Abstract
To investigate the role of Dicer and microRNAs in the mammalian CNS, we used mice in which the second RNase III domain of Dicer was conditionally floxed. Conditional Dicer mice were bred with mice expressing an α-calmodulin kinase II Cre to selectively inactivate Dicer in excitatory forebrain neurons in vivo. Inactivation of Dicer results in an array of phenotypes including microcephaly, reduced dendritic branch elaboration, and large increases in dendritic spine length with no concomitant change in spine density. Microcephaly is likely caused by a 5.5-fold increase in early postnatal apoptosis in these animals as determined by active caspase-3 and TUNEL (terminal deoxynucleotidyl transferase-mediated biotinylated UTP nick end labeling) staining in the cortex. Loss of Dicer function had no measurable effect on cortical lamination as determined by in situ hybridization, suggesting that microcephaly is not caused by defects in neuronal migration. Together, these results illustrate the in vivo significance of Dicer and miRNAs in the mammalian CNS and provide additional support for previous in vitro studies indicating that misregulation of this pathway may result in gross abnormalities in cell number and function that may contribute to a variety of neurological disorders.
Keywords: Dicer, microRNA, CNS, dendritic spine, noncoding RNA, cortex, hippocampus
Introduction
Small RNAs, including microRNAs (miRNAs) have recently emerged as a novel mechanism for post-transcriptional regulation. These noncoding RNAs are naturally occurring, evolutionarily conserved, and are thought to negatively regulate mRNA translation via imperfect base pairing to the 3′ untranslated region (3′ UTR) of messenger RNA (Cao et al., 2006; O'Driscoll, 2006; Vasudevan et al., 2007). To date, over 500 miRNAs have been identified in humans and mice with each individual miRNA potentially regulating many mRNA targets.
In vivo, miRNA production is initiated by the transcription of long primary miRNAs via RNA polymerase II or III (Lee et al., 2004a; Kim, 2005; Borchert et al., 2006). These transcripts are processed by Drosha and DGCR8 (DiGeorge syndrome critical region 8) to pre-miRNAs within the nucleus and exported to the cytoplasm where they are processed by Dicer (Bernstein et al., 2001; Ketting et al., 2001; Knight and Bass, 2001; Lee et al., 2003; Yi et al., 2003; Lund et al., 2004; Kim, 2005). Processing by Dicer results in the production of mature miRNAs that are inserted into an RNA-induced silencing complex resulting in repression of specific gene products by mRNA degradation or translational repression (Kim, 2005).
Although multiple Dicer-like proteins and genes exist in Drosophila (Lee et al., 2004b), fungi, and Arabidopsis, there is a single Dicer gene in Caenorhabditis elegans, mice, and humans (Kadotani et al., 2004), and therefore loss or mutation of Dicer culminates in substantial biological consequences. For example, mutant Dicer alleles in C. elegans demonstrate the role of Dicer in siRNA processing and germ-line development (Knight and Bass, 2001). In mice, Dicer-null embryos arrest at embryonic day 7.5 and lack primitive streak markers, suggesting that Dicer is essential for stem cell maintenance in the early embryo (Bernstein et al., 2003). Loss of Dicer in the developing lung using Cre transgenes results in altered lung epithelial morphogenesis (Harris et al., 2006), whereas loss in the limb bud mesenchyme leads to cell death and improper formation of limb skeletal elements (Harfe et al., 2005). Dicer ablation in zebrafish impairs brain morphogenesis, somitogenesis, and heart development (Giraldez et al., 2005).
Despite these studies, little is understood regarding the global role of Dicer and miRNAs in the CNS of mammalian vertebrates. To date, limited studies have examined the in vivo role of Dicer in the mammalian CNS. In one study, inactivation of Dicer in Purkinje neurons using a Pcp-2 (Purkinje cell protein 2) Cre resulted in a slow progressive neurodegeneration (Schaefer et al., 2007). In a second study, loss of Dicer function in dopaminergic neurons using a dopamine transporter Cre increased apoptosis and neurodegeneration (Hebert and De Strooper, 2007; Kim et al., 2007). Finally, a previous study found that Dicer ablation in the developing olfactory placodes resulted in abnormal terminal differentiation of olfactory neuronal precursor cells as well as the altered maintenance of olfactory progenitor cells (Choi et al., 2008). In the present study, we examine the extent of miRNA function in the cortex and hippocampus using a calmodulin kinase II (CaMKII) Cre in an attempt to understand Dicer's global effect(s) in these tissues. Our results demonstrate that Dicer inactivation in the cortex and hippocampus result in dramatic effects on cellular and tissue morphology, axonal pathfinding, and apoptosis, providing additional evidence that Dicer function is integral to neuronal development.
Materials and Methods
Statistical analysis.
All statistical analysis was performed using GraphPad (San Diego, CA) Prism 4.0 or higher for Macintosh. In experiments in which three or more conditions were analyzed, a one-way ANOVA and Tukey's multiple comparison post test was used which compares each condition versus all conditions for statistical significance. Additional details regarding the N (number of animals) or n (number of cells, sections, dendrites, etc.) are found in the text or pertinent figure legend.
Golgi staining for cellular morphological analysis.
To examine hippocampal morphology in Dicer wild-type (wt) and mutant mice, brains from various genotypes (+/+, +/−, −/−), in the presence and absence of Cre, were impregnated in Golgi solution(s) at postnatal day 21 (P21) using the FD Rapid Golgi staining kit (FD Neurotechnologies, Baltimore, MD) according to the manufactures instructions. Coronal sections of 120 μm were made on a cryostat and hippocampal morphology was examined after staining. For dendritic branch-point analysis, 2–3 apical and basal dendrites were chosen per CA1 hippocampal neuron and two observers blind to genotype determined the number of dendritic branch points emanating from each individual dendrite. For dendritic spine length determination, spines were measured from apical dendrites of Golgi-stained hippocampal CA1 neurons for each Dicer genotype in the presence and absence of Cre expression. For all branch-point experiments results from observers were averaged. Four to five cells were measured per animal and three to four animals were used for each genotype. For Sholl measurements, eight neurons from three animals per genotype were subjected to analysis. Analysis was performed using a customized version of a Sholl analysis plug-in for Image J, which is available at http://www.biology.ucsd.edu/labs/ghosh/software/index.html. Parameters for the Sholl analysis were set as follows in the program: starting radius, 10 μm; step radius, 10 μm; ending radius, 200 μm; radius span, 0.00; span-type median images for dendritic spine and branch-point analysis were acquired at 40×, whereas images for Sholl analysis were captured at a 10× magnification on a Nikon (Melville, NY) 80i microscope.
Electrophysiology from hippocampal neurons.
Membrane currents and potentials were recorded from cultured hippocampal neurons using the whole-cell patch-clamp technique at room temperature. Patch pipettes had resistances of 2.5–4.0 MΩ and were pulled from borosilicate capillary glass (World Precision Instruments, Sarasota, FL). The bath solution had an osmolarity of 300 mOsm and contained (in mm) 120 NaCl, 3 CaCl2, 2 MgCl2, 5 KCl, and 10 HEPES, pH 7.3. The intracellular solution was 290 mOsm and contained (in mm) 120 K-gluconate, 10 KCl, 1 MgCl2, 0.025 CaCl2, 0.2 EGTA, 2 Na2-ATP, 0.2 Na2-GTP, and 10 HEPES, pH 7.2. Spontaneous currents were recorded at a holding potential of −70 mV. Data was collected using a multiclamp 700B amplifier and Clampex acquisition software version 9.2. Traces were visualized using Clampfit 9.2.
Northern blot and qRT-PCR analysis.
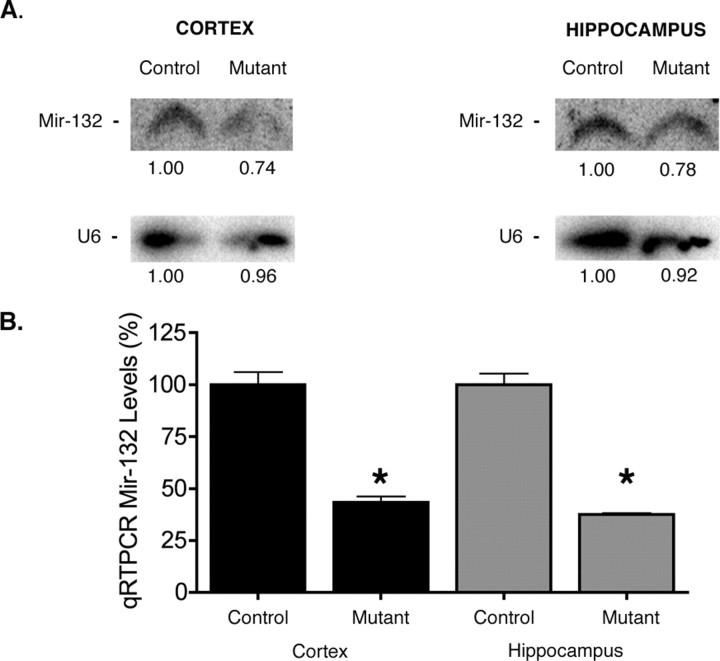
Total RNA was isolated from the cortex, hippocampus and cerebellum of P15 or P21 homozygous Dicer mice in the presence or absence of Cre recombination using Trizol reagent according to manufacturer's instructions. Total RNA (5 μg) was loaded onto 15% denaturing polyacrylamide gels and run at 65 mA for 2.5–3 h at room temperature, and transferred to nitrocellulose at 400 mA for 1 h at room temperature. After crosslinking, membranes were hybridized using antisense DNA oligo probes specific for microRNA 132 (mir-132) or mir-124. The antisense probe used for mir-132 and mir-124, respectively, were 5′gaccatggctgtagactgtta3′ and 5′ggcattcaccgcgtgcctta3′. All probes were 5′ end labeled for 1–2 h using polynucleotide kinase and γ-32P ATP. After overnight hybridization, membranes were washed six times for 10 min in 1× SSC with 1% SDS and exposed to film or a Storm Phosphorimager (General Electric, Piscataway, NJ) was used. Integrated pixel density was calculated using Image J. All images were converted to eight-bit background-subtracted using a rolling radius of 2.5 mm and images were inverted to capture integrated pixel density. Images were then outlined using the freehand tool selection or thresholded to obtain the integrated pixel density. Numbers shown in Figure 2 represent the normalized ratio of mutant and control miRNA expression integrated pixel density.
Figure 2.
MicroRNA levels are decreased in Dicer mutant mice. A, Northern blot analysis for mir-132 at P15 demonstrates a decrease in miRNA levels in the cortex and hippocampus of Dicerflox/flox; CaMKII Cre mice. B, qRT-PCR analysis of mir-132 levels in the cortex and hippocampus of Dicer mutant mice at P21 demonstrates that miRNA levels continue to decrease over time. U6 was used as a loading control in all experiments. Five micrograms of total RNA were loaded per well for cortical and hippocampal mir-132 Northern blot analysis. Twenty nanograms of total RNA were used to generate cDNA for qRT-PCR analysis. Error bars represent the SEM. Values are statistically significant using a one-way ANOVA (*p < 0.001) with a Tukey's multiple comparison-post test.
Taqman quantitative reverse transcriptase (qRT)-PCR analysis was performed on 20 ng of total cortical or hippocampal RNA following the manufacturer's protocol (Applied Biosystems, Foster City, CA) and normalized to U6. Analysis was performed using 2^(ΔCT).
X-gal staining.
Cryosections (30 μm, brain; or 120 μm, whole-mount) for each Dicer genotype were washed three times in a wash solution (0.1 m phosphate buffer with 1% deoxycholate, 2% NP-40 and 1.5 mm MgCl2) and subjected to a 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal) solution (1 mg/ml X-gal and 5 mm potassium ferrocyanide, 5 mm potassium ferricyanide in wash buffer) for 40 min −1 h at 37°C. The reaction was quenched by washing sections three times in wash solution and incubating them in 4% paraformaldehyde for 5 min at room temperature. The sections were washed three times in wash solution and coverslipped using Vectashield mounting medium (Vector Laboratories, Burlingame, CA).
TUNEL staining and immunostaining.
For terminal deoxynucleotidyl transferase-mediated biotinylated UTP nick end labeling (TUNEL) analysis, animals of each genotype were killed at postnatal days 0 and 15 and brains were formalin fixed at 4°C for 2 and 6 h, respectively. Cryosections of 25 μm were made and TUNEL staining was performed using the FragEL DNA detection kit (Calbiochem, La Jolla, CA) according to manufacturer's instructions. Sections from animals used for TUNEL staining were also used to determine the reactivity for the active form of caspase-3. For caspase-3 immunostaining, sections were incubated overnight at 4°C in a rabbit monoclonal antibody directed against the active form of caspase-3 (Cell Signaling Technology, Danvers, MA) at a dilution of 1:500. An AlexaFluor 488-conjugated goat anti-rabbit secondary antibody (Invitrogen, Carlsbad, CA) at a dilution of 1:500 was used for detection. Quantification of apoptotic neurons was assessed by counting the total number of fluorescent (TUNEL or caspase-3 positive) cell bodies in the cortex from mice of each genotype from three to five nonconsecutive cryosections to ensure that neurons were not counted twice. Apoptosis analysis was performed on at least three animals per genotype. Negative controls for TUNEL and caspase-3 staining were slides incubated in the absence of either terminal deoxynucleotidyl transferase enzyme (which catalyzes the reaction for the addition of fluorescent deoxynucleotides to free 3′ OH in TUNEL stain) or caspase-3 antibody. For neuropilin-2 (NP-2) immunostaining, 30 μm coronal sections from P15 brains of wild-type and mutant animals were incubated at a dilution of 1:500 overnight in a rabbit polycolonal antibody directed against neuropilin-2 (Cell Signaling Technology).
Results
Forebrain Dicer inactivation leads to ataxia and reduced life expectancy
To gain a better understanding of the function of Dicer and miRNAs in the developing mammalian CNS, we used a previously established conditional Dicer line in which the fourth exon encoding the second RNase III domain of Dicer is ablated after Cre recombination (Harfe et al., 2005) (Fig. 1 a). Cre recombination was achieved using the R1AG-5 α-CaMKII Cre, which was reported previously to recombine in the cortex and hippocampus beginning at embryonic day 15.5 (Dragatsis and Zeitlin, 2000; Dragatsis et al., 2000). As shown in the Kaplan–Meier curve (Fig. 1 b), inactivation of Dicer using a brain-specific α-CaMKIICre resulted in the early postnatal death of Dicerflox/flox; CaMKII +/− mice (inverted triangles), whereas wild-type and heterozygous animals persist into adulthood. Visual examination indicated that Dicer mutant mice were ataxic with visible tremors during motility. Ataxic gait was normally apparent by postnatal day 14–15, but often occurred as early as postnatal day 12 and was generally characterized by hindleg movement in unison (supplemental Movie 1, available at www.jneurosci.org as supplemental material). Dicer mutant animals also displayed hindlimb crossing prototypical of animals with motor impairments. These results indicate that loss of Dicer function results in a significantly decreased lifespan as well as motor impairments.
Figure 1.
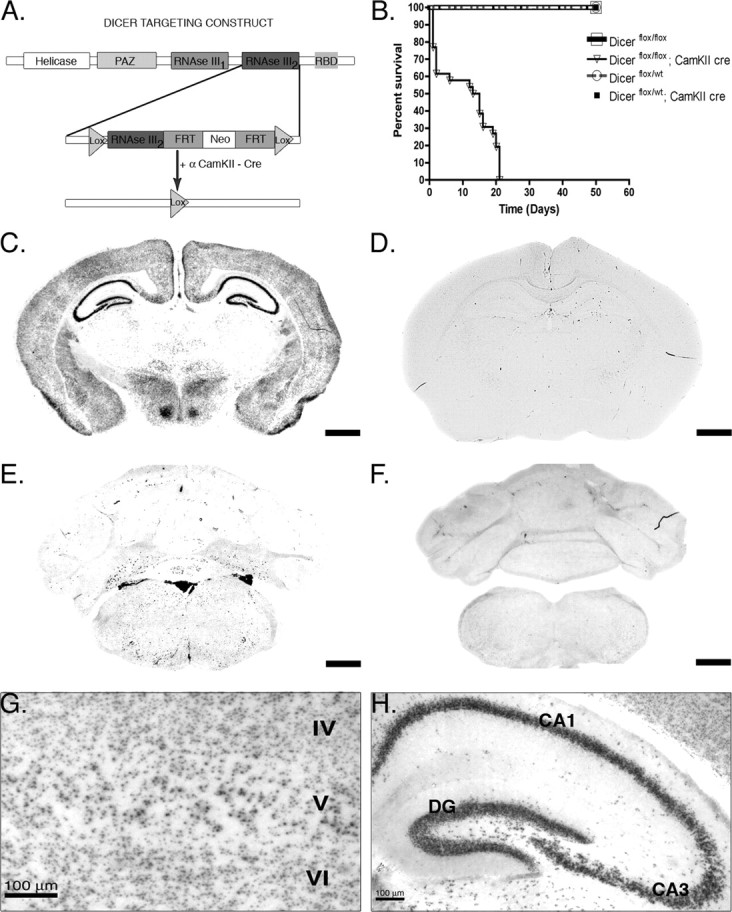
Cre recombination occurs throughout the forebrain and results in early death. A, Dicer targeting construct. The second RNase III domain of Dicer was flanked with Lox P sites. After Cre recombination at embryonic day 15.5, Dicer is rendered inactive, leading to a failure to produce microRNAs in excitatory forebrain postmitotic neurons. B, Kaplan–Meier plot of survival demonstrates that 40% of Dicerflox/flox; CaMKII Cre (n = 26; open triangle) animals die by postnatal day 2 and that the remaining animals die by the end of postnatal day 21, whereas other genotypes persist into adulthood (Dicerflox/flox, n = 20, open square; Dicerflox/wt, n = 20, open circle; Dicerflox/wt; CaMKII Cre, n = 20, closed square). C–H, Floxed Dicer mice were crossed to α-CaMKII Cre- and RosaR26R-expressing mice to visualize brain regions in which Dicer was removed. C, Cre-recombined cells at postnatal day 21 in a Dicerflox/wt; CaMKII Cre; R26R +/− mouse. Recombination is high throughout all cortical layers as well as throughout the pyramidal cell layer of the hippocampus. Little expression/recombination is observed in the thalamus. D, Cre recombination at postnatal day 21 in a Dicerflox/wt; CaMKII Cre; R26R +/+ mouse demonstrating the specificity of X-gal staining. E, X-gal staining at the level of the cerebellum in a Dicerflox/wt; CaMKII Cre; R26R +/− mouse illustrating forebrain specificity of the CaMKII Cre line. F, Similar to the image in D, there is a lack of X-gal staining in the cerebellum in Dicerflox/wt; CaMKII Cre; R26R +/+ mice. G, Higher magnification of cortical layers IV–VI. Recombination occurs in most but not all cortical neurons. H, Higher magnification of the hippocampal formation in a Dicerflox/wt; CaMKII Cre mouse illustrating that recombination occurs throughout the dentate gyrus and CA1–3 regions, but not in strata, where interneurons and glia predominate. Scale bars: C–F, 1 mm. RBD, RNA binding domain; FRT, flipase recognition target.
To verify the neuronal population targeted by the R1AG-5 CaMKIICre as well as investigate the neurons responsible for the observed phenotypes above, floxed Dicer CaMKII mice were crossed to Rosa26R reporter mice to visualize recombined cells/brain regions using β-galactosidase staining (Soriano, 1999). As shown in Figure 1 c–h, Cre recombination is forebrain specific with the highest levels of recombination occurring in the cortex, hippocampus, and caudate–putamen (Fig. 1 c) (data not shown). Closer examination of X-gal staining revealed that Cre recombination was lightly scattered throughout the thalamus, whereas the cortex and all excitatory pyramidal neurons of the CA and dentate region of the hippocampus were X-gal positive (Fig. 1 g,h). Little recombination occurred in caudal or midbrain structures including the cerebellum, medulla, and pons (Fig. 1 e). Similarly, no significant recombination was observed in peripheral organs/tissues (supplemental Fig. 1, available at www.jneurosci.org as supplemental material). Together, these data indicate that the α-calmodulin kinase II Cre is mostly forebrain specific, in agreement with previous studies (Dragatsis et al., 2000).
MiRNA levels are reduced after Dicer inactivation
Because Dicer function is integral to the production of mature miRNAs, we sought to verify that Cre recombination resulted in a loss of Dicer expression in excitatory neurons as well as a loss or reduction in microRNA production (Bernstein et al., 2001; Calabrese et al., 2007). In situ hybridization for Dicer demonstrated loss of transcript in excitatory neurons within the cortex and hippocampus (supplemental Fig. 2B,D, compare insets, E,F, available at www.jneurosci.org as supplemental material). To investigate whether microRNA levels were also reduced, we performed Northern blot analysis on isolated cortical and hippocampal tissue from Dicer mutant and wild-type animals extracted at postnatal day 15. Because the α-calmodulin kinase II Cre is neuron specific and does not target glia or interneurons (Dragatsis and Zeitlin, 2000), we expected only a modest reduction in microRNA expression using Northern blot analysis. Northern blot analysis for mir-132, an miRNA important in dendritic outgrowth, demonstrated that loss of Dicer resulted in a 22% reduction of mir-132 levels in the cortex and 14% decrease in the hippocampus when compared with control animals at postnatal day 15, around the time of onset of motor phenotypes (Fig. 2 a). mir-124a, the most abundant neuronal miRNA (Lagos-Quintana et al., 2002), was also decreased in the cortex of Dicer mutant animals relative to control animals (supplemental Fig. 3, available at www.jneurosci.org as supplemental material). We next asked whether the levels of miR-132 were stable or continued to decline over time. To assure accurate quantification of possibly low miRNA levels, we used quantitative PCR (Jiang et al., 2005; Lee et al., 2007). Quantitative RT-PCR analysis using Taqman probes demonstrated that mir-132 levels continued to decline in the cortex and hippocampus (Fig. 2 b). In Dicer mutant animals, mir-132 levels decreased by >60% in the hippocampus and cortex relative to control animals by P21, corresponding to the time when survival and motor phenotypes were most severe (Fig. 2 b). These data indicate that inactivation of Dicer results in decreased levels of microRNAs in the cortex and hippocampus of Dicer mutant animals and that these levels correlate with the severity of observed phenotypes.
Dicer deficient mice are microcephalic and have increased lateral ventricle size
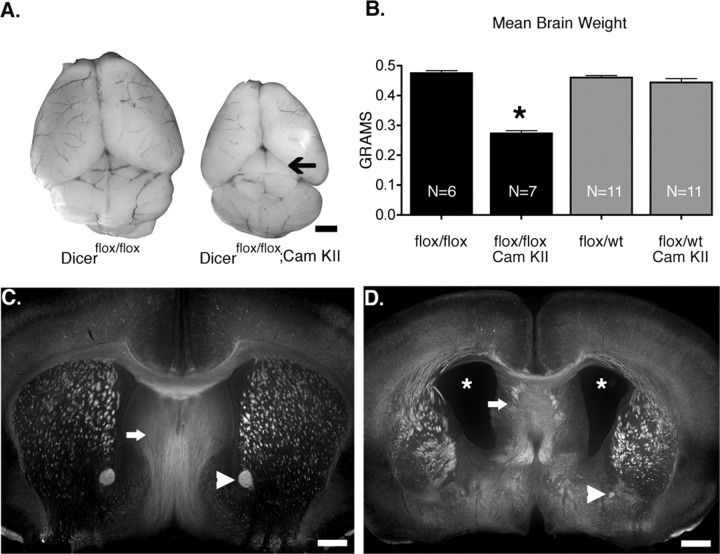
Next we examined gross brain morphology in Dicer mutant and wild-type mice at postnatal day 21. Previous work isolating tissue for Northern blot analysis suggested that vast differences in brain mass existed between mutant and wild-type animals (Fig. 2), and that mutant animals were likely microcephalic. Quantification of brain mass routinely showed an ∼50% reduction in mutant animals (flox/flox, 0.475 ± 0.02 g; flox/flox CaMKII, 0.273 ± 0.02 g; flox/wt, 0.46 ± 0.02 g; flox/wt CaMKII, 0.44 ± 0.04 g), indicating that they were indeed microcephalic (Fig. 3 a,b). Closer examination of gross brain morphology also revealed what appeared to be a smaller cortex leading to increased exposure of the superior colliculus (Fig. 3 a, arrow). Coronal sections demonstrated that mutant animals have abnormally large lateral ventricles, but normal fourth ventricles (Fig. 3 c,d) (data not shown). Coronal sections also indicated that Dicer mutant animals have a smaller anterior commissure (Fig. 3 c,d, arrowhead). Together, these data indicate that loss of Dicer function results in gross brain malformations including microcephaly, increased ventricle size, and reduction in the size of white matter tracts.
Figure 3.
Dicer mutant animals show decreases in brain weight and abnormally large ventricles. A, Brains of Dicer mutant animals are consistently smaller than their wild-type littermates. The arrow highlights increased exposure of superior colliculus in Dicer mutant animals. B, Quantification of brain weight (N indicates number of animals). C, D, Dicerflox/flox (C) and Dicerflox/flox; CaMKII Cre +/− (D) mice immunostained for neuropilin-2 demonstrate substantial differences in ventricular size as well as axonal pathfinding abnormalities (arrows in C and D). Arrowheads illustrate differences in anterior commissure. Scale bars: A, 1 mm; C, D, 500 μm. In B, error bars represent SEM. Values are statistically significant (*p < 0.001) using a one-way ANOVA with a Tukey's multiple comparison post test.
Dicer inactivation results in increased cortical apoptosis, but has no effect on cortical lamination
Because there was a significant decrease in brain mass, increased ventricle size, and a smaller cortex in Dicer mutant mice, we hypothesized that these collective phenotypes may be caused by increased apoptosis. In addition, several in vitro and in vivo studies have demonstrated roles for miRNAs as proapoptotic and antiapoptotic factors (Chan et al., 2005; Karres et al., 2007; Kim et al., 2007; Mott et al., 2007; Raver-Shapira et al., 2007; Xu et al., 2007). To examine whether the observed phenotypes were caused by a loss of neurons through programmed cell death, we performed immunohistochemistry for the cleaved (active) form of caspase-3. Quantification of caspase-3-positive cells in wild-type versus Dicer mutant animals indicates that mutants have a 5.5-fold increase in apoptosis (Dicerflox/+; CaMKII, 13.33 ± 1.02 cells; Dicerflox/flox; CaMKII, 74.32 ± 6.92 cells) with the majority of cell death occurring at or near the ventricular zone of the cortex (Fig. 4 a,b). Because caspase-3 is a marker for apoptosis but may not always be the executioner caspase (Troy et al., 2000, 2001; Prunell and Troy, 2004), we also performed TUNEL staining, which is a better indicator of apoptosis. Figure 4 c illustrates the substantial increase in apoptosis at postnatal day 0 as assessed by TUNEL in Dicer mutant animals. Because mir-21 has been shown to be an antiapoptotic factor in glia (Chan et al., 2005) and there is little available data regarding its presence in neurons, we tested whether levels of this miRNA were decreased in cortex, which would lead to increased levels of neuronal apoptosis. Quantitative RT-PCR demonstrated that levels of mir-21 were only slightly changed (supplemental Fig. 4, available at www.jneurosci.org as supplemental material), suggesting that it is expressed predominately by glia and not neurons. Interestingly, although apoptosis was increased multifold at P0, little to no apoptosis was detected by either TUNEL or caspase-3 immunostaining at postnatal day 15 (data not shown). These data suggest that Dicer is instrumental to the survival of CNS neurons in vivo at early postnatal ages, but does not cause increases in apoptosis at later postnatal ages despite continued reduction of miRNA levels.
Figure 4.
Dicer inactivation leads to increased cortical apoptosis in vivo. A, Quantification of active caspase-3 staining in Dicerflox/wt; CaMKII +/− and Dicerflox/flox; CaMKII +/− at postnatal day 0 (Dicerflox/flox; CaMKII Cre +/−, n = 25, N = 3; Dicerflox/wt; CaMKII Cre +/−, n = 24, N = 3). For negative controls, n = 12 and N = 3. N indicates the number of animals, and n the number of sections analyzed. B, Immunostaining for the active (cleaved) form of caspase-3 illustrating neurons adjacent to the lateral ventricle undergoing programmed cell death. Inset, Apoptotic layer 5 cortical neuron (same section) showing blebbing characteristic of programmed cell death. C, TUNEL staining of Dicerflox/flox (top) and Dicerflox/flox; CaMKII Cre animals (bottom) at postnatal day 0 indicating that Dicer mutant animals have multifold increases in cortical apoptosis with the majority of apoptosis occurring at or around the lateral ventricles in the cortex of Dicer mutant animals. Positive controls were first treated with 1 μg/μl DNase I in 1× TBS-Tween 20/MgSO4 for 20 min at room temperature to cause DNA damage and then processed with other sections. In A and C, negative controls refer to samples untreated with an anti-caspase-3 antibody or terminal deoxynucleotidyl transferase, respectively. Scale bars: B, 10 μm; C, 100 μm. In A, error bars represent SEM. Values are statistically significant using a one-way ANOVA (*p < 0.001) with a Tukey's multiple comparison post test.
Because cortical neurons were lost as a result of increased apoptosis in vivo, we examined whether cortical layering was also perturbed as a consequence of Dicer loss. Under normal conditions, the cortex develops in an inside-out manner, with the first neurons born being found in deeper cortical layers and the last neurons born localizing to more superficial cortical layers. When we performed in situ hybridization for cortical layer-specific markers including CUX1 (layers 2, 3, 4) (Nieto et al., 2004), ER81 (layer 5) (Hevner et al., 2003; Yoneshima et al., 2006), and LDB2 (layers 5, 6) (Arlotta et al., 2005), we found that inactivation of Dicer did not adversely affect the ability of cortical neurons to migrate to their appropriate target layers, consistent with the notion that cortical layering and cell migration is normal in Dicer mutants (supplemental Fig. 5, available at www.jneurosci.org as supplemental material).
Dicer loss in pyramidal neurons leads to changes in dendritic branching, spine morphology, and axonal tracts
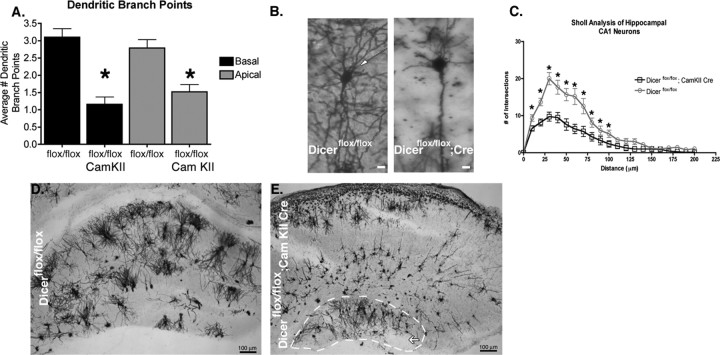
To examine how inactivation of Dicer and miRNAs affects cellular morphology in vivo, we focused our efforts on the hippocampal formation because of its stereotyped architecture, well defined neuronal population, and high recombination efficiency in our system (Fig. 1 c,h) (Dragatsis et al., 2000). Furthermore, because inactivation of Dicer did not severely affect the level of apoptosis in the hippocampus after P0, any effects observed at P21 are likely caused by miRNA loss and not secondary effects associated with apoptosis. Using the Golgi staining technique, we examined several aspects of hippocampal CA1 pyramidal neuron development including dendritic branching, dendritic spine morphology, dendritic spine density, and dendritic spine length. In the absence of Dicer, dramatic effects were observed on apical and basal dendritic branching as measured by branch-point analysis (Fig. 5 a). Basal dendrites from Dicer mutant animals averaged 1.16 ± 0.21 (n = 30) secondary branch points per dendrite, whereas basal dendrites from Dicer wild-type animals averaged 3.1 ± 0.25 (n = 31) secondary branch points per dendrite. Similarly, apical dendrites from Dicer mutant animals averaged 1.51 ± 0.21 (n = 27) branch points per dendrite, whereas Dicer wild-type animals averaged 2.78 ± 0.24 (n = 28) branch points per dendrite. Sholl analysis, which measures dendritic complexity, also confirmed that branching is significantly reduced in mutant versus wild-type animals (Fig. 5 b,c). In addition to reduced dendritic branching, Dicer mutants displayed reduced hippocampal formation size (Fig. 5 d,e, white outline), with a general disorganization of the tissue topology. The dentate gyrus was also affected by loss of miRNAs as it was dramatically reduced in size or completely absent in the most rostral region of the hippocampal formation in a fraction of Dicer mutant animals (Fig. 5 d,e, white arrow).
Figure 5.
Dendritic branching in hippocampal neurons is impaired in Dicer mutant animals. A, Quantification of basal and apical dendritic branch points in Dicerflox/flox and Dicerflox/flox; CaMKII +/− Cre animals demonstrate decreases in the number of dendritic branch points. B, Example of Golgi-stained hippocampal CA1 neurons from Dicerflox/flox and Dicerflox/flox; CaMKII +/− animals illustrating differences in basal and apical branching. C, Sholl analysis of Dicerflox/flox and Dicerflox/flox; CaMKII +/− animals illustrates substantial differences in dendritic complexity (Dicerflox/flox, n = 24, N = 3; Dicerflox/flox; CaMKII +/−, n = 24, N = 3). D, Golgi-stained Dicerflox/flox hippocampus with stereotyped architecture including a well defined dentate gyrus and CA1–3 region. E, Golgi-stained Dicerflox/flox; CaMKII +/− Cre hippocampus outlined in white illustrates decreased size relative to that shown in E (images taken at same magnification). Abnormal hippocampal architecture observed in Dicerflox/flox; CaMKII +/− Cre mice makes distinct layers of the CA region difficult to discern. The white arrow highlights loss of the dentate gyrus. Images were taken at 4× in D and E. n is the number of neurons, N the number of animals. Scale bars: B, 10 μm; E, F, 100 μm. Error bars represent SEM. Values in A are statistically significant using a one-way ANOVA (*p < 0.001) with a Tukey's multiple comparison post test. Values in C are significant using an unpaired two-tailed t test (*p < 0.05).
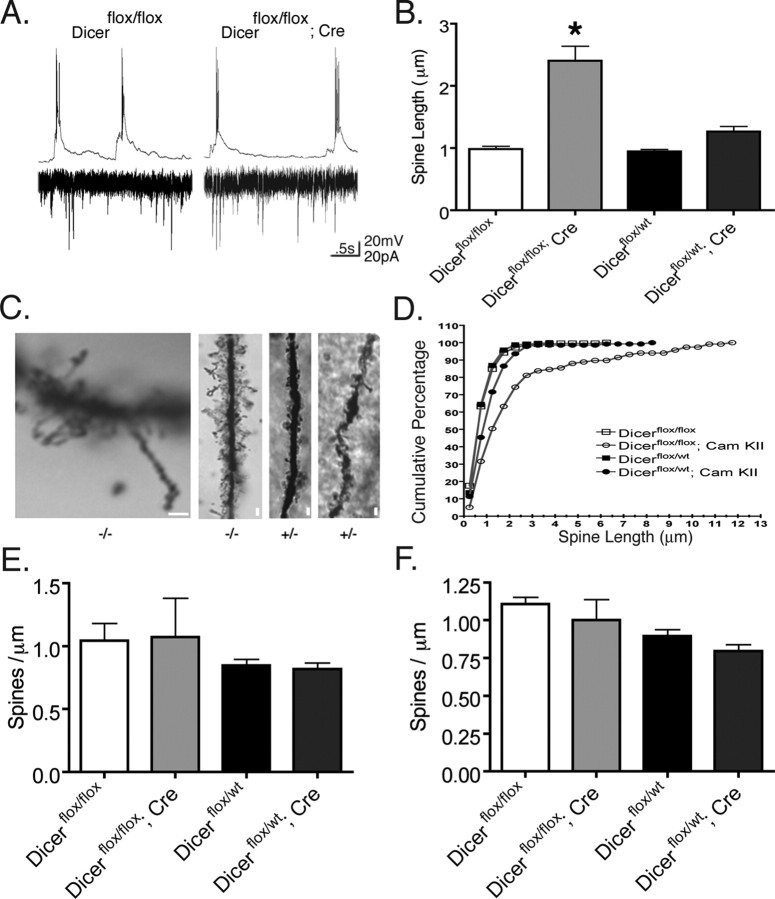
Despite the above cellular and tissue abnormalities in the hippocampus, CA1 neurons from Dicer mutant animals fired spontaneous action potentials and received synaptic input suggesting that dendritic spines were functionally intact (Fig. 6 a). Because mir-134 expression affects dendritic spine width and volume in vitro (Schratt et al., 2006), we tested whether spine morphology was perturbed in Dicer mutant mice. Examination of Golgi-stained apical dendritic spines of CA1 pyramidal neurons from Dicer mutant animals indicated that dendritic spine length is increased by >140% in vivo, with a mean length of 2.4 ± 0.23 μm compared with other genotypes with means ranging from 0.94 to 1.26 μm (Fig. 6 b). Although dendritic spine structures longer than 1.0–1.5 μm are generally considered immature filopodia because of the lack of a spine head, the structures observed in Dicer mutant animals generally retained spine heads. Cumulative distribution plots illustrate that nearly 100% of spines in control animals are less than or equal to 2.0 μm in length, whereas only 70% of knock-out spines measured 2.0 μm or less (Fig. 6 d). Figure 6 c provides examples of dendritic spines found to be 12 μm in length in Dicer mutant animals.
Figure 6.
Inactivation of Dicer in hippocampal CA1 neurons leads to increased apical dendritic spine length but not density in vivo. A, Spontaneous action potentials and synaptic activity from Dicer mutant animals at postnatal day 15 indicate that dendritic spines are functional. B, Mean dendritic spine length for Dicerflox/flox, 0.98 ± 0.04 (n = 245, N = 2); Dicerflox/flox; CaMKII +/−, 2.40 ± 0.23 (n = 117, N = 2); and Dicerflox/wt, 0.94 ± 0.03 (n = 236, N = 3); Dicerflox/wt; CaMKII +/−, 1.26 ± 0.08 (n = 141, N = 3). C, Example of dendritic spines in Dicerflox/flox; CaMKII +/− Cre −/−, and Dicerflox/wt; CaMKII +/− Cre +/− animals. Dendritic spines of Dicerflox/flox; CaMKII +/− Cre (as shown in C, left) were observed up to 12 μm in length. D, Cumulative distribution of apical dendritic spine length in each Dicer genotype. Loss of microRNAs results in one-third of dendritic spines being greater in length than the longest measured dendritic spine in other Dicer genotypes. E, F, Basal and apical dendritic spine density in CA1 hippocampal neurons at postnatal day 21 in Golgi-stained neurons. The number of spines per 50 μm dendrite were counted by three blind observers, and the numbers were averaged. Criterion for inclusion included dendrites of 75 μm or longer. For analysis, spines were counted on basal or apical dendrites starting 25 μm from the cell body for a length of 50 μm. Analysis performed on two to three dendrites per cell in each condition with four to five cells measured. Three to four animals were used per condition. N is the number of animals, and n the number of cells. Scale bars, 2 μm. Error bars represent SEM. Values in B are statistically significant (*p < 0.001) using a one-way ANOVA with a Tukey's post test.
Because such a dramatic effect was observed on dendritic spine length, we also examined the apical and basal dendritic spine density on Golgi-stained hippocampal CA1 neurons at postnatal day 21. Despite occasional segments of high dendritic spine density, there was no significant difference between Dicer mutant and wild-type mice when spine density was measured over 50 μm dendritic lengths (Fig. 6 e,f).
We next asked whether Dicer inactivation affected axonal as well as dendritic structures. In vivo, cortical neurons project to subcortical structures by initially sending processes laterally to the internal capsule; subsequently, these processes turn caudally until they reach their synaptic targets (Bagnard et al., 1998). During these events, neurons are subjected to a multitude of secreted and membrane bound molecules including ephrins and semaphorins. Semaphorins have been demonstrated to act as both attractive and repulsive cues during the development of cortical projections in vivo and in vitro (Bagnard et al., 1998). Because many miRNAs are predicted to target molecules important for axonal pathfinding, we decided to examine whether pathfinding was perturbed in vivo in Dicer mutant animals. Immunohistochemistry for the semaphorin receptor NP-2 in Dicer wild-type and mutant animals revealed pathfinding abnormalities within the lateral septal nucleus, as shown in Figure 3, c and d. In Dicer mutant animals, axons radiated in all directions within the lateral septal nucleus in contrast to the tight, fasciculated organization observed in wild-type animals. These results suggest that loss of Dicer function also affects in vivo axonal pathfinding in mammals.
Discussion
The role of Dicer and small RNAs in the mammalian CNS is currently poorly understood. In the mammalian CNS, Schaefer et al. (2007) examined how Dicer removal influences cerebellar Purkinje neurons in vivo using a conditional system and Pcp-2 Cre. Their study demonstrates that loss of Dicer function results in slow Purkinje cell degeneration. A second study using a conditional system and dopamine transporter Cre similarly demonstrated that Dicer inactivation results in slow cellular death of striatal neurons (Kim et al., 2007). Finally, a recent study revealed that Dicer inactivation in the retina leads to progressive functional and structural degeneration of the retinal architecture (Damiani et al., 2008).
Our study is the first to examine the role of Dicer and miRNAs in excitatory neurons of the neocortex and hippocampus in conditionally floxed mice. In our model system, Dicer is recombined in excitatory neurons as indicated by in situ and x-gal staining (Fig. 1, supplemental Fig. 2, available at www.jneurosci.org as supplemental material). Interestingly, Dicer expression appears to occur both in neuronal cell bodies and in surrounding areas presumably representing non-neuronal cells (supplemental Fig. 2, available at www.jneurosci.org as supplemental material). Because of the continued low levels of signal that are present in the hippocampus and appear to be outside of neuronal cell bodies, we cannot exclude the possibility that low levels of functional Dicer remain in excitatory neurons in this animal model, making it a strong hypomorph. The reduction in miRNAs and the severe phenotypes observed raises the question of whether miRNA dosage plays a key role in brain function as suggested previously (Klein et al., 2007). Nevertheless, our results demonstrate that loss or reduction of Dicer function in excitatory neurons causes significant alterations in dendritic branching, dendritic spine length, apoptosis, and axonal pathfinding in vivo. Furthermore, loss of Dicer function affected the patterning and organization of the hippocampus whereas cortical lamination remained unaltered. Despite the effects on dendritic spine length observed in Dicer mutant animals, loss of Dicer had no measurable effect on dendritic spine density in hippocampal neurons. These studies signify the importance of Dicer, and interfering with the miRNA pathway, in the CNS and demonstrate that misregulation of this pathway results in phenotypes similar to debilitating CNS disorders.
Role of Dicer in brain size and cell survival
After recombination of Dicer, we observed a 5.5-fold increase in apoptosis in the cortex as determined by TUNEL and active caspase-3 staining at postnatal day 0, whereas no increase in apoptosis was detected at P15. Increased cortical apoptosis in early postnatal life is likely the cause of increased third ventricle size and microcephaly in Dicer mutant animals. Dicer is integral to the survival of many cell types including those that contribute to the developing lung epithelium and limb bud (Fukagawa et al., 2004; Harfe et al., 2005; Harris et al., 2006). The observation that Dicer mutants have increased apoptosis early in development (P0), but not later in development (P15) may indicate that a subset of neurons depends on Dicer function for survival. Alternatively, the result may indicate that the timing of Dicer removal determines the outcome, early removal of Dicer may lead to cell death whereas later removal may lead to the other observed phenotypes (e.g., increased spine length). This finding is consistent with a previous report that late inactivation of Dicer does not result in increased apoptosis (Cuellar et al., 2008).
Role of Dicer in dendrite and spine morphology and axon tracts
Given that Dicer loss does not result in further cell death beyond P15, we analyzed the morphological phenotypes associated with Dicer ablation at P21 such as dendrite and spine defects. Abnormalities in dendritic spine density and length are associated with several CNS-related disorders including schizophrenia, fragile X mental retardation, tuberous sclerosis type I, and murine scrapie (Nimchinsky et al., 2002). Furthermore, memory loss associated with aging may be the result of changes in dendritic spine density (Nimchinsky et al., 2002). Previous work has demonstrated that mir-134 is an important determinant in dendritic spine width and volume in hippocampal neurons in vitro (Schratt et al., 2006). In contrast to these findings, inactivation of Dicer resulted in a 140% increase in dendritic spine length with spines observed to be as long as 12 μm. These data indicate that Dicer and the miRNA pathway may regulate multiple aspects of spine morphology in vivo.
It was demonstrated previously that Dicer, miRNAs, and Ago1 all interact with the fragile X mental retardation protein (FMRP), forming an mRNP (messenger ribonuclear protein) complex (Jin et al., 2004). It is therefore interesting that loss of Dicer in hippocampal neurons results in a similar dendritic spine phenotype as that observed in the FMRP-null mice and affected human patients. In neocortical pyramidal neurons of FMRP-null mice, dendritic spine development fails to proceed normally, resulting in longer, phenotypically immature spines (Galvez and Greenough, 2005). In contrast, apical dendritic spines of hippocampal neurons show a global increase in length. Therefore, it appears that in the absence of either FMRP or Dicer, dendritic spines are increased in length, possibly because of the loss of translational suppression on disruption of the miRNA pathway.
In addition to spine phenotypes, inactivation of Dicer results in significant reduction in dendritic complexity (Fig. 5). Interestingly, overexpression of mir-132 has been reported to increase neurite elaboration in vitro (Vo et al., 2005). Using qRT-PCR, we observed a >60% decrease in mir-132 levels in isolated cortical and hippocampal tissue at P21. Thus, downregulation of mir-132 levels may be related to the decrease in dendritic branching observed by branch-point and Sholl analysis (Fig. 5 a–d). Regardless of the specific miRNAs, these results show that loss of Dicer function leads to reduced dendritic branching in vivo.
The axon tract abnormalities observed in our mice phenocopy those of NP-2-null mice, suggesting perturbation of a similar or shared pathway (Giger et al., 2000). In the case of neuropilin-2-null mice, cortical neurons are unable to respond to the repulsive effects of semaphorin 3F culminating in pathfinding abnormalities and smaller anterior commissure, similar to what we observed (Giger et al., 2000). Because our immunostaining indicates the presence of NP-2 on cortical neuron axons, our results suggest that semaphorin 3F may be absent or mislocalized in vivo or that there is altered semaphorin-related intracellular signaling in NP-2-positive cortical neurons in Dicer mutant animals. Regardless of the mechanism, our results mirror observations in zebrafish in which loss of miRNAs caused abnormal pathfinding of trigeminal sensory neurons (Giraldez et al., 2005). Interestingly, the pathfinding of trigeminal neurons is also controlled by the same semaphorin family in vivo, suggesting a conserved regulatory pathway in zebrafish and mice that is disrupted with Dicer loss (Giger et al., 2000).
In summary, our work demonstrates for the first time the in vivo role of Dicer in the neocortex and hippocampal formation of a mammalian species using a conditional Cre-lox system. This work indicates that miRNAs are important regulators of a wide range of cellular and developmental programs necessary for proper CNS function and provide insight into the potential disease mechanisms that are impacted by Dicer.
Footnotes
This work was supported in part by National Institutes of Health (NIH)–National Eye Institute (NEI) Grant T32EY07120 (T.H.D.); NIH Grant R21-MH083090, the Research to Prevent Blindness Career Development Award, Autism Speaks, the March of Dimes, That Man May See, the Sandler Foundation, and the Alfred P. Sloan Foundation (E.M.U.); NIH Grant RO3-DA022201 (M.T.M.); and NIH–NEI Grant EY002162 Core grant for Vision Research. We thank the members of the McManus and Ullian laboratories for careful review of this manuscript. We also thank the Horton Laboratory for aiding in image acquisition. We thank Derrick Southwell for providing the caspase-3 antibody. We thank Dr. Elizabeth Alcamo and Tony Tran for help with the in situ hybridizations.
References
- Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Bagnard D, Lohrum M, Uziel D, Puschel AW, Bolz J. Semaphorins act as attractive and repulsive guidance signals during the development of cortical projections. Development. 1998;125:5043–5053. doi: 10.1242/dev.125.24.5043. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13:1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- Calabrese JM, Seila AC, Yeo GW, Sharp PA. RNA sequence analysis defines Dicer's role in mouse embryonic stem cells. Proc Natl Acad Sci USA. 2007;104:18097–18102. doi: 10.1073/pnas.0709193104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Yeo G, Muotri AR, Kuwabara T, Gage FH. Noncoding RNAs in the mammalian central nervous system. Annu Rev Neurosci. 2006;29:77–103. doi: 10.1146/annurev.neuro.29.051605.112839. [DOI] [PubMed] [Google Scholar]
- Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- Choi PS, Zakhary L, Choi WY, Caron S, Alvarez-Saavedra E, Miska EA, McManus M, Harfe B, Giraldez AJ, Horvitz RH, Schier AF, Dulac C. Members of the miRNA-200 family regulate olfactory neurogenesis. Neuron. 2008;57:41–55. doi: 10.1016/j.neuron.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuellar T, Nelson P, Loeb G, Ullian E, Harfe B, McManus MT. Dicer loss in striatal neurons produces behavioral and neuroanatomical phenotypes in absence of neurodegeneration. Proc Natl Acad Sci USA. 2008;105:5614–5619. doi: 10.1073/pnas.0801689105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiani D, Alexander JJ, O'Rourke JR, McManus MT, Jadhav A, Cepko CL, Hauswirth WW, Harfe BD, Strettoi E. Dicer inactivation leads to progressive functional and structural degereration of the mouse retina. J Neurosci. 2008 doi: 10.1523/JNEUROSCI.0828-08.2008. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragatsis I, Zeitlin S. CaMKIIα-Cre transgene expression and recombination patterns in the mouse brain. Genesis. 2000;26:133–135. doi: 10.1002/(sici)1526-968x(200002)26:2<133::aid-gene10>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- Dragatsis I, Levine MS, Zeitlin S. Inactivation of Hdh in the brain and testis results in progressive neurodegeneration and sterility in mice. Nat Genet. 2000;26:300–306. doi: 10.1038/81593. [DOI] [PubMed] [Google Scholar]
- Fukagawa T, Nogami M, Yoshikawa M, Ikeno M, Okazaki T, Takami Y, Nakayama T, Oshimura M. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat Cell Biol. 2004;6:784–791. doi: 10.1038/ncb1155. [DOI] [PubMed] [Google Scholar]
- Galvez R, Greenough WT. Sequence of abnormal dendritic spine development in primary somatosensory cortex of a mouse model of the fragile X mental retardation syndrome. Am J Med Genet A. 2005;135:155–160. doi: 10.1002/ajmg.a.30709. [DOI] [PubMed] [Google Scholar]
- Giger RJ, Cloutier JF, Sahay A, Prinjha RK, Levengood DV, Moore SE, Pickering S, Simmons D, Rastan S, Walsh FS, Kolodkin AL, Ginty DD, Geppert M. Neuropilin-2 is required in vivo for selective axon guidance responses to secreted semaphorins. Neuron. 2000;25:29–41. doi: 10.1016/s0896-6273(00)80869-7. [DOI] [PubMed] [Google Scholar]
- Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci USA. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KS, Zhang Z, McManus MT, Harfe BD, Sun X. Dicer function is essential for lung epithelium morphogenesis. Proc Natl Acad Sci USA. 2006;103:2208–2213. doi: 10.1073/pnas.0510839103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert SS, De Strooper B. Molecular biology. miRNAs in neurodegeneration. Science. 2007;317:1179–1180. doi: 10.1126/science.1148530. [DOI] [PubMed] [Google Scholar]
- Hevner RF, Daza RA, Rubenstein JL, Stunnenberg H, Olavarria JF, Englund C. Beyond laminar fate: toward a molecular classification of cortical projection/pyramidal neurons. Dev Neurosci. 2003;25:139–151. doi: 10.1159/000072263. [DOI] [PubMed] [Google Scholar]
- Jiang J, Lee EJ, Gusev Y, Schmittgen TD. Real-time expression profiling of microRNA precursors in human cancer cell lines. Nucleic Acids Res. 2005;33:5394–5403. doi: 10.1093/nar/gki863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin P, Zarnescu DC, Ceman S, Nakamoto M, Mowrey J, Jongens TA, Nelson DL, Moses K, Warren ST. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat Neurosci. 2004;7:113–117. doi: 10.1038/nn1174. [DOI] [PubMed] [Google Scholar]
- Kadotani N, Nakayashiki H, Tosa Y, Mayama S. One of the two Dicer-like proteins in the filamentous fungi Magnaporthe oryzae genome is responsible for hairpin RNA-triggered RNA silencing and related small interfering RNA accumulation. J Biol Chem. 2004;279:44467–44474. doi: 10.1074/jbc.M408259200. [DOI] [PubMed] [Google Scholar]
- Karres JS, Hilgers V, Carrera I, Treisman J, Cohen SM. The conserved microRNA miR-8 tunes atrophin levels to prevent neurodegeneration in Drosophila . Cell. 2007;131:136–145. doi: 10.1016/j.cell.2007.09.020. [DOI] [PubMed] [Google Scholar]
- Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans . Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, Hannon G, Abeliovich A. A microRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Klein ME, Lioy DT, Ma L, Impey S, Mandel G, Goodman RH. Homeostatic regulation of MeCP2 expression by a CREB-induced microRNA. Nat Neurosci. 2007;10:1513–1514. doi: 10.1038/nn2010. [DOI] [PubMed] [Google Scholar]
- Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans . Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Lee EJ, Gusev Y, Jiang J, Nuovo GJ, Lerner MR, Frankel WL, Morgan DL, Postier RG, Brackett DJ, Schmittgen TD. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer. 2007;120:1046–1054. doi: 10.1002/ijc.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004a;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004b;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- Mott JL, Kobayashi S, Bronk SF, Gores GJ. mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene. 2007;26:6133–6140. doi: 10.1038/sj.onc.1210436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto M, Monuki ES, Tang H, Imitola J, Haubst N, Khoury SJ, Cunningham J, Gotz M, Walsh CA. Expression of Cux-1 and Cux-2 in the subventricular zone and upper layers II-IV of the cerebral cortex. J Comp Neurol. 2004;479:168–180. doi: 10.1002/cne.20322. [DOI] [PubMed] [Google Scholar]
- Nimchinsky EA, Sabatini BL, Svoboda K. Structure and function of dendritic spines. Annu Rev Physiol. 2002;64:313–353. doi: 10.1146/annurev.physiol.64.081501.160008. [DOI] [PubMed] [Google Scholar]
- O'Driscoll L. The emerging world of microRNAs. Anticancer Res. 2006;26:4271–4278. [PubMed] [Google Scholar]
- Prunell GF, Troy CM. Balancing neuronal death. J Neurosci Res. 2004;78:1–6. doi: 10.1002/jnr.20252. [DOI] [PubMed] [Google Scholar]
- Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, Bentwich Z, Oren M. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- Schaefer A, O'Carroll D, Tan CL, Hillman D, Sugimori M, Llinas R, Greengard P. Cerebellar neurodegeneration in the absence of microRNAs. J Exp Med. 2007;204:1553–1558. doi: 10.1084/jem.20070823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Troy CM, Rabacchi SA, Friedman WJ, Frappier TF, Brown K, Shelanski ML. Caspase-2 mediates neuronal cell death induced by beta-amyloid. J Neurosci. 2000;20:1386–1392. doi: 10.1523/JNEUROSCI.20-04-01386.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troy CM, Rabacchi SA, Hohl JB, Angelastro JM, Greene LA, Shelanski ML. Death in the balance: alternative participation of the caspase-2 and -9 pathways in neuronal death induced by nerve growth factor deprivation. J Neurosci. 2001;21:5007–5016. doi: 10.1523/JNEUROSCI.21-14-05007.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- Vo N, Klein ME, Varlamova O, Keller DM, Yamamoto T, Goodman RH, Impey S. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc Natl Acad Sci USA. 2005;102:16426–16431. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Lu Y, Pan Z, Chu W, Luo X, Lin H, Xiao J, Shan H, Wang Z, Yang B. The muscle-specific microRNAs miR-1 and miR-133 produce opposing effects on apoptosis by targeting HSP60, HSP70 and caspase-9 in cardiomyocytes. J Cell Sci. 2007;120:3045–3052. doi: 10.1242/jcs.010728. [DOI] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneshima H, Yamasaki S, Voelker CC, Molnar Z, Christophe E, Audinat E, Takemoto M, Nishiwaki M, Tsuji S, Fujita I, Yamamoto N. Er81 is expressed in a subpopulation of layer 5 neurons in rodent and primate neocortices. Neuroscience. 2006;137:401–412. doi: 10.1016/j.neuroscience.2005.08.075. [DOI] [PubMed] [Google Scholar]