SUMMARY
The importance of maternal folate consumption for normal development is well established. Yet, the molecular mechanism linking folate metabolism to development remains poorly understood. The enzyme methionine synthase reductase (MTRR) is necessary for utilization of methyl groups from the folate cycle. We found that a hypomorphic mutation of the mouse Mtrr gene results in intrauterine growth restriction, developmental delay and congenital malformations including neural tube, heart and placental defects. Importantly, these defects were dependent upon the Mtrr genotypes of the maternal grandparents. Furthermore, we observed widespread epigenetic instability associated with altered gene expression in the placentas of wildtype grandprogeny of Mtrr-deficient maternal grandparents. Embryo transfer experiments revealed two epigenetic mechanisms of Mtrr deficiency in mice: adverse effects on their wildtype daughters’ uterine environment leading to growth defects in wildtype grandprogeny, and the appearance of congenital malformations independent of maternal environment that persist for five generations, likely through transgenerational epigenetic inheritance.
INTRODUCTION
It is well established that maternal folate deficiency or polymorphisms in genes required for folate metabolism are associated with an increased risk of congenital abnormalities in humans including intrauterine growth restriction, cardiovascular and placental abnormalities and neural tube defects (NTDs). However, conflicting reports have been published showing association and non-association between polymorphisms in folate pathway genes and risk for specific congenital malformations [e.g., NTDs (Rampersaud et al., 2003; van der Put et al., 1995)]. This is reinforced by studies whereby mutations in mouse genes encoding proteins required for folate transport and metabolism do not appear to recapitulate the range of congenital abnormalities produced by maternal folate deficiency in humans (Chen et al., 2001; Deng et al., 2008; Gelineau-van Waes et al., 2008; Pickell et al., 2009; Piedrahita et al., 1999; Swanson et al., 2001). As a result, the underlying mechanism through which folate metabolism acts during development may be more complex than once thought and remains poorly understood.
Folate is a carrier of methyl groups destined for downstream targets including homocysteine for methionine synthesis in a reaction that links the folate and methionine cycles (Figure 1A). The main circulating folate, 5-methyltetrahydrofolate (5-methyl-THF) (Smith et al., 2006), is generated by methylenetetrahydrofolate reductase (MTHFR) (Ghandour et al., 2004; Hart et al., 2002). Its methyl group is exclusively transferred by methionine synthase (MS, encoded by the gene Mtrr) to homocysteine to form methionine and tetrahydrofolate (Shane and Stokstad, 1985). In turn, methionine is a precursor to S-adenosylmethionine (S-AdoMet), which serves as the methyl donor for dozens of cellular substrates including proteins, RNA and DNA (Ghandour et al., 2002; Jacob et al., 1998; Wainfan et al., 1975). Consequently, folate and methionine metabolism are important for successful pregnancy. In mammals, methionine synthase reductase (MTRR, encoded by the Mtrr gene) is responsible for the activation of MS through the reductive methylation of its vitamin B12 cofactor (Yamada et al., 2006). Therefore, MTRR is required for normal progression of the folate and methionine cycles.
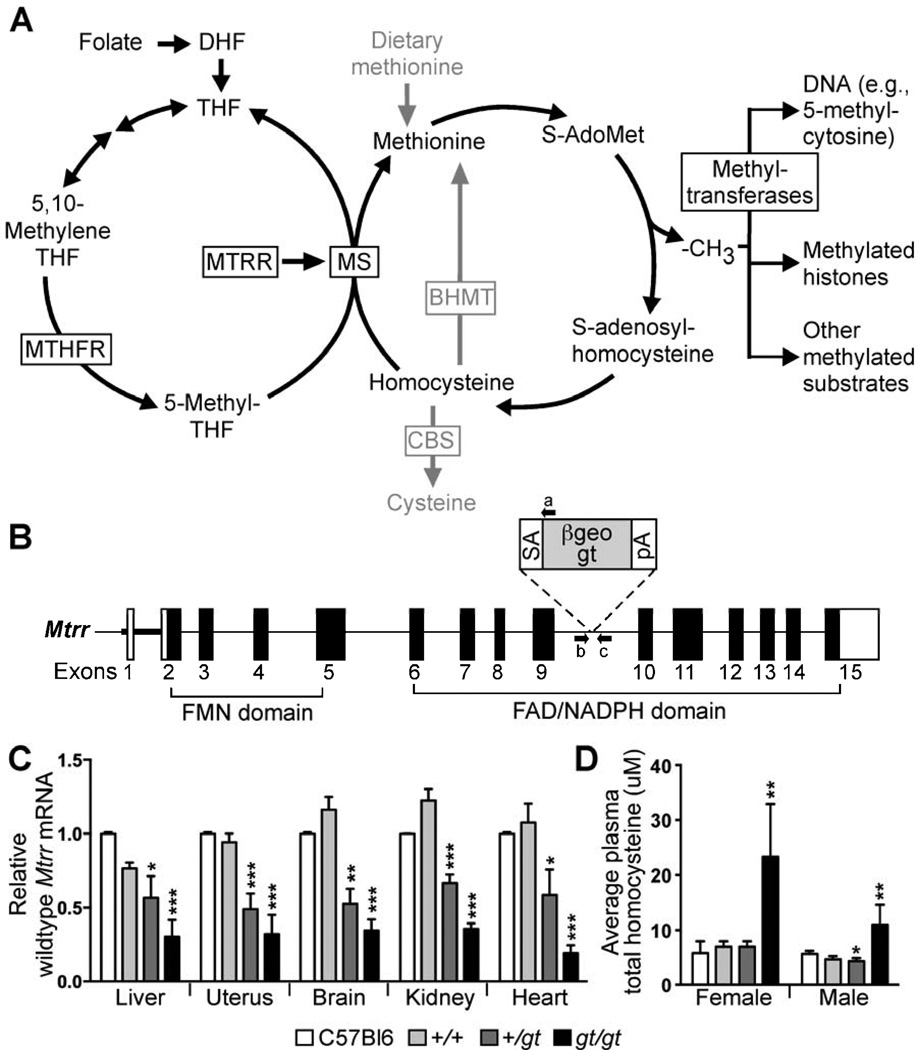
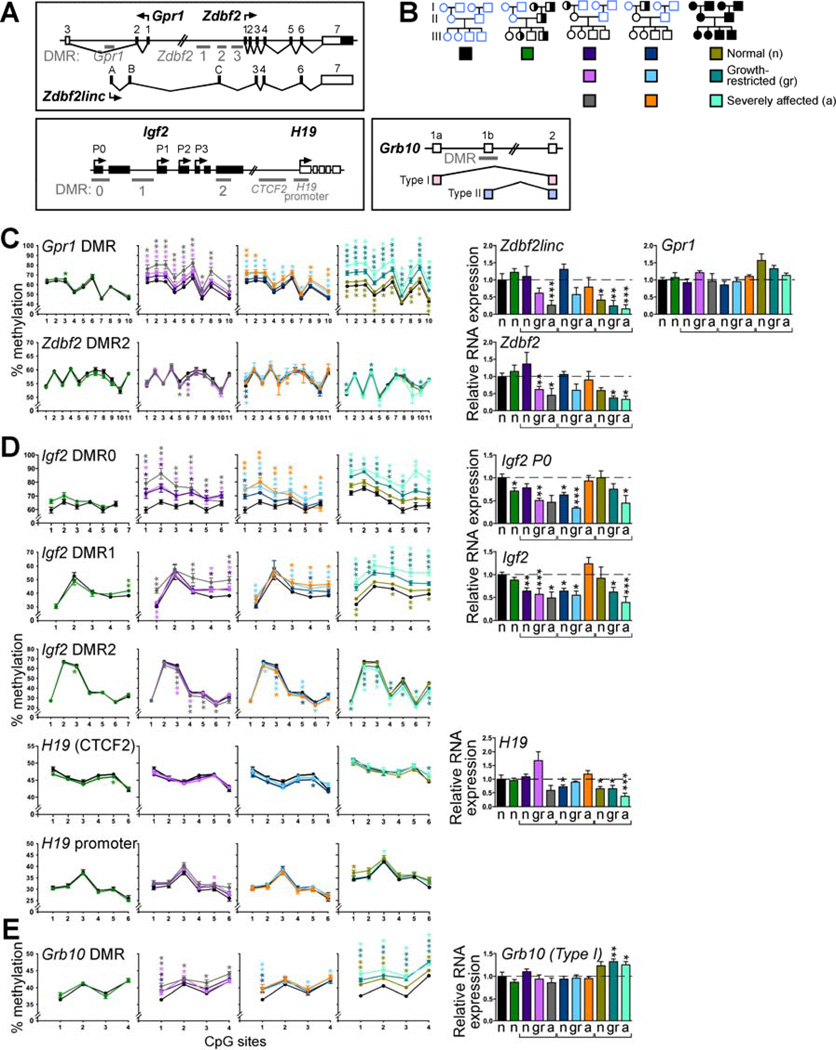
Figure 1. Mtrrgt is a knock-down mutation resulting in hyperhomocysteinemia.
(A) Diagram of folate and methionine metabolisms. (B) β-geo gene-trap (gt) containing a splice-acceptor (SA) and poly-adenylated (pA) tail was randomly inserted into intron 9 of the Mtrr gene to produce the Mtrrgt allele. PCR primers flanking the gene-trap insertion site (arrows b and c) in combination with a primer annealing to the gene-trap (arrow a) discerned the Mtrr+ and Mtrrgt alleles. (C) qRT-PCR analysis of wildtype Mtrr mRNA levels (mean±SEM) in adult organs (N=5 organs/genotype) from Mtrr+/+, Mtrr+/gt and Mtrrgt/gt mice derived from Mtrr+/gt intercrosses compared to C57Bl/6 mice. Data presented as fold difference compared to C57Bl/6 controls (normalized to 1). (D) Plasma total homocysteine concentrations (µm; mean±SD) in Mtrr+/+, Mtrr+/gt and Mtrrgt/gt adult mice compared to C57Bl/6 adults (N=3–8 mice/bar). Statistical analysis: independent comparison of each genotype to C57Bl/6 mice using a t-test. *P<0.05, **P<0.01, ***P<0.005. See also Table S4.
Here, we demonstrate that a hypomorphic mutation in the mouse Mtrr gene (Mtrrgt) disrupts folate metabolism. Remarkably, through a series of highly controlled genetic pedigrees, we show that the Mtrrgt allele is associated with effects on offspring development that are transmitted transgenerationally. In particular, the Mtrr genotype of either maternal grandparent dictates the developmental potential of their wildtype grandprogeny. This occurs through an epigenetic mechanism that leads to distinct phenotypes. Therefore, this is a robust genetic model for studying the epigenetic transmission of developmental disorders between generations.
RESULTS
Mtrrgt mutation causes abnormal folate metabolism in mice
To explore the relationship between folate metabolism and development, we generated Mtrr-deficient mice using embryonic stem cells containing a randomly inserted gene-trap (gt) vector in the Mtrr gene locus (Figure 1B). We verified the insertion of the gene-trap vector into intron 9 of the Mtrr locus by Southern blotting and DNA sequencing (data not shown) and were able to distinguish between Mtrr+/+, Mtrr+/gt and Mtrrgt/gt mice by a three-primer PCR using primers that flanked the gene-trap insertion site combined with a primer specific to the gene-trap vector (Figure 1B).
To test whether transcription of wildtype Mtrr mRNA was disrupted in our model, we performed quantitative reverse transcription (qRT) PCR on RNA collected from adult tissues of Mtrr+/+, Mtrr+/gt and Mtrrgt/gt mice. Despite the gene-trap insertion, we detected wildtype mRNA transcripts in all Mtrrgt/gt tissues tested, albeit at 19.3–35.7% of Mtrr+/+ levels (Figure 1C), indicative of splicing around the gene-trap in some transcripts. Mtrr+/gt tissues contained 49.0–66.8% of Mtrr+/+ mRNA levels (Figure 1C). As a result, Mtrrgt is a knock-down allele.
Increased levels of plasma homocysteine are used as an indicator of folate deficiency (Hague, 2003; Piedrahita et al., 1999). Therefore, using tandem mass spectrometry, we measured plasma total homocysteine concentrations in adult female and male Mtrrgt/gt mice and detected an average of 23.3±5.5 µM (mean±SD) and 11.0±3.6 µM total homocysteine, respectively. These values were 3.3 to 3.7 fold greater than in Mtrr+/+ littermates (females: 7.0±0.6 µM [P<0.005]; males: 4.6±0.3 µM [P<0.05]) (Figure 1D) and are comparable to mouse models with genetic mutations in other folate metabolic enzymes (summarized by Elmore et al., 2007). Elmore et al (2007) also showed increased liver 5-methyl-THF, reduced plasma methionine, and reduced heart AdoMet/AdoHcy ratio in Mtrr-deficient adults compared to wildtype littermates. Altogether, these data reveal that Mtrrgt mice are an appropriate model for studying the impact of abnormal folate metabolism.
Phenotypes of progeny from Mtrr+/gt intercrosses do not correlate with embryonic genotypes
Analysis of the progeny from Mtrr+/gt intercrosses revealed a normal Mendelian ratio at weaning age (36:56:23), indicating that Mtrrgt/gt mice are viable. We observed no obvious phenotypes in Mtrrgt/gt adult mice. However, we noticed that the litter sizes were smaller (5.8 pups/litter, N=20 litters) compared to control C57Bl/6 litters with the same genetic background as the Mtrrgt mouse line (8.3 pups/litter, N=8 litters). Therefore, embryos of Mtrr+/gt intercrosses were analyzed during gestation. At embryonic (E) day 10.5, 55.5% of conceptuses from Mtrr+/gt parents were normal (Figure 2B) in contrast to 95.7% of the conceptuses from C57Bl/6 litters (Figure 2A). Similar numbers of resorptions were present suggesting that there was no difference in embryonic lethality occurring prior to E10.5. However, the remaining conceptuses from the mutant matings showed severe congenital malformations not present in control litters, ranging from developmental delay to NTDs and heart defects. Surprisingly and consistent with Mendelian ratios at weaning, genotyping results indicated that all embryonic genotypes including wildtype were equally affected (Figure 2C).
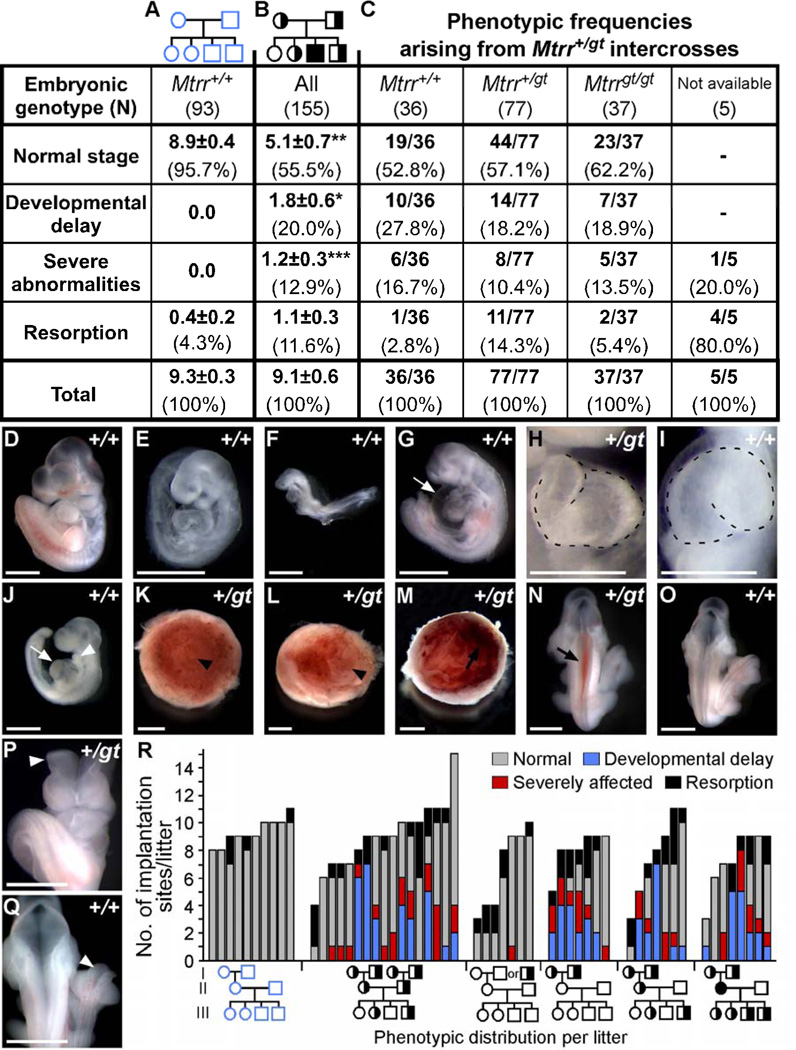
Figure 2. Phenotypes of progeny from Mtrr+/gt intercrosses at E10.5 do not correlate with embryonic or maternal genotypes.
(A-B) Phenotypic frequencies in litters from (B) Mtrr+/gt intercrosses compared to (A) C57Bl/6 crosses denoted by the average number of conceptuses/litter (±SEM) followed by the percentage of total conceptuses in brackets. Statistical analysis: independent comparison of phenotypic frequencies from Mtrr+/gt intercrosses compared to C57Bl/6 using Mann-Whitney test. *P<0.05; **P<0.01; ***P<0.001. (C) Phenotypic frequency in each embryonic genotype derived from Mtrr+/gt intercrosses. No significant difference between embryonic genotypes (Fisher’s exact test). (D-Q) Representative images of conceptuses at E10.5. Embryonic genotype is indicated. (D) Normal-appearing embryo compared to (E-F) developmentally-delayed embryos resembling (E) E9.5- and (F) E8.5-staged embryos. (G-J) Embryos with heart defects including (G) pericardial edema (arrow) or (J) enlarged heart (arrow) and small branchial arches (arrowhead). (H) Normal leftward-cardiac looping vs. (I) abnormal rightward-cardiac looping (dotted line). (K-L) Placentas with (K) normal centered or (L) abnormal off-centered allantois attachment (arrowhead). (M-N) Hemorrhage (arrow) in (M) placenta or (N) embryo. (O) Normal neural tube closure versus (P-Q) open neural tubes (arrowhead) in the (P) cranial or (Q) spinal cord regions. Scale bars: D-H, K-Q, 1 mm; I-J, 0.5 mm. (R) Phenotypic frequencies per litter (generation III) from designated crosses at E10.5. Grandparent genotypes were determined retrospectively. Third pedigree from the left: maternal grandfather was Mtrr+/+ or Mtrr+/gt. Pedigrees: circle, female; square, male; blue outline, C57Bl/6 line; black outline, Mtrr line; white fill, Mtrr+/+; half black/half white, Mtrr+/gt; black fill, Mtrrgt/gt. See also Figure S1 and Table S1.
We found that 20% of embryos at E10.5 were developmentally delayed by approximately 0.5–2.0 days (i.e., contained <30 somite pairs) (Figure 2B, 2E–2F, Table S1), although they otherwise appeared normal. In addition, 12.9% of conceptuses exhibited one or more congenital abnormality (Figure 2, Table S1). Among these, we observed heart defects including pericardial edema (Figure 2G), reversed cardiac looping (Figure 2I) and enlarged hearts that were thicker and disproportionate in size (Figure 2J), abnormal placental development whereby chorioallantoic attachment was eccentric (Figure 2L) or failed to occur (data not shown) and hemorrhaging (Figure 2M–2N). NTDs were also observed including neural tubes that failed to close by the appropriate developmental stage in the cranial region (Figure 2P). These defects are reminiscent of embryos at E10.5 from Mthfr+/− intercrosses (Pickell et al., 2009).
Mtrr deficiency in either maternal grandparent is sufficient to cause defects in their wildtype grandprogeny at E10.5
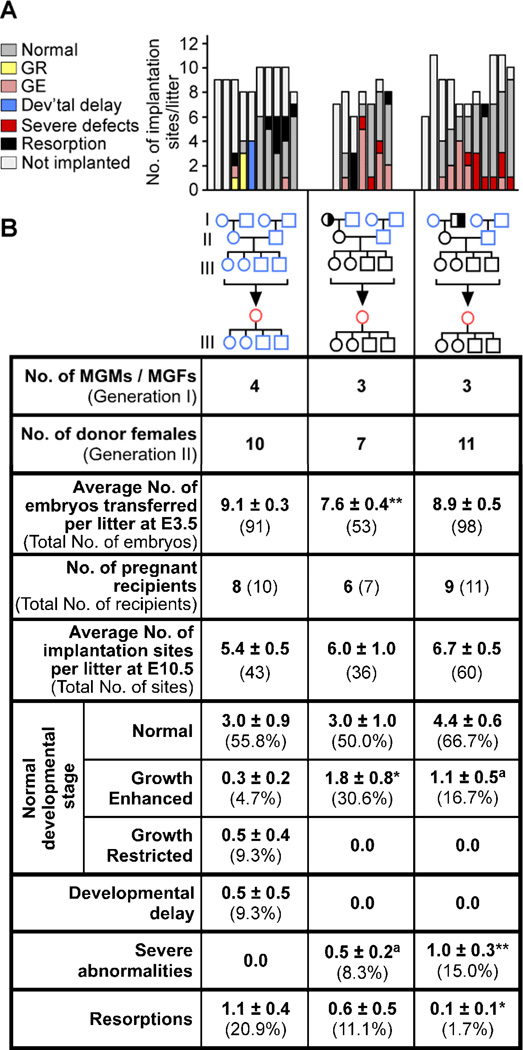
Given that all embryonic genotypes from Mtrr+/gt intercrosses were similarly affected, we suspected that a parental or even grandparental Mtrrgt mutation might affect development. In our initial studies, most of the mice were generated from Mtrr+/gt intercrosses, including Mtrr+/+ controls. This became important when we found that the same types of abnormal conceptuses were observed at a high frequency (44/102 conceptuses) from Mtrr+/+ females crossed to Mtrr+/+ males at E10.5 (Figure S1). Pedigree analysis of these Mtrr+/+ intercrosses revealed that pregnancies with abnormal litters almost always had Mtrr+/gt maternal grandparents (Figures 2R and S1). This implied that normal embryonic development might be associated with a grandparental, and not parental, Mtrrgt mutation.
To verify that a paternal Mtrrgt allele did not contribute to the defects, we crossed Mtrr+/gt males with C57Bl/6 females and assessed the litters at E10.5 (N=10 litters) (Figures 3A–3B and S2). All conceptuses were normal with no obvious developmental delay or defects, except one conceptus with a placenta defect. Since the Mtrr+/gt males were derived from Mtrr+/gt parents, these data indicated that neither the paternal nor the paternal grandparents’ Mtrr genotype significantly affected development at this stage.
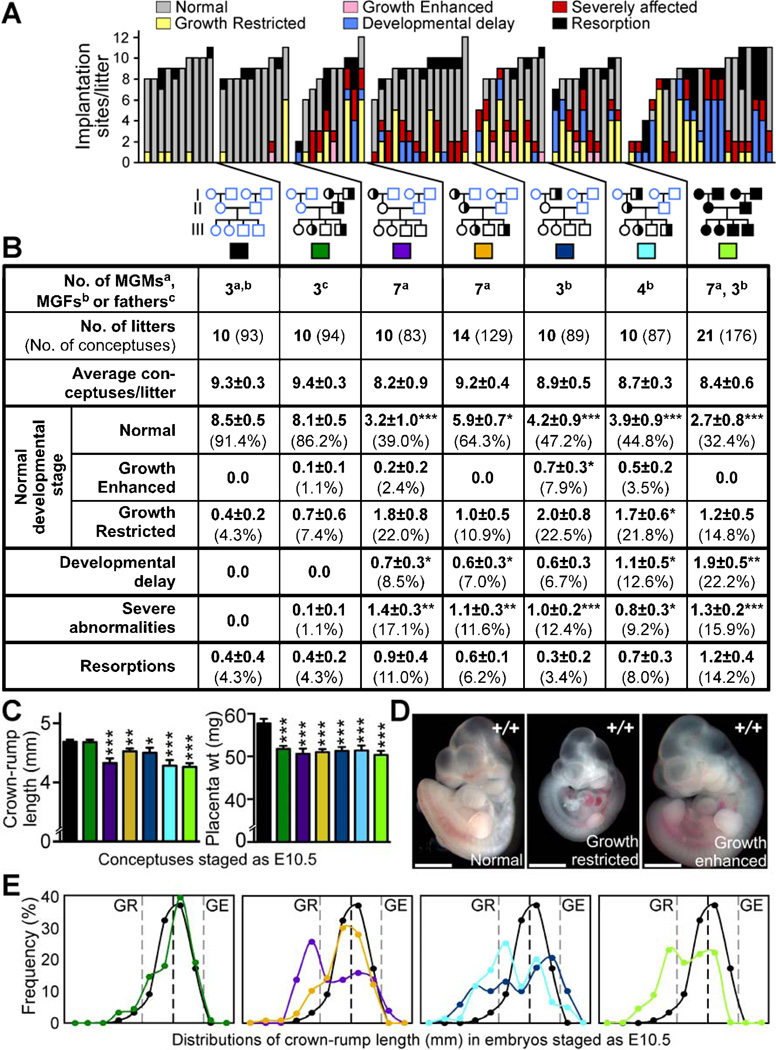
Figure 3. Maternal grandparental Mtrr genotypes are important for a successful pregnancy.
(A-B) Phenotypic frequencies caused by Mtrr+/gt maternal grandmothers (MGMs) or grandfathers (MGFs) at E10.5 compared to C57Bl/6 controls. Mtrr+/gt paternal and Mtrrgt/gt pedigrees were also assessed. Data represented as (A) number of conceptuses/litter with a specific phenotype and (B) average number of phenotypically-affected conceptuses/litter (±SEM) followed by the percentage of total conceptuses in brackets. Colored boxes in (B) indicate the pedigrees assessed in (C) and (E). Pedigrees: circle, female; square, male; blue outline, C57Bl/6 mice; black outline, Mtrr mouse line; white fill, Mtrr+/+; half black/half white fill, Mtrr+/gt; black fill, Mtrrgt/gt. Black asterisks, independent comparison of phenotypic frequencies per litter of each pedigree and C57Bl/6 using Mann-Whitney test. (C) Average embryo crown-rump lengths (±SEM) (left) and average placental weights (±SEM) (right) from conceptuses staged as E10.5 (i.e., 30–40 somite pairs) per pedigree (N=51–89 conceptuses). Statistics: comparison to C57Bl/6 using independent t-test. (D) Representative images of normal, growth-restricted (GR) and growth-enhanced (GE) wildtype embryos at E10.5. Scale bars=1mm. (E) Distribution of crown-rump lengths in E10.5-staged embryos (N=51–89 embryos). Black dotted line, mean; grey dotted lines, two SD from mean. *P<0.05, **P<0.01, ***P<0.005. See also Figure S2 and Table S2.
Next, we examined the specific effect of the maternal grandparental Mtrrgt mutation on embryonic development by crossing Mtrr+/gt females with C57Bl/6 males or C57Bl/6 females with Mtrr+/gt males (generation I). Mtrr+/+ daughters from the resulting litters were then crossed with C57Bl/6 males (generation II). The subsequent Mtrr+/+ litters (generation III) were dissected at E10.5 and the conceptuses were assessed for abnormalities (Figure 3A–3B). Only 39.0% of conceptuses derived from Mtrr+/gt maternal grandmothers and 47.2% of the conceptuses derived from Mtrr+/gt maternal grandfathers appeared normal (Figure 3B). Crown-rump length measurements of embryos within the normal developmental range for E10.5 (30–40 somite pairs) revealed that, on average, these embryos were significantly smaller than C57Bl/6 embryos (Figure 3C). In fact, a large proportion of these so-called ‘normal’ embryos were growth-restricted (22.0% and 22.5% of conceptuses from Mtrr+/gt maternal grandmothers and grandfathers, respectively, as defined as more than two standard deviations below the mean length of C57Bl/6 embryos) (Figure 3D–3E). However, some embryos were actually growth-enhanced, particularly those derived from Mtrr-deficient maternal grandfathers (7.9% growth-enhanced, P<0.05) (Figure 3B, 3D–3E). The presence of growth phenotypes did not correlate with litter size (Table S2). Beyond growth defects, the remaining affected conceptuses were either developmentally delayed or had severe defects (Figure 3A–3B), similar to those described above including NTDs, heart abnormalities, placental defects and hemorrhages (Figure S2). The rates of resorption were not significantly different from C57Bl/6 controls (Figure 3B). Interestingly, some litters were more sensitive to grandparental Mtrr deficiency than others since a proportion of the litters appeared normal whereas up to 100% of conceptuses were affected in other litters (Figure 3A).
Mtrrgt/gt conceptuses derived from pedigrees with Mtrrgt/gt parents and grandparents were also assessed (N=21 litters). These litters showed similar phenotypes but at a higher frequency compared to Mtrr+/+ conceptuses derived from an Mtrr+/gt maternal grandparent (Figures 3 and S2) suggesting a dose-dependent effect of the Mtrrgt allele. The growth phenotype was not assessed in the initial Mtrr+/gt intercrosses or maternal effect pedigrees, which is why the percentage of the conceptuses categorized as phenotypically ‘normal’ is different in these pedigrees (Figures 2 and S1, Table S1) compared to the pedigrees showing a maternal grandparental origin effect (Figures 3 and S2).
Combined, these data implied that an Mtrrgt mutation in either maternal grandparent disrupted the development of their grandprogeny, even when the parents and the conceptus were wildtype. Importantly, the effect was not exacerbated when both the maternal grandmother and mother carried the Mtrrgt allele or when both the maternal grandfather and mother carried the Mtrrgt allele (Figures 3 and S2). This indicates that the presence of the Mtrrgt mutation in either maternal grandparent is sufficient to produce developmental phenotypes in their wildtype grandprogeny.
Mtrr deficiency causes tissue-specific global DNA hypomethylation
Since disruption of the folate cycle is thought to inhibit the transmission of one-carbon methyl groups necessary for DNA methylation, global DNA methylation levels were measured in the Mtrrgt model. Livers and non-pregnant uteri from Mtrr+/gt and Mtrrgt/gt adult mice showed a significant reduction in DNA methylation (44.9–74.2% of C57Bl/6 levels) (Figure 4A). Interestingly, DNA methylation in Mtrr+/gt and Mtrrgt/gt adult brains was not significantly different from controls (Figure 4A) indicating that the brain might be spared from reduced methyl-group availability. DNA hypomethylation was also evident in the livers and uteri of adult wildtype littermates (40–62% of C57Bl/6 levels; Figure 4A), further emphasizing a non-genetic influence in the Mtrrgt model. These data support an epigenetic mechanism for folate metabolism since the dysregulation of DNA methylation precedes the appearance of developmental phenotypes in the Mtrrgt model.
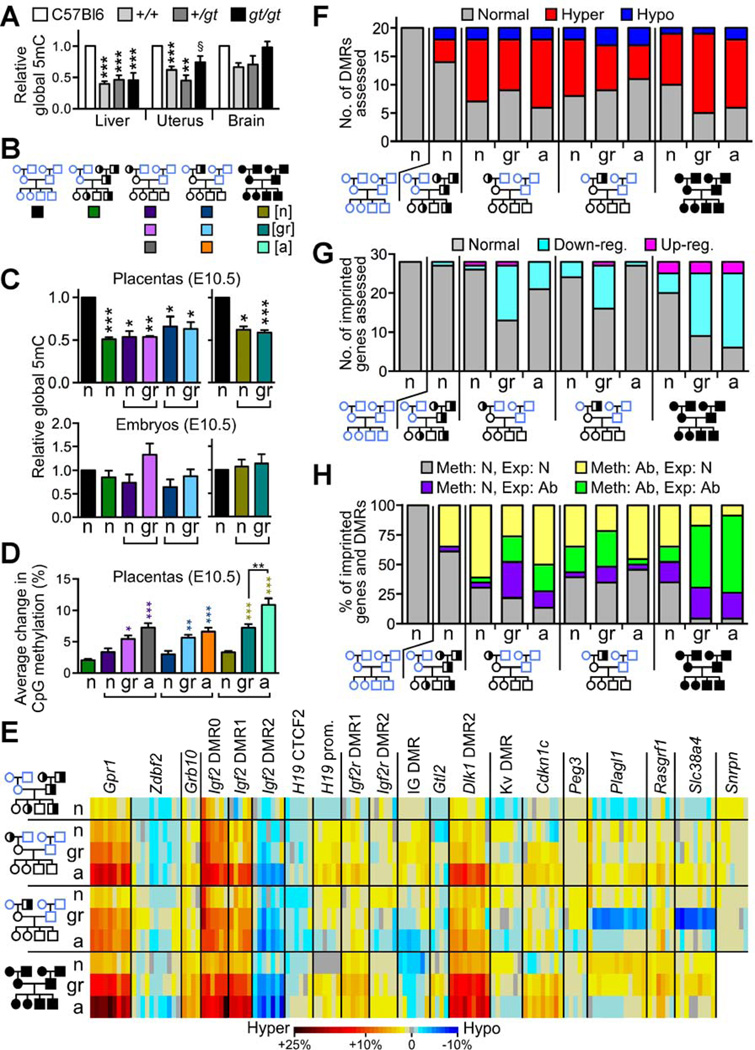
Figure 4. Placentas of wildtype grandprogeny display epigenetic instability at E10.5.
(A) Global 5-methylcytosine (5mC) levels (mean±SEM) in adult tissues from Mtrr+/+, Mtrr+/gt and Mtrrgt/gt mice relative to C57Bl/6 (N=4–18 mice; normalized to 1). (B) Each box denotes the pedigree from which conceptuses analyzed in panels C-D were derived and their phenotype (normal [n], growth-restricted [gr], severely affected [a]). (C) Relative global 5mC levels (mean±SEM) in placentas and embryos derived from the pedigree denoted in (B) compared to C57Bl/6 (N=5–10; normalized to 1). Independent t-tests compared C57Bl/6 to each phenotype/pedigree. (D) Average change in CpG methylation (%) in imprinted DMRs compared to C57Bl/6 controls. Placentas of conceptuses derived from the pedigree denoted in (B) were assessed. Only CpG sites with significant methylation changes were included. Growth-restricted and severely-affected placentas were compared to phenotypically normal placentas within the same pedigree using an independent t-test. (E) A heat-map indicating average methylation change (%) at individual CpG sites in each DMR compared to C57Bl/6. Placentas of phenotypically normal (n), growth-restricted (gr) and severely affected (a) conceptuses (generation III) from each pedigree were assessed. (F-G) The number of (F) DMRs with significant hyper- or hypomethylation and (G) imprinted genes with significant up- or downregulated RNA expression in placentas of conceptuses from the designated pedigree and phenotypic group compared to C57Bl/6. (H) An association analysis between CpG methylation in DMRs and RNA expression of corresponding imprinted genes. Abnormal DMR methylation: ≥1 CpG site with significantly different methylation compared to C57Bl/6. Abnormal RNA expression: significantly different from C57Bl/6. N=3–10 placentas/phenotype/pedigree in panels C-H. Ab, abnormal; Exp, expression; Meth, methylation; N, normal. §P=0.07, *P<0.05, **P<0.01, ***P<0.005. See also Figures S3 and Table S3.
Next, we assessed the effects of maternal grandparental Mtrr deficiency on global DNA methylation in their wildtype grandprogeny at E10.5. Although we did not observe significant differences in the phenotypically normal or growth-restricted embryos derived from an Mtrr+/gt maternal grandparent, there was significant global DNA hypomethylation in the corresponding placentas (53% or 63–66% of C57Bl/6 levels, respectively) (Figure 4C). This implied that the placental epigenetic landscape was altered. Global DNA methylation was also reduced in wildtype placentas derived from Mtrr+/gt fathers (50% of C57Bl/6 levels) (Figure 4C), which further supports an epigenetic mechanism since it shows that altered DNA methylation precedes the appearance of developmental phenotypes. For example, wildtype daughters of an Mtrr+/gt male developed normally despite having hypomethylated placental DNA, and yet in adulthood, these females produce abnormal progeny with hypomethylated placentas (Figures 3A–3B and 4C).
Placentas of wildtype grandprogeny are epigenetically unstable at E10.5
Imprinted loci have previously been used as a model to study epigenetic stability during development (Messerschmidt et al., 2012). It is well known that imprinted genes are regulated by DNA methylation at differentially methylated regions (DMRs) in a parental-origin specific manner. The majority of known imprinted genes are expressed in fetally derived extraembryonic tissue within the placenta and not the maternal decidua, and are required for normal placental development (Coan et al., 2005; Hemberger, 2010; Tunster et al., 2013). To determine the degree of epigenetic instability in grandprogeny of an Mtrr+/gt maternal grandparent, we analyzed imprinted loci for alterations in DNA methylation and gene expression in wildtype placentas at E10.5.
Using the bisulfite pyrosequencing method, we assessed DNA methylation at key CpG sites (N=143) within 20 different DMRs (Table S3). Analysis of placentas of wildtype conceptuses derived from Mtrr+/gt fathers revealed only a few CpG sites (10/143 CpGs in 6/20 DMRs) that were significantly abnormally methylated with an average methylation change (±SEM) of 2.0±0.2% compared to C57Bl/6 levels (Figures 4D–4E and 5, Table S3). This implied that these placentas were epigenetically quite stable, consistent with the absence of phenotypes in these litters (Figure 3). Conversely, a large proportion (45.0%–70.0%) of the DMRs assessed in wildtype placentas of phenotypically normal, growth-restricted and severely affected grandprogeny derived from either Mtrr+/gt maternal grandparent had CpG site methylation levels that were statistically different from C57Bl/6 (Figures 4E and 5, Table S3). The majority of the significantly affected CpG sites were hypermethylated (Figure 4F). The placentas of growth-restricted and severely affected conceptuses had an average CpG methylation change that was significantly larger (5.4–5.5% [P<0.05] and 6.6–7.2% [P<0.005] of C57Bl/6 levels, respectively) compared to phenotypically normal conceptuses from the same pedigree (3.0–3.3% of C57Bl/6 levels) (Figures 4D and 5, Table S3) indicating a positive correlation between epigenetic instability and the severity of the phenotype. Furthermore, some DMRs were more sensitive to grandparental Mtrr deficiency than others since a proportion had only one CpG site with significantly aberrant methylation whereas other DMRs had up to 100% of CpG sites with abnormal methylation that were statistically different from C57Bl/6 controls (Figure S3A). These overall DNA methylation trends were further exacerbated in Mtrrgt/gt placentas (Figures 4D–4F, 5 and S3A, Table S3).
Figure 5. Phenotypic severity is positively correlated with degree of dysregulated DMR methylation and gene expression in placentas at E10.5.
(A) Diagrams of imprinted loci used as examples of epigenetic instability. (B) Each box denotes the pedigree from which generation III placentas (N=3–11) were selected for analysis in panels C-E and their phenotype (e.g., phenotypically normal [n], growth-restricted [gr] or severely affected [a]). (C-E) CpG site-specific methylation (mean±SEM) in the DMRs of (C) Gpr1/Zdbf2, (D) Igf2/H19 and (E) Grb10 loci as determined by pyrosequencing followed by the relative RNA levels (mean±SEM) of the imprinted genes controlled by these DMRs. Values were compared to C57Bl/6 controls. Statistical analysis: for DNA methylation: non-parametric Mann-Whitney test comparing methylation percentages at each CpG site to C57Bl/6; for gene expression: independent t-tests was used to compare each phenotype/pedigree to C57Bl/6. *P<0.05; **P<0.01; ***P<0.005. See also Figure S4 and Tables S3 and S5.
To determine whether the placental epigenetic instability observed was functionally important, we used qRT-PCR to measure the expression of imprinted genes specifically regulated by methylation at these DMRs in the grandprogeny at E10.5 (Table S3). The majority of misexpressed genes were statistically downregulated in a phenotype dependent manner compared to C57Bl/6 placentas (Figure 4G, Table S3), which supports the overall trend of DMR hypermethylation. Importantly, more misexpressed genes were associated with abnormal DMR methylation rather than normal DMR methylation. This occurred regardless of the conceptus’ phenotype and whether it was derived from an Mtrr+/gt maternal grandmother or grandfather (Figures 4H and 5). Together with the observation that there were a considerable number of abnormally methylated DMRs associated with normally expressed genes (Figure 4H), we conclude that dysregulation of DNA methylation likely preceded the changes in gene expression. The instances of gene misexpression associated with normal DMR methylation (Figure 4H) were likely the consequence of secondary effects of Mtrr deficiency including the dysregulation by upstream genes affected by abnormal methylation (e.g., transcription factors or non-coding RNA). Altogether, this suggests a hierarchy of events: an Mtrrgt mutation in either maternal grandparent leads to epigenetic instability in important regulatory regions in placentas of their wildtype grandprogeny, which may in turn cause gene misexpression leading to developmental phenotypes.
As with the global methylation patterns, DMR methylation and gene expression (e.g., the Igf2/H19 locus) in wildtype embryos at E10.5 was not significantly different from controls (Figures 4C and S3B, Table S3). Important changes in DNA methylation and gene expression of specific cell populations might not be apparent when assessing whole embryos. Alternatively, placental cells may be more susceptible than embryonic cells to the intergenerational effects of abnormal folate metabolism. A direct paramutation effect (Rassoulzadegan et al., 2006) is unlikely since normal levels of wildtype Mtrr mRNA were present in Mtrr+/+ placentas and embryos at E10.5 derived from either an Mtrr+/gt parent or grandparent compared to C57Bl/6 controls, regardless of phenotype (Figure S4).
Congenital abnormalities in wildtype grandprogeny are due to epigenetic inheritance
To better understand the mechanism, we performed an embryo transfer experiment (Figures 6 and S5) to distinguish between an epigenetic effect on the ability of females in generation II to carry normal pregnancies (maternal environment) versus the effect on gametes (direct epigenetic inheritance). Wildtype pre-implantation embryos derived from pedigrees with one Mtrr+/gt maternal grandparent and a wildtype mother were collected at E3.25, immediately prior to the initiation of DNA remethylation during epigenetic reprogramming (Saitou et al., 2012). Same stage C57Bl/6 embryos were used as controls. Embryos were transferred into the uteri of pseudopregnant B6D2F1 females and allowed to develop until E10.5. If a phenotype was absent, then an abnormal maternal environment in the original donor mother (i.e., generation II) was the cause of this defect. However, persistence of a phenotype indicates that this defect occurred independent of the maternal environment.
Figure 6. Epigenetic mechanism of Mtrr deficiency leads to distinct phenotypes in their grandprogeny caused by an atypical uterine environment in their daughters or epigenetic inheritance.
Phenotypic frequencies at E10.5 resulting from the transfer of wildtype pre-implantation embryos derived from an Mtrr+/gt maternal grandmother (MGM) and Mtrr+/+ mother (center), or an Mtrr+/gt maternal grandfather (MGF) and Mtrr+/+ mother (right) into a B6D2F1 pseudopregnant recipient female. C57Bl/6 embryos were transferred as a control (left). Data is represented (A) graphically as the number of phenotypes/litter and (B) as the average number of phenotypically-affected conceptuses/litter (±SEM) followed by the percentage of total conceptuses assessed in brackets, unless otherwise indicated. GR, growth-restricted; GE, growth enhanced; Dev’tal, developmental. Independent comparison of phenotypic frequencies from each pedigree to the C57Bl/6 was completed using Mann-Whitney tests. aP=0.05, *P<0.05, **P<0.01. Pedigrees: circle, female; square, male; blue outline, C57Bl/6 mice; black outline, Mtrr mouse line; red outline, B6D2F1 hybrid mice; white fill, Mtrr+/+; half black/half white fill, Mtrr+/gt. See also Figure S5.
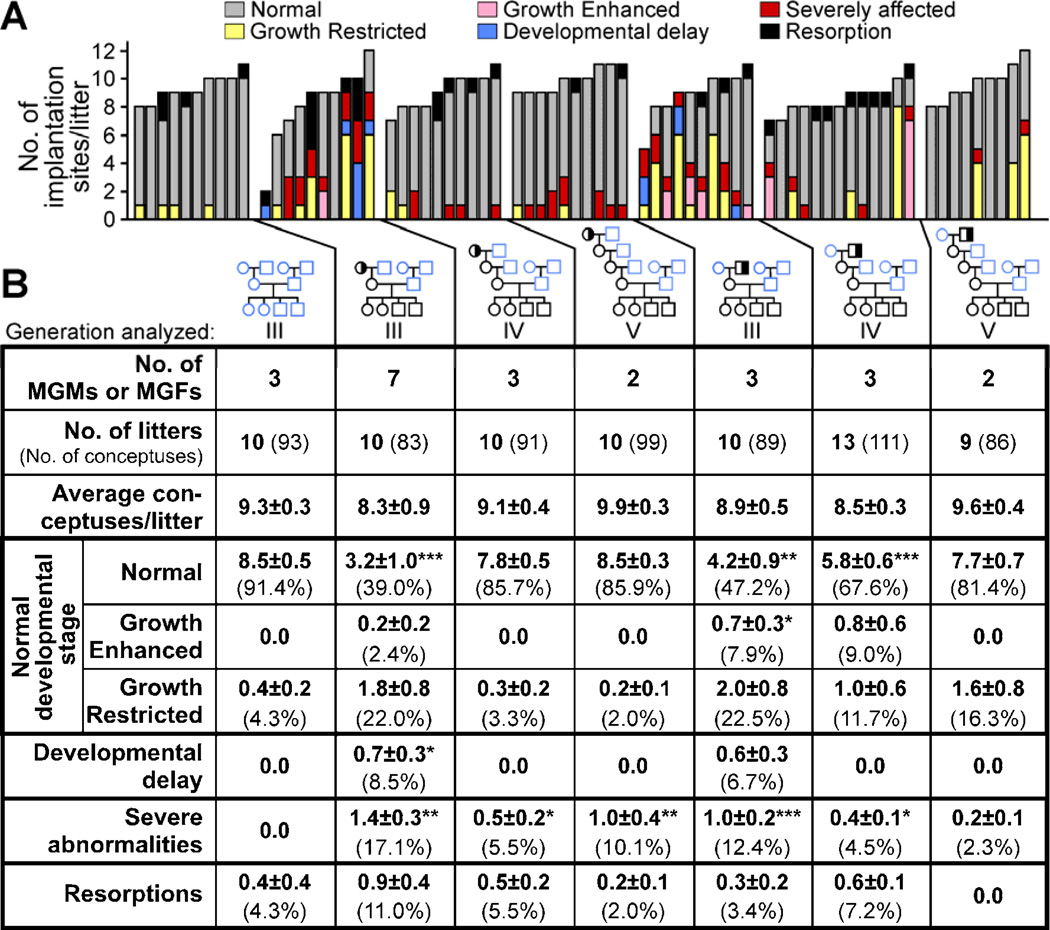
Remarkably, the embryo transfer results suggested that the growth defects and the congenital malformations were due to separate epigenetic mechanisms. Growth restriction and developmental delay phenotypes were not observed in the transferred litters derived from Mtrr+/gt maternal grandmothers (N=36 conceptuses) and Mtrr+/gt maternal grandfathers (N=60 conceptuses) (Figures 6 and S5) indicating that the B6D2F1 maternal environment rescued the growth phenotypes. In fact, we observed an increased frequency of growth enhancement (16.7–30.6% of conceptuses) compared to controls (4.7% [P≤0.05]; N=43 implantation sites) (Figure 6). By contrast, congenital abnormalities, including neural tube, heart and placenta abnormalities, persisted in 8.3–15.0% of the transferred grandprogeny at E10.5 (Figures 6 and S5). No severe abnormalities were apparent in the transferred C57Bl/6 conceptuses. Therefore, Mtrr deficiency results in epigenetic effects both on their daughter’s maternal environment and on their gametes leading to defective fetal and placental development in their grandprogeny. In support of this, we observed the persistence of severe abnormalities but not developmental delay or significant frequencies of growth restriction at E10.5 in generations IV and V derived from an Mtrr+/gt maternal ancestor (Figures 7 and S6). This further signifies that abnormal folate metabolism contributes to epigenetic inheritance through the germline.
Figure 7. Congenital malformations are transmitted at least up to four wildtype generations after an Mtrr+/gt maternal ancestor.
(A-B) The phenotypic frequencies within wildtype litters at E10.5 from generations III, IV and V derived from an Mtrr+/gt maternal grandmother (MGM) or Mtrr+/gt maternal grandfather (MGF) compared to C57Bl/6 pedigrees. Data represented (A) graphically as the number of phenotypes/litter and (B) as the average number of phenotypically affected conceptuses/litter (±SEM) for each pedigree followed by the percentage of total conceptuses in brackets. Phenotypic frequencies/pedigree were independently compared to C57Bl/6 using Mann-Whitney test. *P<0.05, **P<0.01, ***P<0.001. See also Figure S6.
DISCUSSION
We have conducted highly controlled experiments and genetic crosses to show that folate metabolism has distinct transgenerational epigenetic functions that are responsible for specific developmental processes. Remarkably, Mtrr deficiency in either maternal grandparent resulted in a similar spectrum and frequency of phenotypes in their wildtype grandprogeny. Therefore, the initial effect of Mtrr deficiency is likely transmitted through the germline via epigenetic factors, independent of physiological environment and genomic sequence. The epigenetic mechanism behind Mtrr deficiency in mice leads to two distinctive phenotypes: 1) an atypical uterine environment in their wildtype daughters causing growth defects in their wildtype grandprogeny and 2) congenital malformations in their wildtype grandprogeny due to epigenetic inheritance via the germline, the effects of which persist for at least up to four wildtype generations after an Mtrr-deficient maternal ancestor.
Since the folate pathway donates methyl groups necessary for cellular methylation, DNA methylation was a likely epigenetic candidate. Human polymorphisms in folate metabolic enzymes (e.g., MTHFR 677C →T) (Castro et al., 2004; Friso et al., 2002) or the reduction of maternal dietary nutrients (e.g., folate) during periconceptual periods in sheep (Sinclair et al., 2007) also result in hypomethylated or demethylated DNA in the immediate offspring’s genome. However, none of these studies assessed whether these effects were transmitted to subsequent generations. Changes in histone methylation are less likely than changes in DNA methylation to explain the grandparental origin effects simply because histones are largely replaced by protamine proteins during spermatogenesis (Agell et al., 1983).
The widespread dysregulation of DNA methylation in the Mtrrgt model likely explains the wide spectrum of phenotypes observed in the wildtype grandprogeny. Knocking out DNA methyltransferases in conceptuses causes extensive DNA demethylation and phenotypes including growth restriction, developmental delay, NTDs, placental defects and embryonic lethality (Hata et al., 2002; Li et al., 1992; Okano et al., 1999). These phenotypes are reminiscent of those observed in our study and imply that folate plays an important epigenetic role during development. Using imprinted loci as a model for the entire genome, we were able to show extensive epigenetic instability associated with gene misexpression specifically in placentas of the wildtype grandprogeny. At many of the loci assessed, the placentas exhibited locus-specific hypermethylation in the context of global DNA hypomethylation, which is similar to the Mthfr−/− mouse that also has locus-specific methylation trends that do not reflect genome-wide methylation trends (Chan et al., 2010; Chen et al., 2001). The mechanism behind this finding is unknown. Since the Mtrrgt line was extensively backcrossed to sustain genetic robustness, it was impossible to determine the parental origin of imprinted alleles. Regardless, it is unlikely that the alterations in DNA methylation profiles are attributed to a failure to establish parental imprints but rather an inability to maintain genome-wide DNA methylation patterns in imprinted and non-imprinted genes alike. This might be a consequence of reduced methyl group availability or dysregulation of methylation maintenance machinery.
We showed that congenital abnormalities in the Mtrrgt model are caused by transgenerational epigenetic inheritance since these phenotypes persist for four wildtype generations after the initial Mtrrgt ancestor and occur after embryo transfer. The genetic background of the Mtrrgt mouse line is an unlikely cause of the congenital abnormalities since these defects were never observed in the C57Bl/6 control pedigree. Although the mechanism behind epigenetic inheritance remains unknown, we hypothesize that some of the aberrant methylation patterns (epimutations) caused by Mtrr deficiency escape classical epigenetic reprogramming between generations when the genome normally undergoes widespread demethylation. Given the pedigree complexity, inter-individual variation in methylation and wide spectrum of phenotypes observed here, it will be a challenge to identify the precise epimutations that are inherited and that cause specific phenotypes in the Mtrr model. Moreover, it is unclear whether the epigenome is able to stabilize in the generations following an Mtrrgt mutant. In an environmental model of transgenerational epigenetic inheritance (Anway et al., 2005), DNA methylation defects persisted longer than the phenotype (Stouder and Paoloni-Giacobino, 2010). Therefore, once an epigenetic defect is generated, it may never completely revert back to the ancestral state. This concept has important evolutionary implications.
It is unlikely that the transgenerational effects observed here are due to a cytoplasmically-inherited factor, such as a virus, prion or mycoplasma present in the ES cells used to generate the Mtrrgt mouse line. This is because we have preliminary data from two other independently-derived Mtrr mutant mouse lines showing similar developmental effects as Mtrrgt mice at E10.5 including developmental delay, neural tube and heart defects in their wildtype grandprogeny (DJ, X. Zhao, JC and RG, unpublished data). Other mechanisms of transgenerational epigenetic effects include the inheritance of small non-coding RNAs as shown in C. elegans (Grishok et al., 2000; Vastenhouw et al., 2006). The Mtrrgt mouse model will be extremely useful for future studies on the mechanisms of transgenerational inheritance including the inheritance of epimutations and small non-coding RNA.
Our finding of a grandparental but not a direct maternal effect of abnormal folate metabolism appears to differ from another study that used an independently derived Mtrrgt mouse line (Deng et al., 2008). They attributed small embryo size and a ventricular septal defect in embryos at E14.5 to a maternal effect. We ruled out a direct maternal effect at least at E10.5 when we systematically controlled for the grandparental genotypes, since conceptuses from Mtrr+/+, Mtrr+/gt and Mtrrgt/gt mothers were similarly affected. The difference may be due to yet-to-be explained genetic or metabolic differences or to different stages of analysis. Gross assessment of our conceptuses at E14.5 showed few overt embryonic abnormalities suggesting that the severely affected conceptuses die between E10.5 and E14.5 (NP and EW, unpublished data).
Overall, our studies have important consequences for understanding and treating congenital anomalies in humans. First, hypomorphic mutations in the MTRR gene associated with reduced expression may lead to congenital abnormalities even with normal dietary folate. Second, the transgenerational effect of Mtrr deficiency in mice suggests that the full beneficial impact of folate fortification in humans may take more than one generation to observe and that grandparental polymorphisms in folate metabolism pose a risk.
EXPERIMENTAL PROCEDURES
Mice and dissections
ES cells with Mtrrgt allele (XG334) were purchased from BayGenomics (http://baygenomics.ucsf.edu/) and injected into C57Bl/6 blastocysts using standard procedures at BayGenomics. After confirmation of germline transmission of Mtrrgt allele, the progeny were backcrossed to C57Bl/6 mice (http://jaxmice.jax.org) ≥8 generations to become generation I. Mice in the Mtrr+/gt intercross and maternal effect studies (Figures 2A, S1 and Table S1) were backcrossed only 6 generations. Noon of the day the vaginal plug was detected was considered E0.5. Embryonic somite staging was determined according to e-Mouse Atlas Project (http://www.emouseatlas.org). Conceptuses were scored for phenotypes, photographed, weighed and snap frozen for storage. All experiments were performed in accordance with the Canadian Council on Animal Care and the University of Calgary Committee on Animal Care (Protocol No. M06109) and UK Government Home Office licensing procedures.
Diet
Mice were fed a normal breeding diet ad libitem from weaning onwards. For more information see the Extended Experimental Procedures and Table S4.
Genotyping
A three-primer PCR reaction was used to detect the presence or absence of the gene-trap in the Mtrr allele. Two primers, a (5’-GAGATTGGGTCCCTCTTCCAC) and b (5’-GCTGCGCTTCTGAATCCACAG), flanked the gene-trap insertion site and detected the wildtype band (272 bp) (Figure 1B). Primer a together with primer c (5’-CGACTTCCGGAGCGGATCTC), which annealed to the gene-trap, detected the mutant band (383 bp). PCR parameters: 30 seconds at 95ºC, 1 minute at 62ºC and 1 minute at 72ºC for 30 cycles. DNA samples were obtained from ear or yolk sac tissue.
DNA and RNA extraction
For Mtrr transcript analysis, total RNA extraction from adult organs was completed using TRIzol reagent according to manufacturer’s instructions (Invitrogen). For transcript analysis of imprinted genes, whole placentas and embryos at E10.5 were homogenized using lysing matrix D beads (MP Biomedicals). Total RNA extraction was completed using the GenElute™ Mammalian Total RNA Miniprep Kit (Sigma) or AllPrep DNA/RNA Mini Kit (Qiagen). All extracts were treated with DNAse I (Fermentas).
qRT-PCR
Reverse-transcription reactions were performed with the RevertAid™ H Minus First Strand cDNA Synthesis Kit (Fermentas) by using 2–5 µg of RNA in a 20-µL reaction according to manufacturer’s instructions. PCR amplification was conducted using MESA SYBR® Green qPCR MasterMix Plus (Eurogentec) on an Applied Biosystems 7500 system or a Roche LightCycler® 480 System. Transcript levels were normalized to Actb and/or Hprt RNA levels and the fold change was quantified using the standard curve method. cDNA levels in C57Bl/6 placentas were normalized to 1. Experiments were conducted in triplicate with at least three biological replicates. For primer sequences, refer to Table S5.
Total homocysteine concentrations
Total homocysteine in plasma was measured by the Biochemical Genetics Unit, Dept of Clinical Biochemistry, Cambridge University Hospitals NHS Foundation Trust using underivatized LC-MS/MS (Waters Acquity LC system and Quattro Premiere mass spectrometer) operated in electrospray ionization positive mode. A deuterated internal standard, d8-homocystine, was added to the plasma, followed by reduction of disulfides with dithiothreitol (Magera et al., 1999; Weaving et al., 2006). Calibrators were prepared by spiking normal plasma with a range of known homocysteine concentrations. Fifty µL of d8-homocystine was added to 50 µL of plasma, which when reduced gave twice as much homocysteine. The mixture was reduced with dithiothreitol and proteins precipitated with acetonitrile and 0.1% formic acid. The samples were centrifuged, transferred to 96-well plates and the supernatant was injected into the tandem mass spectrometer via a cyano LC column (Supelcosil LC-CN). Multiple reaction monitoring (MRM) was used to quantify the total homocysteine against a standard curve. Quality control samples were run at two levels in every batch, which had designed acceptance limits before the results were accepted.
DNA methylation analysis
Tissue was snap frozen immediately after phenotypic scoring and stored at −80°C. DNA extraction from all samples occurred simultaneously to reduce sample variability. Global DNA methylation was measured using the SuperSense™ Methylated DNA Quantification Kit (Epigentek) or the MethylFlash Methylated DNA Quantification Kit (Fluorometric) (Epigentek) according to manufacturer’s instructions. Briefly, genomic DNA extracted from tissue was exposed to a fluorescently-labeled antibody against 5-methylcytosine (5mC) and the fluorescence intensity was quantified with a FLUOstar OPTIMA fluorescence plate reader (BMG LABTECH) at 540EX/590EMnm. Global 5mC levels were calculated as a percentage of total cytosine in DNA and normalized to 1. 4–8 biological replicates were used per category tested.
To measure relative CpG methylation at imprinted DMRs, genomic DNA was bisulphite treated using an Imprint® DNA Modification Kit (Sigma). 50 ng of bisulphite-converted DNA was used as a template for PCR together with 0.2 µM of each biotinylated primer and 0.25 units of HotStarTaq DNA Polymerase (Qiagen) or TrueStart* Taq DNA Polymerase (Fermentas). Refer to Table S5 for primer sequences. PCR parameters: 30 seconds at 95°C, 30 seconds at the annealing temperature (Table S5) and 45 seconds at 72°C for 40 cycles. PCR products were purified using Streptavidin Sepharose™ High Performance columns (GE Healthcare) and hybridized to the sequencing primer using a PyroMark® Q96 Vacuum Prep Workstation (Qiagen). Further sequencing was performed using the PyroMark® Gold Reagents kit (Qiagen) on a PyroMark MD Pyrosequencing System (Qiagen). The mean CpG methylation was calculated using at least four biological replicates and two technical replicates.
Embryo transfers
Using M2 media (M7167, Sigma), embryos were flushed at E3.25 from uteri of Mtrr+/+ females (derived from one Mtrr+/gt parent) mated with C57Bl/6 males. No hormones were used. The embryos were cultured in KSOM (MR-121-D, Millipore) micro-drops covered with mineral oil (ES-005-C, Millipore) at 37°C for no more than 30 minutes before transferring them into uteri of E2.5 pseudopregnant [C57Bl/6 × DBA/2] F1 hybrid (B6D2F1) recipient females (http://www.criver.com). C57Bl/6 embryos were transferred as controls. B6D2F1 females, which have 50% genetic similarity to C57Bl/6 mice, were used since C57Bl/6 mice are notoriously poor recipients. Donor and recipient females were 7–10 weeks old. Embryos were transferred regardless of appearance and litters were never pooled. Transferred litters were dissected at E10.5, the timing of which corresponded to the staging of the recipient female (Ueda et al., 2003).
Statistical analysis
Statistical analyses were performed using GraphPad Prism® 6 software. Parametric data (e.g., litter sizes, mRNA expression, homocysteine concentrations, embryo lengths, placenta weights and global DNA methylation levels) were analyzed using independent unpaired t-tests. Global DNA methylation percentages were normalized by arc sine transformation prior to t-tests. Nonparametric data (e.g., phenotypic frequencies and bisulfite pyrosequencing data) was analyzed using a Mann-Whitney or Kruskal-Wallis test. P values <0.05 were considered significant.
Equipment and Software
A Zeiss SteREO Discovery.V8 microscope with a AxioCam MRc5 camera and AxioVision 4.7.2 software (Carl Zeiss) imaging software program were used to obtain embryo images and embryo crown-rump lengths. PCR and pyrosequencing primers were designed using Primer3 v0.4.0 software (Rozen and Skaletsky, 2000) and Pyromark Assay Design 2.0 software (Qiagen), respectively. Graphs were generated using GraphPad Prism® 6 software. Figures were constructed using Canvas X software.
Supplementary Material
RESEARCH HIGHLIGHTS.
Mutation in the Mtrr gene causes abnormal folate metabolism in mice
Mtrr genotype of maternal grandparent affects development of wildtype grandprogeny
Placentas of wildtype grandprogeny display widespread epigenetic instability
Mtrr mutation impacts both the in utero environment and epigenetic inheritance
ACKNOWLEDGEMENTS
We thank Cameron Fielding, Xiang Zhao, Frances Snider and Nuala Daw for their technical assistance. This work was supported by grants from the CIHR (to RG, JC), NIH-NHLBI (to RG) and Centre for Trophoblast Research (CTR) (to EW). NP was funded by a CTR graduate studentship. AFS was supported by FP7 grants from EpiHealth and Epigenesys, MRC and Wellcome Trust. EW was funded by the CIHR Training Program in Genetics, Child Development & Health and fellowships from the CTR, NSERC and CIHR.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Agell N, Chiva M, Mezquita C. Changes in nuclear content of protein conjugate histone H2A–ubiquitin during rooster spermatogenesis. FEBS Lett. 1983;155:209–212. doi: 10.1016/0014-5793(82)80604-2. [DOI] [PubMed] [Google Scholar]
- Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science. 2005;308:1466–1469. doi: 10.1126/science.1108190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro R, Rivera I, Ravasco P, Camilo ME, Jakobs C, Blom HJ, de Almeida IT. 5,10-methylenetetrahydrofolate reductase (MTHFR) 677CT and 1298AC mutations are associated with DNA hypomethylation. J Med Genet. 2004;41:454–458. doi: 10.1136/jmg.2003.017244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan D, Cushnie DW, Neaga OR, Lawrance AK, Rozen R, Trasler JM. Strain-specific defects in testicular development and sperm epigenetic patterns in 5,10-methylenetetrahydrofolate reductase-deficient mice. Endocrinology. 2010;151:3363–3373. doi: 10.1210/en.2009-1340. [DOI] [PubMed] [Google Scholar]
- Chen Z, Karaplis AC, Ackerman SL, Pogribny IP, Melnyk S, Lussier-Cacan S, Chen MF, Pai A, John SW, Smith RS, et al. Mice deficient in methylenetetrahydrofolate reductase exhibit hyperhomocysteinemia and decreased methylation capacity, with neuropathology and aortic lipid deposition. Hum Mol Genet. 2001;10:433–443. doi: 10.1093/hmg/10.5.433. [DOI] [PubMed] [Google Scholar]
- Coan PM, Burton GJ, Ferguson-Smith AC. Imprinted genes in the placenta--a review. Placenta. 2005;26(Suppl A):S10–S20. doi: 10.1016/j.placenta.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Deng L, Elmore CL, Lawrance AK, Matthews RG, Rozen R. Methionine synthase reductase deficiency results in adverse reproductive outcomes and congenital heart defects in mice. Mol Genet Metab. 2008;94:336–342. doi: 10.1016/j.ymgme.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore CL, Wu X, Leclerc D, Watson ED, Bottiglieri T, Krupenko NI, Krupenko SA, Cross JC, Rozen R, Gravel RA, et al. Metabolic derangement of methionine and folate metabolism in mice deficient in methionine synthase reductase. Mol Genet Metab. 2007;91:85–97. doi: 10.1016/j.ymgme.2007.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friso S, Choi SW, Girelli D, Mason JB, Dolnikowski GG, Bagley PJ, Olivieri O, Jacques PF, Rosenberg IH, Corrocher R, et al. A common mutation in the 5,10-methylenetetrahydrofolate reductase gene affects genomic DNA methylation through an interaction with folate status. Proc Natl Acad Sci U S A. 2002;99:5606–5611. doi: 10.1073/pnas.062066299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelineau-van Waes J, Heller S, Bauer LK, Wilberding J, Maddox JR, Aleman F, Rosenquist TH, Finnell RH. Embryonic development in the reduced folate carrier knockout mouse is modulated by maternal folate supplementation. Birth Defects Res A Clin Mol Teratol. 2008;82:494–507. doi: 10.1002/bdra.20453. [DOI] [PubMed] [Google Scholar]
- Ghandour H, Chen Z, Selhub J, Rozen R. Mice deficient in methylenetetrahydrofolate reductase exhibit tissue-specific distribution of folates. J Nutr. 2004;134:2975–2978. doi: 10.1093/jn/134.11.2975. [DOI] [PubMed] [Google Scholar]
- Ghandour H, Lin BF, Choi SW, Mason JB, Selhub J. Folate status and age affect the accumulation of L-isoaspartyl residues in rat liver proteins. J Nutr. 2002;132:1357–1360. doi: 10.1093/jn/132.6.1357. [DOI] [PubMed] [Google Scholar]
- Grishok A, Tabara H, Mello CC. Genetic requirements for inheritance of RNAi in C. elegans. Science. 2000;287:2494–2497. doi: 10.1126/science.287.5462.2494. [DOI] [PubMed] [Google Scholar]
- Hague WM. Homocysteine and pregnancy. Best Pract Res Clin Obstet Gynaecol. 2003;17:459–469. doi: 10.1016/s1521-6934(03)00009-9. [DOI] [PubMed] [Google Scholar]
- Hart DJ, Finglas PM, Wolfe CA, Mellon F, Wright AJ, Southon S. Determination of 5-methyltetrahydrofolate (13C–labeled and unlabeled) in human plasma and urine by combined liquid chromatography mass spectrometry. Anal Biochem. 2002;305:206–213. doi: 10.1006/abio.2002.5662. [DOI] [PubMed] [Google Scholar]
- Hata K, Okano M, Lei H, Li E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development. 2002;129:1983–1993. doi: 10.1242/dev.129.8.1983. [DOI] [PubMed] [Google Scholar]
- Hemberger M. Genetic-epigenetic intersection in trophoblast differentiation: implications for extraembryonic tissue function. Epigenetics. 2010;5:24–29. doi: 10.4161/epi.5.1.10589. [DOI] [PubMed] [Google Scholar]
- Jacob RA, Gretz DM, Taylor PC, James SJ, Pogribny IP, Miller BJ, Henning SM, Swendseid ME. Moderate folate depletion increases plasma homocysteine and decreases lymphocyte DNA methylation in postmenopausal women. J Nutr. 1998;128:1204–1212. doi: 10.1093/jn/128.7.1204. [DOI] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Magera MJ, Lacey JM, Casetta B, Rinaldo P. Method for the determination of total homocysteine in plasma and urine by stable isotope dilution and electrospray tandem mass spectrometry. Clin Chem. 1999;45:1517–1522. [PubMed] [Google Scholar]
- Messerschmidt DM, de Vries W, Ito M, Solter D, Ferguson-Smith A, Knowles BB. Trim28 is required for epigenetic stability during mouse oocyte to embryo transition. Science. 2012;335:1499–1502. doi: 10.1126/science.1216154. [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Pickell L, Li D, Brown K, Mikael LG, Wang XL, Wu Q, Luo L, Jerome-Majewska L, Rozen R. Methylenetetrahydrofolate reductase deficiency and low dietary folate increase embryonic delay and placental abnormalities in mice. Birth Defects Res A Clin Mol Teratol. 2009;85:531–541. doi: 10.1002/bdra.20575. [DOI] [PubMed] [Google Scholar]
- Piedrahita JA, Oetama B, Bennett GD, van Waes J, Kamen BA, Richardson J, Lacey SW, Anderson RG, Finnell RH. Mice lacking the folic acid-binding protein Folbp1 are defective in early embryonic development. Nat Genet. 1999;23:228–232. doi: 10.1038/13861. [DOI] [PubMed] [Google Scholar]
- Rampersaud E, Melvin EC, Siegel D, Mehltretter L, Dickerson ME, George TM, Enterline D, Nye JS, Speer MC. Updated investigations of the role of methylenetetrahydrofolate reductase in human neural tube defects. Clin Genet. 2003;63:210–214. doi: 10.1034/j.1399-0004.2003.00043.x. [DOI] [PubMed] [Google Scholar]
- Rassoulzadegan M, Grandjean V, Gounon P, Vincent S, Gillot I, Cuzin F. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature. 2006;441:469–474. doi: 10.1038/nature04674. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Saitou M, Kagiwada S, Kurimoto K. Epigenetic reprogramming in mouse pre-implantation development and primordial germ cells. Development. 2012;139:15–31. doi: 10.1242/dev.050849. [DOI] [PubMed] [Google Scholar]
- Shane B, Stokstad EL. Vitamin B12-folate interrelationships. Annu Rev Nutr. 1985;5:115–141. doi: 10.1146/annurev.nu.05.070185.000555. [DOI] [PubMed] [Google Scholar]
- Sinclair KD, Allegrucci C, Singh R, Gardner DS, Sebastian S, Bispham J, Thurston A, Huntley JF, Rees WD, Maloney CA, et al. DNA methylation, insulin resistance, and blood pressure in offspring determined by maternal periconceptional B vitamin and methionine status. Proc Natl Acad Sci U S A. 2007;104:19351–19356. doi: 10.1073/pnas.0707258104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DE, Kok RM, Teerlink T, Jakobs C, Smulders YM. Quantitative determination of erythrocyte folate vitamer distribution by liquid chromatography-tandem mass spectrometry. Clin Chem Lab Med. 2006;44:450–459. doi: 10.1515/CCLM.2006.085. [DOI] [PubMed] [Google Scholar]
- Stouder C, Paoloni-Giacobino A. Transgenerational effects of the endocrine disruptor vinclozolin on the methylation pattern of imprinted genes in the mouse sperm. Reproduction. 2010;139:373–379. doi: 10.1530/REP-09-0340. [DOI] [PubMed] [Google Scholar]
- Swanson DA, Liu ML, Baker PJ, Garrett L, Stitzel M, Wu J, Harris M, Banerjee R, Shane B, Brody LC. Targeted disruption of the methionine synthase gene in mice. Mol Cell Biol. 2001;21:1058–1065. doi: 10.1128/MCB.21.4.1058-1065.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunster SJ, Jensen AB, John RM. Imprinted genes in mouse placental development and the regulation of fetal energy stores. Reproduction. 2013;145:R117–R137. doi: 10.1530/REP-12-0511. [DOI] [PubMed] [Google Scholar]
- Ueda O, Yorozu K, Kamada N, Jishage K, Kawase Y, Toyoda Y, Suzuki H. Possible expansion of “Window of Implantation” in pseudopregnant mice: time of implantation of embryos at different stages of development transferred into the same recipient. Biol Reprod. 2003;69:1085–1090. doi: 10.1095/biolreprod.103.017608. [DOI] [PubMed] [Google Scholar]
- van der Put NM, Steegers-Theunissen RP, Frosst P, Trijbels FJ, Eskes TK, van den Heuvel LP, Mariman EC, den Heyer M, Rozen R, Blom HJ. Mutated methylenetetrahydrofolate reductase as a risk factor for spina bifida. Lancet. 1995;346:1070–1071. doi: 10.1016/s0140-6736(95)91743-8. [DOI] [PubMed] [Google Scholar]
- Vastenhouw NL, Brunschwig K, Okihara KL, Muller F, Tijsterman M, Plasterk RH. Gene expression: long-term gene silencing by RNAi. Nature. 2006;442:882. doi: 10.1038/442882a. [DOI] [PubMed] [Google Scholar]
- Wainfan E, Moller ML, Maschio FA, Balis ME. Ethionine-induced changes in rat liver transfer RNA methylation. Cancer Res. 1975;35:2830–2835. [PubMed] [Google Scholar]
- Weaving G, Rocks BF, Iversen SA, Titheradge MA. Simultaneous quantitation of homocysteine, cysteine and methionine in plasma and urine by liquid chromatography-tandem mass spectrometry. Ann Clin Biochem. 2006;43:474–480. doi: 10.1258/000456306778904605. [DOI] [PubMed] [Google Scholar]
- Yamada K, Gravel RA, Toraya T, Matthews RG. Human methionine synthase reductase is a molecular chaperone for human methionine synthase. Proc Natl Acad Sci U S A. 2006;103:9476–9481. doi: 10.1073/pnas.0603694103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.