Abstract
Functionalized magnetic nanoparticles (mNPs) have shown promise in biosensing and other biomedical applications. Here we use functionalized mNPs to develop a highly sensitive, versatile sensing strategy required in practical biological assays and potentially in vivo analysis. We demonstrate a new sensing scheme based on magnetic spectroscopy of nanoparticle Brownian motion (MSB) to quantitatively detect molecular target. MSB uses the harmonics of oscillating mNPs as a metric for the freedom of rotational motion, thus reflecting the bound state of the mNP. The harmonics can be detected in vivo from nanogram quantities of iron within 5 seconds. Using a streptavidin-biotin binding system, we show that the detection limit of the current MSB technique is lower than 150 pM (0.075 picomole), which is much more sensitive than previously reported techniques based on mNP detection. Using mNPs conjugated with two anti-thrombin DNA aptamers, we show that thrombin can be detected with high sensitivity (4 nM or 2 picomole). A DNA-DNA interaction was also investigated. The results demonstrated that sequence selective DNA detection can be achieved with 100 pM (2 nM) sensitivity. The results of using MSB to sense these interactions, verified that the MSB based sensing technique can achieve rapid measurement (within 10 sec), and is suitable for detecting and quantifying a wide range of biomarkers or analytes. It has the potential to be applied in variety of biomedical applications or diagnostic analyses.
Keywords: magnetic nanoparticles, Brownian motion, relaxation time, biosensing
1. Introduction
One of the major challenges in biosensing, diagnostics and pharmaceutical drug development is the rapid, accurate and sensitive monitoring of the concentration of biomarkers, drugs or pathogens in biological samples. Driven by this challenge, a number of new biosensing platforms have been reported using a variety of detecting techniques. Some of these techniques provide high sensitivity. For example, an optical biosensor based on wavelength shifts using porous silicon, has demonstrated sensitivity for streptavidin and DNA at pico- and femtomolar concentrations. (Lin et al. 1997) An electrochemical biosensor based on silicon nanowire field-effect transistors has achieved sensitivity for streptavidin down to hundreds of femtomolar concentrations.(Duan et al. 2012) An aptamer sensor based on silicon microring resonators showed the detection limit of 1.4 nM for thrombin.(Park et al. 2013) An electrochemical aptamer biosensor using chitosan-Au nanocomposites has demonstrated the capability to detect thrombin with a detection limit of 5.5 fM.(Zhao et al. 2012) However, the disadvantages of these methods limit their biomedical application. For example, the traditional techniques are usually time-consuming, require large sample volumes and multiple washing steps.(Adler et al. 2008; Homola 2003) In addition, optical sensors based on fluorescence detection usually suffer from high background signals.(Owicki 2000)
In comparison, sensing methods based on magnetic nanoparticles (mNPs) have advantages including biocompatibility, environmentally safety, and low cost to synthesize; moreover, mNP-based sensing methods provide less background noise, because there is little or no magnetic signal from biological samples (Haun et al. 2010; Shao et al. 2012). Hence, they have received considerable attention for developing biosensing and diagnostic tools. To date, various mNPs based detection methods have been reported, including AC susceptometry (Park et al. 2011), Hall effect measurements (Mihajlovic et al. 2005), magnetoresistance measurements (Baselt et al. 1998), superconducting quantum interference devices (SQUIDs)(Kotitz et al. 1999), and MRI spin-spin relaxation time (T2) assay(Perez et al. 2002). However, many of these established techniques suffer from extreme conditions requiring low temperatures or long measurement times while admitting low sensitivity and limited types of target molecules. The most important limitation is that all are incapable of in vivo use with high sensitivity. (Haun et al. 2010; Shao et al. 2012)
A relatively new mNP sensing method, magnetic spectroscopy of nanoparticle Brownian motion (MSB), has been shown to be capable of sensing the change in any property influencing the rotational Brownian motion of the mNPs within seconds.(Rauwerdink et al. 2010; Rauwerdink and Weaver 2010b; Weaver and Kuehlert 2012; Weaver et al. 2009) Particularly of interest to biosensing applications, MSB is able to quantitatively measure the bound fraction (Rauwerdink and Weaver 2011) and relaxation times (Weaver and Kuehlert 2012). The measurements rely on the fact that mNPs tend to align with the applied magnetic field, but that tendency is countered by Brownian motion that randomizes the mNPs’ alignment. The extent of disorder caused by Brownian motion is linked directly to environmental conditions including temperature (Weaver et al. 2009), bound state (Rauwerdink and Weaver 2010a, 2011), and viscosity (Rauwerdink and Weaver 2010b). Higher harmonics were used instead of the fundamental frequency to avoid picking up the signal of the applied field. The odd harmonics are of interest in describing the shape of the magnetization and its saturation. The detection using harmonics can be achieved with high sensitivity and it has been shown that in imaging applications (Gleich and Weizenecker 2005), the harmonics can be measured in vivo with ng of NPs (Weizenecker et al. 2007; Weizenecker et al. 2009). The harmonics increase linearly with the number of mNPs so the ratio of the harmonics was used as a concentration-independent metric to characterize the relaxation time. (Weaver and Kuehlert 2012) The ratio of 5th over the 3rd harmonics is an appropriate parameter to reflect the bound state of the mNPs.
In this study, we first verified the feasibility of MSB sensing the mNPs’ restricted rotation secondary to the analyte binding multiple mNPs together. Linking mNPs together produce larger changes in the MSB signal than simply binding the analyte to the mNP. (Rauwerdink and Weaver 2010a) The well characterized biotin and streptavidin system with high affinity was chosen as the first model system demonstrating the validity of the technique and allowing us to explore some of the parameters affecting the measurement. We then tested DNA aptamer- thrombin system, and a DNA-DNA interaction system. The results show that MSB can achieve specific detection with high sensitivity, and capable to detect analyte directly in biological sample. Thus there is potential for MSB biosensing to be utilized for a wide range of biomarker in a variety of biomedical applications or diagnostic analyses, both in vitro and in vivo.
2. Materials and Methods
2.1 Material
Iron oxide nanoparticles functionalized with amine groups or streptavidin were obtained from MicroMod (Micromod Partikeltechnologie GmbH, Germany). EZ-link Sulfo-NHS-LC-Biotin obtained from Thermo. Streptavidin protein and human thrombin protein were purchased from Sigma Aldrich (St. Louis, MO) and Abcam (Cambridge, MA), respectively. All the chemicals used in this research were analytical grade reagents. Previously reported 15-mer (Bock et al. 1992) and 29-mer (Tasset et al. 1997) anti-thrombin aptamer sequences with biotin modification at 5′ were used in the thrombin targeted system. For the DNA-DNA binding system, two 12 base single-strand DNAs (ssDNA), S1 and S2, with biotin modification at 5′ and 3′ respectively were designed. Complementary strands were synthesized so half of each was complementary to S1 and the other half to S2. All the aptamer and DNA sequences in this study were custom synthesized by Integrated DNA technologies, Inc. (Coralville, IA) Spacers of 10 Ts were added between the biotin and the aptamer sequence to enhance the accessibility of the aptamer towards its target protein by reducing steric effects. The DNA sequences are listed in supplementary material.
2.2 Preparation of Functional mNPs
Before conjugation, mNPs were washed using a magnetic separator, and reconstituted in PBS buffer with 0.005% tween 20 (pH 7.4). To prepare biotin modified mNP, EZ-link Sulfo-NHS-LC-Biotin was added to the amine labeled mNP solution and incubated at room temperature for 2 hours. Aptamer and ssDNA modified mNPs were achieved by conjugating the terminal biotin moieties on the aptamer/ssDNA to the streptavidin labeled mNPs. The molar ratio of the DNA aptamer to the streptavidin groups was 50:1. This reaction mixture was incubated at room temperature in PBS at pH 7.4 for 2 hours. After conjugation, the modified mNPs were washed 3 times to remove all the free biotin, aptamers or ssDNA, and resuspended in PBS or Tris-HCl, respectively. The mNPs were stable in solution without precipitation for months.
Each sample contained approximately 150 μg mNP diluted in 500 μl buffer. PBS (pH 7.4) buffer was used for streptavidin detection; buffer containing 50 mM Tris-HCl, 140 mM NaCl, 1 mM MgCl2 (pH 7.4) was used for thrombin detection; 20 mM Tris-HCl was used for DNA detection. All the samples are prepared in triplicates. MSB measurements were taken before and 10 min after the addition of the analyte to allow binding to occur.
2.3 Characterization of amine surface density on mNPs
The number of amine groups on each mNP was evaluated using the biotin quantitation kit purchased from thermo pierce (Rockford, IL). Briefly, the biotinylated NP was added to a mixture of 4′-hydroxyazobenzene-2-carboxylic acid (HABA) and avidin. Because of its higher affinity for avidin, biotin displaces the HABA and results in a decrease of absorbance at 500 nm, which is proportional to the concentration of biotin in the sample.
2.4 Detection of biomolecule interactions via MSB
mNPs suspended in solution have a dynamic response to a magnetic field that depends on the balance between magnetic forces tending to align the mNPs and thermal effects that hinder the alignment of the mNPs. Relaxation times are used to characterize these mechanisms. We employed larger mNPs where Neel relaxation is minimized and Brownian relaxation is dominant (Connolly and St Pierre 2001) to maximize the sensitivity to binding because binding only affects the Brownian relaxation. In this study, mNPs with 50 nm iron oxide composite cores and 113 nm hydrodynamic radii were used. MSB is most sensitive to a given biomarker when the biomarker links multiple mNPs together restricting rotation motion and increasing the relaxation time. When the biomarker has multiple binding sites, ligands from two different mNPs can bind the same biomarker molecule, assembling the mNPs into clusters or aggregates. Such an interaction leads to an increasing effective size of the mNP cluster resulting in increased the Brownian relaxation time, which can be sensed through the MSB measurement. The harmonics were immediately recorded at 290, 510, 737, 1050, 1270, 1740 and 2110 Hz and 10 mT, and data acquired at 1270 Hz was used to analyze the detection sensitivity. The ratio of the 5th over the 3rd harmonics (R53) at each frequency was used as a concentration-independent metric.(Weaver and Kuehlert 2012).
The mth measured harmonic, Hm, can be estimated from the harmonics of the aggregated NPs, HA, and of the free NPs, Hf, assuming that the signal from the two types of free NPs is approximately the same:
| (1) |
where [A] is the concentration of the aggregate, [NPT] is the total concentration of both types of NPs. Equation 1 can be used to make a least squares estimate the concentrations directly if the individual harmonics for both the bound and free NPs are known. More generally, Eq. 1 shows that the harmonics are monotonically related to the concentrations. The ratio is more complicated but it is still monotonic with concentration. For relatively low biomarker concentrations, the biomarker concentration is linearly related to the concentration of aggregates:
| (2) |
where the Kd are the disassociation constants for the reactions binding the NP targeting agents to the biomarker, N1 and N2 are the number of ligands immobilized on the NPs, and [NP1] and [NP2] are the concentrations of the two types of unbound NPs. The salient point is that the biomarker concentration is linear with the measured harmonics. The relationship becomes nonlinear at higher biomarker concentrations when more than one biomarker molecule is bound to the same NP and the number of three and four NP aggregates becomes significant.
3. Results and Discussion
3.1 MSB detection Scheme
Streptavidin and biotin binding pairs were used for a proof of concept of the detection scheme. The streptavidin and biotin interaction is well characterized and has the highest affinity (Kd=10−14 ~ −15M) among all known biological noncovalent binding pairs.(Weber et al. 1989) This system has also been frequently used as a model system for mNP based sensing strategies, which allows us to compare results. Streptavidin is a homotetramer with four biotin binding sites, which can interaction with multiple biotin moieties on different mNPs, and results to the increased mNP cluster size. In order to verify the correlation between the analyte concentration, mNP cluster size and the MSB signal, we first compared the MSB measurement of mNP assembly with size analysis of the hydrodynamic mNP cluster size as shown in Figure 1.
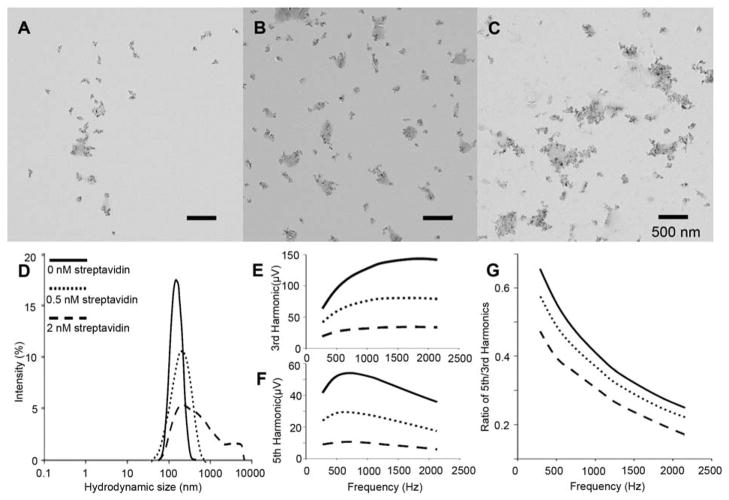
Fig. 1.
Characterization of mNP in three different bound states. A–C, TEM images of mNP with the addition of 0, 0.5 and 2 nM streptavidin, respectively; D, hydrodynamic size analysis of mNP in different bound states; E and F are MSB measurement of 3rd and 5th harmonics of mNP samples of different bound states; and G is the ratio of 5th and 3rd harmonics in F and E.
Increased streptavidin concentration produced larger mNP aggregations as observed on TEM images (Figure 1, A–C) and as determined by hydrodynamic size analysis (Figure 1D). The average hydrodynamic sizes were 142.6, 169.7 and 308.3 nm in the presence of 0, 0.5 and 2 nM streptavidin respectively, and the width of the size distribution also increased with the streptavidin concentration. These size increases correlate well to the streptavidin amount; the linear correlation coefficient is 0.996 with a P-value of 5.7%. MSB measurements were taken of the same samples; the 3rd (Figure 1E), 5th (Figure 1F) harmonics and the ratio of 5th and 3rd harmonics (R53) dropped with increasing streptavidin concentration. The results confirm that the raw MSB signal as well as the concentration independent parameter (R53) accurately reflect the mNP cluster size. Therefore, MSB can be used as a sensing technique capable of detecting the protein via binding between the protein and its targeting reagent immobilized on the mNPs. Moreover, the results confirmed the reliability and convenience of using the R53 to indicate the level of binding and as a parameter for sensing the biomarker concentration.
3.2 Optimization of the sensing scheme
Using the MSB technique to sense biomolecule interactions is based on the quantification of the rotational Brownian motion of the biomolecule conjugated mNP aggregates compared to that of free mNPs. The sensing efficiency of this strategy characterized in Eq. 2, depends on several critical parameters including, N, the number of biomarker targeting agents on the mNP. Nonspecific binding is the other critical parameter.
First of all, nonspecific interaction between other molecules and the targeted mNPs is a critical problem in the design of sensing systems. Several generally applied strategies for eliminating nonspecific interaction were tested, including the addition of tween 20, bovine serum albumin (BSA) and polyethylene glycol (PEG) 2000. The result shows that with the addition of 0.001%~0.005% tween 20, nonspecific interaction was effectively reduced. (Figure S1) The addition of tween 20 produced diluted mNP samples that were stable in solution for more than two weeks, and exhibited the least inhibition on the specific binding between biotin-conjugated mNP and streptavidin. (Figure S2)
We also examined the effect of the surface density of biotin conjugated on the mNP surface on the MSB signal. Interestingly, there is an optimal surface density of about 4600 amine groups (~40%) on each mNP converted to biotin. Less or more conversion resulted in less signal response. (Figure S3) It is important to note that the biotin conversion ratio had unobservable effect on the hydrodynamic size of the mNPs; it was always 135 ± 4 nm. This indicates that the different signal response was not caused by differences in the size distribution. We believe the impact of the surface ligand density on the signal drop is related with whether the binding events happened between biotins on different mNPs or between biotin groups on the same mNP. More specifically, the streptavidin may prefer to bind to multiple biotin groups on the same mNP instead of between different mNPs, when mNP was densely immobilized with biotin, a phenomenon we term “nonaggregate binding”. Nonaggregate binding produces a much smaller signal drop than that observed from assembly of multiple mNPs. This result indicates that the ligand surface density is a crucial parameter for analytes with two or more binding sites for the targeting reagent.
3.3 Sensitivity and Selectivity of the Streptavidin Detection
MSB signal response upon increasing streptavidin concentration, ranging from 50 to 2000 pM (0.025 to 1 picomole) was investigated, including both of the harmonic amplitude (Figure S4) and the R53 (Figure 2). Two sets of the control samples were applied in this experiment to verify the specificity of this detection strategy. 1) Plain mNP has no biotin conjugated with streptavidin added, and 2) biotin conjugated mNP with the addition of BSA. As shown Figure 2, 1.1% and 2.0%, which were caused by nonspecific interaction between mNPs, and between mNPs and protein. At the same concentration of streptavidin added, the specific binding between the biotin mNP and the streptavidin caused 20.6% signal drop.
Fig. 2.
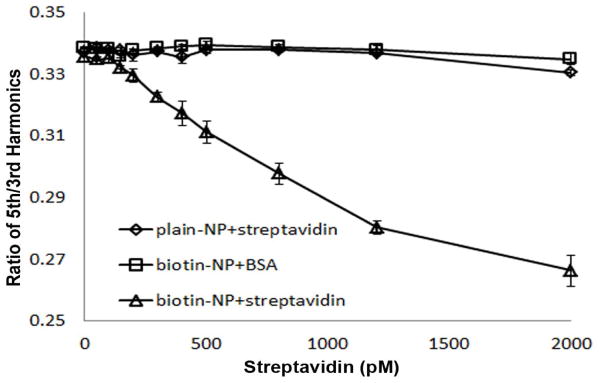
Ratio of 5th and 3rd harmonics of biotin conjugated mNP determined by MSB measurement as a function of streptavidin concentration. Two control samples are applied in this experiment to verify the specificity of the detection: 1) mNP without biotin functionalized in presence of increasing concentration of streptavidin; 2) biotin functionalized mNP sample with the addition of BSA.
The MSB signal change that was statistically significant was estimated using the probability that a given measurement was in the distribution characterized by the mean and standard deviation of the control measurements. In this case, 150 pM is significantly different from the controls with a p-value less than 5%. The MSB signal is linearly proportional to the concentration range from 150 pM to 1200 mM streptavidin with correlation coefficients of 0.994. At higher streptavidin concentration, the relationship becomes nonlinear, because more streptavidin bind to the aggregate NPs instead of free NPs. In the nonlinear range, the signal is monotonic with the concentration, which still can be interpreted, but is less accurate.
The demonstrated sensitivity of 150 pM (0.075 picomole) is much better than previous methods have demonstrated. Previously, Kotitz and colleagues used relaxation measurements and showed that the lowest avidin amount that can be detected is 200 nM. (Kotitz et al. 1999) Yang and colleagues reported a superconducting quantum interference device (SQUID), and demonstrated 1.5 μM or 300 mg avidin as the sensitivity. (Yang et al. 2006) Park and colleagues developed an AC suspetometer to measure complex AC magnetic susceptibility of magnetic mNP, and showed that the detection limit of biotin horseradish peroxidase (HRP) can be as low as 1.0 mg/ml (about 20 μM). (Park et al. 2011) In this study, we show MSB sensing scheme has the detection limit <0.1 picomole streptavidin (6 ng in term of mass). The detecting sensitivity obtained by MSB measurement is about 10~100 times better than the previous reports listed above.
3.4 Sensing DNA Aptamer/Protein Binding
We further explored an aptamer/protein binding system using the MSB detection system. Because of the advantages over antibodies, aptamers emerged as a group of competitive biorecognition reagents and have shown great potential for biomedical sensing in a wide variety of detecting methods.(Nimjee et al. 2005; Zhou et al. 2010) However, the aptamer as targeting reagent has not been applied with a mNP sensing system. In this study, we tested the thrombin/aptamer system to verify the feasible detecting proteins using an aptamer/protein interaction. Two previously reported anti-thrombin aptamers (15 mer and 29 mer) were used.
These two aptamers have distinct tertiary conformations that can selectively bind two distinct specific epitopes (Fibrinogen binding site for 15 mer aptamer and heparin binding domain for 29 mer aptamer).(Bock et al. 1992; Tasset et al. 1997) mNPs conjugated to 15 mer and 29 mer anti-thrombin aptamer (15 aTa and 29 aTa) were prepared separately, so that each mNP was conjugated to only one type of aptamer. The two mNPs were mixed in equal numbers for each sample. The R53 significantly dropped (P<0.001) in the presence of 4 nM thrombin, and was linearly proportional to thrombin concentration in the range of 4–20 nM. (Figure 3)
Fig. 3.
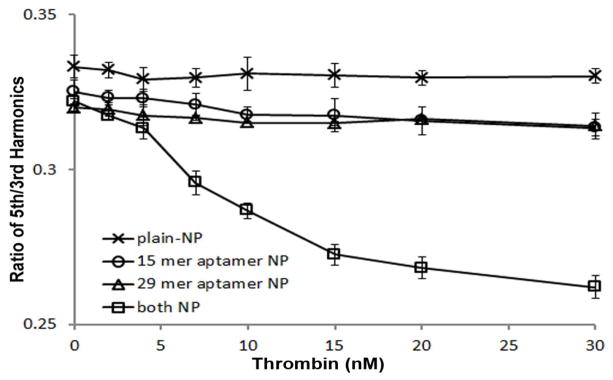
Ratio of 5th and 3rd harmonics of two populations of mNPs, conjugated with 15 mer and 29 mer anti-thrombin aptamer respectively, determined by MSB measurement as a function of thrombin concentration. Three control samples are applied in this experiment to verify the specificity of the detection: mNP without aptamer functionalized, and only one population of mNP functionalized with either 15 mer or 29 mer anti-thrombin aptamer with the increasing concentration of thrombin added.
Thrombin has hetero-binding domain, which requires the binding of thrombin to two mNPs with different aptamers in order to assemble the mNPs into larger clusters. This minimizes nonaggregate binding. Addition of the highest concentration of thrombin to either the 15 mer aptamer mNP or the 29 mer aptamer mNP alone produced a relatively small MSB signal change (3.9% and 3.2%, respective) compared to the change produced when both mNPs were present (26.3%), as shown in Figure 3. This result confirmed that the signal drop was indeed caused by a cooperative effect of two binding events linking two mNPs. The undecorated mNP without any aptamer conjugated produced a significantly smaller effect (0.9%).
3.5 Sensing DNA Hybridization
Besides proteins, nucleic acids are also important biomarkers, including microRNA, siRNA, mRNA circulating DNA and so on.(Schwarzenbach et al. 2011) In order to extend the application of MSB based sensing schemes, an ssDNA sensing target was tested in this study. mNPs were divided into two populations, and each was functionalized with either 12 mer DNA strand 1 (S1) or 12 mer DNA strand 2 (S2). 6 Ts was added between the ssDNA sensing moiety and the biotin to enhance the flexibility and improve the accessibility of the ssDNA. The analyte molecule was a 24 mer DNA with two 12 mer complementary regions S1′ and S2′ for both of the S1 and S2 strands. As shown in Figure 4A, the MSB detection showed a linear range of 200–2000 pM (R=0.995) with a detection limit of 100 pM (0.05 picomole) of analyte DNA (P<0.001), and the linear range is 200–2000 pM. Three control samples were prepared: first, mNPs with no DNA conjugated, showed only 0.5% signal change with the addition of 10 nM complementary DNA; the other two are the samples with either S1-NP or S2-NP alone, which show insignificant MSB signal drop (1.3% and 1.4%) caused by nonaggregate binding, compared with the signal changed (17.3%) caused by the sample with both S1 and S2-NP as shown in Figure 4A.
Fig. 4.
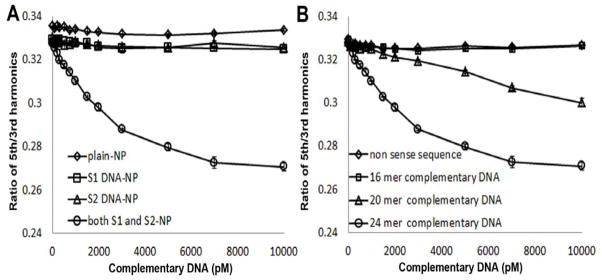
(A) ssDNA detection determined by MSB measurement. Ratio of 5th and 3rd harmonics of two populations of mNPs, conjugated with two distinct DNA sequence respectively, determined by MSB measurement as a function of complementary DNA strand concentration. Three control samples were tested to confirm the specificity of the detection. (B) ssDNA with different length (24 mer, 20 mer and 16 mer) was investigated. A 24 mer ssDNA with non-complementary sequence as a control sample was tested.
A non-complementary, nonsense 24-mer DNA strand, tested with identical experimental conditions, produced no MSB signal change (0.7% signal drop at 10 nM concentration), indicating the sequence selectivity of this assay. (Figure 4) Two shorter complementary strands-16-mer and 20-mer were also investigated to compare with the 24-mer (complete length) strand. The result showed that a significant MSB signal drop was observed in presence of 1 nM (0.5 picomole) of the 20-mer strand. With the addition of 16-mer strand complementary strand, no signal change can be detected at 0–100 nM concentration range at the identical sensing time (10 min).(Figure 4) However, after 2 days, the signal of sample with 100 nM 16-mer complementary ssDNA dropped 3.2% (data not shown). These results indicate that the reduced affinity of the shorter strands decreased the sensitivity of the method, and the low affinity target with 16-mer length can barely be detected for the concentration range employed.
Finally, to verify that MSB can function as a diagnostic tool in the biological environment, we tested the capability of detecting ssDNA directly in serum using MSB.(Figure S5) Although the detection limit we show here is about 400 pM, which is not as sensitive as in PBS (100 pM), it can be concluded that MSB is capable of estimating the ssDNA level directly in serum. This result confirmed that it is feasible to apply MSB in biological samples, and even in in vivo analysis.
4. Conclusion
In this study, we demonstrated the feasibility of using MSB for biosensing applications. The analyte links mNPs together inhibiting their rotational freedom, which reduces their MSB signal. The streptavidin and biotin system was used as a model system. A size analysis of the aggregated mNPs showed that the hydrodynamic size of the mNP is consonant with the MSB measurement. The sensitivity of the streptavidin system was 150 pM or 0.075 picomole streptavidin, which is much lower than previously reported sensing techniques based on mNP detection. We used an aptamer targeting system to detect thrombin, and the sensitivity was about 4 nM. We tested a sequence specific ssDNA sensing scheme, and the results demonstrate the impact of affinity on the sensitivity and the potential specificity of these systems. The sensitivity was 100 pM. As discussed above, several reports about optical sensors and electrochemical sensors have demonstrated better sensitivity than we have demonstrated here. However, MSB is a less cumbersome approach: requiring minimal chemical functionalization/fabrication; measurements are relatively fast, ~10 sec, and can be made longitudinally over time. Further development is focus on improving the sensitivity of magnetization detection, as well as the signal to noise ratio. We do not know the ultimate sensitivity but the demonstrated sensitivity is already in the range previously demonstrated by other mNP based techniques. Most importantly, we have shown that MSB sensing can be achieved in biological sample, and it has the potential for in vivo detection: the harmonics used have been measured from small quantities of mNPs in vivo.(Weizenecker et al. 2009)
In short, the approach we have demonstrated here is a rapid and versatile technique, suitable for wide range of bio-markers. MSB is a promising new biosensing method, and has the potential to be applied in broad array of biomedical applications, including both in vitro and in vivo molecular detection.
Supplementary Material
Scheme 1.
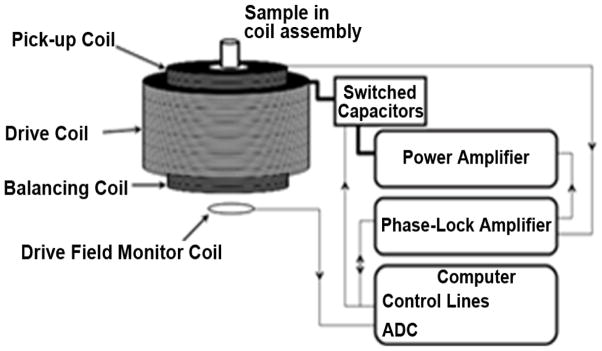
Diagram of the apparatus used to measure the MSB signal from mNP samples. A sinusoidal voltage of a given frequency and amplitude are generated by phase-lock amplifier controlled by computer. This voltage is output to the power amplifier, which drives the current into the resonant drive coil. The computer switches the capacitor to create variant resonant frequency of the drive coil. The output voltage of the phase-lock amplifier is adjusted by monitoring the current generated in a field measurement coil to obtain the correct drive field. The pickup coil and balancing coil were fixed inside the drive field coil in series. The output is measured by the phase-lock amplifier, which output the measured voltage in pickup coil at each harmonic to the computer.
The freedom of rotational motion of mNP can be measured by MSB within seconds.
The MSB signal of mNP decreased with increasing analyte concentration.
We optimized sensing scheme using streptavidin/biotin as model system.
MSB can detect protein and DNA with high specificity and sensitivity.
MSB is a versatile technique, suitable for both in vitro and in vivo detection.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baselt DR, Lee GU, Natesan M, Metzger SW, Sheehan PE, Colton RJ. Biosens Bioelectron. 1998;13:731–739. doi: 10.1016/s0956-5663(98)00037-2. [DOI] [PubMed] [Google Scholar]
- Bock LC, Griffin LC, Latham JA, Vermaas EH, Toole JJ. Nat. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- Connolly J, St Pierre TG. J Magn Magn Mater. 2001;225:156–160. [Google Scholar]
- Duan XX, Li Y, Rajan NK, Routenberg DA, Modis Y, Reed MA. Nat Nanotechnol. 2012;7:401–407. doi: 10.1038/nnano.2012.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleich B, Weizenecker R. Nat. 2005;435:1214–1217. doi: 10.1038/nature03808. [DOI] [PubMed] [Google Scholar]
- Haun JB, Yoon TJ, Lee H, Weissleder R. Wiley Interdiscip Rev-Nanomed Nanobiotechnol. 2010;2:291–304. doi: 10.1002/wnan.84. [DOI] [PubMed] [Google Scholar]
- Kotitz R, Weitschies W, Trahms L, Brewer W, Semmler W. J Magn Magn Mater. 1999;194:62–68. [Google Scholar]
- Lin VSY, Motesharei K, Dancil KPS, Sailor MJ, Ghadiri MR. Sci. 1997;278:840–843. doi: 10.1126/science.278.5339.840. [DOI] [PubMed] [Google Scholar]
- Mihajlovic G, Xiong P, von Molnar S, Ohtani K, Ohno H, Field M, Sullivan GJ. Appl Phys Lett. 2005:87. [Google Scholar]
- Nimjee SM, Rusconi CP, Sullenger BA. Annu Rev Med. 2005;56:555–583. doi: 10.1146/annurev.med.56.062904.144915. [DOI] [PubMed] [Google Scholar]
- Park K, Harrah T, Goldberg EB, Guertin RP, Sonkusale S. Nanotechnol. 2011:22. doi: 10.1088/0957-4484/22/8/085501. [DOI] [PubMed] [Google Scholar]
- Park MK, Kee JS, Quah JY, Netto V, Song J, Fang Q, La Fosse EM, Lo GQ. Sens Actuators, B. 2013;176:552–559. [Google Scholar]
- Perez JM, Josephson L, O’Loughlin T, Hogemann D, Weissleder R. Nat Biotechnol. 2002;20:816–820. doi: 10.1038/nbt720. [DOI] [PubMed] [Google Scholar]
- Rauwerdink AM, Giustini AJ, Weaver JB. Nanotechnol. 2010;21:455101. doi: 10.1088/0957-4484/21/45/455101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauwerdink AM, Weaver JB. Appl Phys Lett. 2010a;96:033702. [Google Scholar]
- Rauwerdink AM, Weaver JB. J Magn Magn Mater. 2010b;322:609–613. [Google Scholar]
- Rauwerdink AM, Weaver JB. Med Phys. 2011;38:1136–1140. doi: 10.1118/1.3549762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzenbach H, Hoon DSB, Pantel K. Nat Rev Cancer. 2011;11:426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- Shao HL, Min C, Issadore D, Liong M, Yoon TJ, Weissleder R, Lee H. Theranostics. 2012;2:55–65. doi: 10.7150/thno.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasset DM, Kubik MF, Steiner W. J Mol Biol. 1997;272:688–698. doi: 10.1006/jmbi.1997.1275. [DOI] [PubMed] [Google Scholar]
- Weaver JB, Kuehlert E. Med Phys. 2012;39:2765–2770. doi: 10.1118/1.3701775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver JB, Rauwerdink AM, Hansen EW. Med Phys. 2009;36:1822–1829. doi: 10.1118/1.3106342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber PC, Ohlendorf DH, Wendoloski JJ, Salemme FR. Sci. 1989;243:85–88. doi: 10.1126/science.2911722. [DOI] [PubMed] [Google Scholar]
- Weizenecker J, Borgert J, Gleich B. Phys Med Biol. 2007;52:6363–6374. doi: 10.1088/0031-9155/52/21/001. [DOI] [PubMed] [Google Scholar]
- Weizenecker J, Gleich B, Rahmer J, Dahnke H, Borgert J. Phys Med Biol. 2009;54:L1–L10. doi: 10.1088/0031-9155/54/5/L01. [DOI] [PubMed] [Google Scholar]
- Yang HC, Yang SY, Fang GL, Huang WH, Liu CH, Liao SH, Horng HE, Hong CY. J Appl Phys. 2006;99:124701. [Google Scholar]
- Zhao J, Lin F, Yi Y, Huang Y, Li H, Zhang Y, Yao S. Analyst. 2012;137:3488–3495. doi: 10.1039/c2an35340g. [DOI] [PubMed] [Google Scholar]
- Zhou J, Battig MR, Wang Y. Anal Bioanal Chem. 2010;398:2471–2480. doi: 10.1007/s00216-010-3987-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.