Abstract
Polyketide synthases (PKSs) are responsible for synthesizing a myriad of natural products with agricultural, medicinal relevance. The PKSs consist of multiple functional domains of which each can catalyze a specified chemical reaction leading to the synthesis of polyketides. Biochemical studies showed that protein-substrate and protein-protein interactions play crucial roles in these complex regio-/stereo- selective biochemical processes. Recent developments on X-ray crystallography and protein NMR techniques have allowed us to understand the biosynthetic mechanism of these enzymes from their structures. These structural studies have facilitated the elucidation of sequence-function relationship of PKSs and will ultimately contribute to the prediction of product structure. This review will focus on the current knowledge of type I PKS structures and the protein-protein interactions in this system.
Keywords: Natural products, biosynthesis, antibiotics, fatty acid synthase, X-ray crystallography, protein NMR, mutagenesis, macromolecular docking
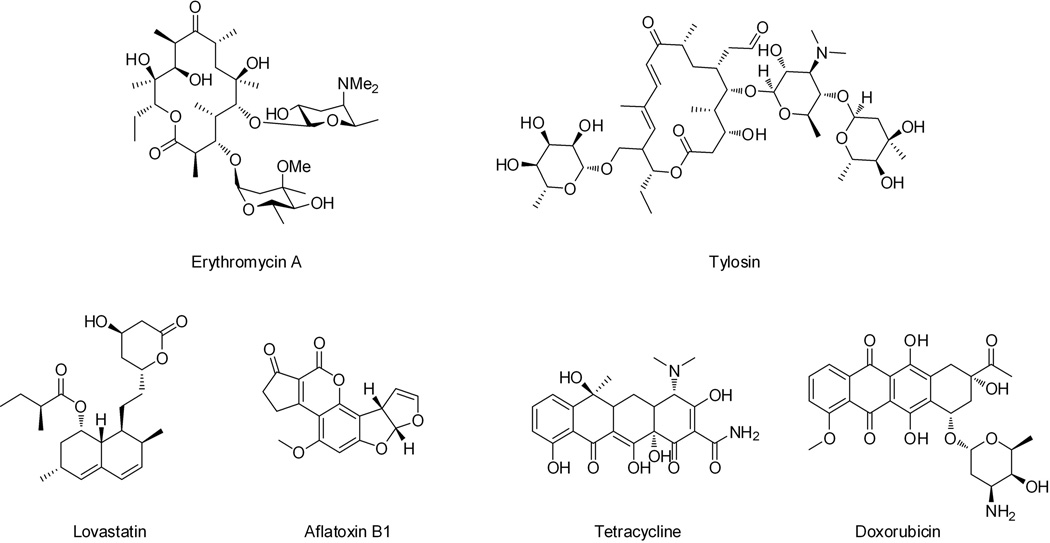
Polyketides constitute a major family of secondary metabolites that have diverse structures and broad biological activities (Walsh, 2008; Staunton et al., 2001). As a result, polyketides are ample sources for the development of frontline therapeutics towards different diseases (Newman et al., 2012). Many polyketide natural products have become important drug molecules, including human antibiotics erythromycin A and tetracycline; veterinary antibiotic tylosin; anti-cancer agent doxorubicin; immunosuppressant rapamycin and cholesterol-lowering drug lovastatin (Figure 1).
Figure 1.
Examples of polyketides synthesized by different types of polyketide synthases: type I modular PKS (erythromycin A and tylosin), type I iterative PKS (lovastatin, aflatoxin B1), and type II PKS (tetracycline and doxorubicin). 107×53mm (300 × 300 DPI).
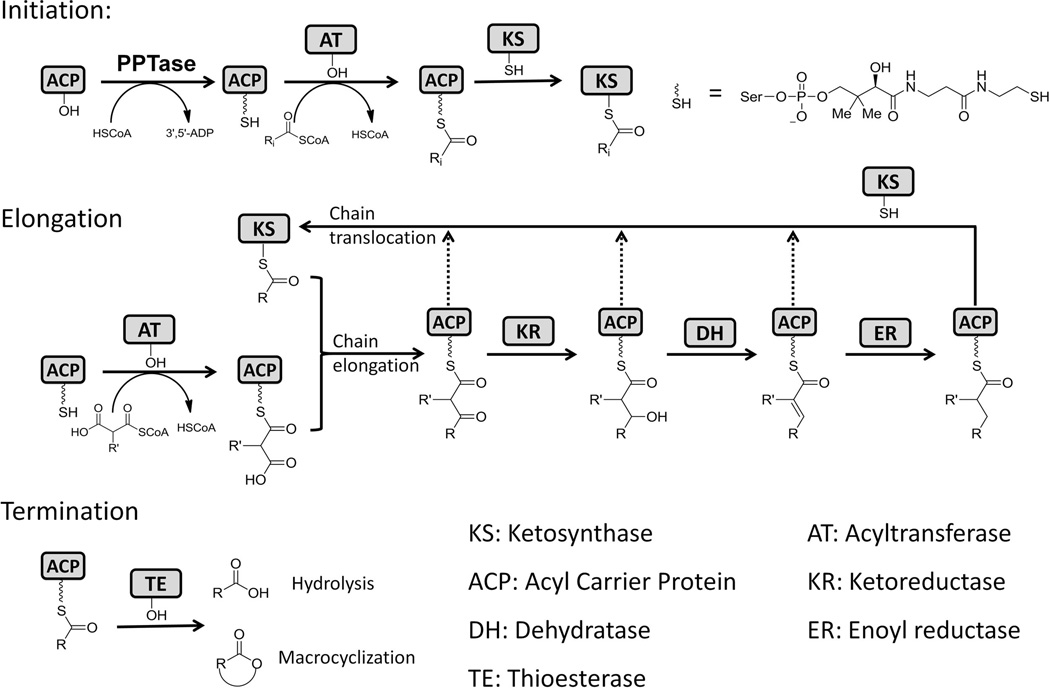
Notwithstanding the diversity displayed by the structures, all polyketides are assembled from simple cellular building blocks, such as acetate, by a group of enzymes called polyketide synthases (PKSs) (Khosla, 2009). Resembling the biosynthetic mechanism of fatty acid synthases (FASs), polyketide biosynthesis by PKSs can be divided into three steps: initiation, elongation and termination. Both FASs and PKSs are composed of catalytic domains that work in coordination to polymerize the simple building blocks (mostly in the form of acyl-CoAs). In the initiation step, the acyl carrier protein (ACP) must first be posttranslationally phosphopantetheinylated at the active site serine within the conserved DSL motif by a phosphopantetheinyl transferase (PPTase). The PPTase is either encoded in the PKS gene cluster as a dedicated enzyme, or is shared with FASs or other PKSs in the same host (Lambalot et al., 1996). The acyl-CoA:ACP transacylase (AT) is responsible for transferring the monomeric acyl groups, such as malonyl or methylmalonyl, from the coenzyme A carrier to the free thiol of the phosphopantetheine (pPant) arm on the modified (holo-) ACP. The ketosynthase (KS) catalyzes the successive decarboxylative Claisen condensations between the growing polyketide chain and the monomeric/extender unit supplied by ACP to elongate the nascent polyketide backbone. AT, KS and ACP are the minimal domains required for the PKS synthesis hence are called minimal PKSs. Following each chain extension step in which the polyketide backbone is increased in size by one ketide, tailoring domains such as β-ketoreductase (KR), dehydratase (DH), and enoylreductase (ER) can selectively reduce the newly formed polyketide. Whereas in FASs, the β-keto positions are completely reduced to methylenes and yield a saturated fatty acyl product, the tailoring domains in PKS can be used in different combinations at each β-position to yield products of different oxidation states. Also unique to PKSs, a methyl group may be introduced at the α position of the growing chain by C-methyltransferase (MT) domain (Hertweck, 2009). Finally, when the polyketide chain reaches the desired size, the product can be offloaded from the PKSs via different release mechanisms, such as hydrolysis or macrocyclization catalyzed by a thioesterase (TE) (Du et al., 2010) (Figure 2).
Figure 2.
General chemical reactions catalyzed by PKSs. 215×145mm (300 × 300 DPI).
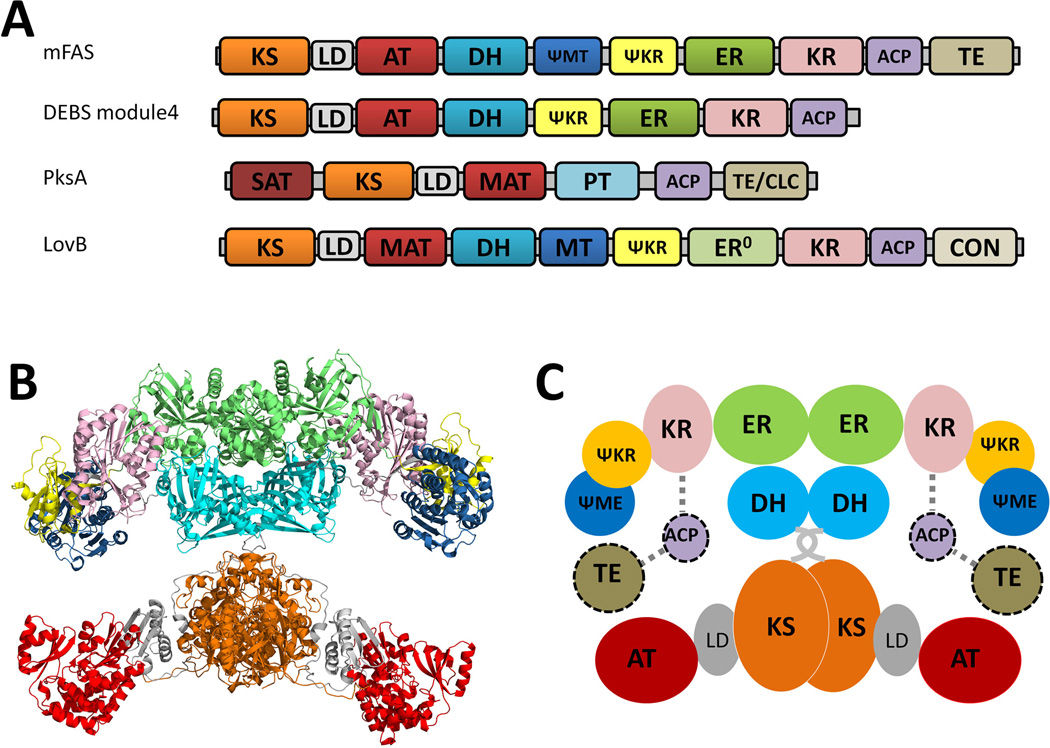
Based on the enzyme primary sequence, catalytic domain organization and product structure, PKSs can be classified into three distinct types (Weissman, 2009; Hertweck, 2009): type I PKSs are large multidomain enzymes that can function either modularly or iteratively; type II PKSs consist of dissociated enzymes that function iteratively, and are typically associated with aromatic polyketides from bacteria; Types III PKSs contain single KS domains that catalyze condensations between various acyl-CoAs directly without the use of ACP domains. This classification is also supported by the phylogenetic analysis based on the amino acid sequence of KS domains (Jenke-Kodama et al., 2005; Kroken et al., 2003). Among the three classes of PKSs, type I PKSs have the highest complexity in their protein structures and product diversities. These megasynthases are featured by having multiple catalytic domains juxtaposed on a single polypeptide chain. These covalently joined domains result in increased reaction rates and protection of labile intermediates. Depending on whether the polyketide backbone extension is performed using distinct sets of domains (each set contains minimal PKS domains and is referred as a module), or repeatedly on a single set of domains, type I PKSs can be divided into two subcategories: modular (non-iterative) and iterative PKSs (Weissman, 2009; Hertweck, 2009; Cox, 2007).
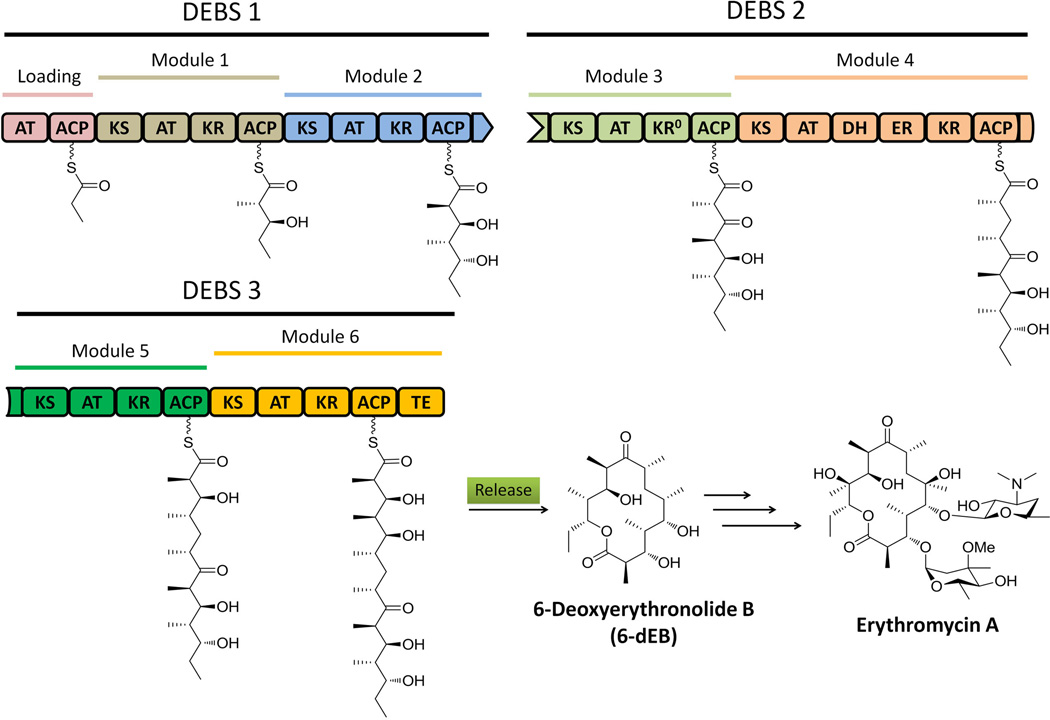
Modular type I PKSs, which are usually found in bacteria, synthesize polyketides in an assembly line fashion: the growing polyketide chain is delivered from one module to the next and each catalytic domain in a module is typically used only once (Meier et al., 2009). The number of the modules present in a PKS defines the number of chain extension steps, while the presence of KR, DH and ER domains in each module determine the degree of β-keto reduction after each elongation (Cane et al., 1999; Fischbach et al., 2006). Therefore, the modular nature of the PKSs and the associated structures of the polyketide products are colinear, which can be used to predict product from PKS sequence and vice versa (Fischbach et al., 2006). The best studied modular PKS is the 6-deoxyerythronide B synthase (DEBS) involved in the synthesis of 6-deoxyerythronide B (6-dEB), which is the aglycon of the antibiotic erythromycin A. In DEBS, one loading module and six extending modules are required to process one propionate and six methylmalonyl-CoA units to produce 6-dEB (Figure 3). The colinear feature of type I PKSs has greatly enabled the rational manipulation of the domains and modules towards the generation of structurally altered products. In the case of DEBS, over fifty different macrolides have been synthesized through the engineering of AT and β-processing domains (McDaniel et al., 1999).
Figure 3.
Biosynthesis of 6-deoxyerythronolide B on type I modular PKS DEBSs. The domain with a “0” notation indicate nonreactive domain which misses important catalytic/binding site. 215×150mm (300 × 300 DPI).
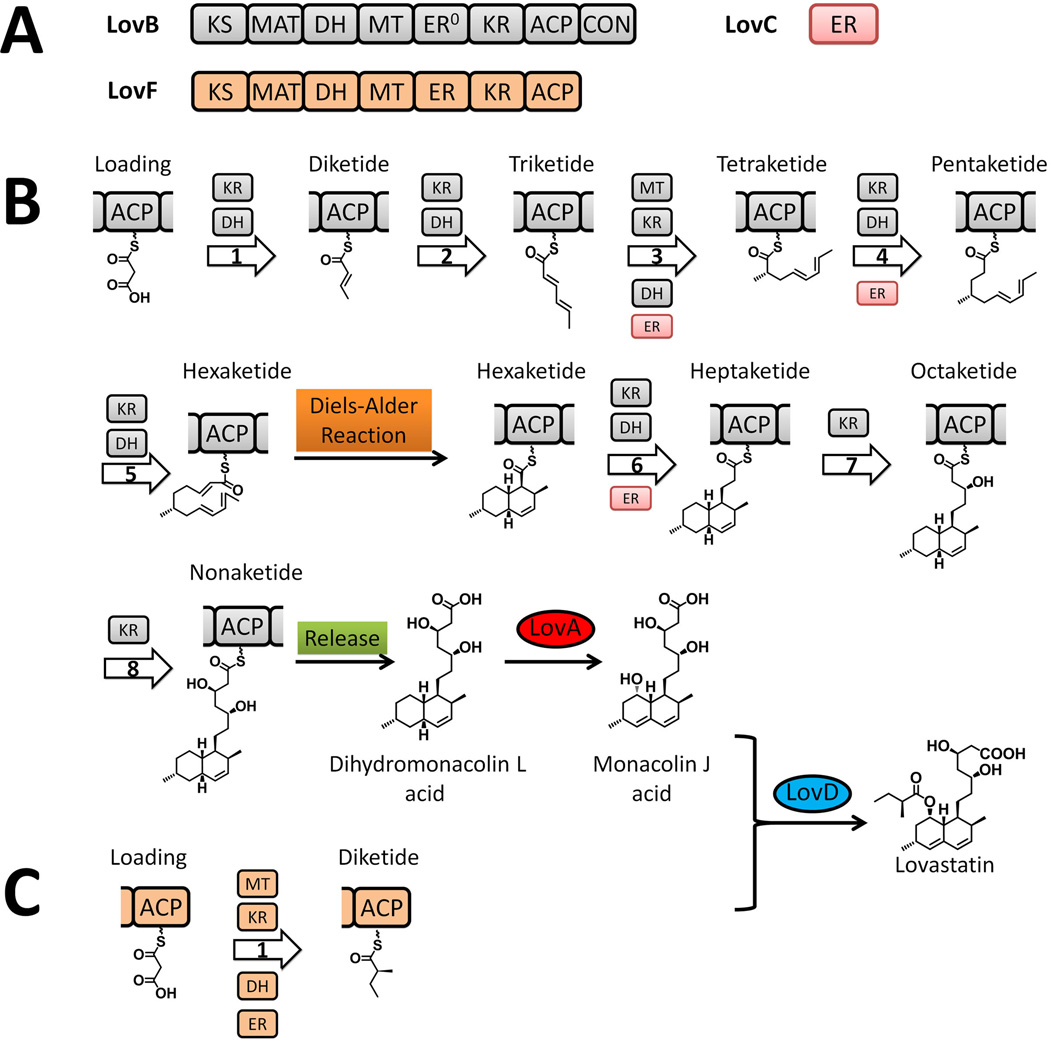
Iterative type I PKSs, such as the lovastatin nonaketide synthase LNKS (or LovB) from Aspergillus terreus, are mostly found in filamentous fungi, with some recent discoveries of bacterial examples, including the PKSs that synthesize the polyketide core of enediynes (Shen, 2003). Each iterative type I PKS consists of a single set of catalytic domains, which can vary and define different extent of reductive modification in the product (Figure 4-A). While the linear organization and the iterative use of the domains resemble those of mammalian fatty acid synthase (mFAS), the programming rules dictating the permutative use of catalytic domains in iterative type I PKSs are significantly more complicated and mysterious. In the LovB case, different combinations of MT-KR-DH-ER usage are involved in the eight cycles of chain elongation, resulting in the generation of different β-oxidation states of different chain length intermediates, by the same set of domains. The necessity of such complex programming rule is evident in the product of LovB, dihydromonacolin L, in which the decalin core is formed through the intramolecular Diels-Alder cyclization of a precisely prepared triene at the hexaketide stage (Auclair et al., 2000; Kennedy et al., 1999; Ma et al., 2009) (Figure 4-B). In sharp contrast, a different type I PKS in the lovastatin pathway, LovF, which has nearly the same domain architecture as LovB, performs only a single iteration of chain extension and tailoring to yield the 2-methylbutyrate side chain in lovastatin (Figure 4-C) (Xie et al., 2009; Kennedy et al., 1999). The contrast in programming rules between LovB and LovF highlights the complexity and perhaps engineering potential of this subclass of type I PKSs.
Figure 4.
Biosynthesis of lovastatin. (A) Domain organization of type I iterative PKSs LovB and LovF. (B) Biosynthesis of dihydromonacolin L on LovB in presence of LovC. (C) Biosynthesis of 2-methylbutyrate side chain on LovF. 215×209mm (300 × 300 DPI).
Because of the similarity in the domain organization between mFAS and type I PKSs (Figure 5-A), the X-ray crystal structures of mFAS have provided the first look into the organization of the catalytic units in the multidomain type I PKS megasynthases (Maier et al., 2008; Maier et al., 2006). Much of the finding in mFASs can be extended to that of type I PKSs, although significant differences are expected. While the crystal structure of an intact module (from modular PKS) or an entire iterative PKS is not yet available, significant structural information of individual PKS domains have been obtained in recent years. These include the X-ray structures of excised KS-AT (Tang et al., 2006; Tang et al., 2007), KR (Keatinge-Clay et al., 2006; Keatinge-Clay, 2007; Zheng et al., 2010; Zheng et al., 2011), DH (Keatinge-Clay, 2008; Akey et al., 2010), KR-ER (Zheng et al., 2012) and TE (Tsai et al., 2001; Tsai et al., 2002; Akey et al., 2006; Akey et al., 2010) domains from type I modular PKSs. Structures of ACPs from different families of PKSs have been obtained via high-resolution, liquid-state NMR analysis. These high resolution structures have greatly increased our understanding of the structural biology and enzymology of the PKSs components and domains. A number of publications have reviewed the various aspects of structural enzymology of FASs and PKSs (Weissman et al., 2008; Tsai et al., 2009; Smith et al., 2007; Keatinge-Clay, 2012).
Figure 5.
Structural overview of mammalian FAS. (A) Comparison of the linear organizations of mFAS and type I PKSs. (B) The crystal structure of mFAS with a resolution of 3.2 Å. PDB code: 2VZ9 (C) Cartoon representation of the mFAS organization with predicted ACP-TE domains. 215×157mm (300 × 300 DPI).
1. Biochemical and structural analysis of mammalian FASs
The head-to-head model of FASs was proposed through sets of mutation complementation experiments (Witkowski et al., 1996; Joshi et al., 1997; Joshi et al., 1998b). Inactivation mutations can be introduced to different catalytic domains without disrupting the folding of the domains, thereby allowing the combination of different mutant monomers to generate homodimers and heterodimers (Witkowski et al., 1996). The ACP domains were shown to crosstalk with the KS domain from either subunits to accomplish chain elongations in FAS (Joshi et al., 1998b), but could only interact with the DH domain from the same subunit (Joshi et al., 1997; Joshi et al., 1998a). Heterodimers containing one subunit with either an inactive KS or AT domain, when combined with the other subunit inactivated in any one of the other five domains, including ACP, KR, DH, ER and TE domain, retained the ability to synthesize fatty acids(Joshi et al., 1998a). Additionally, a null mutant FAS with all seven active site-compromised domains was capable of synthesizing fatty acid product in vitro after pairing with a wild-type FAS subunit (Joshi et al., 2003). In the head-to-head model, the ACP is able to interact with KS-AT from both subunits and can only interact with DH-ER-KR-TE domains within the same subunit (Rangan et al., 2001). The model was further supported by the cross-linking of KS and ACP via 1, 3-dibromopropanone (DBP), in which intra-subunit linked heterodimers were recovered (Witkowski et al., 1999). The KS domains were proposed to be positioned at the interface of the two subunits (Witkowski et al., 2004). This is motivated by biochemical evidences that KS domains from type I and type II FASs are always found as dimers (Moche et al., 1999; Price et al., 2003; Witkowski et al., 2004), and that the two subunits can be covalently crosslinked via bis-maleimido at low concentrations (Witkowski et al., 2004). Therefore the KS domains must play a key role in maintaining the dimeric structures of FAS, and by inference, PKSs.
The 4.5 Å resolution X-ray structure of porcine FAS, which was subsequently refined to 3.2 Å resolution, showed the overall structure of mFAS adopts an X-shaped organization that is consistent with the head-to-head model (Maier et al., 2008; Maier et al., 2006; Asturias et al., 2005). The individual catalytic domains were mapped out by fitting the density diagram with previously-solved type II FAS components, including FabB (KS) (Olsen et al., 2001), FabA (DH) (Leesong et al., 1996), FabZ (DH) (Kimber et al., 2004), and FabG (KR) (Price et al., 2001) from E. coli; FabD (MAT, malonyl-CoA:ACP transacylase) from S. coelicolor (Keatinge-Clay et al., 2003); and a zinc-free quinone reductase (ER) from Thermus thermophilus (Shimomura et al., 2003). Starting from the N-terminus of the mFAS, the dimeric KS domains are located at the center of the lower body where each is connected to the monomeric ‘leg’ MAT domain (Figure 5-B and Figure 5-C). Dimeric DH, ER and monomeric KR together form the upper body of the FAS, in which KR domains are the ‘arms’. The ‘chest’ DH adopts a ‘double-hot-dog’ fold containing portion from core domain of FAS. KR domain plays a central role in connecting the ‘shoulder’ ER and DH domains as well as two newly identified non-enzymatic domains consisting of the rest of the core domain. The first non-catalytic domain was named ‘pseudo-methyltransferase’ (ψ-MT) due to its structural similarity toward methyltransferase family, while the second domain was designated as ‘pseudo-ketoreductase’ (ψ-KR) resembling a truncated KR scaffold that interacts with the KR. In the lower body, the linker region between KS and MAT adopts a α2β3-fold (see more discussion of this region in the PKS part). The upper body and lower body of the mFAS are barely contact with each other with a contact surface area of 230 Å. In contrast, the KS, DH and the ER domains contribute to ~5000 Å2 dimer interface of the mFAS. The linker region between the MAT and the DH domains interact with the KS domain from the other subunit, also contributing to dimer formation.
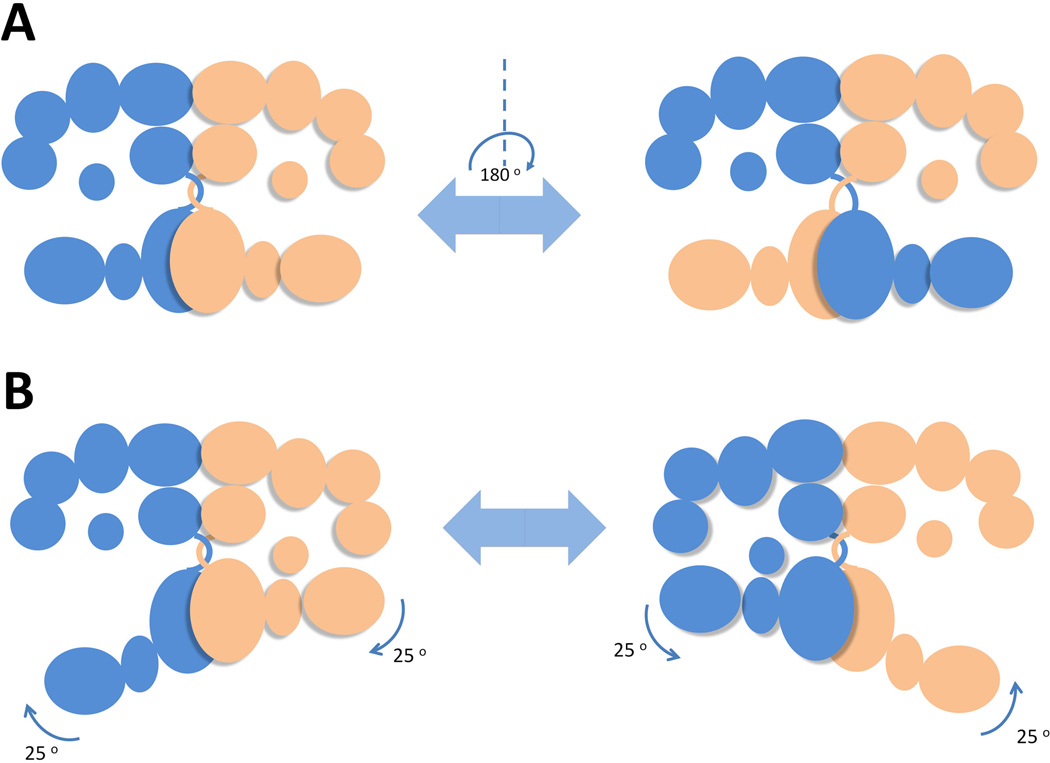
Collectively, the dimeric mFAS clearly contains two reaction chambers, in which fatty acid assembly could take place independently. Interestingly, the two reaction chambers appeared to be asymmetrical: the distance between the active sites of MAT and KR domains is 15 Å shorter in the narrower chamber comparing to that in the wider chamber, suggesting that the fatty acid synthesis in each of reaction chamber might not proceed at the same pace and conformation changes. The anchor point of ACP was revealed at the center of the upper portion of the reaction chambers. The ACP and TE domains were unsolved in the reported mFAS crystal due to the inherent domain flexibility, but structures of standalone human TE (Chakravarty et al., 2004) and rat FAS ACP (Reed et al., 2003) are available. The ψ-MT domain is located at vicinity of ACP anchor point, confining the movement of ACP within the region of the active sites of the catalytic domains. These structure features enable the holo-ACP to dock at different locations and shuttle the substrate to the specific active site from one chamber but not active sites from the other. Considering that the upper and lower bodies of the FAS is connected by the flexible MAT-DH linker and the limited protein-protein interactions between the two halves, it is highly possible that the lower body of one subunit could swivel up to 180 degrees with respects to the upper body to interact with the ACP from the other subunit (Figure 6-A). This hypothesis is verified by the electron microscopic analysis, in which an 80–100 degree rotation of the lower body with respects to the upper body was clearly present in reconstructed structures. The combined X-ray and EM structures explain the biochemical findings in mutant complementation and crosslinking experiments. This swiveling mechanism also injects FASs with additional catalytic route that contributes to approximately 20 % of fatty acid synthesis rate, as estimated from in vitro mutant complementation (Rangan et al., 2001).
Figure 6.
Schematic configurations adopted by mFAS to position catalytic domains into the proximity of ACP domain for domain-domain interactions. (A) A full 180o swiveling of lower body relative to the upper body permits the ACP to crosstalk with KS-AT domains from both subunits. (B) a 25o rotation of the lower body against the upper body leads to the two states of the reaction chambers-open and closed. 215×157mm (300 × 300 DPI).
To examine the dynamics of the flexible FAS that permit the functional interactions of the ACP with all the catalytic domains, Asturias and Smith utilized single-particle electron microscopy (EM) to continuously capture a series of configurations of FAS dimer from Rattus norvegicus (Brignole et al., 2009). The FAS mutant (Δ22) was constructed by the removal of the 22-residue ACP-TE linker to restrict the mobility of the TE domain. When monitored by electron microscope in the absence of substrates, both ACP and TE domains were visible in EM reconstructions. In the presence of substrates, asymmetrical arrangement of the upper β-processing domains was observed in both Δ22 and DH0 mutant FASs. Specifically, a swinging motion (up to 25 degrees) of the lower body relative to the upper body resulted in the opening of one of the reaction chambers and closing of the other (Figure 6-B). While the open configuration of one of the chamber is required by the β-process domains to catalyze the reduction steps, the closure of the reaction chamber is proposed to facilitate the substrate loading and condensation reactions by the KS and AT domains. Additionally, since the TE domain is completely ‘locked’ outside upon the closure of the chamber, the chain releasing step likely happens in the open chamber.
2. Comparison between Type I PKSs and mFAS
The mammalian FASs are evolutionarily closely related to the type I iterative and modular PKSs (Smith et al., 2008). In addition to the highly similar sequences of catalytic domains and linear positioning of the domains (Figure 5-A), the parallel between mFAS and PKSs can be extended to the linker regions and nonenzymatic domains ψ-MT and ψ-KR. In particular, the presence of the ψ-MT domain in mFAS and the catalytic MT domain in the iterative fungal PKSs indicate shared evolutionary ancestor. Despite the domain similarities, the mFAS and type I PKSs are functionally very different. In the case of modular PKSs, each module only performs one chain elongation and one defined set of β-processing, and the chain is passed unidirectionally down the assembly line to downstream modules. The different substrate specificities of the domains, different combinations of KR-DH-ER cassette, and the juxtaposition of different mFAS-like modules, all contribute to the diverse structures that can be generated from type I PKSs. So far, no X-ray structure of an intact PKS module has been solved, although the structures of nearly all the individual domains have been obtained. From a structural point of view, the fascinating aspects of modular PKSs include the spatial arrangement of different modules with respect to each other, and how protein-protein interactions between the modules are facilitated through ACPs and linker regions to achieve the assembly line like biosynthetic logic.
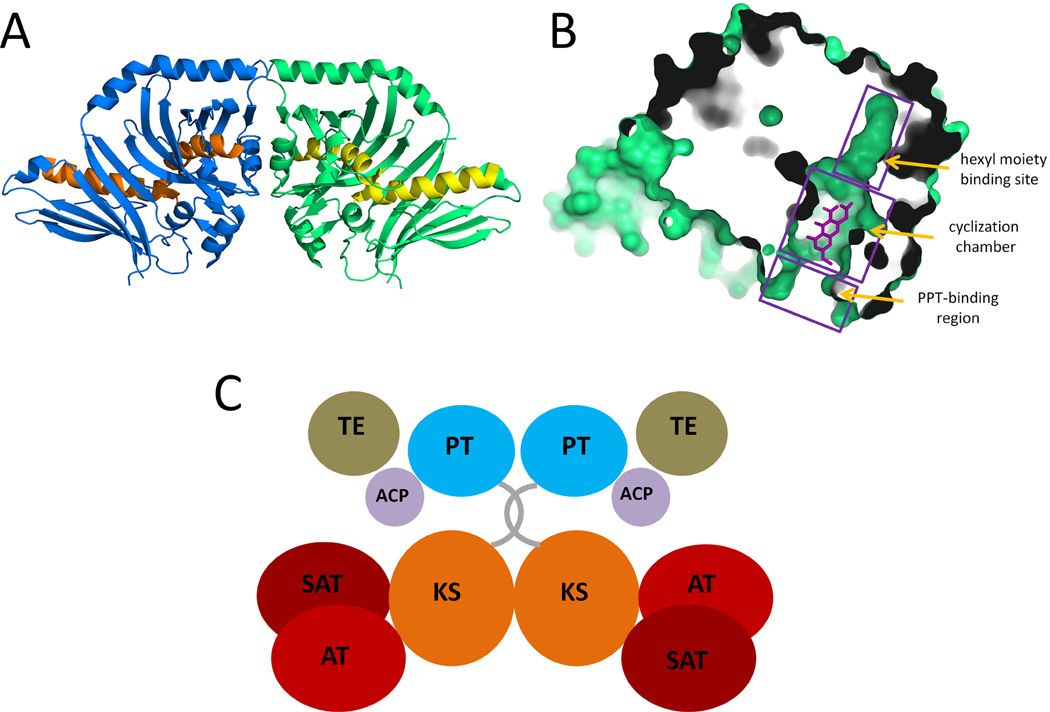
While the modular PKSs is certainly Nature’s example of “more is better” when it comes to generating chemical diversity, the iterative type I PKSs can be considered a more compact version in which modules of different functions are condensed into a single set of domains. Although iterative PKSs are functionally and structurally more closely related to mFAS, there are significant differences that make this subclass of PKS fascinating from a biochemical and structural perspective. Depending on the type of product, different tailoring domains are present in the PKS. For nonreduced polyketides in which β-processing is absent, additional domains such as product template (PT) and Claisen-like cyclase (TE/CLC) are present to fix the regioselectivity of intramolecular cyclization reactions. For reduced polyketides, the more familiar KR, DH, ER and MT domains can be found in the PKS, although it is common that some domains lost their functions during evolution. The current structural understanding of the domains from iterative type I PKSs is not as extensive as in the modular PKS, mostly due to the difficulties in isolating functional and soluble domains. In recent years, however, numerous structures, including that of AT (Liew et al., 2012), PT (Crawford et al., 2009), TE/CLC (Korman et al., 2010) have been solved, which have provided insights into how some of these distinct domains can fit into the overall dimeric structure of the PKSs (Figure 11-C, also see detailed discussion in the section 3.4). Another distinction between mFAS and iterative PKS is the recruitment of smaller, in trans enzymes for product tailoring and product release. This can be exemplified in the lovastatin biosynthesis, in which a dissociated ER enzyme (LovC) is required for enoylreduction of three intermediates on LovB (Figure 4-B), and a dissociated acyltranferase (LovD) is required for product release/transfer from LovF (Figure 4). From a limited number of experiments, the interaction between the iterative PKS and these dissociated enzymes appear to be specific. Hence understanding this unique protein-protein interaction is important for fully decoding the programming rules of iterative PKSs.
Figure 11.
The crystal structure of the PT domain from PksA. (A) Ribbon representation of the PT dimer showing the double hot dog (DHD) fold. PDB code: 3HRR. (B) Surface representation of the PT monomer substrate pocket. The product analogue HC8 is co-crystallized and is shown as sticks (C) Predicted domain organization of PksA. 215×147mm (300 × 300 DPI).
Finally, the most intriguing differences between mFAS and iterative PKSs are undoubtedly the ability of the tailoring domains in the reducing iterative PKSs to only function on selected intermediates during the different cycles of chain extension, and how overall chain length is determined. Currently, nearly no information is known regarding how the selectivity is so precisely maintained. As a result of this cryptic selectivity, highly similar iterative PKSs can produce drastically different products. Recent genome sequences of different fungi species have revealed that each contains dozens of such PKSs with unknown products, yet unlike the modular PKSs, no product can be predicted from these enzymes due to the utter lack of understanding of their sequence-function relationships. It is expected that crystal structures of the iterative PKS domains, including KS, KR, DH, ER, MT and ACP, will provide significant breakthroughs needed for decoding these machineries.
3. Structures and catalytic mechanisms of PKS domains
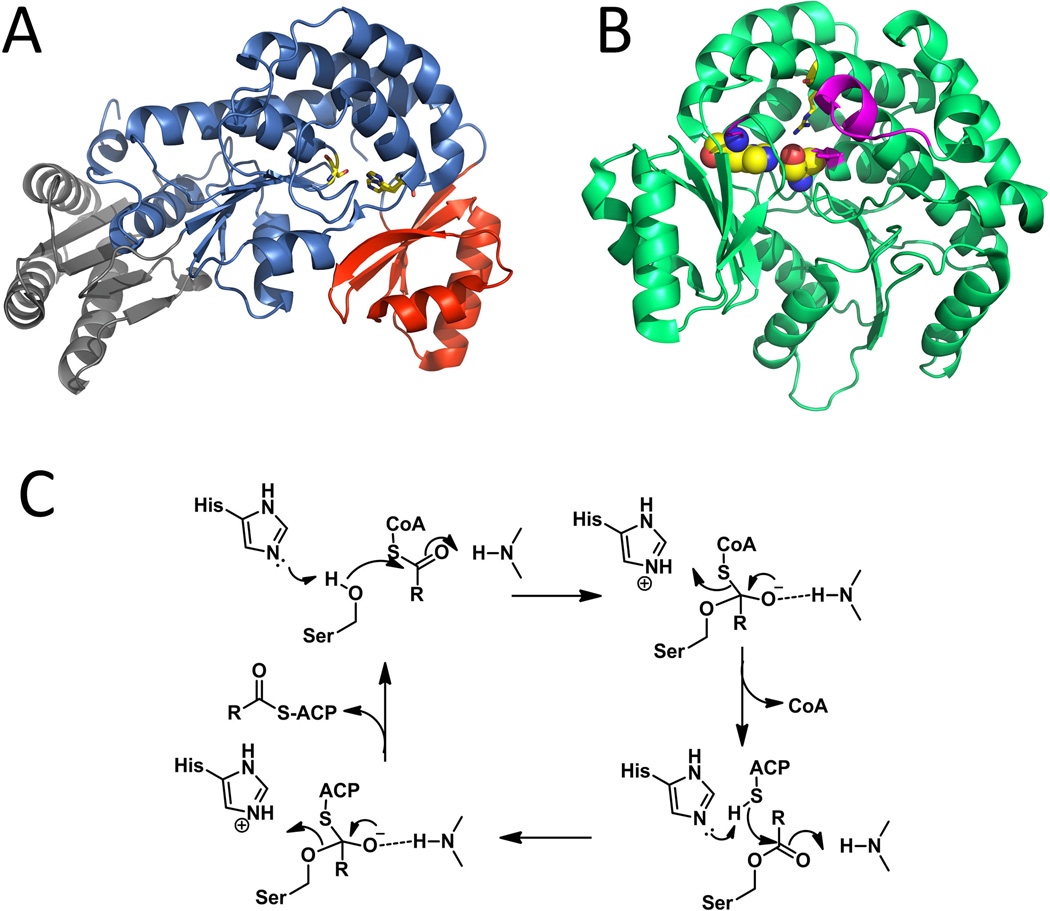
3.1. Ketosynthase (KS)
A KS domain accepts the polyketide chain from upstream (or cognate) ACP and catalyzes the decarboxylative Claisen condensation between the growing polyketide in the form of an acyl-S-KS complex and an extender unit acyl-S-ACP to give an elongated product (Figure 7-B). Several KS crystal structures from type I PKS system have been solved in the context of standalone KS-AT didomain, including DEBS KS-AT3 (Tang et al., 2007) and KS-AT5 (Tang et al., 2006). The KS forms homodimer and each monomer adopts a thiolase-like fold with αβαβα topology (Figure 7-A, blue regions). The homodimeric nature of KS domains is recognized to be crucial in maintaining the dimeric organizations of the entire FASs and PKSs (Tang et al., 2006). The KS-KS interactions are mainly via the backbone hydrogen bonds between two anti-parallel β-strands. A dimeric coiled-coil N-terminal peptide, which is present exclusively in modular PKSs, offers additional contact between the two KS monomers (Tang et al., 2006). This coiled dimer not only helps to stabilize the overall KS structure but also acts as a docking site for the upstream module during module-module communication (Figure 7-A, orange regions). A ~100 residue linker is found connecting the KS and AT domains and has unique αβα fold with α helices close to the KS side and the β strand interacting with the C-terminal α helix on the AT domain (Figure 7-A, yellow regions). The highly conserved hydrophobic interactions between KS and KS-AT linker, as well as AT and KS-AT linker are exclusively found in type I PKSs and are proposed to anchor the relative positions of KS and AT domains. Lastly, a ~ 30 residue post-AT linker loop has significant interactions with the KS domain through a number of hydrogen bonds (Figure 7-A, red regions).
Figure 7.
Selected PKS KS structures. (A) KS-AT dimer from DEBS module 5. Orange: dimeric N-terminal coiled-coil peptide; Blue: KS domain; Yellow: KS-AT linker; Green: AT domain; Red: post AT linker. PDB code: 2HG4. (B) Mechanism of KS catalyzed chain elongation. (C) KS-CLF heterodimer. Blue: KS domain; Green: CLF domain; Orange: grasping loop; The reactive cysteine, Cys169, and a gating residue that regulates chain length, Phe116′, are shown in sticks to the ~17-A polyketide tunnel. PDB code: 1TQY. 215×164mm (300 × 300 DPI).
Analogous to the well-studied FAS KSs, the KSs from type I PKSs have the same catalytic triad Cys-His-His, in which the cysteine is the nucleophile involved in the trans-thioesterification reactions. The cysteine is buried in the substrate pocket and can only be reached by pPant arm attached to the ACP domain (Tang et al., 2006). The active sites of KS from the different crystal structures can be perfectly superimposed, indicating the arrangement of the triad is likely optimum. The two histidine residues locate in the vicinity of the cysteine contribute to the formation of an oxyanion hole that stabilize the tetrahedron intermediate and presumably promote the decarboxylation of the extender unit (Figure 7-B).
While FAS KSs have higher affinity toward saturated acyl chains, the type I PKS KS domains can exhibit varying degrees of substrate specificity. The DEBS KSs show promiscuity toward polyketide chains ranging from diketide to decaketide, and those with different oxidation states on the β-carbons (Khosla et al., 1999a). On the other hand, some type I PKSs KS domains appeared to be more selective towards the functional groups present on cognate intermediates (Khosla et al., 2007; Khosla et al., 1999a). For example, KSs from trans-AT type I PKSs are highly specific towards the growing intermediates. Phylogenetic analysis of those KSs allows predicting structure features on the β/γ position of their substrates without knowing information on the reductive domains in the previous modules (Nguyen et al., 2008b). Crystal structures of KS domains show that the size and the shape of the substrate binding pocket determine specificity. The binding channel can be divided into a well-conserved phosphopantetheinyl binding region and a more variable acyl-binding region (Tang et al., 2006; Tang et al., 2007). The acyl-binding pockets of FAS KSs are mainly formed by hydrophobic residues and as a result, specific towards saturated fatty acyl groups. In contrast, the acyl-binding regions of PKS KSs are generally amphipathic, allowing the formation of hydrogen bond and salt bridge to stabilize the acyl intermediates with different levels of reduction (from unreduced to completely reduced). The substrate binding channel of type I PKS KSs are also much wider compared to that of FAS KSs, which also contribute to the observed increased substrate tolerance.
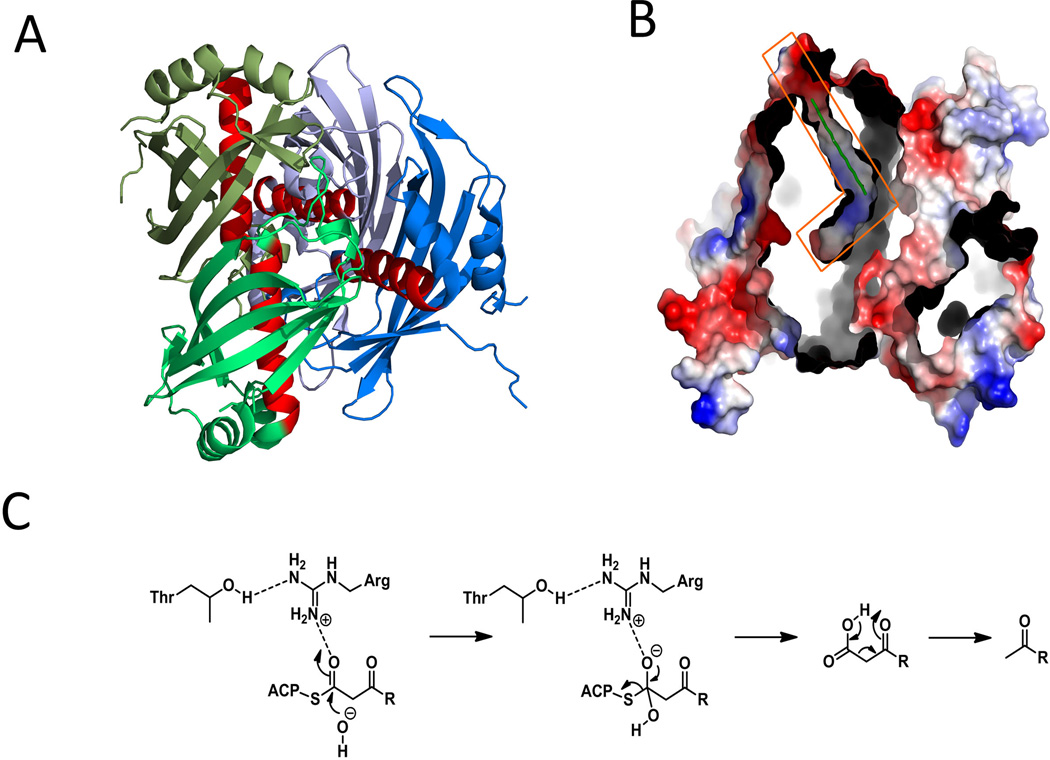
The KS domains in PKSs have been associated with the control of polyketide chain length. This is particularly important in the iterative PKSs, such as the iterative type I and bacterial type II systems. KS domain swapping experiments between two fungal iterative PKSs that synthesize polyketides of different length have led to the biosynthesis of new polyketides of different chain length (Zhu et al., 2007). Although the swapping experiment did not afford products of expected sizes, the experiment nevertheless hinted that KS is at least partially involved in controlling chain length. Direct insight into chain length control was obtained from both mutagenesis and structural studies (Keatinge-Clay et al., 2004) on the KS-CLF heterodimer in type II PKSs. The chain-length factor (CLF) is a catalytically inactive version of the KS domain and the interface of the KS-CLF dimer defines the size of the substrate binding channel. An amphipathic polyketide binding tunnel is clearly visible, originating from the active site cysteine to the KS-CLF interface (Figure 7-C). When larger gatekeeping residues at the interface of the act KS-CLF were changed to smaller residues, the mutant KS-CLF were able to synthesize longer polyketides. The opposite effect was observed when smaller gatekeeping residues were mutated to larger ones in the tcm KS-CLF (Tang et al., 2003). The mechanism of chain length control in both iterative type I and type II PKSs is based on size of the polyketide rather than the number of elongation cycles. This is both supported by the crystal structure and through various precursor directed biosynthesis experiments (Tang et al., 2004; Watanabe et al., 2003).
3.2 Acyltransferase (AT)
Acyltransferase (AT) is the catalytic domain that selects and loads the extender acyl group onto the ACP domains during the elongation cycle of FAS and PKSs. AT domains therefore serve as the gatekeeper in building block selection and play a key role in maintaining the fidelity of fatty acid and polyketide biosynthesis. AT domains from FASs and iterative type I PKSs use malonyl-CoA as extender units. In comparison, AT domains from modular type I PKSs have a much broader variation in substrate specificity. AT domains associated with the loading of starter units have been shown to select acetyl-CoA, propionyl-CoA, isobutyryl-CoA, crotonyl-CoA or isopentyl-CoA, etc (Liou et al., 2003; Lau et al., 2000). Extender AT domains in PKSs have been shown to select a variety of extender units in addition to malonyl-CoA, such as the most common propionate-derived 2S–methylmolanyl-CoA. The biogenesis and incorporation of different acyl-CoAs by the AT domain as a means to introduce structural diversity into polyketides has been covered in several recent reviews (Chan et al., 2009; Wilson et al., 2012).
In modular type I PKSs, instead of a dedicated AT domain built in within each module, there are numerous examples of PKSs in which a disassociated trans-acting AT enzyme are involved in the loading of the ACP domains with, in most cases, malonyl-CoA (Cheng et al., 2003; Piel et al., 2004; Nguyen et al., 2008a; Kopp et al., 2005; Zhao et al., 2010). These standalone AT enzymes interact with nearly all the ACPs domains in the PKS assembly line, thereby exhibiting broader ACP substrate specificities. For example, the trans-AT enzyme from disorazole pathway shows higher activity toward heterologous ACP domains than that of typical cis-acting AT domains (Wong et al., 2011). The malonyl-CoA:ACP transacylase (MAT) involved in fatty acid biosynthesis in Streptomyces, which is commonly shared by different type II PKSs, can also act as a trans-AT enzyme on type I PKSs. When the cis-acting AT domain in the sixth module of DEBSs is inactivated, MAT complemented the otherwise stalled pathway, loaded a malonyl group onto the ACP, and led to the effective production of 2-desmethyl-6-dEB (Kumar et al., 2003).
Over the last decade, the structures of ATs from different biosynthetic systems have been determined, including MATs from E. coli (Serre et al., 1995) and Streptomyces coelicolor (Keatinge-Clay et al., 2003), modular type I PKS ATs from DEBS (Tang et al., 2006; Tang et al., 2007), trans-acting AT from disorazole pathway (Wong et al., 2011) and iterative type I PKS AT from dynemicin pathway (DYN10) (Liew et al., 2012). The monomeric ATs all share the same two-subdomain structure: a core subdomain contains α/β hydrolases-like core with a smaller ferredoxin-like subdomain (Figure 8-A). Both cis- and trans-acting ATs use the highly conserved catalytic Ser-His dyad and ping-pong bi-bi mechanism to transfer the acyl group from CoA to the ACP domains (Ruch et al., 1973). The nucleophilicity of catalytic serine hydroxyl group is strengthened through hydrogen bonding with the histidine, as evidenced by the ~ 3 Å distance between the Nε2 atom of the histidine and the Oγ atom of the serine (Figure 8-B) (Keatinge-Clay et al., 2003). The oxyanion hole that stabilizes the tetrahedron intermediate is formed by the backbone amides of a glutamine from the conserved PGQGXQ motif and a residue adjacent upstream to the catalytic serine (Keatinge-Clay et al., 2003; Serre et al., 1995) and the exiting CoA is protonated by the catalytic histidine. A second tetrahedral intermediate is formed when the ACP phosphopantetheine thiol attacks the ester carbonyl. The acyl binding cleft is relatively hydrophobic, thus preventing the binding of water molecules and the hydrolysis of acylated enzyme oxyesters in the absence of free thiol acceptors (Serre et al., 1995). In cases where hydrolysis is noted, the hydrolyzed acyl group binds to the pocket non-covalently and greatly inhibit the AT efficiency, as seen in dynemicin AT DYN10 (Liew et al., 2012).
Figure 8.
Selected AT structures and typical catalytic mechanism. (A) Structure of ATDYN10 from dynemicin pathway. KS-AT linker (gray), α/β hydrolases-like core (Blue) and ferrodoxin-like smaller subdomain (Red). The catalytic His-Ser dyad is shown as sticks. PDB code: 4AMP. (B) Residues and region important to substrate binding or selection in DEBS AT5: catalytic dyad are shown as spheres; the arginine stabilizes the substrate 3-carboxylate group is shown as sticks; important region that defines the substrate specificity are labeled in magentas. PDB code: 2HG4. (C) catalytic mechanism of AT. 215×188mm (300 × 300 DPI).
Co-crystallization of MAT with either acetate or malonyl-CoA showed that the presence of a positively charged side chain of the highly conserved arginine (for instance, Arg122 in S. coelicolor MAT) can stabilize the 3-carboxylate group via a salt bridge (Keatinge-Clay et al., 2003; Rangan et al., 1997; Serre et al., 1995). Molecular docking and bioinformatics studies of different AT domains have led to the identification of several conserved sequences motifs that play key roles in determining the substrate specificity. First, a motif that is located about 30 residues upstream of the catalytic serine forms part of the substrate-binding pocket and influences the selection between malonyl-CoA and methylmalonyl-CoA. After analyzing over two hundred domains, Yadav and his coworkers mapped a conserved ZTXX[AT][QE] motif to malonyl-specific ATs and [RQSED]V[DE]VVQ to methylmalonyl–specific ATs (Z represents hydrophilic residue, while X represents hydrophobic residue) (Yadav et al., 2003). This specificity code for AT was partially verified in motif swapping experiment with DEBS AT4 (Reeves et al., 2001). Second, the residue beyond the catalytic serine, which contributes to the formation of oxyanion hole, is also sterically important in substrate binding. A branched hydrophobic residue (Leu or Val) is present in malonyl-specific ATs, while a residue with more linear sides chain (Glu or Met) is found in this position for methylmalonyl-specific ATs (Haydock et al., 1995). Third, a YASH motif that includes the catalytic histidine is present exclusively in the methylmalonyl-specific DEBS ATs (Reeves et al., 2001), while a HAFH motif is found in malonyl-specific ATs. Lastly, in the DEBS KS-AT5 structure, the first α helix of α-β-α loop of the C-terminal linker region was positioned at the entrance of the substrate pocket and was shown to be essential in determining the substrate specificity through swapping between rapamycin AT2 and DEBS AT2 (Lau et al., 1999) (Figure 8-C).
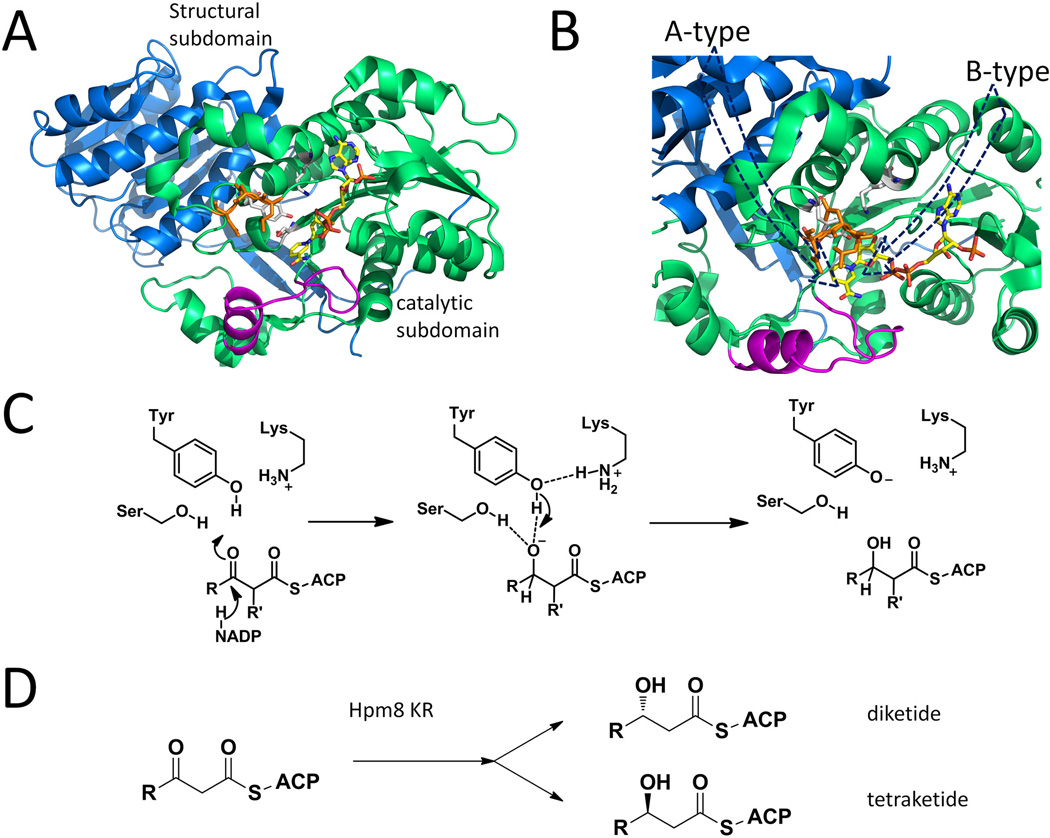
3.3 Ketoreductase (KR)
Ketoreductases (KRs) in PKSs and FASs stereospecifically reduce the carbonyl group of the β-ketoacyl-ACP intermediates. They belong to the short-chain dehydrogenase/reductases (SDR) family, which are well known to be NAD(P)+/NAD(P)H dependent oxidoreductases. All of the reported KRs contain at least one highly conserved Rossmann fold, which is composed of a central, 6 or 7 strands β-sheet flanked with 3 or 4 α-helices on each side. This structural moiety creates a characteristic groove that binds the NADPH cofactor (Kavanagh et al., 2008). KR can be a standalone protein (type II) or a built-in domain in type I PKSs. Depending on the stereochemistry (D/L) on the OH-bearing β-carbon in the product, a type I KR can be classified as either an “A-type” (for L- configuration) or a “B-type” KR (for D- configuration) (Keatinge-Clay, 2007). When the substrate is an α-substituted β-ketoacyl intermediate, some KR may catalyze epimerization on the α-carbon before the ketoreduction (Valenzano et al., 2009; Keatinge-Clay, 2007). Based on the configuration on the accepted α-carbon, the A- and B-type KRs can be further classified into A1, A2, B1 and B2: those reducing an unepimerized 2R substrate are denoted with a “1” and those accept an epimerized 2S substrate are denoted with a “2”. Reduction-incompetent KRs are called C type: C1-type for KRs that lack both the catalytic tyrosine residue and NADPH binding motif; C2-type for KRs that present the complete catalytic triad but miss the NADPH binding motif. A protocol was developed to assign substituent stereochemistry accordingly by combining the KR fingerprints with the presence of other reductive domains (Keatinge-Clay, 2007). Four type I KR domain crystal structures have been reported, including one A-type KR (Amphotericin KR2 (Zheng et al., 2010)), two B-type KRs (DEBS KR1 (Keatinge-Clay et al., 2006) and tylosin KR1 (Keatinge-Clay, 2007)) and one C2-type KR (pikromycin KR3 (Zheng et al., 2011)).
Type I PKS KRs show pseudodimeric structures comprise two SDR subdomains: a structural subdomain followed by a catalytic subdomain in sequence. In modules that contain the ER domain, sequence for ER domain is inserted into those of the structural and catalytic subdomains. The NADPH binding site is missing in the structural subdomain and the catalytic subdomain is stabilized by the structural subdomain (exemplified by DEBS KR1, Figure 9-A). The conserved Ser-Tyr-Lys triad in the catalytic subdomain is responsible for activation of the target β-carbonyl group on the substrate and to stabilize the resulting oxyanion after the hydride addition (Cane, 2010) (Figure 9-C). Usually an inserted “lid” consists of a helix and a loop helps to form the substrate binding pocket. Variations in residues between different KR types have been found to dictate stereochemical outcome. In B-type KR, an “LDD” motif is located in the loop that precedes the active site (Figure 9-A), while a tryptophan is found in at a key position between catalytic serine and tyrosine in an A-type KR. These features result to two alternative substrate entry modes for A-type and B-type KRs. In A-type KRs, the phosphopantetheinyl arm slips into the active site from the groove behind the lid helix, while in B-type KRs, that entry mode is prevented because of the LDD motif. Instead, the substrate enters the active site from the opposite side relative to the lid (Figure 9-B). The different positioning of the substrate lead to the hydride from the NADPH attacking from different prochiral faces of the thioester carbonyl and generating different ketoreduction stereochemistry.
Figure 9.
Selected KR structures and typical catalytic mechanism. (A) DEBS KR1 with cofactor binding, catalytic triads are shown as sticks. Blue: structural subdomain; Green: catalytic subdomain; Orange: “LDD” moiety shown as sticks. Magenta: Lid helix and loop. PDB code: 2FR0. (B) Different paths of entrance result in the different product stereochemistries in A and B type KRs. The presence of orange DLL loop in B-type KR will prevent substrate from entering behind the magenta lid. (C) Mechanism of KR catalyzed reduction. (D) Substrate-tuned stereoselectivity in Hpm8KR. 215×176mm (300 × 300 DPI).
Based on sequences comparison, KRs from iterative PKSs should share similar two-subdomain structure as modular type I KRs. One would expect that since a single KR is utilized during the different iterations of chain elongation, all the β-carbonyls that are reduced by KR would display the same stereochemistry. Interesting, careful stereochemistry analysis of the hypothemycin PKS Hpm8 shows that the KR domain can switch the stereoselectivity when processing substrates of different lengths (Figure 9-D). Mutations of A- and B-type KR “fingerprint residues” in Hpm8 did not alter the differential stereoselectivity, which indicates additional structural features are necessary to direct substrate binding mode. Through swapping sequences with another iterative type I PKS (Rdc5), Zhou et al. pointed out that the β5α5α6a motif should be the determinant region that modulates this novel substrate-tuned stereoselectivity (Zhou et al., 2012).
3.4 Dehydratase (DH) and DH-Like Domains
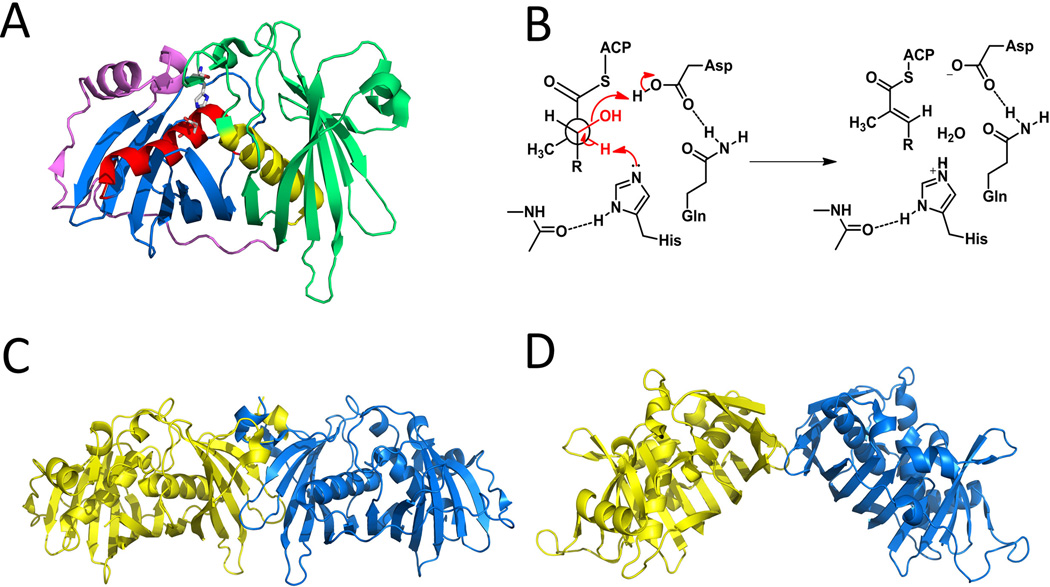
The PKS dehydratase domain (DH) catalyze reversible dehydration reactions on the β-hydroxyl-acyl intermediate bound to the ACP, resulting in an α,β-unsaturated acyl-ACP intermediate in either the cis or trans configuration. The downstream reactions catalyzed by ER, KS or TE will pull the equilibrium toward the direction of dehydration. Both the mFAS and PKS DHs are hypothesized to have been evolved from a common type II dehydratase ancestor through gene duplication and fusion (Akey et al., 2010; Keatinge-Clay, 2008). Up to date, X-ray structures of five DH domains from modular PKS have been reported, including DEBS DH4 (Keatinge-Clay, 2008) and the four DHs from curacin A biosynthetic pathway (Akey et al., 2010). As shown in the reported structures, the DH domains exist as a dimer, with each monomer itself forms a pseudodimer of two ‘‘hot-dog” folds, resulting in a “double hot-dog” fold. Each “hot-dog” fold composes of a “sausage-like” long helix wrapped around by a “bun” that consists of a curved β-sheet (Figure 10-A, sausages are in red and yellow, buns are in blue and green). The ‘‘catalytic dyad’’ active site in each monomer is located at a tunnel formed between the N- and C-terminal “hot-dog” folds, comprising of a histidine from the first “hot-dog” fold, and an aspartate from the other (Figure 10-A, both residues are shown as sticks). Dimerization may not be necessary for catalysis as the complete active site is formed without participation from the other monomer, which is supported by the functional mFAS DH monomers (Smith et al., 2003). In the reported crystals, the substrate tunnel in DH is covered by a flexible region formed by less ordered elements. Specific protein-protein interaction with ACP domains may be necessary for opening of the cover and delivery of the substrate.
Figure 10.
Selected DH structures and typical catalytic mechanism. (A) Double hot-dog structure in DEBS DH4. The catalytic histidine and aspartate (shown as sticks) are provided from two different monomers. PDB code:3EL6. (B) A typical catalytic mechanism of DH domain. (C) An extended DH dimer conformation exemplified by DEBS DH4. PDB code:3EL6. (D) “V” shaped DH dimer in mFAS. PDB code: 2VZ9. 215×127mm (300 × 300 DPI).
The dehydration proceeds through deprotonation of the α-carbon by the histidine and abstraction of β-hydroxyl by the aspartic acid. The configuration of the double bond formed is determined mainly by the chirality of the β-hydroxyl group following ketoreduction by the KR domain in the same module. Products of A-type KRs are dehydrated to cis- (or Z) double-bond products and B-type products to trans- (or E) double-bonds. The critical constraint that results in this specificity is that the substrate α-hydrogen and β-hydroxyl groups need to be adjacent to the prepositioned histidine and aspartic acid, respectively, while within the confines of the substrate tunnel. In vitro biochemistry assays have shown that all of the (E)-2-ene-acyl-ACP products in modular PKSs share a common syn dehydration mechanism (Guo et al., 2010; Valenzano et al., 2010) (Figure 10-B).
In contrast to the highly similar structures for PKS and mFAS DH monomers, the corresponding dimeric structures are notably different in the angle formed by the two monomers. All reported PKS DH dimers have similar flat contour but the mFAS DH dimer is V-shaped (Keatinge-Clay, 2008; Akey et al., 2010; Maier et al., 2008) (Figure 10-C and 10-D). The PKS DH dimer interface is formed by complementary contacts mainly between the residues on the β strands of N-terminal hot-dog folds in each monomer. The interface area accounts only for 4%–6% of the total monomer surface area, which is similar to that at the mFAS DH dimer interface. The elongated DH dimer seems to be common in modular PKSs because the hydrophobic residues involved in dimerization are conserved. Such conformations in the PKS DH dimers may be a requirement in spatially assembling multiple modules of PKSs, and suggests that the multi-domain architecture of the reducing portion of modular PKSs (Top portion in 3D structure) should be different from that of the mFAS. (See the discussion in ER section)
DH-like double hot-dog fold is also found in the reported structure of product template domain (PT) (Figure 11-A) from a fungal iterative, nonreducing PKS (PksA) involved in aflatoxin biosynthesis (Crawford et al., 2009). Phylogenetic analysis (Kroken et al., 2003) suggests that iterative nonreducing PKSs are evolved from ancient reducing PKSs. Combining this with the fact that the PksA PT was found to be dimeric both in the crystal and in solution, and has a double “hot-dog” fold like the DH domain, it is reasonable to infer that PTs are evolved from the DH and are located at the positions of DH domains in the three dimensional structure (Figure 11-C). PTs catalyze regio-specific aldol cyclization of the unreduced poly-β-ketoacyl:ACP intermediate synthesized by the iterative PKS. The PT domain of PksA catalyzes stepwise first- and second-ring cyclizations to yield the ACP-tethered bicyclic intermediate (Crawford et al., 2008). A deep pocket that can accommodate the phosphopantetheinyl tethered intermediate chain is found in PksA PT crystal. The pocket can be divided into three regions: a pPant-binding region close to the surface, a cyclization chamber that can accommodate the bicyclic product core and a hexyl moiety binding site sitting in the innermost part of the pocket that is specific for the hexanoyl starter unit (Figure 11-B). Notably, a ‘wet’ side of the cyclization chamber, which contains a network of crystallographic water molecules and nearby hydrophilic residues, is proposed to keep C9 in the substrate in the electrophilic keto tautomeric form for the intramolecular aldol cyclizations (Crawford et al., 2009). The catalytic dyad, located at the surface of the cyclization chamber, is also composed of a histidine and an aspartate from two different “hot-dog” folds like in the DH domain. The catalytic histidine acts as base to deprotonate the methylene and generates the nucleophile. The aspartate is involved in polarizing the catalytic histidine, instead of helping removing a hydroxyl as in DH domain. PT domains of different cyclization regioselectivity have been functionally characterized and can be grouped phylogenetically (Li et al., 2010). It is expected that the active site geometry will dictate the positioning of the unreduced polyketide chain with respect to the histidine residue, which can then lead to different cyclization patterns.
3.5 Enoylreductase (ER)
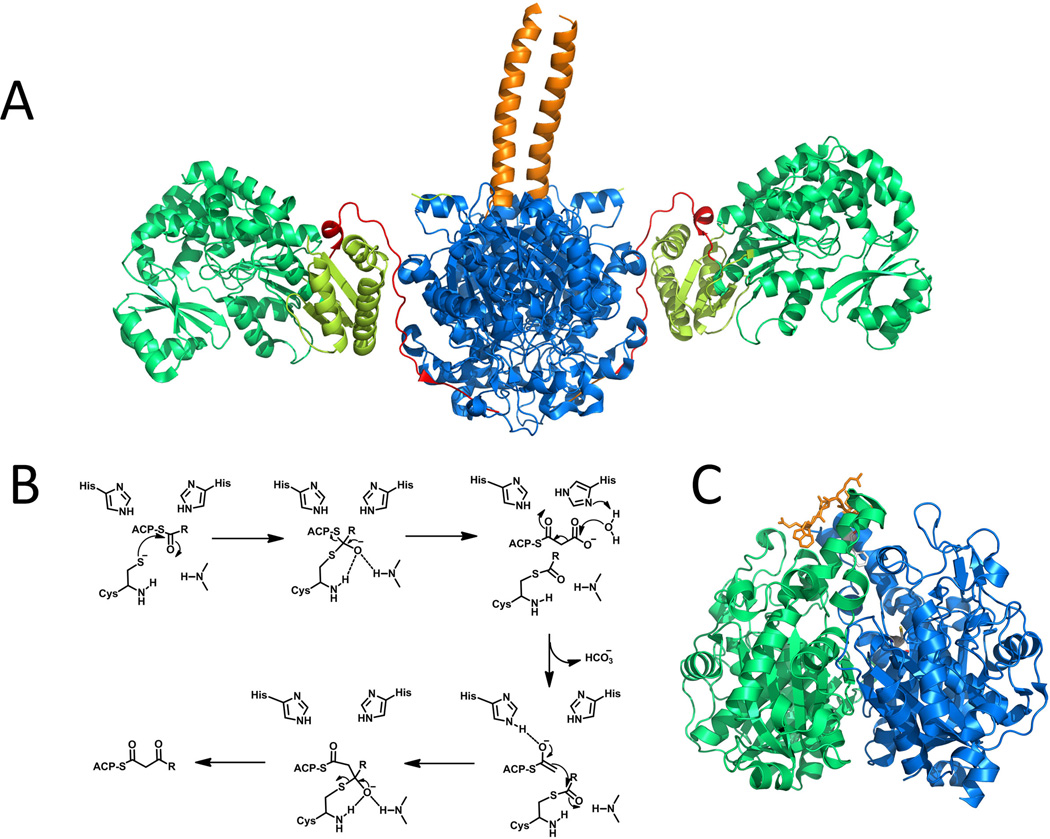
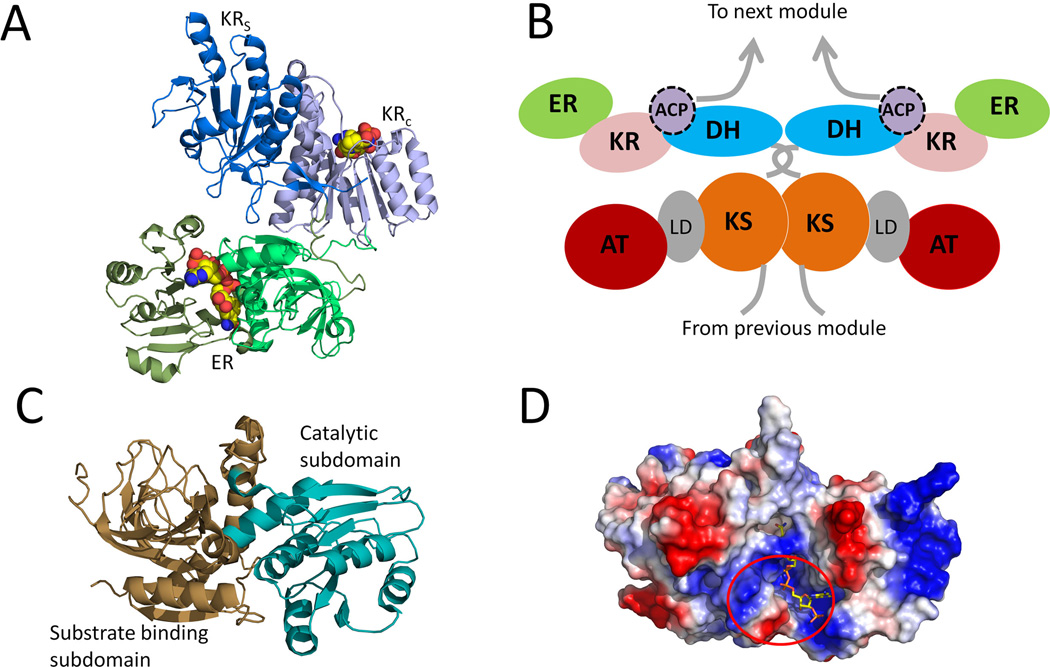
The enoyl-acyl carrier protein reductases (ERs) catalyze the reduction of an enoyl-ACP to an α, β-saturated acyl-ACP. Different from KRs, ERs use the 4-pro-R hydride instead of 4-pro-S hydride of NADPH for enoylreduction (Yin et al., 2001; Ames et al., 2012). Compared to other type I PKS domains, ER has the least structural and functional information. Very recently, structures of ER from both the modular (cis) and iterative (trans) PKSs were reported, including SpnER2 (solved in the form of ER-KR didomain) from spinosyn PKS module 2 (Zheng et al., 2012) and the trans-acting ER LovC involved in lovastatin biosynthesis(Ames et al., 2012). SpnER2 reduces a tetraketide, while LovC interacts with LovB (containing a null ER) and selectively reduces the tetra-, penta-, and heptaketides intermediates (Kennedy et al., 1999; Ma et al., 2009). Like the ER domain in mFAS, PKS ERs also belong to Medium-chain dehydrogenases/reductases (MDR) superfamily (Nordling et al., 2002), which typically comprise of two subdomains: a C-terminal Rossmann-fold NADPH binding subdomain and a catalytic N-terminal substrate-binding subdomain (Figure 12-A and Figure 12-C). The NADPH binding site is located in a cleft between the two subdomains. In the LovC-crotonoyl-CoA complex, the four-carbon crotonoyl group extends into a hydrophobic putative substrate-binding pocket that is adjacent to the nicotinamide ring of the bound NADP+. In contrast to most enzymes in the MDR family that form homodimers, both SpnER2 and LovC were shown to exist as monomers. The monomeric form of LovC was rationalized by the presence of additional loop (~10 residues) in the region between the catalytic and cofactor-binding domains. Computer modeling shows that the extended loop prevents homodimer formation because of significant steric clash with the modeled dimeric partner. This additional loop in LovC is conserved among other trans-acting ERs that are found in fungal iterative type I PKSs. The monomeric form of these ER may be required for interaction with the ACP partners in the PKSs. Indeed, gel-filtration interaction study indicates that LovC forms a complex with the LovB. A docking simulation using a homology model of LovB ACP and the solved structure of LovC suggests that a positively charged surface patch adjacent to the active site of LovC may be required for LovB-LovC interactions (Figure 12-D).
Figure 12.
The crystal structures of the ER domains. (A) structure of Spn(KR+ER)2 didomain with cofactor (as spheres) binding. PDB code: 3SLK. (B) Proposed schematic of a module in a multimodular PKS. (C) Structure of LovC. Brown: substrate binding subdomain; palegreen: catalytic subdomain. PDB code: 3B70. (D) surface charge on LovC and predicted docking region with LovB ACP. Bound NADP is shown in sticks. 215×143mm (300 × 300 DPI).
Our current understanding of enoylreduction process is that a hydride is transferred from NADPH to the β-carbon of the substrate, followed by protonation of the α-carbon by either a general acid or solvent. Structural guided mutagenesis studies on both SpnER2 and LovC show that a conserved lysine (K422 in SpnER2 and K54 in LovC) is critical for the enoyl reduction activity. Compared to other positions, mutations to this lysine lowers the activity more significantly, which agrees with the recent mutational study on the ER domain from module 13 of rapamycin PKS (RAPS), which also concluded that no single residue alone serves as oxyanion hole or Brønsted acid (Kwan et al., 2010). Tyr241 in SpnER2, located at the opposite side of the conserved lysine in the active site cleft, is proposed to protonate the α-carbon following hydride addition to generate the S configuration on the product. In vivo mutagenesis shows that changing the corresponding tyrosine to valine in DEBS ER4 switches the product stereochemistry at the α-position from S to R (Kwan et al., 2008). Upon mutation of the tyrosine, the aforementioned lysine is proposed carry out an additional function in protonating the α-carbon from the opposite side and generates the R configuration.
Crystallization of Spn(ER-KR)2 also allowed investigation of the ER-KR interface in modular type I PKS, which is larger than that observed for mFAS. ER-KR contacts are mostly found between the nonconserved hydrophilic residues in the catalytic subdomain of SpnKR2 and those in the substrate-binding subdomain of SpnER2. The linkers that connect ER and two KR subdomains help mediate this interface. The C-terminal end of the insertion loop is shorter than that in mFAS, leading to the observed differences in ER-KR orientation. Divergent evolution of multimodular PKSs and mFASs based on the monomeric state of SpnER2 is proposed (Zheng et al., 2012). In the multimodular PKSs, the monomeric ER is necessary to enable sufficient flexibility of the ACP domain in the same module to interact with both the intramodular domains and the downstream, dimeric catalytic domain (either KS or TE) (Figure 12-B).
3.6 Thioesterase (TE)
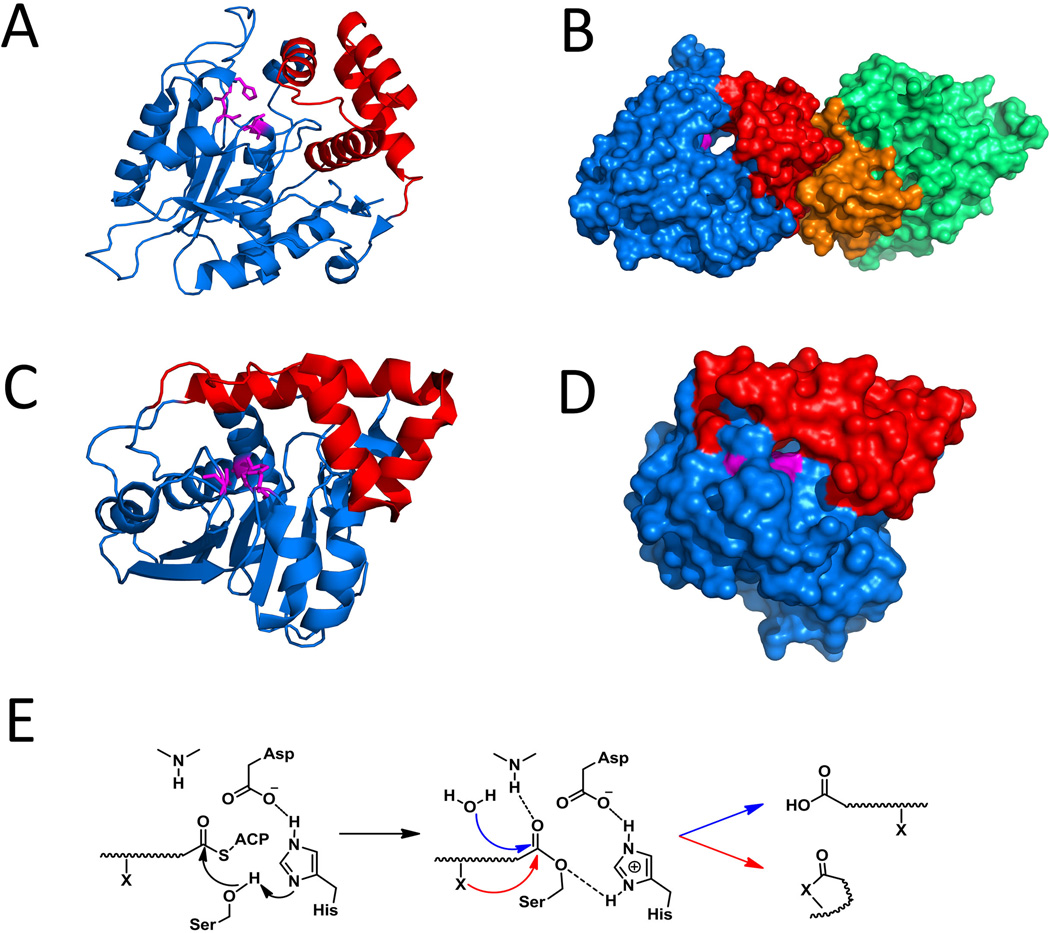
Like in mFAS, thioesterase (TE) in PKS system is involved in the product release. PKS TEs can break ACP thioester linkage by catalyzing cyclization or hydrolysis of the acyl substrate. Two types of TEs with distinct structures are found in PKSs: α/β hydrolase-fold TEs and “hot-dog” fold TEs. The α/β hydrolase-fold TEs can be cis-acting domains located at end of the modular PKS assembly line or at the C-terminus of the iterative PKS (type I TEs), or dissociated enzymes responsible for the hydrolytic release of stalled, aberrantly loaded chains on PKSs (types II TEs). They facilitate covalent catalysis in which the acyl chain is first transferred to the active site serine, generating an acyl-O-TE intermediate. Inter- or intramolecular nucleophilic attack on the carbonyl of acyl-O-TE oxyester then releases the product from these TEs (Figure 13-E). In contrast, “hot-dog” fold TEs feature a five-stranded anti-parallel β-sheet wrapping around a long α-helix and several shorter helices (Figure 14-A) and utilize a different catalytic mechanism to cleave the thioester bond, in which no covalent enzyme-substrate intermediate is formed (Figure 14-C). To date, numerous TE structures have been reported, including type I TEs from modular PKSs DEBS (Tsai et al., 2001), pikromycin PKS (Akey et al., 2006; Tsai et al., 2002; Giraldes et al., 2006), tautomycetin PKS (Scaglione et al., 2010) and curacin A PKS (Gehret et al., 2011); from the iterative norsolorinic acid synthase (Korman et al., 2010); type II TE from the pathways of rifamycin (Claxton et al., 2009); “hot-dog” fold TEs from the biosynthetic pathways of the enediynes calicheamicin (Kotaka et al., 2009) and dynemicin (Liew et al., 2010).
Figure 13.
Selected structures of α/β-hydrolases TE domains in PKS system. Lid region are shown in red and catalytic residues are shown in magentas. (A) structure of pikromycin TE (type I). PDB code: 1MNQ. (B) surface representation of pikromycin TE dimer. (C) structure of Rifamycin TE (Type II). PDB code: 3FLA. (D) a surface representation of rifamycin TE. (E) Catalytic mechanism of α/β-hydrolases TE (Hydrolysis vs. macrocyclization). 215×190mm (300 × 300 DPI).
Figure 14.
Selected structures of hot-dog fold TE domains in PKS system. (A) hot-dog fold dynemicin TE DynE7 tetramer structures (“sausages” are in red). PDB code: 2XEM. (B) Surface representation of DynE7 tetramer and the L-shaped substrate binding channel in orange box. (C) Mechanism of decarboxylative hydrolysis in hot-dog fold TEs. 215×152mm (300 × 300 DPI).
Whereas TE from the mFAS (Chakravarty et al., 2004) is monomeric, all type I TEs from modular PKSs have been crystalized as dimers. The active-site triad (Ser-His-Asp/Glu) and most of the important secondary structure features of α/β-hydrolases are conserved among the four available structures. The α/β-hydrolase core contains a central seven-stranded β-sheet connected by α-helices, with β2 antiparallel to the remaining strands. The characteristic αD helix between β6 and β7 in the α/β-hydrolase family is replaced by a less-conserved, subdomain that functions as a “lid”. The substrate binding region is formed between the α/β hydrolase core and the lid subdomain (Figure 13-A and Figure 13-C). Depending on orientation of the lid subdomain with respect to the core, TE can adopt either a closed or open conformation with respect to bulk solvent. The DEBS, pikromycin and tautomycetin TEs are partially closed and the dimer interface is formed through hydrophobic interaction between helices in the lid subdomain. A long substrate channel is visible in the structure and spans the entire width of the protein, with the active site triad located at the center of the channel (Figure 13-B). The acyl substrate is delivered into the active site serine from one end of the channel by the upstream ACP and is exported from the other end at the completion of the reaction. The different residues that form the substrate channel in the four TEs, mostly on the interior side of the lid subdomain, account for the highly variable channel shape and the substrate specificities. In the macrolactone forming DEBS and pikromycin TEs, the wide channel can accommodate the acyl chain in a cyclized conformation, while in the hydrolytic tautomycetin TE, a narrower and more constricted channel favoring a linear product is found. The size of the substrate channel can be affected by pH and increases upon increasing pH in DEBS and pikromycin TEs (Tsai et al., 2002). Curacin TE is an example of an open-conformation TE. This domain can catalyze multiple reactions including the combination of hydrolysis, elimination of sulfate group and decarboxylation to generate alkene products from β-sulfated acyl-ACP substrates (Gehret et al., 2011).
Type II TEs remove incorrect acyl chains from carrier domains. This is to ensure no stalling of the PKS function occurs due to incorrectly loaded or tailoring moieties. Disruption of type II TE usually results in a significant decrease in product yield. Type II TEs also show the α/β hydrolase fold similar to that of type I TEs (Figure 13-C). In the reported crystal structure of rifamycin TE, several forms with different conformations of the lid were captured. The flexibility is caused by a “lid loop” that is 16 residues long. Movement of the lid helices and the flexible lid loop affects the size and shape of the substrate chamber, which allows the TE to bind a variety of different substrates. The interior surface of the substrate chamber is hydrophilic, which explains the board substrate specificity towards charged and uncharged substrates, short-or medium-chain length, or branched acyl thioesters (Figure 13-D). The slightly positive charged surface surrounding the phosphopantetheine entrance is involved in the protein-protein interactions with ACP.
Unlike typical hot-dog fold TEs that utilize acyl-CoA substrates (Dillon et al., 2004), enediyne hot-dog fold TEs release polyketide products from the ACPs. Important residues for ester bond activation and oxyanion intermediate stabilization in other hot-dog fold TEs are missing in enediyne hot-dog fold TEs, which suggests a different catalytic mechanism. Actually these enediyne TEs, including the characterized CalE7 and DynE7, form a new TEBC subfamily (Liew et al., 2010) among the hot-dog fold TEs (Dillon et al., 2004). They show as tetramers in solution and in reported crystal structures (Figure 14-A). Contacts within the primary dimer create an L-shaped substrate binding channel, comprising of a hydrophilic outer phosphopantetheinyl binding channel and a hydrophobic inner polyene binding channel (Figure 14-B). Mutagenesis study (Kotaka et al., 2009) shows that the presence of a highly conserved arginine residue in the central helix is essential to the catalyzed decarboxylic hydrolysis and may function as an oxyanion hole (Figure 14-C).
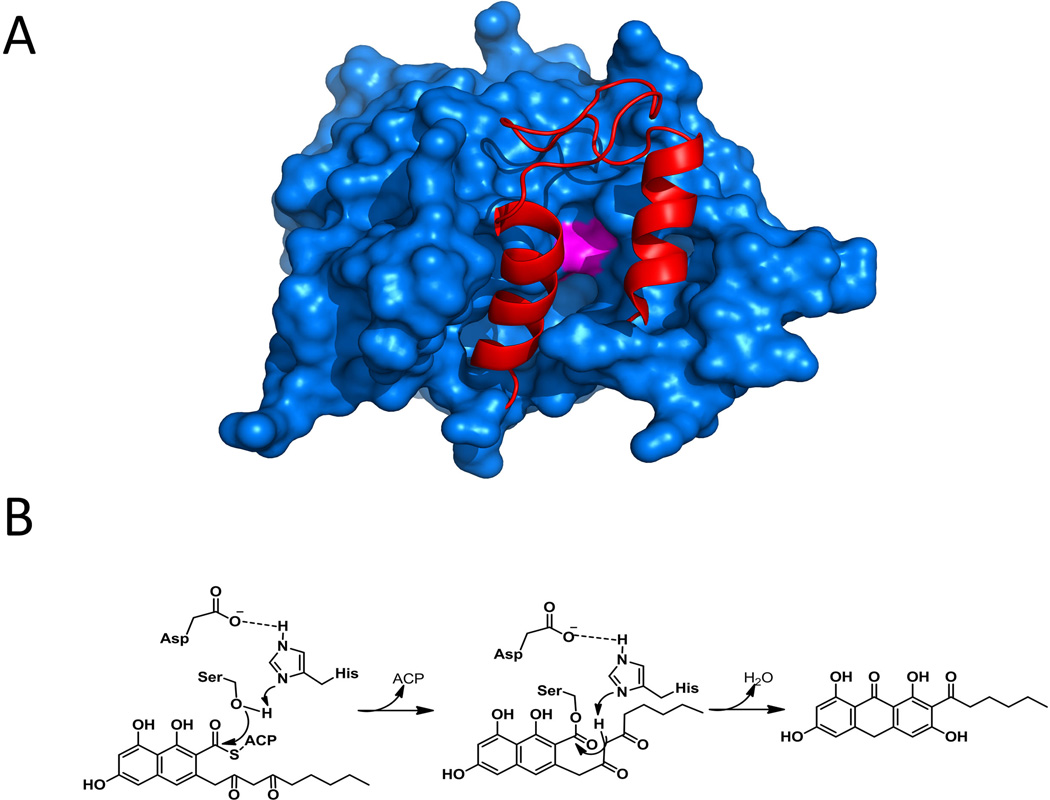
Some C-terminal TE domains of nonreducing iterative PKSs from fungi catalyze the regioselective Claisen condensation using thioester carbonyl as the electrophile and release of the aromatic polyketide, and hence are also referred to as TE/CLC domain (Fujii et al., 2001). In the absence of the TE/CLC domain, the polyketide intermediate can be released in as α-pyrone shunt products by nucleophilic attack from an enolate oxygen (Crawford et al., 2008; Fujii et al., 2001; Ma et al., 2008). The monomeric TE/CLC domain from the norsolorinic acid PKS, PksA, shares similar α/β-hydrolase fold with the typical modular PKS TEs (Korman et al., 2010). The substrate-binding region is similarly located between the core and the lid, and contains hydrophobic residues for binding of the aromatic substrate. A loop region in the lid is highly flexible, and is proposed to be the entrance to the binding pocket (Figure 15-A). Once delivered, the polyketide acyl intermediate is transesterified from the pPant arm of the ACP to the active site serine. After release of the freed ACP, the lid can close and the diketo portion of the substrate can rotate into a conformation that favors enolate formation and subsequent attack on the carbonyl of the oxyester (Figure 15-B). Therefore, ACP-TE/CLC protein-protein interaction and reorganization of the TE/CLC pocket are prerequisites for substrate delivery and product formation. In support to this proposed interaction, NMR titration experiments have shown interaction between analogous region of the NRPS TE domain and the peptidyl carrier protein partner (Koglin et al., 2008; Frueh et al., 2008).
Figure 15.
Structure and mechanism of TE/CLC domain. (A) Surface representation of closed PksA TE. Red: lid region; Magentas: catalytic residues. PDB code: 3ILS. (B) Mechanism of TE catalyzed Claisen condensation. 215×163mm (300 × 300 DPI).
3.7 Acyl carrier proteins (ACPs)
Acyl carrier protein (ACP) is a small (~80–100 residues) non-catalytic domain that tethers the growing polyketide and building block on the pPant arm. The posttranslational modification that transforms apo ACP domains into its holo form is catalyzed by a phosphopantetheinyl transferase (PPTase), such as sfp from Bacillus subtillis (Quadri et al., 1998). Using the 20Å pPant arm, the ACP delivers substrates to the different catalytic domains during PKS catalysis. To facilitate the assortment of protein-protein interactions, ACP binds to the specific enzyme partners through different structural features on the small protein. Therefore, obtaining the structures of ACPs and decoding the residues that are responsible for the different (and orthogonal) interactions is the key in understanding PKS function. This is especially important in type I PKSs, in which fine-tuned KS:ACP interactions play key roles in conferring the unidirectional substrate transfer of modular PKSs (Kapur et al., 2010; Kapur et al., 2012), and controlling the permutation of tailoring domain functions in iterative PKSs. Due to the inherent flexibility of the ACP domains and the adjacent linker regions, ACPs are typically absent in the crystallographic snapshots of intact megasynthases. However, the solution structures of ACPs have been obtained via solution-phase NMR spectroscopy. To date, several solution structures of ACPs have been characterized, including: DEBS ACP2 (Alekseyev et al., 2007), holo and apo DEBS ACP6 (Tran et al., 2010), calicheamicin ACP (Lim et al., 2011), curacin ACPI (Busche et al., 2012), apo- (Crump et al., 1997), holo- (Evans et al., 2008), derivatized forms (Evans et al., 2009; Haushalter et al., 2011) of actinorhodin ACP, frenolicin holo-ACP (Li et al., 2003), oxytetracycline apo-ACP (Findlow et al., 2003) and norsolorinic PKS holo-ACP (Wattana-amorn et al., 2010).
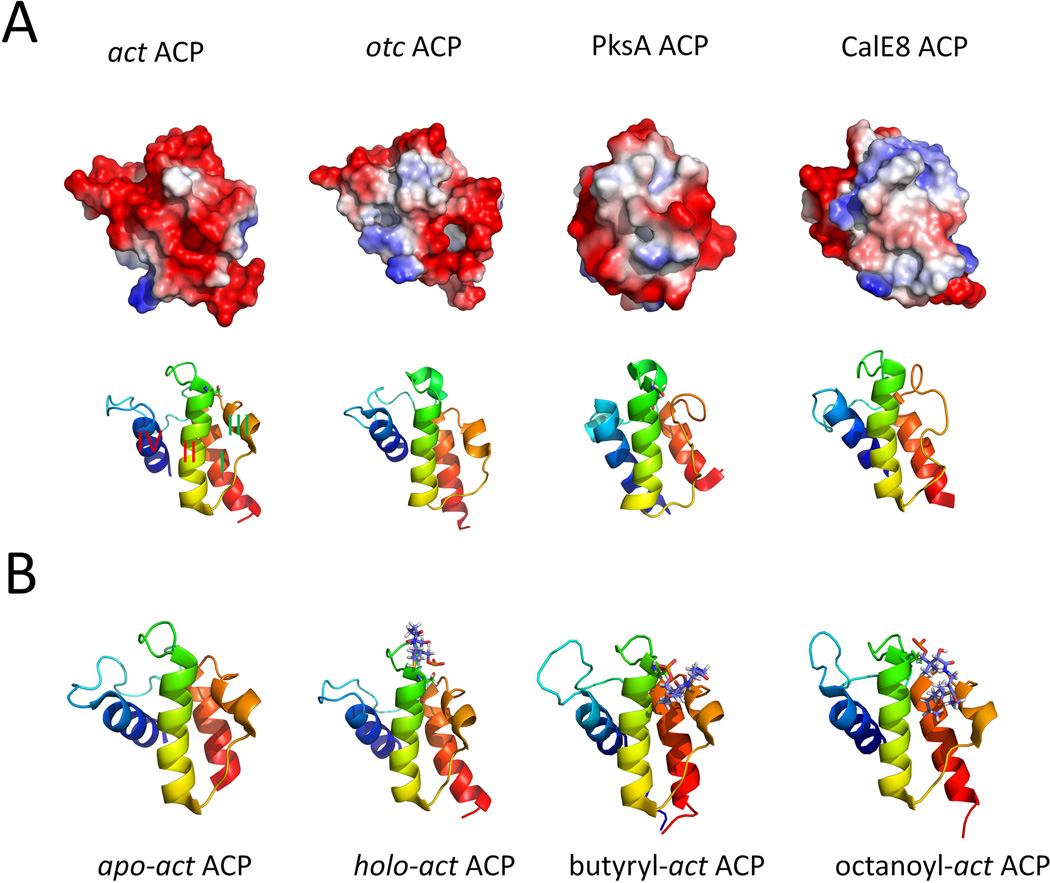
Despite the overall low sequence similarity among ACPs from different PKSs, nearly all ACPs exhibit a similar right-handed twisted bundle consisting of three α-helices (helix I, helix II and helix IV) connected by two loops. In some structures an additional short helix (helix III) is found in the second loop depends on its helix-loop equilibrium conformations (Sharma et al., 2006; Cantu et al., 2012). The longer helix-I is positioned anti-parallel to helix II and helix IV, while helix I and helix IV are arranged at a small angle relative to each other (Figure 16-A). The overall helical bundle is stabilized via inter-helical hydrophobic interactions. The well-conserved “DSL” motif containing the active serine is located at the N-terminus of helix II, which is identified as the universal protein-protein ‘recognition helix’ (Zhang et al., 2003; Weissman et al., 2006; Tsai et al., 2009). This recognition helix is featured by a large number of negatively charged residues and is demonstrated by mutagenesis studies to play a crucial role for ACPs to communicate with other catalytic domains (Chen et al., 2006).
Figure 16.
Selected structures of PKS ACPs. (A) Ribbon representation and electrostatic potential surface of selected ACPs. PDB codes: act ACP: 2K0X; oct ACP: 1NQ4; PksA ACP: 2KR5; CalE8 ACP: 2L9F. (B) Type II ACP sequesters the hydrophobic polyketide chains. Comparison of different forms of act ACP. PDB codes: apo: 2K0Y; holo: 2K0X; butyryl: 2KG9; octanoyl: 2KGC. 215×180mm (300 × 300 DPI).
Type II PKS ACPs sequester bound acyl chains within an internal cavity (Zornetzer et al., 2006; Upadhyay et al., 2009; Roujeinikova et al., 2002; Evans et al., 2009), which was proposed to protect reactive intermediates from unintended intramolecular cyclization and bulk solvent. Actinorhodin ACP (actACP) represents the first PKS component in which the three dimensional structure is determined, and is the most thoroughly studied ACP. NMR studies identified the possible interactions between the tethered substrate and ACP. The holo-form of the ACP exhibits detectable conformational changes compared to the apo-ACP, with helix III moving closer to helix II. Leu43 on helix II was reported to interact with the pantothenate methyls on the pPant arm. This conformational change is used to account for the dissociation of the ACP-PPTase complex (Evans et al., 2008), in which the ACP and PPTase mainly interact along helix II (Weissman et al., 2006). Structures of several short-chain acyl actACPs were determined. Development of nonhydrolyzable thioether or pantetheinamide analogues of the thioester acylACPs substrate is necessary for some substrates because of spontaneous hydrolysis in water. High-resolution NMR structures revealed dramatic conformational changes of actACP when nonpolar acyl chains including butyryl, hexanoyl, and octanoyl are attached (Evans et al., 2009). Binding cavity within the ACP is created by the movement of helix III (Figure 16-B). Although the resulting cleft formed between helix II and helix III is sufficiently deep, the acyl chains still occupy only the front portion to keep the thioester part availability for other enzymes. Another study probed the interaction between act ACP and the tethered substrate (Haushalter et al., 2011), using chemoenzymatically synthesized emodic-act ACP (Haushalter et al., 2008; La Clair et al., 2004; Worthington et al., 2006a). The emodyl moiety was used to mimic the late-stage polyaromatic intermediates in the actinorhodin pathway. Although the 3D structure of emodic-act ACP was not solved, comparison of NMR signals between holo- and emodic-act ACP allowed identification of interacting residues, which are also located near the acyl binding cavity between helix II and helix III.
In contrast to type II PKS ACPs, ACPs from type I PKSs are unable to sequester the hydrophobic chains in an acyl binding cavity (Wattana-amorn et al., 2010; Lim et al., 2011; Busche et al., 2012). Usually no conformation changes of type I PKS ACP are observed when acylated with polyketide chains. The type I PKS ACPs contain relatively fewer negative charges on the “recognition helix” compared to the freestanding type II PKS ACPs. Only very weak interactions between the relatively more hydrophobic surface and the polyketide chain have been observed in a few cases (Lim et al., 2011). The lack of substrate binding in type I PKS ACP is consistent with similar observation with the type I rat FAS ACP (Ploskon et al., 2008). The absence of a protective role in type I PKS ACP has been attributed to these ACPs are less exposed to solvent due to crowding from adjacent domains; as well as the expected higher efficiency of substrate delivery in type I PKSs.
4. Protein-Protein interactions involving ACP
During each cycle for polyketide extension, the ACP is involved in all of the catalytic steps, including extender-unit loading by AT, chain elongation and translocation by the KS, reductive modifications by KR, DH, ER and/or other domains. For the multidomain type I PKSs, the mobility and flexibility of the ACP domain facilitates the shuttling of substrates to and between all the different active sites. The binding of ACP with other functional domains are either modulated by the recognition of the acyl substrates or a specific region on the ACP or both. However, these dynamic domain-domain interactions have to be weak and transient to ensure efficient product synthesis. Studying the determinants for efficient transport or delivery of carrier protein-tethered substrates is essential for understanding the complex catalytic process on PKSs.
Because of its transient nature, detailed insights into ACP based interactions are less likely to come from single crystallographic snapshot of intact multidomain enzymes. Currently, several approaches are involved to study the ACP-centered protein-protein interactions, including computer simulation of docking models based on reported structures, mutagenesis followed by biochemical assays and solution-phase NMR spectroscopy to probe the dynamic behaviors. ACP centered protein-protein interaction in type I modular PKSs are focused in the following discussion.
4.1. KS-ACP interactions
In type I modular PKS, KS shows broad polyketide substrate specificity (Khosla et al., 1999b) and KS-ACP protein-protein interactions play an important role in both catalytic steps involving KS. The chain elongation step involves intramodular KS-ACP interactions (KSn-ACPn) in which the KS catalyzes the decarboxylative condensation and transfers the extended polyketide product to the ACP within the same module. The chain translocation step involves intermodular KS-ACP interactions (ACPn-1-KSn), in which the KS is acylated with the polyketide intermediate from the ACP of the previous module prior to the elongation step. The polyketide chain transfer in type I modular PKS is unidirectional: from the upstream ACPn-1 to KSn during the chain translocation and then to ACPn during the chain elongation. Specific ACPn-KSn interactions prevent the back transfer of elongated polyketide between the pair. This duel substrate specificity of the KS in the two separate catalytic events is clearly supported by in vitro assays using dissected minimal PKS domains from the DEBS PKS (Gokhale et al., 1999; Tsuji et al., 2001; Wu et al., 2002; Chen et al., 2006).
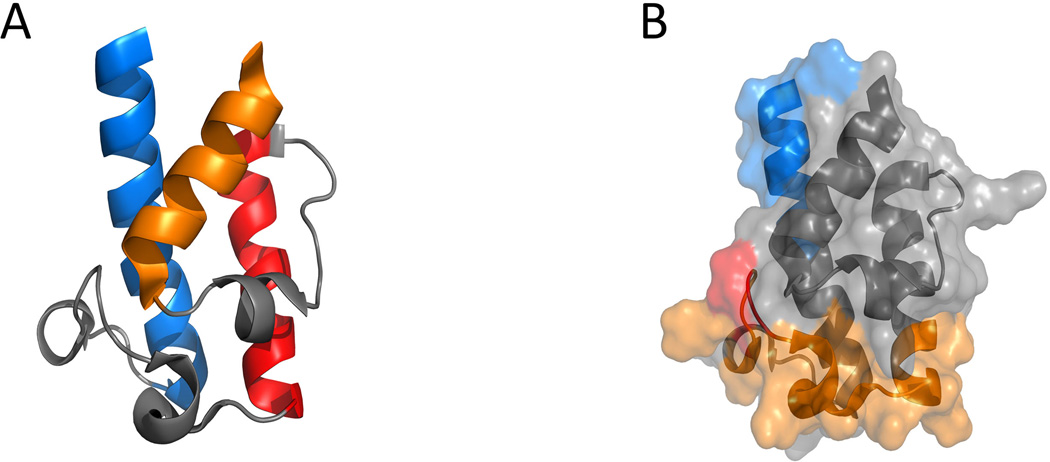
Recent mechanistic investigations discovered that the docking sites on ACP for intermodular chain translocation and intramodular chain elongation are located at separate regions in the ACP domain. Loop I of the ACP domain is involved in the KSn-ACPn recognition during the chain elongation step (Kapur et al., 2010), while the first ten residues of helix I are responsible for controlling the ACPn-1-KSn interactions during the chain translocation step (Kapur et al., 2012) (Figure 17). The orthogonal recognition regions were identified from separate kinetic assays that measure the rate of elongation (Chen et al., 2006) or back-transfer reaction (Wu et al., 2001) using dissected DEBS KS-AT didomain and a large series of chimeric DEBS ACPs created from swapping secondary structural elements between ACPs.
Figure 17.
Structure features of DEBS ACP2. PDB code: 2JU2. (A) Ribbon structure of DEBS ACP2, helix I in blue, helix II in red, helix IV in orange, loops in gray. (B) Regions in DEBS ACP2 that involved in ACP-KS/AT interactions. Docking interface for chain translocation is shown in blue; Docking interface for chain elongation is shown in Orange and key residues at position 44, 45 are shown in red; The rest of the protein is shown in gray. 215×103mm (300 × 300 DPI).
In a molecular docking model that models protein-protein interaction during chain elongation, homology structure of ACP5 is docked into a deep cleft defined by KS5, KS5-AT5 linker and AT5 from the same polypeptide strand (Kapur et al., 2010). Interestingly, no significant contact is found between ACP5 and KS5 domain from the other subunit. Two regions of KS5-AT5 linker were identified to interact electrostatically with residues 44 and 45 in loop I of ACP5. Mutation of these two ACP residues led to 90% reduction in chain elongation activity.
The molecular basis for intermodular chain transfer were explained by another docking study using the homology structure of DEBS ACP4 and the crystal structure of KS5-AT5 (Kapur et al., 2012). ACP4 was found to dock in the same cleft that ACP5 docks, but the position and orientation of the ACPs are very different. The position of ACP4 in the docking model is consistent with the NMR structure of the dimeric complex formed between DEBS2 C-terminal (downstream to ACP4) and DEBS3 N-terminal linker domains (Broadhurst et al., 2003). The ACP4 interacts with KS5-AT5 linker mainly by electrostatic interaction between E23 on ACP4 helix I and R551 on KS5-AT5 linker. This proposed interaction site between ACPn-1 and KSn is supported by the NMR titration experiments using labeled ACP2 and unlabeled KS3-AT3, in which 1H-15N HSQC signals for some residues in helix I disappeared in the presence of the KS3-AT3 didomain (Charkoudian et al., 2011).
This dual mode of binding by two ACPs on the same KS to achieve unidirectional chain transfer can be extended to other type I modular PKS systems because these motifs discussed above are highly conserved as revealed by the primary sequence alignments (Kapur et al., 2010). In iterative PKSs, the main difference is that the ACP can dock to both elongation and transfer sites. Indeed, constructing of a mutant DEBS ACP that contains both ACPn-1 and ACPn recognition features reprogrammed the non-iterative DEBS module 3 to catalyze two successive rounds of chain elongation (Kapur et al., 2012).
4.2. AT-ACP interactions
The AT domains in PKSs not only need to select the correct extender unit acyl CoA, but must also recognize the cognate ACP in the acyl transfer reaction. For example, the interaction between type II FAS HpMCAT and HpACP is moderate with a binding affinity of KD=4.31 ×10−5 M (Zhang et al., 2007). Biochemical analysis also indicated that ATs from modular PKSs show moderate preference (10–20:1) toward their cognate ACPs compared to noncogate ACPs (Wong et al., 2010). Deletion of KS-AT linker or post-AT linker results in lower AT activity and ACP specificity, which suggests that these linkers play important roles in catalysis and ACP recognition (Wong et al., 2010).
Molecular docking studies using DEBS ACP3-AT3 (Wong et al., 2010) and dynemicin PKS ACP-AT (Liew et al., 2012) showed that the ACP is docked in the cleft between KS-AT linker and the α/β hydrolases-like core (larger subdomain of AT), and is from the side accessible to the AT active site. In the dynemicin ACP-AT complex, the fully extended pPant arm points toward the AT active site, and the ACP-AT interface is mainly formed through electrostatic interactions in two regions: the first is between a region close to the DSL motif on ACP and the entrance of the substrate tunnel on the AT larger subdomain; the second is between loops I, II of ACP and the KS-AT linker. Salt bridges formed between the two domains contribute to a majority of the contacts. Some of salt- bridge forming residues (Arg, Asp and Glu) are conserved among AT and ACP sequences. The importance of these sites was confirmed by alanine-scanning mutagenesis using the trans-acting disorazole ACP1. The AT showed 50% decreased transacylation activity towards disorazole ACP1 when Asp45 is mutated into an alanine (Wong et al., 2011).
4.3. KR-ACP Interactions
Detailed biochemical studies using standalone KRs from DEBS modules 1, 2 and 6 have shown that KRs are highly specific toward the β-ketoacyl substrates and portions of the pPant arm, but are more tolerant toward the ACP carrier (Chen et al., 2007; Siskos et al., 2005). The molecular docking model of DEBS-KR1 and its cognate diketide acyl-ACP was built up in a stepwise fashion: first an apo-ACP/KR complex is generated from the crystal structure of DEBS-KR1 and a homology model of apo-ACP1 based on solution structure of DEBS ACP2; then the pPant arm is attached to the ACP1; finally the 2S–methyl-3-oxopentanoyl substrate is appended to the pPant arm (Anand et al., 2012). In the final docking model, the acyl unit and the pPant arm were immobilized in the shallow cavity of KR1 through hydrophilic and hydrophobic interactions. An arginine and phenylalanine on KR1 interact with the aspartate and leucine, respectively, in the conserved “DSL” moiety of ACP1. In another docking model, a homology model of amphotericin ACP2 was generated from solution structure of DEBS ACP2 and was docked onto the A-type KR2 (Zheng et al., 2010). The active site of ACP is found facing the KR catalytic groove in an orientation that favors formation of the L-β-hydroxyl product. Protein-protein interactions are found in two regions: between the ACP2 helix II and the KR4 catalytic groove; and between the ACP2 loop II and the KR lid helix. A pair of hydrophobic interaction is found between the valine immediately following the catalytic serine in ACP2 and a conserved leucine at the C-terminal end of α4 in the KR structural subdomain. A salt bridge is also found in this model between a conserved ACP2 helix II arginine and an aspartate at the C-terminal end of α4. Similar docking model between ACP and KR is observed using the B-type tylosin KR1. In this model, the pPant-bound substrate enters from the opposite side of the active site groove to result in opposite stereospecific reduction.
4.4. DH-ACP interactions
The crystal structure of DEBS DH4 revealed several conserved hydrophobic residues that can interact with the cognate ACP4. Comparing to type II FAS DH, the ACP docking region on DEBS-DH4 is only slightly positive charged (Keatinge-Clay, 2008; Anand et al., 2012). As shown in FAS DH-ACP complex (Kimber et al., 2004; Zhang et al., 2001), a highly conserved phenylalanine and arginine pair near the DH active site entrance can interact with the leucine and aspartate residues, respectively, in the conserved “DSL” motif of ACP. This key role of the arginine was confirmed by the mutagenesis study, in which mutation of this residue to aspartic acid in DH4 significantly decreased the catalytic efficiency of DEBS PKS (Keatinge-Clay, 2008). The acyl-ACP4/DH complex, which was built stepwise from the apo-ACP/DH complex, also confirmed role of Phe-Arg pair in ACP docking, and showed details of substrate binding. The entire pentaketide acyl substrate and a portion of the pPant arm enter the deep cavity of the DH domain (Anand et al., 2012).
4.5. TE-ACP Interactions
A type I TE usually shows broad specificities towards the acyl substrates (Sharma et al., 2007; Weissman et al., 1998; Aggarwal et al., 1995), while interacts with ACP domains weakly. In DEBS, ACP6 in the last module is jointed to the TE domains through a linker of eleven residues. The interactions between ACP6 and TE were studied using in vitro biochemical assays and solution NMR (Tran et al., 2008; Tran et al., 2010). The covalent linkage between the ACP6 and DEBSTE has been shown to be highly important for efficient catalysis, as separating the two domains into standalone proteins significantly reduced the activity of the TE domain (Tran et al., 2008). This result hinted that protein-protein interactions between the two domains may be minimal. This was further supported by titration of TE into an ACP6 solution in a solution NMR study. Similar results were also observed in a titration experiment using the calicheamicin ACP and TE domains. Therefore, for most TE domains, only the acyl substrate and part of the pPant arm is required for TE recognition, while the identity of the ACP substrate is not critical (Tran et al., 2010; Lim et al., 2011). This has been widely used in construction of hybrid and nonnatural type I PKS assembly lines, in which the TE domain can be placed after different ACP domains to facilitate product release or cyclization (Bedford et al., 1996). In addition, the TE has been shown to catalyze hydrolysis or cyclization reaction using acyl substrates attached as N-acetylcysteamine thioesters (Wang et al., 2009). Iterative type I PKS TEs have also been found to be broadly specific towards acyl substrates, while not requiring an ACP partner for catalysis. This is demonstrated in using the Gibberella zeae PKS13TE domain (Zhou et al., 2008) as a biocatalytic tool in the release of dihydromonacolin L from the iterative PKS LovB (Ma et al., 2009).
4.6. Halogenase-ACP Interactions
In the modular PKS CurA involved in the biosynthesis of curacin A, a set of three ACPs (ACPI-ACPII-ACPIII) are found in tandem. The three ACPs are highly identical in sequence and function synergistically with each other (Busche et al., 2012; Busche et al., 2009). This module also contains a nonheme Fe2+/α-ketoglutarate-dependent halogenase domain, of which the crystal structure was reported previously (Khare et al., 2010). The ACP-halogenase interaction in CurA is specific, as the γ-chlorination only occurs when the acyl substrate is loaded on cognate ACPI. The protein-protein interactions between ACPI and the halogenase domain was investigated using solution NMR spectroscopy (Busche et al., 2012). Similar to all type I modular ACPs, ACPI adopts the four-helix fold and the 3-hydroxyl-3-methyl-glutaryl substrate is only weakly bound to the surface of ACPI. The interactions between ACPI and the halogenase is also weak, as titration of the halogenase into ACPI solution did not result in any notable chemical shift perturbations in the NMR signals. Mutagenesis study of the non-conserved, solvent exposed residues on ACPI led to the identification of several residues that have the strongest impact on chlorination efficiency, which included an asparagine and an isoleucine on helix II and an alanine on helix III. These residues form a surface patch surrounding the ACPI pPant arm attachment site, thereby providing specific contacts for halogenase-ACPI recognition (Busche et al., 2012).
Summary and perspective
Recent development on X-ray crystallography, electron microscopic analysis and NMR techniques has promoted significant advancement in our understanding of the structural and mechanism basis of multidomain PKSs. Almost all the modular PKS catalytic domains, both as standalone domains and as multi-domain fragments have solved three-dimensional structures, which have provided detailed information on the mechanism of catalysis and possible interactions with neighboring domains. However, the detailed information regarding many domain-domain interactions are still unknown. Crystallization of a complete module of modular PKS, or an intact iterative type I PKS will be extremely informative towards complete understanding of these nanomachines and is the next grand challenge in this field. To date, most studies of protein-protein interaction involving ACPs are still based on molecular docking between reported crystal structures or homology structures. Because of the dynamic nature of such protein-protein interactions during polyketide biosynthesis, it is difficult for X-ray crystallography to capture the weak or transient interactions. Recently, researchers are pursuing a strategy to install mechanism-based crosslinkers onto the pPant arm of the ACP and covalently capture these proteins in action (Meier et al., 2010; Worthington et al., 2010; Worthington et al., 2006b; Kapur et al., 2008). Using similar linkers, X-ray structure that captured the TE-PCP (peptidyl carrier protein) interactions in the related nonribosomal peptide synthetase was recently reported (Liu et al., 2011). The PCP-TE didomain from E. coli enterobactin synthetase was captured in the crystal structure where the PCP domain forms extensive hydrophobic interactions with the TE domain. The substrate bound pPant moiety fits in a channel leading to the TE catalytic triad, which provides detailed insight into how the pPant arm of holo carrier protein interacts with its partner enzyme to stabilize domain/domain interactions. Such tools will also be highly useful for the PKS system. In parallel, development of new NMR techniques, such as paramagnetic relaxation enhancement (Wang et al., 2012), for investigating transient processes will also be highly beneficial.
Acknowledgements
We gratefully acknowledge Dr. Yit-Heng Chooi for critical reading of this manuscript.
Research in our laboratory on related topics are supported by the National Institute of Health (1R01GM085128).
Footnotes
Declaration of interest
This manuscript is submitted to Critical Reviews in Biochemistry and Molecular Biology exclusively. The authors report no declarations of interest.
References
- Aggarwal R, Caffrey P, Leadlay PF, Smith CJ, Staunton J. The Thioesterase of the Erythromycin-Producing Polyketide Synthase - Mechanistic Studies in-Vitro to Investigate Its Mode of Action and Substrate-Specificity. J Chem Soc, Chem Commun. 1995:1519–1520. [Google Scholar]
- Akey DL, Kittendorf JD, Giraldes JW, Fecik RA, Sherman DH, Smith JL. Structural basis for macrolactonization by the pikromycin thioesterase. Nat Chem Biol. 2006;2:537–542. doi: 10.1038/nchembio824. [DOI] [PubMed] [Google Scholar]
- Akey DL, Razelun JR, Tehranisa J, Sherman DH, Gerwick WH, Smith JL. Crystal Structures of Dehydratase Domains from the Curacin Polyketide Biosynthetic Pathway. Structure. 2010;18:94–105. doi: 10.1016/j.str.2009.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alekseyev VY, Liu CW, Cane DE, Puglisi JD, Khosla C. Solution structure and proposed domain domain recognition interface of an acyl carrier protein domain from a modular polyketide synthase. Protein Sci. 2007;16:2093–2107. doi: 10.1110/ps.073011407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames BD, Nguyen C, Bruegger J, Smith P, Xu W, Ma S, Wong E, Wong S, Xie X, Li JW, Vederas JC, Tang Y, Tsai SC. Crystal structure and biochemical studies of the trans-acting polyketide enoyl reductase LovC from lovastatin biosynthesis. Proc Nat Acad Sci USA. 2012;109:11144–11149. doi: 10.1073/pnas.1113029109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand S, Mohanty D. Modeling holo-ACP:DH and holo-ACP:KR complexes of modular polyketide synthases: a docking and molecular dynamics study. BMC Struct Biol. 2012;12:10. doi: 10.1186/1472-6807-12-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asturias FJ, Chadick JZ, Cheung IK, Stark H, Witkowski A, Joshi AK, Smith S. Structure and molecular organization of mammalian fatty acid synthase. Nat Struct Mol Biol. 2005;12:225–232. doi: 10.1038/nsmb899. [DOI] [PubMed] [Google Scholar]
- Auclair K, Sutherland A, Kennedy J, Witter DJ, Van den Heever JP, Hutchinson CR, Vederas JC. Lovastatin nonaketide synthase catalyzes an intramolecular Diels-Alder reaction of a substrate analogue. J Am Chem Soc. 2000;122:11519–11520. [Google Scholar]
- Bedford D, Jacobsen JR, Luo G, Cane DE, Khosla C. A functional chimeric modular polyketide synthase generated via domain replacement. Chem Biol. 1996;3:827–831. doi: 10.1016/s1074-5521(96)90068-x. [DOI] [PubMed] [Google Scholar]
- Brignole EJ, Smith S, Asturias FJ. Conformational flexibility of metazoan fatty acid synthase enables catalysis. Nat Struct Mol Biol. 2009;16:190–197. doi: 10.1038/nsmb.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadhurst RW, Nietlispach D, Wheatcroft MP, Leadlay PF, Weissman KJ. The structure of docking domains in modular polyketide synthases. Chem Biol. 2003;10:723–731. doi: 10.1016/s1074-5521(03)00156-x. [DOI] [PubMed] [Google Scholar]
- Busche A, Gottstein D, Hein C, Ripin N, Pader I, Tufar P, Eisman EB, Gu L, Walsh CT, Sherman DH, Lohr F, Guntert P, Dotsch V. Characterization of molecular interactions between ACP and halogenase domains in the Curacin A polyketide synthase. ACS Chem Biol. 2012;7:378–386. doi: 10.1021/cb200352q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busche AE, Aranko AS, Talebzadeh-Farooji M, Bernhard F, Dotsch V, Iwai H. Segmental isotopic labeling of a central domain in a multidomain protein by protein trans-splicing using only one robust DnaE intein. Angew Chem Int Ed Engl. 2009;48:6128–6131. doi: 10.1002/anie.200901488. [DOI] [PubMed] [Google Scholar]
- Cane DE. Programming of erythromycin biosynthesis by a modular polyketide synthase. J Biol Chem. 2010;285:27517–27523. doi: 10.1074/jbc.R110.144618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cane DE, Walsh CT. The parallel and convergent universes of polyketide synthases and nonribosomal peptide synthetases. Chem Biol. 1999;6:R319–R325. doi: 10.1016/s1074-5521(00)80001-0. [DOI] [PubMed] [Google Scholar]
- Cantu DC, Forrester MJ, Charov K, Reilly PJ. Acyl carrier protein structural classification and normal mode analysis. Protein Sci. 2012;21:655–666. doi: 10.1002/pro.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarty B, Gu Z, Chirala SS, Wakil SJ, Quiocho FA. Human fatty acid synthase: structure and substrate selectivity of the thioesterase domain. Proc Nat Acad Sci USA. 2004;101:15567–15572. doi: 10.1073/pnas.0406901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YA, Podevels AM, Kevany BM, Thomas MG. Biosynthesis of polyketide synthase extender units. Nat Prod Rep. 2009;26:90–114. doi: 10.1039/b801658p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charkoudian LK, Liu CW, Capone S, Kapur S, Cane DE, Togni A, Seebach D, Khosla C. Probing the interactions of an acyl carrier protein domain from the 6-deoxyerythronolide B synthase. Protein Sci. 2011;20:1244–1255. doi: 10.1002/pro.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AY, Cane DE, Khosla C. Structure-based dissociation of a type I polyketide synthase module. Chem Biol. 2007;14:784–792. doi: 10.1016/j.chembiol.2007.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AY, Schnarr NA, Kim CY, Cane DE, Khosla C. Extender unit and acyl carrier protein specificity of ketosynthase domains of the 6-deoxyerythronolide B synthase. J Am Chem Soc. 2006;128:3067–3074. doi: 10.1021/ja058093d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YQ, Tang GL, Shen B. Type I polyketide synthase requiring a discrete acyltransferase for polyketide biosynthesis. Proc Nat Acad Sci USA. 2003;100:3149–3154. doi: 10.1073/pnas.0537286100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claxton HB, Akey DL, Silver MK, Admiraal SJ, Smith JL. Structure and functional analysis of RifR, the type II thioesterase from the rifamycin biosynthetic pathway. J Biol Chem. 2009;284:5021–5029. doi: 10.1074/jbc.M808604200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox RJ. Polyketides, proteins and genes in fungi: programmed nano-machines begin to reveal their secrets. Org Biomol Chem. 2007;5:2010–2026. doi: 10.1039/b704420h. [DOI] [PubMed] [Google Scholar]
- Crawford JM, Korman TP, Labonte JW, Vagstad AL, Hill EA, Kamari-Bidkorpeh O, Tsai SC, Townsend CA. Structural basis for biosynthetic programming of fungal aromatic polyketide cyclization. Nature. 2009;461:1139–1143. doi: 10.1038/nature08475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford JM, Thomas PM, Scheerer JR, Vagstad AL, Kelleher NL, Townsend CA. Deconstruction of iterative multidomain polyketide synthase function. Science. 2008;320:243–246. doi: 10.1126/science.1154711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump MP, Crosby J, Dempsey CE, Parkinson JA, Murray M, Hopwood DA, Simpson TJ. Solution structure of the actinorhodin polyketide synthase acyl carrier protein from Streptomyces coelicolor A3(2) Biochemistry. 1997;36:6000–6008. doi: 10.1021/bi970006+. [DOI] [PubMed] [Google Scholar]
- Dillon SC, Bateman A. The Hotdog fold: wrapping up a superfamily of thioesterases and dehydratases. BMC Bioinformatics. 2004;5:109. doi: 10.1186/1471-2105-5-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L, Lou L. PKS and NRPS release mechanisms. Nat Prod Rep. 2010;27:255–278. doi: 10.1039/b912037h. [DOI] [PubMed] [Google Scholar]
- Evans SE, Williams C, Arthur CJ, Burston SG, Simpson TJ, Crosby J, Crump MP. An ACP structural switch: conformational differences between the apo and holo forms of the actinorhodin polyketide synthase acyl carrier protein. ChemBioChem. 2008;9:2424–2432. doi: 10.1002/cbic.200800180. [DOI] [PubMed] [Google Scholar]
- Evans SE, Williams C, Arthur CJ, Ploskon E, Wattana-amorn P, Cox RJ, Crosby J, Willis CL, Simpson TJ, Crump MP. Probing the Interactions of early polyketide intermediates with the Actinorhodin ACP from S. coelicolor A3(2) J Mol Biol. 2009;389:511–528. doi: 10.1016/j.jmb.2009.03.072. [DOI] [PubMed] [Google Scholar]
- Findlow SC, Winsor C, Simpson TJ, Crosby J, Crump MP. Solution structure and dynamics of oxytetracycline polyketide synthase acyl carrier protein from Streptomyces rimosus. Biochemistry. 2003;42:8423–8433. doi: 10.1021/bi0342259. [DOI] [PubMed] [Google Scholar]
- Fischbach MA, Walsh CT. Assembly-line enzymology for polyketide and nonribosomal Peptide antibiotics: logic, machinery, and mechanisms. Chem Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- Frueh DP, Arthanari H, Koglin A, Vosburg DA, Bennett AE, Walsh CT, Wagner G. Dynamic thiolation-thioesterase structure of a non-ribosomal peptide synthetase. Nature. 2008;454:903–906. doi: 10.1038/nature07162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii I, Watanabe A, Sankawa U, Ebizuka Y. Identification of Claisen cyclase domain in fungal polyketide synthase WA, a naphthopyrone synthase of Aspergillus nidulans. Chem Biol. 2001;8:189–197. doi: 10.1016/s1074-5521(00)90068-1. [DOI] [PubMed] [Google Scholar]
- Gehret JJ, Gu L, Gerwick WH, Wipf P, Sherman DH, Smith JL. Terminal alkene formation by the thioesterase of curacin A biosynthesis: structure of a decarboxylating thioesterase. J Biol Chem. 2011;286:14445–14454. doi: 10.1074/jbc.M110.214635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldes JW, Akey DL, Kittendorf JD, Sherman DH, Smith JL, Fecik RA. Structural and mechanistic insights into polyketide macrolactonization from polyketide-based affinity labels. Nat Chem Biol. 2006;2:531–536. doi: 10.1038/nchembio822. [DOI] [PubMed] [Google Scholar]
- Gokhale RS, Tsuji SY, Cane DE, Khosla C. Dissecting and exploiting intermodular communication in polyketide synthases. Science. 1999;284:482–485. doi: 10.1126/science.284.5413.482. [DOI] [PubMed] [Google Scholar]
- Guo X, Liu T, Valenzano CR, Deng Z, Cane DE. Mechanism and stereospecificity of a fully saturating polyketide synthase module: nanchangmycin synthase module 2 and its dehydratase domain. J Am Chem Soc. 2010;132:14694–14696. doi: 10.1021/ja1073432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haushalter RW, Filipp FV, Ko KS, Yu R, Opella SJ, Burkart MD. Binding and pK(a) Modulation of a Polycyclic Substrate Analogue in a Type II Polyketide Acyl Carrier Protein. ACS Chem Biol. 2011;6:413–418. doi: 10.1021/cb200004k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haushalter RW, Worthington AS, Hur GH, Burkart MD. An orthogonal purification strategy for isolating crosslinked domains of modular synthases. Bioorg Med Chem Lett. 2008;18:3039–3042. doi: 10.1016/j.bmcl.2008.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haydock SF, Aparicio JF, Molnar I, Schwecke T, Khaw LE, Konig A, Marsden AF, Galloway IS, Staunton J, Leadlay PF. Divergent sequence motifs correlated with the substrate specificity of (methyl)malonyl-CoA:acyl carrier protein transacylase domains in modular polyketide synthases. FEBS Lett. 1995;374:246–248. doi: 10.1016/0014-5793(95)01119-y. [DOI] [PubMed] [Google Scholar]
- Hertweck C. The biosynthetic logic of polyketide diversity. Angew Chem Int Ed Engl. 2009;48:4688–4716. doi: 10.1002/anie.200806121. [DOI] [PubMed] [Google Scholar]
- Jenke-Kodama H, Sandmann A, Muller R, Dittmann E. Evolutionary implications of bacterial polyketide synthases. Mol Biol Evol. 2005;22:2027–2039. doi: 10.1093/molbev/msi193. [DOI] [PubMed] [Google Scholar]
- Joshi AK, Rangan VS, Smith S. Differential affinity labeling of the two subunits of the homodimeric animal fatty acid synthase allows isolation of heterodimers consisting of subunits that have been independently modified. J Biol Chem. 1998a;273:4937–4943. doi: 10.1074/jbc.273.9.4937. [DOI] [PubMed] [Google Scholar]
- Joshi AK, Rangan VS, Witkowski A, Smith S. Engineering of an active animal fatty acid synthase dimer with only one competent subunit. Chem Biol. 2003;10:169–173. doi: 10.1016/s1074-5521(03)00023-1. [DOI] [PubMed] [Google Scholar]
- Joshi AK, Witkowski A, Smith S. Mapping of functional interactions between domains of the animal fatty acid synthase by mutant complementation in vitro. Biochemistry. 1997;36:2316–2322. doi: 10.1021/bi9626968. [DOI] [PubMed] [Google Scholar]
- Joshi AK, Witkowski A, Smith S. The malonyl/acetyltransferase and beta-ketoacyl synthase domains of the animal fatty acid synthase can cooperate with the acyl carrier protein domain of either subunit. Biochemistry. 1998b;37:2515–2523. doi: 10.1021/bi971886v. [DOI] [PubMed] [Google Scholar]
- Kapur S, Chen AY, Cane DE, Khosla C. Molecular recognition between ketosynthase and acyl carrier protein domains of the 6-deoxyerythronolide B synthase. Proc Nat Acad Sci USA. 2010;107:22066–22071. doi: 10.1073/pnas.1014081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapur S, Lowry B, Yuzawa S, Kenthirapalan S, Chen AY, Cane DE, Khosla C. Reprogramming a module of the 6-deoxyerythronolide B synthase for iterative chain elongation. Proc Nat Acad Sci USA. 2012;109:4110–4115. doi: 10.1073/pnas.1118734109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapur S, Worthington A, Tang Y, Cane DE, Burkart MD, Khosla C. Mechanism based protein crosslinking of domains from the 6-deoxyerythronolide B synthase. Bioorg Med Chem Lett. 2008;18:3034–3038. doi: 10.1016/j.bmcl.2008.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanagh K, Jornvall H, Persson B, Oppermann U. The SDR superfamily: functional and structural diversity within a family of metabolic and regulatory enzymes. Cell Mol Life Sci. 2008;65:3895–3906. doi: 10.1007/s00018-008-8588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keatinge-Clay A. Crystal structure of the erythromycin polyketide synthase dehydratase. J Mol Biol. 2008;384:941–953. doi: 10.1016/j.jmb.2008.09.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keatinge-Clay AT. A tylosin ketoreductase reveals how chirality is determined in polyketides. Chem Biol. 2007;14:898–908. doi: 10.1016/j.chembiol.2007.07.009. [DOI] [PubMed] [Google Scholar]
- Keatinge-Clay AT. The structures of type I polyketide synthases. Nat Prod Rep. 2012;29:1050–1073. doi: 10.1039/c2np20019h. [DOI] [PubMed] [Google Scholar]
- Keatinge-Clay AT, Maltby DA, Medzihradszky KF, Khosla C, Stroud RM. An antibiotic factory caught in action. Nat Struct Mol Biol. 2004;11:888–893. doi: 10.1038/nsmb808. [DOI] [PubMed] [Google Scholar]
- Keatinge-Clay AT, Shelat AA, Savage DF, Tsai SC, Miercke LJ, O'Connell JD, 3rd, Khosla C, Stroud RM. Catalysis, specificity, and ACP docking site of Streptomyces coelicolor malonyl-CoA:ACP transacylase. Structure. 2003;11:147–154. doi: 10.1016/s0969-2126(03)00004-2. [DOI] [PubMed] [Google Scholar]
- Keatinge-Clay AT, Stroud RM. The structure of a ketoreductase determines the organization of the beta-carbon processing enzymes of modular polyketide synthases. Structure. 2006;14:737–748. doi: 10.1016/j.str.2006.01.009. [DOI] [PubMed] [Google Scholar]
- Kennedy J, Auclair K, Kendrew SG, Park C, Vederas JC, Hutchinson CR. Modulation of polyketide synthase activity by accessory proteins during lovastatin biosynthesis. Science. 1999;284:1368–1372. doi: 10.1126/science.284.5418.1368. [DOI] [PubMed] [Google Scholar]
- Khare D, Wang B, Gu LC, Razelun J, Sherman DH, Gerwick WH, Hakansson K, Smith JL. Conformational switch triggered by alpha-ketoglutarate in a halogenase of curacin A biosynthesis. Proc Nat Acad Sci USA. 2010;107:14099–14104. doi: 10.1073/pnas.1006738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosla C. Structures and mechanisms of polyketide synthases. J Org Chem. 2009;74:6416–6420. doi: 10.1021/jo9012089. [DOI] [PubMed] [Google Scholar]
- Khosla C, Gokhale RS, Jacobsen JR, Cane DE. Tolerance and specificity of polyketide synthases. Annu Rev Biochem. 1999a;68:219–253. doi: 10.1146/annurev.biochem.68.1.219. [DOI] [PubMed] [Google Scholar]
- Khosla C, Gokhale RS, Jacobsen JR, Cane DE. Tolerance and specificity of polyketide synthases. Annu Rev Biochem. 1999b;68:219–253. doi: 10.1146/annurev.biochem.68.1.219. [DOI] [PubMed] [Google Scholar]
- Khosla C, Tang Y, Chen AY, Schnarr NA, Cane DE. Structure and mechanism of the 6-deoxyerythronolide B synthase. Annu Rev Biochem. 2007;76:195–221. doi: 10.1146/annurev.biochem.76.053105.093515. [DOI] [PubMed] [Google Scholar]
- Kimber MS, Martin F, Lu Y, Houston S, Vedadi M, Dharamsi A, Fiebig KM, Schmid M, Rock CO. The structure of (3R)-hydroxyacyl-acyl carrier protein dehydratase (FabZ) from Pseudomonas aeruginosa. J Biol Chem. 2004;279:52593–52602. doi: 10.1074/jbc.M408105200. [DOI] [PubMed] [Google Scholar]
- Koglin A, Lohr F, Bernhard F, Rogov VV, Frueh DP, Strieter ER, Mofid MR, Guntert P, Wagner G, Walsh CT, Marahiel MA, Dotsch V. Structural basis for the selectivity of the external thioesterase of the surfactin synthetase. Nature. 2008;454:907–911. doi: 10.1038/nature07161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp M, Irschik H, Pradella S, Muller R. Production of the tubulin destabilizer disorazol in Sorangium cellulosum: Biosynthetic machinery and regulatory genes. ChemBioChem. 2005;6:1277–1286. doi: 10.1002/cbic.200400459. [DOI] [PubMed] [Google Scholar]
- Korman TP, Crawford JM, Labonte JW, Newman AG, Wong J, Townsend CA, Tsai SC. Structure and function of an iterative polyketide synthase thioesterase domain catalyzing Claisen cyclization in aflatoxin biosynthesis. Proc Nat Acad Sci USA. 2010;107:6246–6251. doi: 10.1073/pnas.0913531107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotaka M, Kong R, Qureshi I, Ho QS, Sun H, Liew CW, Goh LP, Cheung P, Mu Y, Lescar J, Liang ZX. Structure and catalytic mechanism of the thioesterase CalE7 in enediyne biosynthesis. J Biol Chem. 2009;284:15739–15749. doi: 10.1074/jbc.M809669200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroken S, Glass NL, Taylor JW, Yoder OC, Turgeon BG. Phylogenomic analysis of type I polyketide synthase genes in pathogenic and saprobic ascomycetes. Proc Nat Acad Sci USA. 2003;100:15670–15675. doi: 10.1073/pnas.2532165100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Koppisch AT, Cane DE, Khosla C. Enhancing the modularity of the modular polyketide synthases: transacylation in modular polyketide synthases catalyzed by malonyl-CoA:ACP transacylase. J Am Chem Soc. 2003;125:14307–14312. doi: 10.1021/ja037429l. [DOI] [PubMed] [Google Scholar]
- Kwan DH, Leadlay PF. Mutagenesis of a modular polyketide synthase enoylreductase domain reveals insights into catalysis and stereospecificity. ACS Chem Biol. 2010;5:829–838. doi: 10.1021/cb100175a. [DOI] [PubMed] [Google Scholar]
- Kwan DH, Sun Y, Schulz F, Hong H, Popovic B, Sim-Stark JC, Haydock SF, Leadlay PF. Prediction and manipulation of the stereochemistry of enoylreduction in modular polyketide synthases. Chem Biol. 2008;15:1231–1240. doi: 10.1016/j.chembiol.2008.09.012. [DOI] [PubMed] [Google Scholar]
- La Clair JJ, Foley TL, Schegg TR, Regan CM, Burkart MD. Manipulation of carrier proteins in antibiotic biosynthesis. Chem Biol. 2004;11:195–201. doi: 10.1016/j.chembiol.2004.02.010. [DOI] [PubMed] [Google Scholar]
- Lambalot RH, Gehring AM, Flugel RS, Zuber P, LaCelle M, Marahiel MA, Reid R, Khosla C, Walsh CT. A new enzyme superfamily - the phosphopantetheinyl transferases. Chem Biol. 1996;3:923–936. doi: 10.1016/s1074-5521(96)90181-7. [DOI] [PubMed] [Google Scholar]
- Lau J, Cane DE, Khosla C. Substrate specificity of the loading didomain of the erythromycin polyketide synthase. Biochemistry. 2000;39:10514–10520. doi: 10.1021/bi000602v. [DOI] [PubMed] [Google Scholar]
- Lau J, Fu H, Cane DE, Khosla C. Dissecting the role of acyltransferase domains of modular polyketide synthases in the choice and stereochemical fate of extender units. Biochemistry. 1999;38:1643–1651. doi: 10.1021/bi9820311. [DOI] [PubMed] [Google Scholar]
- Leesong M, Henderson BS, Gillig JR, Schwab JM, Smith JL. Structure of a dehydratase-isomerase from the bacterial pathway for biosynthesis of unsaturated fatty acids: two catalytic activities in one active site. Structure. 1996;4:253–264. doi: 10.1016/s0969-2126(96)00030-5. [DOI] [PubMed] [Google Scholar]
- Li Q, Khosla C, Puglisi JD, Liu CW. Solution structure and backbone dynamics of the holo form of the frenolicin acyl carrier protein. Biochemistry. 2003;42:4648–4657. doi: 10.1021/bi0274120. [DOI] [PubMed] [Google Scholar]
- Li Y, Xu W, Tang Y. Classification, Prediction, and Verification of the Regioselectivity of Fungal Polyketide Synthase Product Template Domains. J Biol Chem. 2010;285:22762–22771. doi: 10.1074/jbc.M110.128504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew CW, Nilsson M, Chen MW, Sun H, Cornvik T, Liang ZX, Lescar J. Crystal structure of the acyltransferase domain of the iterative polyketide synthase in enediyne biosynthesis. J Biol Chem. 2012;287:23203–23215. doi: 10.1074/jbc.M112.362210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew CW, Sharff A, Kotaka M, Kong R, Sun HH, Qureshi I, Bricogne G, Liang ZX, Lescar J. Induced-fit upon Ligand Binding Revealed by Crystal Structures of the Hot-dog Fold Thioesterase in Dynemicin Biosynthesis. J Mol Biol. 2010;404:291–306. doi: 10.1016/j.jmb.2010.09.041. [DOI] [PubMed] [Google Scholar]
- Lim J, Kong R, Murugan E, Ho CL, Liang ZX, Yang D. Solution structures of the acyl carrier protein domain from the highly reducing type I iterative polyketide synthase CalE8. PLoS One. 2011;6:e20549. doi: 10.1371/journal.pone.0020549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou GF, Lau J, Cane DE, Khosla C. Quantitative analysis of loading and extender acyltransferases of modular polyketide synthases. Biochemistry. 2003;42:200–207. doi: 10.1021/bi0268100. [DOI] [PubMed] [Google Scholar]
- Liu Y, Zheng T, Bruner SD. Structural basis for phosphopantetheinyl carrier domain interactions in the terminal module of nonribosomal peptide synthetases. Chem Biol. 2011;18:1482–1488. doi: 10.1016/j.chembiol.2011.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma SM, Li JW, Choi JW, Zhou H, Lee KK, Moorthie VA, Xie X, Kealey JT, Da Silva NA, Vederas JC, Tang Y. Complete reconstitution of a highly reducing iterative polyketide synthase. Science. 2009;326:589–592. doi: 10.1126/science.1175602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma SM, Zhan J, Xie X, Watanabe K, Tang Y, Zhang W. Redirecting the cyclization steps of fungal polyketide synthase. J Am Chem Soc. 2008;130:38–39. doi: 10.1021/ja078091o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier T, Jenni S, Ban N. Architecture of mammalian fatty acid synthase at 4.5 A resolution. Science. 2006;311:1258–1262. doi: 10.1126/science.1123248. [DOI] [PubMed] [Google Scholar]
- Maier T, Leibundgut M, Ban N. The crystal structure of a mammalian fatty acid synthase. Science. 2008;321:1315–1322. doi: 10.1126/science.1161269. [DOI] [PubMed] [Google Scholar]
- McDaniel R, Thamchaipenet A, Gustafsson C, Fu H, Betlach M, Betlach M, Ashley G. Multiple genetic modifications of the erythromycin polyketide synthase to produce a library of novel "unnatural" natural products. Proc Nat Acad Sci USA. 1999;96:1846–1851. doi: 10.1073/pnas.96.5.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier JL, Burkart MD. The chemical biology of modular biosynthetic enzymes. Chem Soc Rev. 2009;38:2012–2045. doi: 10.1039/b805115c. [DOI] [PubMed] [Google Scholar]
- Meier JL, Haushalter RW, Burkart MD. A mechanism based protein crosslinker for acyl carrier protein dehydratases. Bioorg Med Chem Lett. 2010;20:4936–4939. doi: 10.1016/j.bmcl.2010.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moche M, Schneider G, Edwards P, Dehesh K, Lindqvist Y. Structure of the complex between the antibiotic cerulenin and its target, beta-ketoacyl-acyl carrier protein synthase. J Biol Chem. 1999;274:6031–6034. doi: 10.1074/jbc.274.10.6031. [DOI] [PubMed] [Google Scholar]
- Newman DJ, Cragg GM. Natural Products As Sources of New Drugs over the 30 Years from 1981 to 2010. J Nat Prod. 2012;75:311–335. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T, Ishida K, Jenke-Kodama H, Dittmann E, Gurgui C, Hochmuth T, Taudien S, Platzer M, Hertweck C, Piel J. Exploiting the mosaic structure of trans-acyltransferase polyketide synthases for natural product discovery and pathway dissection. Nat Biotechnol. 2008a;26:225–233. doi: 10.1038/nbt1379. [DOI] [PubMed] [Google Scholar]
- Nguyen T, Ishida K, Jenke-Kodama H, Dittmann E, Gurgui C, Hochmuth T, Taudien S, Platzer M, Hertweck C, Piel J. Exploiting the mosaic structure of trans-acyltransferase polyketide synthases for natural product discovery and pathway dissection. Nat Biotechnol. 2008b;26:225–233. doi: 10.1038/nbt1379. [DOI] [PubMed] [Google Scholar]
- Nordling E, Jornvall H, Persson B. Medium-chain dehydrogenases/reductases (MDR). Family characterizations including genome comparisons and active site modeling. Eur J Biochem. 2002;269:4267–4276. doi: 10.1046/j.1432-1033.2002.03114.x. [DOI] [PubMed] [Google Scholar]
- Olsen JG, Kadziola A, von Wettstein-Knowles P, Siggaard-Andersen M, Larsen S. Structures of beta-ketoacyl-acyl carrier protein synthase I complexed with fatty acids elucidate its catalytic machinery. Structure. 2001;9:233–243. doi: 10.1016/s0969-2126(01)00583-4. [DOI] [PubMed] [Google Scholar]
- Piel J, Hui DQ, Fusetani N, Matsunaga S. Targeting modular polyketide synthases with iteratively acting acyltransferases from metagenomes of uncultured bacterial consortia. Environ Microbiol. 2004;6:921–927. doi: 10.1111/j.1462-2920.2004.00531.x. [DOI] [PubMed] [Google Scholar]
- Ploskon E, Arthur CJ, Evans SE, Williams C, Crosby J, Simpson TJ, Crump MP. A mammalian type I fatty acid synthase acyl carrier protein domain does not sequester acyl chains. J Biol Chem. 2008;283:518–528. doi: 10.1074/jbc.M703454200. [DOI] [PubMed] [Google Scholar]
- Price AC, Rock CO, White SW. The 1.3-Angstrom-resolution crystal structure of beta-ketoacyl-acyl carrier protein synthase II from Streptococcus pneumoniae. J Bacteriol. 2003;185:4136–4143. doi: 10.1128/JB.185.14.4136-4143.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AC, Zhang YM, Rock CO, White SW. Structure of beta-ketoacyl-[acyl carrier protein] reductase from Escherichia coli: negative cooperativity and its structural basis. Biochemistry. 2001;40:12772–12781. doi: 10.1021/bi010737g. [DOI] [PubMed] [Google Scholar]
- Quadri LE, Weinreb PH, Lei M, Nakano MM, Zuber P, Walsh CT. Characterization of Sfp, a Bacillus subtilis phosphopantetheinyl transferase for peptidyl carrier protein domains in peptide synthetases. Biochemistry. 1998;37:1585–1595. doi: 10.1021/bi9719861. [DOI] [PubMed] [Google Scholar]
- Rangan VS, Joshi AK, Smith S. Mapping the functional topology of the animal fatty acid synthase by mutant complementation in vitro. Biochemistry. 2001;40:10792–10799. doi: 10.1021/bi015535z. [DOI] [PubMed] [Google Scholar]
- Rangan VS, Smith S. Alteration of the substrate specificity of the malonyl-CoA/acetyl-CoA:acyl carrier protein S-acyltransferase domain of the multifunctional fatty acid synthase by mutation of a single arginine residue. J Biol Chem. 1997;272:11975–11978. doi: 10.1074/jbc.272.18.11975. [DOI] [PubMed] [Google Scholar]
- Reed MAC, Schweizer M, Szafranska AE, Arthur C, Nicholson TP, Cox RJ, Crosby J, Crump MP, Simpson TJ. The type I rat fatty acid synthase ACP shows structural homology and analogous biochemical properties to type II ACPs. Org Biomol Chem. 2003;1:463–471. doi: 10.1039/b208941f. [DOI] [PubMed] [Google Scholar]
- Reeves CD, Murli S, Ashley GW, Piagentini M, Hutchinson CR, McDaniel R. Alteration of the substrate specificity of a modular polyketide synthase acyltransferase domain through site-specific mutations. Biochemistry. 2001;40:15464–15470. doi: 10.1021/bi015864r. [DOI] [PubMed] [Google Scholar]
- Roujeinikova A, Baldock C, Simon WJ, Gilroy J, Baker PJ, Stuitje AR, Rice DW, Slabas AR, Rafferty JB. X-ray crystallographic studies on butyryl-ACP reveal flexibility of the structure around a putative acyl chain binding site. Structure. 2002;10:825–835. doi: 10.1016/s0969-2126(02)00775-x. [DOI] [PubMed] [Google Scholar]
- Ruch FE, Vagelos PR. The isolation and general properties of Escherichia coli malonyl coenzyme A-acyl carrier protein transacylase. J Biol Chem. 1973;248:8086–8094. [PubMed] [Google Scholar]
- Scaglione JB, Akey DL, Sullivan R, Kittendorf JD, Rath CM, Kim ES, Smith JL, Sherman DH. Biochemical and structural characterization of the tautomycetin thioesterase: analysis of a stereoselective polyketide hydrolase. Angew Chem Int Ed Engl. 2010;49:5726–5730. doi: 10.1002/anie.201000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serre L, Verbree EC, Dauter Z, Stuitje AR, Derewenda ZS. The Escherichia-Coli Malonyl-Coa-Acyl Carrier Protein Transacylase at 1.5-Angstrom Resolution - Crystal-Structure of a Fatty-Acid Synthase Component. J Biol Chem. 1995;270:12961–12964. doi: 10.1074/jbc.270.22.12961. [DOI] [PubMed] [Google Scholar]
- Sharma AK, Sharma SK, Surolia A, Surolia N, Sarma SP. Solution structures of conformationally equilibrium forms of holo-acyl carrier protein (PfACP) from Plasmodium falciparum provides insight into the mechanism of activation of ACPs. Biochemistry. 2006;45:6904–6916. doi: 10.1021/bi060368u. [DOI] [PubMed] [Google Scholar]
- Sharma KK, Boddy CN. The thioesterase domain from the pimaricin and erythromycin biosynthetic pathways can catalyze hydrolysis of simple thioester substrates. Bioorg Med Chem Lett. 2007;17:3034–3037. doi: 10.1016/j.bmcl.2007.03.060. [DOI] [PubMed] [Google Scholar]
- Shen B. Polyketide biosynthesis beyond the type I, II and III polyketide synthase paradigms. Curr Opin Chem Biol. 2003;7:285–295. doi: 10.1016/s1367-5931(03)00020-6. [DOI] [PubMed] [Google Scholar]
- Shimomura Y, Kakuta Y, Fukuyama K. Crystal structures of the quinone oxidoreductase from Thermus thermophilus HB8 and its complex with NADPH: implication for NADPH and substrate recognition. J Bacteriol. 2003;185:4211–4218. doi: 10.1128/JB.185.14.4211-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siskos AP, Baerga-Ortiz A, Bali S, Stein V, Mamdani H, Spiteller D, Popovic B, Spencer JB, Staunton J, Weissman KJ, Leadlay PF. Molecular basis of Celmer’s rules: stereochemistry of catalysis by isolated ketoreductase domains from modular polyketide synthases. Chem Biol. 2005;12:1145–1153. doi: 10.1016/j.chembiol.2005.08.017. [DOI] [PubMed] [Google Scholar]
- Smith JL, Sherman DH. Biochemistry. An enzyme assembly line. Science. 2008;321:1304–1305. doi: 10.1126/science.1163785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, Tsai SC. The type I fatty acid and polyketide synthases: a tale of two megasynthases. Nat Prod Rep. 2007;24:1041–1072. doi: 10.1039/b603600g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, Witkowski A, Joshi AK. Structural and functional organization of the animal fatty acid synthase. Prog Lipid Res. 2003;42:289–317. doi: 10.1016/s0163-7827(02)00067-x. [DOI] [PubMed] [Google Scholar]
- Staunton J, Weissman KJ. Polyketide biosynthesis: a millennium review. Nat Prod Rep. 2001;18:380–416. doi: 10.1039/a909079g. [DOI] [PubMed] [Google Scholar]
- Tang Y, Chen AY, Kim CY, Cane DE, Khosla C. Structural and mechanistic analysis of protein interactions in module 3 of the 6-deoxyerythronolide B synthase. Chem Biol. 2007;14:931–943. doi: 10.1016/j.chembiol.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Kim CY, Mathews II, Cane DE, Khosla C. The 2.7-Angstrom crystal structure of a 194-kDa homodimeric fragment of the 6-deoxyerythronolide B synthase. Proc Nat Acad Sci USA. 2006;103:11124–11129. doi: 10.1073/pnas.0601924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Lee TS, Khosla C. Engineered biosynthesis of regioselectively modified aromatic polyketides using bimodular polyketide synthases. PLoS Biol. 2004;2:227–238. doi: 10.1371/journal.pbio.0020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Tsai SC, Khosla C. Polyketide chain length control by chain length factor. J Am Chem Soc. 2003;125:12708–12709. doi: 10.1021/ja0378759. [DOI] [PubMed] [Google Scholar]
- Tran L, Broadhurst RW, Tosin M, Cavalli A, Weissman KJ. Insights into protein-protein and enzyme-substrate interactions in modular polyketide synthases. Chem Biol. 2010;17:705–716. doi: 10.1016/j.chembiol.2010.05.017. [DOI] [PubMed] [Google Scholar]
- Tran L, Tosin M, Spencer JB, Leadlay PF, Weissman KJ. Covalent linkage mediates communication between ACP and TE domains in modular polyketide synthases. ChemBioChem. 2008;9:905–915. doi: 10.1002/cbic.200700738. [DOI] [PubMed] [Google Scholar]
- Tsai SC, Ames BD. Structural enzymology of polyketide synthases. Methods Enzymol. 2009;459:17–47. doi: 10.1016/S0076-6879(09)04602-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SC, Lu HX, Cane DE, Khosla C, Stroud RM. Insights into channel architecture and substrate specificity from crystal structures of two macrocycle-forming thioesterases of modular polyketide synthases. Biochemistry. 2002;41:12598–12606. doi: 10.1021/bi0260177. [DOI] [PubMed] [Google Scholar]
- Tsai SC, Miercke LJ, Krucinski J, Gokhale R, Chen JC, Foster PG, Cane DE, Khosla C, Stroud RM. Crystal structure of the macrocycle-forming thioesterase domain of the erythromycin polyketide synthase: versatility from a unique substrate channel. Proc Nat Acad Sci USA. 2001;98:14808–14813. doi: 10.1073/pnas.011399198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji SY, Cane DE, Khosla C. Selective protein-protein interactions direct channeling of intermediates between polyketide synthase modules. Biochemistry. 2001;40:2326–2331. doi: 10.1021/bi002463n. [DOI] [PubMed] [Google Scholar]
- Upadhyay SK, Misra A, Srivastava R, Surolia N, Surolia A, Sundd M. Structural insights into the acyl intermediates of the Plasmodium falciparum fatty acid synthesis pathway: the mechanism of expansion of the acyl carrier protein core. J Biol Chem. 2009;284:22390–22400. doi: 10.1074/jbc.M109.014829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzano CR, Lawson RJ, Chen AY, Khosla C, Cane DE. The biochemical basis for stereochemical control in polyketide biosynthesis. J Am Chem Soc. 2009;131:18501–18511. doi: 10.1021/ja908296m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzano CR, You YO, Garg A, Keatinge-Clay A, Khosla C, Cane DE. Stereospecificity of the dehydratase domain of the erythromycin polyketide synthase. J Am Chem Soc. 2010;132:14697–14699. doi: 10.1021/ja107344h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh CT. The chemical versatility of natural-product assembly lines. Acc Chem Res. 2008;41:4–10. doi: 10.1021/ar7000414. [DOI] [PubMed] [Google Scholar]
- Wang M, Zhou H, Wirz M, Tang Y, Boddy CN. A thioesterase from an iterative fungal polyketide synthase shows macrocyclization and cross coupling activity and may play a role in controlling iterative cycling through product offloading. Biochemistry. 2009;48:6288–6290. doi: 10.1021/bi9009049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Schwieters CD, Tjandra N. Parameterization of solvent-protein interaction and its use on NMR protein structure determination. J Magn Reson. 2012;221:76–84. doi: 10.1016/j.jmr.2012.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Wang CC, Boddy CN, Cane DE, Khosla C. Understanding substrate specificity of polyketide synthase modules by generating hybrid multimodular synthases. J Biol Chem. 2003;278:42020–42026. doi: 10.1074/jbc.M305339200. [DOI] [PubMed] [Google Scholar]
- Wattana-amorn P, Williams C, Ploskon E, Cox RJ, Simpson TJ, Crosby J, Crump MP. Solution structure of an acyl carrier protein domain from a fungal type I polyketide synthase. Biochemistry. 2010;49:2186–2193. doi: 10.1021/bi902176v. [DOI] [PubMed] [Google Scholar]
- Weissman KJ. Introduction to Polyketide Biosynthesis. Complex Enzymes in Microbial Natural Product Biosynthesis, Part B: Polyketides, Aminocoumarins and Carbohydrates. 2009;459:3–16. doi: 10.1016/S0076-6879(09)04625-4. [DOI] [PubMed] [Google Scholar]
- Weissman KJ, Hong H, Popovic B, Meersman F. Evidence for a protein-protein interaction motif on an acyl carrier protein domain from a modular polyketide synthase. Chem Biol. 2006;13:625–636. doi: 10.1016/j.chembiol.2006.04.010. [DOI] [PubMed] [Google Scholar]
- Weissman KJ, Muller R. Protein-protein interactions in multienzyme megasynthetases. ChemBioChem. 2008;9:826–848. doi: 10.1002/cbic.200700751. [DOI] [PubMed] [Google Scholar]
- Weissman KJ, Smith CJ, Hanefeld U, Aggarwal R, Bycroft M, Staunton J, Leadlay PF. The thioesterase of the erythromycin-producing polyketide synthase: Influence of acyl chain structure on the mode of release of substrate analogues from the acyl enzyme intermediates. Angew Chem Int Edit. 1998;37:1437–1440. doi: 10.1002/(SICI)1521-3773(19980605)37:10<1437::AID-ANIE1437>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Wilson MC, Moore BS. Beyond ethylmalonyl-CoA: the functional role of crotonyl-CoA carboxylase/reductase homologs in expanding polyketide diversity. Nat Prod Rep. 2012;29:72–86. doi: 10.1039/c1np00082a. [DOI] [PubMed] [Google Scholar]
- Witkowski A, Ghosal A, Joshi AK, Witkowska HE, Asturias FJ, Smith S. Head-to-head coiled arrangement of the subunits of the animal fatty acid synthase. Chem Biol. 2004;11:1667–1676. doi: 10.1016/j.chembiol.2004.09.016. [DOI] [PubMed] [Google Scholar]
- Witkowski A, Joshi A, Smith S. Fatty acid synthase: In vitro complementation of inactive mutants. Biochemistry. 1996;35:10569–10575. doi: 10.1021/bi960910m. [DOI] [PubMed] [Google Scholar]
- Witkowski A, Joshi AK, Rangan VS, Falick AM, Witkowska HE, Smith S. Dibromopropanone cross-linking of the phosphopantetheine and active-site cysteine thiols of the animal fatty acid synthase can occur both inter- and intrasubunit - Reevaluation of the side-by-side, antiparallel subunit model. J Biol Chem. 1999;274:11557–11563. doi: 10.1074/jbc.274.17.11557. [DOI] [PubMed] [Google Scholar]
- Wong FT, Chen AY, Cane DE, Khosla C. Protein-Protein Recognition between Acyltransferases and Acyl Carrier Proteins in Multimodular Polyketide Synthases. Biochemistry. 2010;49:95–102. doi: 10.1021/bi901826g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong FT, Jin X, Mathews II, Cane DE, Khosla C. Structure and Mechanism of the trans-Acting Acyltransferase from the Disorazole Synthase. Biochemistry. 2011;50:6539–6548. doi: 10.1021/bi200632j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthington AS, Burkart MD. One-pot chemo-enzymatic synthesis of reporter-modified proteins. Org Biomol Chem. 2006a;4:44–46. doi: 10.1039/b512735a. [DOI] [PubMed] [Google Scholar]
- Worthington AS, Porter DF, Burkart MD. Mechanism-based crosslinking as a gauge for functional interaction of modular synthases. Org Biomol Chem. 2010;8:1769–1772. doi: 10.1039/b925966j. [DOI] [PubMed] [Google Scholar]
- Worthington AS, Rivera H, Torpey JW, Alexander MD, Burkart MD. Mechanism-based protein cross-linking probes to investigate carrier protein-mediated biosynthesis. ACS Chem Biol. 2006b;1:687–691. doi: 10.1021/cb6003965. [DOI] [PubMed] [Google Scholar]
- Wu N, Cane DE, Khosla C. Quantitative analysis of the relative contributions of donor acyl carrier proteins, acceptor ketosynthases, and linker regions to intermodular transfer of intermediates in hybrid polyketide synthases. Biochemistry. 2002;41:5056–5066. doi: 10.1021/bi012086u. [DOI] [PubMed] [Google Scholar]
- Wu N, Tsuji SY, Cane DE, Khosla C. Assessing the balance between protein-protein interactions and enzyme-substrate interactions in the channeling of intermediates between polyketide synthase modules. J Am Chem Soc. 2001;123:6465–6474. doi: 10.1021/ja010219t. [DOI] [PubMed] [Google Scholar]
- Xie X, Meehan MJ, Xu W, Dorrestein PC, Tang Y. Acyltransferase mediated polyketide release from a fungal megasynthase. J Am Chem Soc. 2009;131:8388–8389. doi: 10.1021/ja903203g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav G, Gokhale RS, Mohanty D. Computational approach for prediction of domain organization and substrate specificity of modular polyketide synthases. J Mol Biol. 2003;328:335–363. doi: 10.1016/s0022-2836(03)00232-8. [DOI] [PubMed] [Google Scholar]
- Yin YF, Gokhale R, Khosla C, Cane DE. Erythromycin biosynthesis. The 4-pro-S hydride of NADPH is utilized for ketoreduction by both module 5 and module 6 of the 6-deoxyerythronolide B synthase. Bioorg Med Chem Lett. 2001;11:1477–1479. doi: 10.1016/s0960-894x(00)00529-1. [DOI] [PubMed] [Google Scholar]
- Zhang L, Liu W, Xiao J, Hu T, Chen J, Chen K, Jiang H, Shen X. Malonyl-CoA: acyl carrier protein transacylase from Helicobacter pylori: Crystal structure and its interaction with acyl carrier protein. Protein Sci. 2007;16:1184–1192. doi: 10.1110/ps.072757307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YM, Rao MS, Heath RJ, Price AC, Olson AJ, Rock CO, White SW. Identification and analysis of the acyl carrier protein (ACP) docking site on beta-ketoacyl-ACP synthase III. J Biol Chem. 2001;276:8231–8238. doi: 10.1074/jbc.M008042200. [DOI] [PubMed] [Google Scholar]
- Zhang YM, Wu B, Zheng J, Rock CO. Key residues responsible for acyl carrier protein and beta-ketoacyl-acyl carrier protein reductase (FabG) interaction. J Biol Chem. 2003;278:52935–52943. doi: 10.1074/jbc.M309874200. [DOI] [PubMed] [Google Scholar]
- Zhao C, Coughlin JM, Ju J, Zhu D, Wendt-Pienkowski E, Zhou X, Wang Z, Shen B, Deng Z. Oxazolomycin biosynthesis in Streptomyces albus JA3453 featuring an "acyltransferase-less" type I polyketide synthase that incorporates two distinct extender units. J Biol Chem. 2010;285:20097–20108. doi: 10.1074/jbc.M109.090092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Gay DC, Demeler B, White MA, Keatinge-Clay AT. Divergence of multimodular polyketide synthases revealed by a didomain structure. Nat Chem Biol. 2012;8:615–621. doi: 10.1038/nchembio.964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Keatinge-Clay AT. Structural and functional analysis of C2-type ketoreductases from modular polyketide synthases. J Mol Biol. 2011;410:105–117. doi: 10.1016/j.jmb.2011.04.065. [DOI] [PubMed] [Google Scholar]
- Zheng J, Taylor CA, Piasecki SK, Keatinge-Clay AT. Structural and functional analysis of A-type ketoreductases from the amphotericin modular polyketide synthase. Structure. 2010;18:913–922. doi: 10.1016/j.str.2010.04.015. [DOI] [PubMed] [Google Scholar]
- Zhou H, Gao ZZ, Qiao KJ, Wang JJ, Vederas JC, Tang Y. A fungal ketoreductase domain that displays substrate-dependent stereospecificity. Nat Chem Biol. 2012;8:331–333. doi: 10.1038/nchembio.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Zhan J, Watanabe K, Xie X, Tang Y. A polyketide macrolactone synthase from the filamentous fungus Gibberella zeae. Proc Nat Acad Sci USA. 2008;105:6249–6254. doi: 10.1073/pnas.0800657105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Yu F, Li XC, Du L. Production of dihydroisocoumarins in Fusarium verticillioides by swapping ketosynthase domain of the fungal iterative polyketide synthase Fum1p with that of lovastatin diketide synthase. J Am Chem Soc. 2007;129:36–37. doi: 10.1021/ja0672122. [DOI] [PubMed] [Google Scholar]
- Zornetzer GA, Fox BG, Markley JL. Solution structures of spinach acyl carrier protein with decanoate and stearate. Biochemistry. 2006;45:5217–5227. doi: 10.1021/bi052062d. [DOI] [PMC free article] [PubMed] [Google Scholar]