Abstract
Dyskeratosis congenita (DC) is a telomere-biology disorder characterized by a mucocutaneous triad, aplastic anemia, and predisposition to cancer. Mutations in a narrow segment of TINF2 exon 6 have been recognized to cause often-severe DC that is either sporadic or autosomal dominant. We describe three children with very early presentations of DC, including one with the severe variant known as Revesz syndrome. Whereas most TINF2 mutations reported to date are missense changes, each of our patients carried a novel heterozygous nonsense or frameshift mutation, revealing a new 5’ boundary to the affected gene segment in patients with DC. Examination of patient-derived lymphoblastoid cell lines revealed stable expression of the predicted truncated TIN2 proteins. In co-immunoprecipitation assays, the ability of a truncation mutant to interact with TRF1 was severely impaired, whereas the ability of the most common DC-associated mutant was much less affected. This suggests that disruption of TIN2-TRF1 interaction may contribute to the severe clinical phenotype observed in the context of the TIN2 truncation mutation but is unlikely to be the primary cause of telomere shortening associated with the more prevalent TIN2 missense mutations. Telomere flow-FISH analysis of one pedigree demonstrated the dramatic effect a de novo nonsense TINF2 mutation had on telomere length in early development. These cases underscore the severe manifestations of truncating TINF2 mutations.
Keywords: Aplastic anemia, bone marrow failure, dyskeratosis congenita, Revesz syndrome, TIN2, TINF2, telomere
INTRODUCTION
Dyskeratosis congenita (DC; MIM #s 127550, 305000, 224230) is an inherited bone marrow failure and cancer predisposition syndrome that classically is defined by a triad of oral leukoplakia, nail dystrophy, and abnormal skin pigmentation (1, 2). DC is heterogeneous in presentation and can have variable additional manifestations, such as pulmonary fibrosis, liver disease, enteropathy, and urogenital abnormalities (3). Two rare, severe variants of DC have been described, Revesz syndrome (RS; MIM #268130) and Hoyeraal-Hreidarsson syndrome (HHS; MIM #300240) (1, 2). Individuals with RS manifest the additional defining feature of bilateral exudative retinopathy. Other features of RS include intrauterine growth retardation, intracranial calcification and cerebellar hypoplasia (4). HHS is characterized by cerebellar hypoplasia along with microcephaly, developmental delay, immunodeficiency, intrauterine growth retardation, and bone marrow failure (5, 6).
At the molecular level, most patients with DC have very short telomeres, best characterized in leukocytes, and defined as a length less than the first percentile for age (7-9). To date, approximately 50% of affected individuals have genetic alterations in one of six genes affecting telomere biology (1). Five of those genes affect components of telomerase, the enzyme responsible for the maintenance of telomere length. The sixth gene implicated in DC, TINF2, encodes TIN2, a member of the telomere-associated shelterin complex, which plays integral roles in the structure and function of telomeres (10). Interestingly, patients harboring TINF2 mutations are reported to have extremely short telomeres, correlating clinically with the frequently early age of presentation (less than 10 years) and severe manifestations of the disease (11, 12). The described mutations are all heterozygous, and map to a short, highly conserved segment of exon 6 of unknown function (Fig. 1A). Of the 40 reported probands with TINF2-associated DC or one of its variants, 35 harbored missense mutations, whereas only five probands had a truncating mutation (Fig. 1A and Table 1) (10-13). Two different TINF2 mutations have been reported in five individuals with RS (11, 12). Here, we describe three children with novel nonsense/frameshift TINF2 mutations who developed severe aplastic anemia (SAA) and other manifestations of DC at an early age. We demonstrate expression of the expected truncated TIN2 proteins, which, with the severe telomere shortening observed in the patients, indicates that loss of the TIN2 C-terminus results in significant deleterious effects. We further show that one of the truncation mutants has markedly reduced interaction with the shelterin complex member TRF1, whereas the most commonly found missense mutation does not, revealing a specific loss of function of the TIN2 truncation.
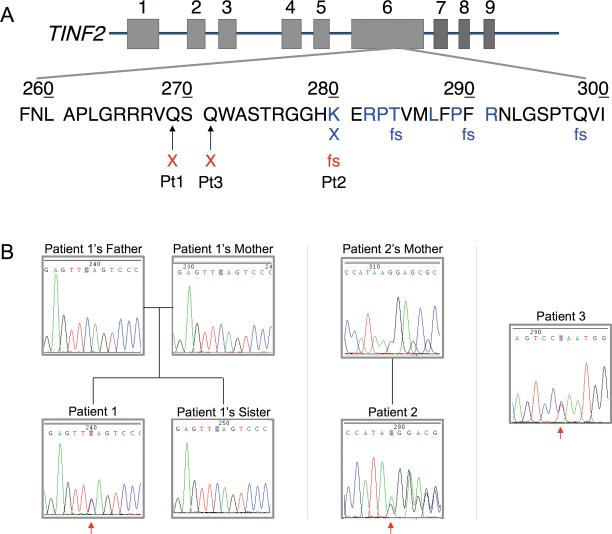
Figure 1.
A) Localization of TINF2 mutations. The genomic TINF2 structure is depicted with exons as rectangles. The TIN2 short isoform contains exons 1-6, whereas the long isoform includes exons 1-9 (29). Residues previously reported to be affected by missense mutations are in blue (10, 11, 13, 30). X and fs are placed below sites of known nonsense and frameshift mutations, respectively. Previously reported nonsense (X) and frameshift (fs) mutations are in blue, and new mutations in red. B) TINF2 sequence chromatograms. Chromatograms are of the described patients and some family members, as labeled. An arrow points to the reported heterozygous change in each patient. (Chromatograms of Patient 2's mother and Patient 3 are courtesy of Ambry Genetics, Aliso Viejo, CA)
Table 1.
DC-associated TINF2 mutations
| TINF2 base change | TIN2 amino acid change | Mutation type | Associated phenotype(s) | Number of reported probands (ref) |
|---|---|---|---|---|
| Previously reported mutations | ||||
| c.845G>A | R282H | Missense | DC DC/RS DC/HH/RS HH Ataxia-pancytopenia |
12 (10, 11) 2 (10, 11) 1 (11) 2 (11) 1 (13) |
| c.844C>T | R282C | Missense | DC | 7 (11) |
| c.838A>G* | K280E | Missense | DC | 1 (10) |
| c.844C>A† | R282S | Missense | DC | 1 (10) |
| c.847C>T | P283S | Missense | DC/HH | 1 (11) |
| c.847C>G | P283A | Missense | DC | 1 (11) |
| c.848C>A | P283H | Missense | DC | 1 (11) |
| c.850A>G | T284A | Missense | DC | 1 (11) |
| c.860T>C | L287P | Missense | DC | 1 (11) |
| c.865_866delinsAG | P289S | Missense | DC | 1 (11) |
| c.871A>G | R291G | Missense | DC | 1 (11) |
| c.838A>T | K280X | Nonsense | DC/HH/RS HH/RS |
1 1 (11) |
| c.867_868insC | F290LfsX2 | Frameshift | DC | 1 (11) |
| c.892delC | Q298RfsX19 | Frameshift | DC | 1 (11) |
| c.849_850insC | T284HfsX8 | Frameshift | DC | 1 (11) |
| Mutations reported in this series | ||||
| c.805C>T | Q269X | Nonsense | DC | 1 |
| c.811C>T | Q271X | Nonsense | DC | 1 |
| c.839delA | K280RfsX36 | Frameshift | DC/RS | 1 |
METHODS
Ethics and patient ascertainment
All described probands and family members were enrolled on a research protocol that was approved by the Baylor College of Medicine Institutional Review Board. The subjects were ascertained through clinical encounters over a two year period at Texas Children's Cancer Center and Hematology Service, a tertiary referral center. The probands represent three of the four patients found to have TINF2 mutations, the fourth patient having the most common mutation, TIN2p.R282H.
Molecular genetic studies
Peripheral blood was obtained from Patients 1 and 2 and the sister of Patient 1, and DNA extracted using Puregene Blood Core Kit B (Qiagen). Saliva was collected from the parents of Patient 1 and DNA extracted using the Oragene-DNA sample collection kit (DNA Genotek). We performed polymerase chain reaction amplification of TINF2 exon 6 using published primers (11), followed by bidirectional sequencing and comparison to the coding sequence of TINF2 isoform 1 (NM_001099274.1) (14). The patients’ mutations were independently confirmed through clinically certified labs (GeneDx, Gaithersburg, MD, or Ambry Genetics, Aliso Viejo, CA). Biologic samples of Patient 3 (skin fibroblast culture) and Patient 2's mother (peripheral blood) were obtained and subjected to TINF2 exon 6 sequencing solely by Ambry Genetics.
Telomere length analysis
Telomere flow-FISH (15) analyses of Patient 1 and his family and Patient 2 were performed by Repeat Diagnostics (Vancouver, BC).
Western blotting for TIN2
EBV-transformed lymphoblastoid cell lines (LCLs) were generated from Patient 1, Patient 1's sister, and Patient 2 by the tissue culture core laboratory within the Department of Molecular and Human Genetics, Baylor College of Medicine. We prepared small scale whole cell lysates by combining cytoplasmic and nuclear fractions as previously described (16). Protein was measured using Pierce BCA protein assay kit (Thermo Scientific), and specific protein expression analyzed by western blotting under the following conditions: 50 μg of total protein was loaded onto NuPage 4-12% bis-tris gel (Invitrogen), and TIN2 probed using a 1:2000 dilution of polyclonal rabbit anti-TIN2 antibody #865, which targets full-length TIN2 (kindly provided by Dr. Titia De Lange, Rockefeller University) (17). Beta actin served as the loading control, and was assayed using monoclonal antibody AC-15 (Sigma-Aldrich) at 1:5000 dilution. Analysis of TIN2 protein in Patient 3's skin fibroblasts was not possible because of poor growth of the culture
Plasmids
TINF2 short isoform cDNA sequence derived from pLPC-N-FH2-TIN2 plasmid (17) (kindly provided by Titia De Lange, Rockefeller University) was amplified and cloned into pcDNA3.1, creating pAB625, which was subsequently used for mutagenesis and co-immunoprecipitation experiments. Myc-TRF1 plasmid was generously provided by Ming Lei (University of Michigan) (18).
Mutagenesis
TINF2 mutations, c.805C>T/p.Q269X and c.845G>A /p.R282H, were introduced into pAB625 using oligonucleotide single-stranded mutagenesis (19) to give rise to pAB631 and pAB629, respectively. Mutations were confirmed by sequencing.
Co-immunoprecipitation assays
HEK293T cells (2 × 106) were transfected with 4 μg of either Myc-TRF1 plasmid, pAB625 (FLAG-TINF2), pAB629 (FLAG-TINF2-R282H) or pAB631(FLAG-TINF2-Q269X) using Lipofectamine LTX with PLUS Reagent (Invitrogen) according to the manufacturer's protocols. Extracts were made by adding 500 μl RIPA buffer (50 mM Tris-HCL pH 8.0, 150 mM NaCl, 1% Triton X-100, 0.5% Na deoxycholate, 0.1% SDS) and 100 μl of Protease Inhibitor cocktail III (Calbiochem) to a frozen pellet for 30 min on ice. Immunoprecipitations were performed by mixing 750 μg of Myc-TRF1 plasmid transfected HEK293T cell extracts with 750 μg of either pAB625, pAB629 or pAB631 transfected cell extracts. Unmixed controls of 750 μg of Myc-TRF1 plasmid or pAB625 transfected cell extracts were also prepared. Mixtures were rotated 2 hrs at 4°C and subsequently 10 μg of α-myc antibody 9E-10 (Sigma-Aldrich) was added to each reaction for 1 hr. One hundred fifty microliters of RIPA washed Protein G Plus-Agarose beads were added to each reaction and were rotated overnight at 4°C. Beads were collected by mild centrifugation, washed 4 times with 500 μl of RIPA buffer and finally resuspended in 40 μl of RIPA buffer and 20 μl of SDS-PAGE buffer. Forty microliters of each reaction were separated on 10% SDS-polyacrylamide gels and proteins analyzed by sequential western blotting using α-FLAG M2 monoclonal antibody (Sigma-Aldrich; 1:1000) and α-Myc antibody 9E-10 (Sigma-Aldrich; 1:5000). Input extract western blots were performed by loading 50 μg of each reaction collected after the 2 hr mixing period. In addition to α-FLAG and α-Myc antibodies, the membrane was probed with β-actin AC-15 antibody (Sigma-Aldrich; 1:5000).
PATIENTS AND RESULTS
Patient 1
A 4-year-old Hispanic male was referred to us for evaluation of pancytopenia with macrocytic anemia [white blood count (WBC) 2,970/μl; absolute neutrophil count (ANC) 1,160/μl; hemoglobin 8.3 gm/dl; platelet count 36,000/μl; mean corpuscular volume (MCV) 96.8 fl]. Bone marrow aspiration and biopsy revealed 15 to 30% cellularity, with maturing trilineage hematopoiesis and normal cytogenetics. Past medical history was significant for an undescended testis and phimosis. His family history was noncontributory. Physical findings of dystrophic nails, oral leukoplakia, and subtle lacy hyperpigmentation involving the neck, inguinal, and scrotal areas established the diagnosis of classical DC. Within weeks of presentation, the patient became dependent on platelet transfusions, and his ANC decreased to 240/μl, at which point he received matched related donor hematopoietic stem cell transplantation (HSCT).
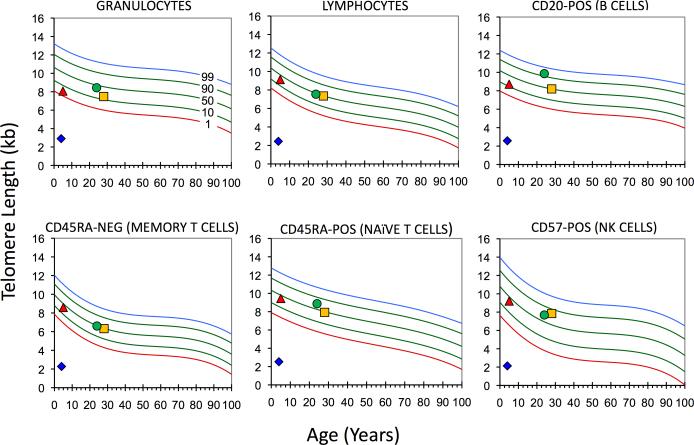
Consistent with the clinical diagnosis of DC, telomere flow-FISH analysis demonstrated telomere lengths well below the first percentile for all WBC subsets tested (Fig. 2) (7). TINF2 exon 6 sequence analysis revealed a novel heterozygous mutation, TINF2c.805C>T, TIN2 p.Q269X (Fig. 1). TINF2 sequences and telomere lengths of the parents and sibling were normal (Figs. 1B and 2), indicating that the patient carried a de novo TINF2 mutation.
Figure 2.
Dramatic telomere shortening in one generation in the presence of a TINF2c.805C>T: TIN2p.Q269X heterozygous mutation. WBC subset telomere flow-FISH analysis (15) performed on Patient 1, his parents and sibling, as indicated. Proband, diamond; Sister, triangle; Mother, circle; and Father, square. (Courtesy of Repeat Diagnostics, Vancouver, BC)
Patient 2
A 21-month-old Hispanic male, with an unremarkable family history, was referred for further evaluation of SAA. He was a product of a normal pregnancy but at age 9 months was diagnosed with bilateral exudative retinopathy requiring multiple laser surgeries and resulting in loss of vision in one eye. He was found to be thrombocytopenic (platelets of 41,000 /μl) at 13 months of age and, during the following five months, developed macrocytic anemia (hemoglobin nadir 5.8 gm/dl, MCV 102 fl) and neutropenia (ANC 390/μl). His platelets concurrently decreased to 12,000/μl. His bone marrow showed 20 to 40% cellularity with marked myeloid and megakaryocytic hypoplasia, and normal cytogenetics. At that time, he was noted to have nail dystrophy and oral leukoplakia. On presentation to our clinic at age 21 months, his weight and head circumference were below the third percentile. In addition, he was delayed in gross motor and language areas, and he had a wide-based gait with poor coordination suggestive of cerebellar dysfunction. Brain magnetic resonance imaging confirmed cerebellar hypoplasia. Given the above findings, he was diagnosed with RS, a severe variant of DC. Due to the severity of his bone marrow failure, the patient underwent matched umbilical cord blood transplantation following reduced intensity conditioning (RIC). He experienced multiple complications resulting in his death on day +92 after transplant.
Telomere length analysis prior to umbilical cord transplant demonstrated very short telomeres in all WBC subsets, with median lengths of 2.3 to 2.8 kilobases. TINF2 exon 6 sequencing revealed a novel heterozygous deletion, TINF2c.839delA (Fig. 1B), resulting in a frameshift and a premature stop codon, which could result in the expression of a truncated variant of TIN2, p.K280RfsX36 (Fig. 1A). His mother's TINF2 sequence analysis was normal (Fig. 1B); his father was unavailable for testing but reportedly had no medical problems.
Patient 3
A Caucasian female, with an unremarkable family history, was first noted to have leukopenia, macrocytic anemia, and thrombocytopenia at 21 months of age (WBC 3,600/μl, ANC 1,800/μl, hemoglobin 10.1 gm/dl, MCV 100 fl, and platelets 31,000/μl). Over the next two years, her anemia and thrombocytopenia progressed (hemoglobin 4.1 gm/dl and platelets 6,000/μl) and she developed neutropenia (ANC 400/μl). She was referred to our clinic at age 3.5 years for further management. Her examination was normal with no mucocutaneous findings. Bone marrow evaluation revealed 5% cellularity with no dysplastic or cytogenetic abnormalities. Testing for Fanconi anemia and paroxysmal nocturnal hemoglobinuria were negative. Telomere length testing was not clinically available at that time. She was diagnosed with SAA and underwent a 6/6 HLA-matched sibling donor HSCT using RIC. Shortly after undergoing HSCT, she developed skin hyperpigmentation, nail dystrophy progressing to vanishing nails, and oral leukoplakia. These changes initially were thought to be graft-versus-host disease, but they did not improve with multiple immunosuppressive therapies. A diagnosis of DC was entertained. DKC1 and TERC sequencing of a buccal sample were normal. Additional features of DC, such as epiphora, esophageal stricture, and osteopenia-related fractures, developed. Other complications of DC became apparent when the patient turned 10 years old (seven years post HSCT). She had progressive interstitial lung disease with fibrosis, as well as gastrointestinal bleeding secondary to enteropathy and non-cirrhotic portal hypertension. She eventually developed hepatopulmonary syndrome and died at the age of 12 years from multi-organ failure.
After identification of TINF2's involvement in DC, a skin biopsy was obtained from the patient and TINF2 exon 6 sequencing from a skin fibroblast culture was performed, demonstrating a novel heterozygous mutation, TINF2c.811C>T (Fig. 1B), which would potentially lead to expression of a truncated protein, p.Q271X (Fig. 1A). Parental testing was not performed.
Mutant TIN2 proteins are expressed in cells
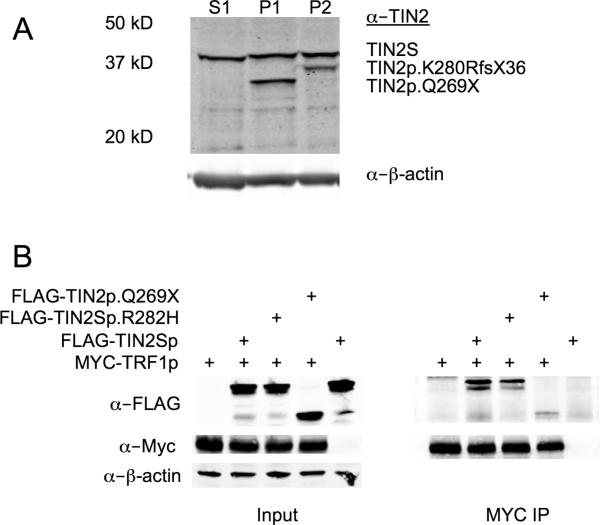
To address the question of whether the expected truncated TIN2 proteins were expressed, western blotting of LCL whole cell lysates from Patients 1 and 2 was performed. This revealed stable expression of truncated TIN2 proteins of the predicted sizes of approximately 30 kD and 35 kD, respectively, in addition to expression of full length TIN2 (~39 kD) (Fig. 3A). It was consistently noted that the amount of the mutant protein was less than the full length TIN2 (see also Supplemental Figure 1), possibly indicating that the truncated proteins were less stable. Alternatively, this could be due to the lack of TIN2's normal C terminus, reducing polyclonal antibody recognition of the truncated protein.
Figure 3.
A) Stable expression of truncated TIN2 proteins. Western blotting of whole cell extracts from LCLs of Patient 1's sister, Patient 1, and Patient 2 (Lanes S1, P1, and P2, respectively). The positions of the full length short isoform of TIN2 in all, and the truncated TIN2 in Patients 1 and 2 are denoted by arrows. Beta actin served as the loading control. (n.b., The slight difference in migration in S1 as compared to P1 and P2 was not consistently seen; e.g., see Supplemental Figure 1) B) TIN2p.Q269X has markedly reduced binding to TRF1. Co-immunoprecipitations of mixtures of extracts from transfected HEK293T cells analyzed by western blotting (MYC IP). Mixtures prior to immunoprecipitation are shown (Input).
TIN2p.Q269X has much reduced binding to TRF1
TIN2 has four known binding partners, three members of the shelterin complex, TRF1, TRF2, and TPP1, and a component of cohesin, SA1. TIN2 interacts with TRF2, TTP1 and SA1 via its N terminal domain, whereas the region that interacts with TRF1 is more central (20-23). Crystal structural analysis of an internal TIN2 peptide (256-276) bound to TRF1 demonstrated that the C-terminal boundary of interaction on TIN2 is aa 267 (18), the penultimate residue in TIN2p.Q269X. Therefore, we reasoned that the truncation mutants, and possibly previously described TIN2 missense mutants, might have allosteric effects on TRF1 binding. To address this question, we incubated extracts prepared from HEK293T cells transfected with Myc-TRF1 with extracts from cells transfected with FLAG-TIN2S, FLAG-TIN2p.Q269X or FLAG-TIN2p.R282H (the most common missense mutant found in patients) and then immunoprecipitated TRF1 with α-Myc antibody. We found TIN2p.Q269X was markedly impaired for its ability to interact with TRF1, whereas TIN2p.R282H retained substantial binding activity (Fig. 3B). Similar results were obtained with extracts from co-transfected cells and when TIN2 was immunoprecipitated (data not shown).
DISCUSSION
TINF2 mutations have been found to account for 11 to 24% of cases with DC (24). Missense mutations are most common, both in terms of distinct mutations and number of affected probands, whereas only one nonsense mutation and three frameshift mutations have previously been identified (Table 1) (10-13). Our patients carried novel nonsense and frameshift mutations, thereby expanding the number of distinct DC-associated TINF2 mutations to a total of 18. RS is a severe and rare variant of DC associated with the additional morbidities of retinopathy and central nervous system involvement (1, 2, 4). TINF2 is the only gene discovered to be implicated in this subset of patients with DC, and to date, two mutations (namely, TINF2c.838A>T; TIN2p.K280X and TINF2c.845G>A; TIN2p.R282H) have been reported (Table 1). Patient 2's mutation (TINF2c.839delA; TIN2p.K280RfsX36) constitutes the third mutation causing RS. Therefore, we propose that TINF2 should be the first DC-related gene sequenced in such patients.
The severity of DC has not been specifically graded, but has been generally linked to the age of onset of aplastic anemia and the number of associated somatic abnormalities. Patients manifesting prior to 10 years of age are considered to have the severest form (25). Alternatively, patients with the two variants of DC, RS and HHS, are referred to as having severe disease (1, 2). DC has been known to show phenotypic variability even within groups carrying the same genetic mutation (26), possibly reflecting added genetic and/or environmental modifiers on the dysfunctional telomere maintenance. Hence, establishing genotype-phenotype correlations has been difficult and complex. However, Walne, et al., observed that most individuals with TINF2 mutations tended to develop severe DC, with approximately two-thirds developing SAA in the first decade of life (11). Focusing on the small subgroup of probands with nonsense or frameshift TINF2 mutations (including those in our series), we noted that all patients uniformly presented with very early DC, as early as 1 year of age. In addition to Patient 2 presenting with RS, Patient 1 displayed evidence of organ involvement at birth (undescended testis, phimosis), suggesting a pivotal role of TINF2 in early development.
Heretofore, all DC-associated TINF2 mutations map to a short segment of exon 6 that would consequently affect amino acids (aa) 280-298 of the transcribed protein, TIN2 (Fig. 1A) (10, 11). Two of our described mutations (TINF2c.805C>T; TIN2p.Q269X and TINF2c.811C>T; TIN2p.Q271X) lie closer to the N-terminus and would, therefore, extend the affected segment to aa 269. The function of this segment remains unidentified. Notably, however, the TIN2p.Q269X truncation immediately abuts the residues known to mediate an interaction between TIN2 and TRF1 (18). We found the TIN2p.Q269X mutant is unable to bind TRF1 efficiently, whereas the TIN2p.R282H mutant retains substantial binding. Because patients harboring a TIN2p.R282H mutation have markedly short telomeres and manifest all of the features of DC, it is unlikely that disrupted TRF1 binding is the main factor driving disease pathogenesis. Supporting this notion is the observation that missense mutation of residues known to directly interact with TRF1 has not been reported. Reduction in TIN2-TRF1 binding may, however, contribute to the severe phenotype observed in some patients such as our patient bearing the TIN2p.Q269X truncation mutation.
It is notable that N-terminal nonsense or frameshift mutations have not been reported in patients, suggesting that haploinsufficiency of an N terminal function (e.g., TRF2 binding) does not underlie the telomere dysfunction. Moreover, in our patient LCLs bearing more centrally located nonsense or frameshift mutations, truncated proteins were readily observed (Fig. 3A). Whether the result in solely haploinsufficiency of C terminal functions or dominant negative effects remains to be determined.
Finally, telomere length is inherited as a quantitative trait and correlates significantly with paternal, but not maternal, telomere length (27). Patient 1 had a documented de novo mutation (Fig. 1B), and his extremely short telomeres (Fig. 2), in contrast to his father's, demonstrate the drastic negative impact that a heterozygous nonsense TINF2 mutation can have on telomere length in a single generation, similar to what has been reported with missense TINF2 mutations (10, 11). This raises the possibility that TINF2 mutations result in telomere shortening through increased telomere attrition rather than or in addition to altered lengthening mechanisms. Further research is needed to elucidate the exact pathophysiology underlying TINF2's deleterious effects.
Supplementary Material
Acknowledgements
We are grateful to our patients and their families for participating in our research study, and being gracious with their time and effort. We would like to acknowledge Drs. Robert Krance, Juan Carlos Bernini and Alana Kennedy-Nasser for their referral of patients with DC to our care and study, as well as for their dedicated clinical effort with each reported patient. We would also like to thank the personnel at the Baylor College of Medicine tissue culture core lab for generation of the LCLs; Dr. Titia de Lange (Rockefeller University) for the anti-TIN2 antibody and the TIN2 plasmid; Dr. Ming Lei (University of Michigan) for the TRF1 plasmid; and our Baylor College of Medicine colleagues Dr. Catherine Bollard for technical advice, Christopher Williams for technical assistance, and Drs. Monica Gramatges and Lee Ligon-Borden for critical review of the manuscript.
Footnotes
Conflict of interest statements
Ghadir S. Sasa, M.D. denies presence of any potential conflict.
Albert Ribes-Zamora denies presence of any potential conflict.
Nya D. Nelson denies presence of any potential conflict.
Alison A. Bertuch. M.D., Ph.D. denies presence of any potential conflict.
During the submission of our manuscript, the mutation harbored by Patient 1 was reported by Vulliamy, et al. (28), who received our patient's sample for mutation testing while we were independently sequencing TINF2.
REFERENCES
- 1.Walne AJ, Dokal I. Advances in the understanding of dyskeratosis congenita. Br J Haematol. 2009 doi: 10.1111/j.1365-2141.2009.07598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Savage SA, Alter BP. Dyskeratosis congenita. Hematol Oncol Clin North Am. 2009;23(2):215–231. doi: 10.1016/j.hoc.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kirwan M, Dokal I. Dyskeratosis congenita: a genetic disorder of many faces. Clin Genet. 2008;73(2):103–112. doi: 10.1111/j.1399-0004.2007.00923.x. [DOI] [PubMed] [Google Scholar]
- 4.Revesz T, Fletcher S, al-Gazali LI, et al. Bilateral retinopathy, aplastic anaemia, and central nervous system abnormalities: a new syndrome? J Med Genet. 1992;29(9):673–675. doi: 10.1136/jmg.29.9.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hreidarsson S, Kristjansson K, Johannesson G, et al. A syndrome of progressive pancytopenia with microcephaly, cerebellar hypoplasia and growth failure. Acta Paediatr Scand. 1988;77(5):773–775. doi: 10.1111/j.1651-2227.1988.tb10751.x. [DOI] [PubMed] [Google Scholar]
- 6.Berthet F, Tuchschmid P, Boltshauser E, et al. The Hoyeraal-Hreidarsson syndrome: don't forget the associated immunodeficiency. Eur J Pediatr. 1995;154(12):998. doi: 10.1007/BF01958649. [DOI] [PubMed] [Google Scholar]
- 7.Alter BP, Baerlocher GM, Savage SA, et al. Very short telomere length by flow fluorescence in situ hybridization identifies patients with dyskeratosis congenita. Blood. 2007;110(5):1439–1447. doi: 10.1182/blood-2007-02-075598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Westin ER, Chavez E, Lee KM, et al. Telomere restoration and extension of proliferative lifespan in dyskeratosis congenita fibroblasts. Aging Cell. 2007;6(3):383–394. doi: 10.1111/j.1474-9726.2007.00288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gourronc FA, Robertson M, Herrig AK, et al. Proliferative defects in dyskeratosis congenita skin keratinocytes are corrected by expression of the telomerase reverse transcriptase, TERT, or by activation of endogenous telomerase through expression of papillomavirus E6/E7 or the telomerase RNA component, TERC. Exp Dermatol. 19(3):279–288. doi: 10.1111/j.1600-0625.2009.00916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Savage SA, Giri N, Baerlocher GM, et al. TINF2, a component of the shelterin telomere protection complex, is mutated in dyskeratosis congenita. Am J Hum Genet. 2008;82(2):501–509. doi: 10.1016/j.ajhg.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walne AJ, Vulliamy T, Beswick R, et al. TINF2 mutations result in very short telomeres: analysis of a large cohort of patients with dyskeratosis congenita and related bone marrow failure syndromes. Blood. 2008;112(9):3594–3600. doi: 10.1182/blood-2008-05-153445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sarper N, Zengin E, Kilic SC. A child with severe form of dyskeratosis congenita and TINF2 mutation of shelterin complex. Pediatr Blood Cancer. 2010;55(6):1185–1186. doi: 10.1002/pbc.22624. [DOI] [PubMed] [Google Scholar]
- 13.Tsangaris E, Adams SL, Yoon G, et al. Ataxia and pancytopenia caused by a mutation in TINF2. Hum Genet. 2008;124(5):507–513. doi: 10.1007/s00439-008-0576-7. [DOI] [PubMed] [Google Scholar]
- 14.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35(Database issue):D61–65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baerlocher GM, Vulto I, de Jong G, et al. Flow cytometry and FISH to measure the average length of telomeres (flow FISH). Nat Protoc. 2006;1(5):2365–2376. doi: 10.1038/nprot.2006.263. [DOI] [PubMed] [Google Scholar]
- 16.Smeaton MB, Miller PS, Ketner G, et al. Small-scale extracts for the study of nucleotide excision repair and non-homologous end joining. Nucleic Acids Res. 2007;35(22):e152. doi: 10.1093/nar/gkm974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ye JZ, de Lange T. TIN2 is a tankyrase 1 PARP modulator in the TRF1 telomere length control complex. Nat Genet. 2004;36(6):618–623. doi: 10.1038/ng1360. [DOI] [PubMed] [Google Scholar]
- 18.Chen Y, Yang Y, van Overbeek M, et al. A shared docking motif in TRF1 and TRF2 used for differential recruitment of telomeric proteins. Science. 2008;319(5866):1092–1096. doi: 10.1126/science.1151804. [DOI] [PubMed] [Google Scholar]
- 19.Kunkel TA, Roberts JD, Zakour RA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 20.Kim SH, Kaminker P, Campisi J. TIN2, a new regulator of telomere length in human cells. Nat Genet. 1999;23(4):405–412. doi: 10.1038/70508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Canudas S, Houghtaling BR, Kim JY, et al. Protein requirements for sister telomere association in human cells. Embo J. 2007;26(23):4867–4878. doi: 10.1038/sj.emboj.7601903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Houghtaling BR, Cuttonaro L, Chang W, et al. A dynamic molecular link between the telomere length regulator TRF1 and the chromosome end protector TRF2. Curr Biol. 2004;14(18):1621–1631. doi: 10.1016/j.cub.2004.08.052. [DOI] [PubMed] [Google Scholar]
- 23.Ye JZ, Donigian JR, van Overbeek M, et al. TIN2 binds TRF1 and TRF2 simultaneously and stabilizes the TRF2 complex on telomeres. J Biol Chem. 2004;279(45):47264–47271. doi: 10.1074/jbc.M409047200. [DOI] [PubMed] [Google Scholar]
- 24.Savage SA. Dyskeratosis congenita. In: Pagon RA, Bird TC, Dolan CR, et al., editors. GeneReviews. Vol. 2010. University of Washington; Seattle: 2009. [Google Scholar]
- 25.Dokal I. Dyskeratosis congenita in all its forms. Br J Haematol. 2000;110(4):768–779. doi: 10.1046/j.1365-2141.2000.02109.x. [DOI] [PubMed] [Google Scholar]
- 26.Vulliamy TJ, Marrone A, Knight SW, et al. Mutations in dyskeratosis congenita: their impact on telomere length and the diversity of clinical presentation. Blood. 2006;107(7):2680–2685. doi: 10.1182/blood-2005-07-2622. [DOI] [PubMed] [Google Scholar]
- 27.Njajou OT, Cawthon RM, Damcott CM, et al. Telomere length is paternally inherited and is associated with parental lifespan. Proc Natl Acad Sci U S A. 2007;104(29):12135–12139. doi: 10.1073/pnas.0702703104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vulliamy T, Beswick R, Kirwan M, et al. Telomere length measurement can distinguish pathogenic from non-pathogenic variants in the shelterin component, TIN2. Clin Genet. 2011 doi: 10.1111/j.1399-0004.2010.01605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaminker PG, Kim SH, Desprez PY, et al. A novel form of the telomere-associated protein TIN2 localizes to the nuclear matrix. Cell Cycle. 2009;8(6):931–939. doi: 10.4161/cc.8.6.7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Du HY, Mason PJ, Bessler M, et al. TINF2 mutations in children with severe aplastic anemia. Pediatr Blood Cancer. 2009;52(5):687. doi: 10.1002/pbc.21903. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.