 days post surgery mice were bled to ensure equilibration and 13–17 days post surgery tissues were harvested from both mice. The number of non-vascular-associated P14 memory T cells was calculated in “donor” and “parabiont” (see Supplementary Fig. 3). A minimum threshold of 25 events was applied for calculation of P14 T-cell KLF2GFP and CD69 MFI.
Retroviral construct generation
To generate the S1PR1 expressing retrovirus vector (MiT-S1PR1), a BglII/NotI fragment including FLAG-tagged mouse S1PR1 was PCR amplified from a MSCV-S1PR1-hCD4 vector27,28 (a gift from J. Cyster, University of California, San Francisco, San Francisco, CA) and was subsequently subcloned into the retroviral vector, MSCV-IRES-Thy1.1 (MiT) which was kindly provided from J. Chen (Massachusetts Institute of Technology, Cambridge, MA)17,18. A similar construct was generated with PCR amplified mouse KLF2 cDNA17. MiT-S1PR1, MiT-KLF2 and MiT (“empty” vector) plasmids were co-transfected with the retroviral packaging vector, pCL-Eco into 293T cells using Lipofectamine 2000 (Life Technology). The 293T line was established from a mycoplasma-free culture, but was not routinely assayed for mycoplama contamination. Supernatants were collected at 48 h after transfection.
Retroviral transduction approaches and calculations
P14 CD8+ T cells were activated either in vivo or in vitro prior to retroviral transduction. For in vivo activation, P14 mice were infected iv with 1 × 106 LCMV Armstrong and harvested as in previous studies52. In vitro activation involved culture of 2–3 × 106 bulk P14 splenocytes with either plate-bound anti-CD3 (2 μg/ml) and soluble anti-CD28 (0.5 μg/ml) per well of a 24-well plate, or with 250nM gp33–41 peptide. At 24 h post activation, 3–6 × 106 activated splenocytes were incubated per well of a 24-well tissue culture plate with 10 μg/ml polybrene and 20 ng/ml hIL-2 and incubated at 37 °C for 30 min. After centrifugation of the plates (at 411g for 5 min at 21°C), the culture media was removed and 1 ml of S1PR1, KLF2 or empty retroviral supernatant was added to each well including 10μg/ml polybrene and 20ng/ml IL-2. Spin infection (1141g for 90 min at 21°C) was followed by a 1 h 37 °C incubation to rest the cells before in vivo transfer. In some experiments, retronectin was used to enhance transduction efficiency by coating 6-well plates with 25 μg/ml retronectin followed by a 2000g spin of 4 ml retrovirus onto the plate for 90 min at 4 °C. Virus supernatant was aspirated off and 6 × 106 activated P14 splenocytes and 20 ng/ml hIL-2 were added to the plate and incubated for 30 min at 37 °C. The plates were then spun at 600g for 30 min at 21°C. For adoptive transfer, 2.5–5 × 104 S1PR1 transduced P14 cells (together with the same number of congenically distinct empty-vector transduced P14 cells) were cotransferred into host animals that were infected with LCMV the following day.
At 3–60 days post transfer, mice were treated with anti-CD8 antibody iv (as described above) and lymphocytes were harvested from indicated tissues. Percent transduction of live non-vascular-associated P14 cells was calculated, based on the Thy-1.1 marker. For normalization, the percent transduction for P14 CD8+ T cells within a given tissue was normalized to the percent transduction found in the spleen of the same animal (Supplementary Fig. 5a).
To characterize transduction efficiency, and monitor proliferation after transduction, splenocytes were set at 1 × 106 cells in 2 ml complete media/well in a 24-well plate with 10–20 ng/ml hIL-2. Every 2 days, live cells were counted and re-set at 1 × 106 cells in 2 ml/well and replenished with new media containing hIL-2. Transduction efficiency over time was also monitored (using the Thy-1.1 transduction marker). Some experiments included 250 nm gp33–41 peptide for the first 2 days after transduction, with similar results.
For DNFB model studies, P14 splenocytes were activated in vitro for 24 h (with 1 μM gp33–41 peptide) then transduced with S1PR1, KLF2 or empty retroviral vectors (as described above) and then maintained in vitro for 4 to 5 days with hIL-2 (20 ng/ml). From these cultures, 2 × 106 cells Thy-1.1 expressing cells transduced with S1PR1 or KLF2 vectors were co-transferred with an equal number of Thy-1.1 expressing P14 cells transduced with empty vector into normal hosts (that were subsequently treated with DNFB, as described below).
In all cases, hosts and both donor populations used for co-transfers differed in CD45 alleles (being CD45.1/.1, CD45.2/.2 or CD45.1/.2 - the combinations differing between experiments) to allow for identification of donor populations using CD45 allele-specific antibodies.
In vitro activated effector P14 transfer into uninfected hosts
For analysis of KLF2 expression in in vitro activated effector P14 cells transferred into uninfected hosts, KLF2GFP and congenic WT P14 were activated in vitro with 1 μM gp33–41 peptide for 24 h and then cultured for 2 additional days in vitro with 20 ng/ml hIL-2. Next, 1 × 106 cells from both cultures were co-transferred into uninfected hosts, and 12–15 days later the KLF2GFP status of cells in different tissues were analyzed.
2,4-Dinitro-1-Fluorobenzene (DNFB) treatment model
In vitro activated P14 T cells were transduced with S1PR1 or KLF2 (and congenic P14 T cells transduced with the empty retroviral vector) followed by culture in IL-2, as described above. Cells transduced with S1PR1 or KLF2 were co-transferred with empty-vector transduced cells (2 × 106 of each population) into normal congenic hosts. Alternatively, KLF2GFP P14 CD8+ T cells and congenic WT P14 cells were activated in a similar way (but without transduction) and 2 × 106 cells of each population co-transferred into normal congenic hosts. At 1 h post-transfer, host mice were shaved and treated with 20 μl of 0.5% DNFB (Sigma) (dissolved in acetone/oil (4:1)) to 1 cm2 area of flank skin, essentially as described8. The other side of the flank skin (contralateral) was treated with vehicle without DNFB. On day 2, 3, 5 and 8 post treatment, skin tissues were harvested (as above) and number of transduced P14 cells was determined.
Ex vivo cytokine and inhibitors assay
WT and KLF2GFP P14 CD8+ T cells were co-transferred into C57BL/6 mice and infected with LCMV the following day. At 4.5 days post infection, bulk splenocytes were harvested and added to 96-well flat bottom plates at 7 × 105 cells/well in 200 μl complete RPMI media, in the presence of cytokines at the following concentrations: TGF-β (10 ng/ml), IL-33 (100 ng/ml), IL-6 (100 ng/ml), IL-15 (20 ng/ml), TNF (125 ng/ml), IL-12 (10 ng/ml), IL-18 (10 ng/ml), IFN-α (50 U/ml) in the indicated combinations, similar to previous studies13,21,40. In some experiments, the Akt inhibitor AKTi (0.25–2 μM, Millipore) or the PI(3)K inhibitor LY294002 (1.25–10 μM, Millipore) were also included from the beginning of the culture period. At 10–40 h of culture, cells were stained with appropriate antibodies and analyzed by flow cytometry.
In vitro cytokine and inhibitors assay and RNA isolation
WT and KLF2GFP P14 splenocytes (5 × 106 cells/ml) were primed in vitro culture in a 10 cm cell culture dish with 250 nM gp33–41 peptide for 48 h. Activated WT and KLF2GFP P14 splenocytes were mixed 1:1 and 2 × 105 cells of the mixed splenocytes were cultured in 96-well flat bottom plates, with no additional cytokines or with TGF-β (10 ng/ml) and IL-33 (100 ng/ml), in the presence or absence of LY294002 (10 μM) or AKTi (1 μM) inhibitors for 48 h. Cells were analyzed by flow cytometry, or, to determine gene expression changes, live P14 CD8+ T cells were sorted using a FACS aria (BD Biosciences) (gating on live, CD3+, CD8+ events), and quantitative RT PCR was performed with the primers indicated above.
In vivo LY294002 administration
Congenically distinct WT and KLF2GFP P14 CD8+ T cells (2.5–5 × 104 cells of each type) were co-transferred into C57BL/6 and infected with LCMV. Four days post infection, 50 mg/kg LY294002 (LC Laboratories) in 50μl DMSO, or 50μl DMSO only (as a vehicle control) was administered ip twice (injections 12 h apart). On day 5 (12 h after the last injection), tissues were harvested and KLF2GFP expression and cell counts determined (see Fig. 7 and S7).
Statistical analysis
Data were analyzed using Prism software 4.0 (GraphPad). For standard data sets, an unpaired two-tailed Student’s t-test was used (with Welch’s correction utilized when variances were found to be different, using the F-test). For values that differed by >10-fold, the data was log10 transformed prior to t-test analysis. When data was normalized (on indicated control samples), normalization involved division of all values by the overall mean of the control values, in order to avoid type I and II errors during calculation of significance through the t-test53. Asterisks indicate obtained p-values: unless otherwise noted in the figure legend, *** indicates P < 0.001; **, P < 0.01; *, P < 0.05 and “NS” indicates P > 0.05. Data sets (in Prism format) are available upon request.
Supplementary Material
Acknowledgments
We thank J. Cyster (University of California-San Francisco) and J. Chen (MIT) for providing retroviral constructs, K. Walkowiak and J. Schenkel for input on parabiosis, L. Mackay (University of Melbourne, Australia) and D. Kaplan for advice on DNFB studies, and the Jamequist lab for intellectual support. This work was supported by the NIH through the following awards: R37 AI38903 to SCJ; R37 AI39560 to KAH; Predoctoral training grant T32 AI07313 to CNS and T90 DE022732 to KGA).
Footnotes
Author contributions
CNS, JYL, DM, KAH and SCJ designed the experiments; CNS, JYL, and KGA performed experiments; and CNS, JYL and SCJ wrote the manuscript.
References
- 1.Mackay CR, Marston WL, Dudler L. Naive and memory T cells show distinct pathways of lymphocyte recirculation. J Exp Med. 1990;171:801–817. doi: 10.1084/jem.171.3.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klonowski KD, et al. Dynamics of blood-borne CD8 memory T cell migration in vivo. Immunity. 2004;20:551–562. doi: 10.1016/s1074-7613(04)00103-7. [DOI] [PubMed] [Google Scholar]
- 3.Masopust D, et al. Dynamic T cell migration program provides resident memory within intestinal epithelium. J Exp Med. 2010;207:553–564. doi: 10.1084/jem.20090858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wakim LM, Woodward-Davis A, Bevan MJ. Memory T cells persisting within the brain after local infection show functional adaptations to their tissue of residence. Proc Natl Acad Sci (USA) 2010;107:17872–17879. doi: 10.1073/pnas.1010201107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hofmann M, Pircher H. E-cadherin promotes accumulation of a unique memory CD8 T-cell population in murine salivary glands. Proc Natl Acad Sci (USA) 2011;108:16741–16746. doi: 10.1073/pnas.1107200108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gebhardt T, et al. Memory T cells in nonlymphoid tissue that provide enhanced local immunity during infection with herpes simplex virus. Nat Immunol. 2009;10:524–530. doi: 10.1038/ni.1718. [DOI] [PubMed] [Google Scholar]
- 7.Jiang X, et al. Skin infection generates non-migratory memory CD8+ T(RM) cells providing global skin immunity. Nature. 2012;483:227–231. doi: 10.1038/nature10851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mackay LK, et al. Long-lived epithelial immunity by tissue-resident memory T (TRM) cells in the absence of persisting local antigen presentation. Proc Natl Acad Sci (USA) 2012;109:7037–7042. doi: 10.1073/pnas.1202288109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schenkel JM, Fraser KA, Vezys V, Masopust D. Sensing and alarm function of resident memory CD8(+) T cells. Nat Immunol. 2013;14:509–513. doi: 10.1038/ni.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu L, et al. Epidermal injury and infection during poxvirus immunization is crucial for the generation of highly protective T cell-mediated immunity. Nat Med. 2010;16:224–227. doi: 10.1038/nm.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bevan MJ. Memory T cells as an occupying force. Eur J Immunol. 2011;41:1192–1195. doi: 10.1002/eji.201041377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sheridan BS, Lefrancois L. Regional and mucosal memory T cells. Nat Immunol. 2011;12:485–491. doi: 10.1038/ni.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ariotti S, Haanen JB, Schumacher TN. Behavior and function of tissue-resident memory T cells. Adv Immunol. 2012;114:203–216. doi: 10.1016/B978-0-12-396548-6.00008-1. [DOI] [PubMed] [Google Scholar]
- 14.Masopust D, Picker LJ. Hidden memories: frontline memory T cells and early pathogen interception. J Immunol. 2012;188:5811–5817. doi: 10.4049/jimmunol.1102695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shin H, Iwasaki A. A vaccine strategy that protects against genital herpes by establishing local memory T cells. Nature. 2012;491:463–467. doi: 10.1038/nature11522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Casey KA, et al. Antigen-independent differentiation and maintenance of effector-like resident memory T cells in tissues. J Immunol. 2012;188:4866–4875. doi: 10.4049/jimmunol.1200402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carlson CM, et al. Kruppel-like factor 2 regulates thymocyte and T-cell migration. Nature. 2006;442:299–302. doi: 10.1038/nature04882. [DOI] [PubMed] [Google Scholar]
- 18.Bai A, Hu H, Yeung M, Chen J. Kruppel-like factor 2 controls T cell trafficking by activating L-selectin (CD62L) and sphingosine-1-phosphate receptor 1 transcription. J Immunol. 2007;178:7632–7639. doi: 10.4049/jimmunol.178.12.7632. [DOI] [PubMed] [Google Scholar]
- 19.Matloubian M, et al. Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature. 2004;427:355–360. doi: 10.1038/nature02284. [DOI] [PubMed] [Google Scholar]
- 20.Grayson JM, Murali-Krishna K, Altman JD, Ahmed R. Gene expression in antigen-specific CD8+ T cells during viral infection. J Immunol. 2001;166:795–799. doi: 10.4049/jimmunol.166.2.795. [DOI] [PubMed] [Google Scholar]
- 21.Schober SL, et al. Expression of the transcription factor lung Kruppel-like factor is regulated by cytokines and correlates with survival of memory T cells in vitro and in vivo. J Immunol. 1999;163:3662–3667. [PubMed] [Google Scholar]
- 22.Cyster JG, Schwab SR. Sphingosine-1-phosphate and lymphocyte egress from lymphoid organs. Ann Rev Immunol. 2012;30:69–94. doi: 10.1146/annurev-immunol-020711-075011. [DOI] [PubMed] [Google Scholar]
- 23.Weinreich MA, et al. KLF2 transcription-factor deficiency in T cells results in unrestrained cytokine production and upregulation of bystander chemokine receptors. Immunity. 2009;31:122–130. doi: 10.1016/j.immuni.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Odumade OA, Weinreich MA, Jameson SC, Hogquist KA. Kruppel-like factor 2 regulates trafficking and homeostasis of gammadelta T cells. J Immunol. 2010;184:6060–6066. doi: 10.4049/jimmunol.1000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anderson KG, et al. Cutting Edge: Intravascular Staining Redefines Lung CD8 T Cell Responses. J Immunol. 2012;189:2702–2706. doi: 10.4049/jimmunol.1201682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masopust D, Vezys V, Wherry EJ, Barber DL, Ahmed R. Cutting edge: gut microenvironment promotes differentiation of a unique memory CD8 T cell population. J Immunol. 2006;176:2079–2083. doi: 10.4049/jimmunol.176.4.2079. [DOI] [PubMed] [Google Scholar]
- 27.Kuo CT, Veselits ML, Leiden JM. LKLF: a transcriptional regulator of single-positive T cell quiescence and survival. Science. 1997;277:1986–1990. doi: 10.1126/science.277.5334.1986. [DOI] [PubMed] [Google Scholar]
- 28.Shiow LR, et al. CD69 acts downstream of interferon-alpha/beta to inhibit S1P1 and lymphocyte egress from lymphoid organs. Nature. 2006;440:540–544. doi: 10.1038/nature04606. [DOI] [PubMed] [Google Scholar]
- 29.Bankovich AJ, Shiow LR, Cyster JG. CD69 suppresses sphingosine 1-phosophate receptor-1 (S1P1) function through interaction with membrane helix 4. J Biol Chem. 2010;285:22328–22337. doi: 10.1074/jbc.M110.123299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Masopust D, Vezys V, Marzo AL, Lefrancois L. Preferential localization of effector memory cells in nonlymphoid tissue. Science. 2001;291:2413–2417. doi: 10.1126/science.1058867. [DOI] [PubMed] [Google Scholar]
- 31.Sallusto F, Geginat J, Lanzavecchia A. Central memory and effector memory T cell subsets: function, generation, and maintenance. Annu Rev Immunol. 2004;22:745–763. doi: 10.1146/annurev.immunol.22.012703.104702. [DOI] [PubMed] [Google Scholar]
- 32.Lo CG, Xu Y, Proia RL, Cyster JG. Cyclical modulation of sphingosine-1-phosphate receptor 1 surface expression during lymphocyte recirculation and relationship to lymphoid organ transit. J Exp Med. 2005;201:291–301. doi: 10.1084/jem.20041509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Graler MH, Huang MC, Watson S, Goetzl EJ. Immunological effects of transgenic constitutive expression of the type 1 sphingosine 1-phosphate receptor by mouse lymphocytes. J Immunol. 2005;174:1997–2003. doi: 10.4049/jimmunol.174.4.1997. [DOI] [PubMed] [Google Scholar]
- 34.Ledgerwood LG, et al. The sphingosine 1-phosphate receptor 1 causes tissue retention by inhibiting the entry of peripheral tissue T lymphocytes into afferent lymphatics. Nat Immunol. 2008;9:42–53. doi: 10.1038/ni1534. [DOI] [PubMed] [Google Scholar]
- 35.Hart GT, Hogquist KA, Jameson SC. Kruppel-like factors in lymphocyte biology. J Immunol. 2012;188:521–526. doi: 10.4049/jimmunol.1101530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moran AE, et al. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J Exp Med. 2011;208:1279–1289. doi: 10.1084/jem.20110308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sinclair LV, et al. Phosphatidylinositol-3-OH kinase and nutrient-sensing mTOR pathways control T lymphocyte trafficking. Nat Immunol. 2008;9:513–521. doi: 10.1038/ni.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schluns KS, Lefrancois L. Cytokine control of memory T-cell development and survival. Nat Rev Immunol. 2003;3:269–279. doi: 10.1038/nri1052. [DOI] [PubMed] [Google Scholar]
- 39.Takada K, et al. Kruppel-like factor 2 is required for trafficking but not quiescence in postactivated T cells. J Immunol. 2011;186:775–783. doi: 10.4049/jimmunol.1000094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Freeman BE, Hammarlund E, Raue HP, Slifka MK. Regulation of innate CD8+ T-cell activation mediated by cytokines. Proc Natl Acad Sci (USA) 2012;109:9971–9976. doi: 10.1073/pnas.1203543109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hedrick SM. The cunning little vixen: Foxo and the cycle of life and death. Nat Immunol. 2009;10:1057–1063. doi: 10.1038/ni.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang YE. Non-Smad pathways in TGF-beta signaling. Cell Res. 2009;19:128–139. doi: 10.1038/cr.2008.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Plas DR, Thompson CB. Akt activation promotes degradation of tuberin and FOXO3a via the proteasome. J Biol Chem. 2003;278:12361–12366. doi: 10.1074/jbc.M213069200. [DOI] [PubMed] [Google Scholar]
- 44.Mackay LK, et al. The development pathway for CD103+CD8+ tissue-resident memory T cells of skin. Nat Immunol. 2013 doi: 10.1038/ni.2744. In Press. [DOI] [PubMed] [Google Scholar]
- 45.Zhu J, et al. Immune surveillance by CD8alphaalpha+ skin-resident T cells in human herpes virus infection. Nature. 2013;497:494–497. doi: 10.1038/nature12110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Debes GF, et al. Chemokine receptor CCR7 required for T lymphocyte exit from peripheral tissues. Nat Immunol. 2005;6:889–894. doi: 10.1038/ni1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bromley SK, Thomas SY, Luster AD. Chemokine receptor CCR7 guides T cell exit from peripheral tissues and entry into afferent lymphatics. Nat Immunol. 2005;6:895–901. doi: 10.1038/ni1240. [DOI] [PubMed] [Google Scholar]
- 48.Finlay D, Cantrell DA. Metabolism, migration and memory in cytotoxic T cells. Nat Rev Immunol. 2011;11:109–117. doi: 10.1038/nri2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ohkura N, Kitagawa Y, Sakaguchi S. Development and maintenance of regulatory T cells. Immunity. 2013;38:414–423. doi: 10.1016/j.immuni.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 50.Zhu J, Paul WE. Peripheral CD4+ T-cell differentiation regulated by networks of cytokines and transcription factors. Immunol Rev. 2010;238:247–262. doi: 10.1111/j.1600-065X.2010.00951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rutishauser RL, Kaech SM. Generating diversity: transcriptional regulation of effector and memory CD8 T-cell differentiation. Immunol Rev. 2010;235:219–233. doi: 10.1111/j.0105-2896.2010.00901.x. [DOI] [PubMed] [Google Scholar]
- 52.Araki K, et al. mTOR regulates memory CD8 T-cell differentiation. Nature. 2009;460:108–112. doi: 10.1038/nature08155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Valcu M, Valcu CM. Data transformation practices in biomedical sciences. Nat Methods. 2011;8:104–105. doi: 10.1038/nmeth0211-104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.