Abstract
Background:
The detection of V600E BRAF mutations has fundamental clinical consequences as the treatment option with BRAF inhibitors such as vemurafenib or dabrafenib yields response rates of ∼48%. Heterogeneity with respect to BRAF mutation in different metastases has been described in single cases. As this has important implications for the determination of BRAF status and treatment of patients, it is essential to acquire more data.
Methods:
A total of 300 tumour samples from 187 melanoma patients were analysed for BRAF mutations by pyrosequencing. Equivocal results were confirmed by capillary sequencing. Clinical data with respect to melanoma type, tumour site and survival were summarised for 53 patients with multiple analysed tumour samples (2–13 per patient).
Results:
BRAF mutations were found in 84 patients (44.9%) and 144 tumour samples (48%) with BRAF mutations in 45.5% of primary tumours and 51.3% of metastases, respectively. In 10 out of 53 patients (18.9%) where multiple samples were analysed results were discordant with respect to mutation findings with wild-type and mutated tumours in the same patient. Mutations did not appear more frequently over the course of disease nor was its occurrence associated with a specific localisation of metastases.
Conclusion:
As heterogeneity with respect to BRAF mutation status is detected in melanoma patients, subsequent testing of initially wild-type patients can yield different results and thus make BRAF inhibitor therapy accessible. The role of heterogeneity in testing and for clinical response to therapy with a BRAF inhibitor needs to be further investigated.
Keywords: detection method, BRAF mutation, melanoma, pyrosequencing, immunohistochemistry, repeated testing
Discovery of the activating (oncogenic) V600E BRAF mutation that is present in ∼41–50% of melanomas (Houben et al, 2004; Curtin et al, 2005) has paved the way to targeted therapy with BRAF inhibitors. The first BRAF inhibitor to gain approval, vemurafenib (Zelboraf), has demonstrated improvement of survival in patients with metastatic melanoma who present with the V600E mutation (Chapman et al, 2011) and another BRAF inhibitor, dabrafenib, has also shown to be effective (Hauschild et al, 2012). Other variant BRAF mutations (V600K) have been described and were shown to be associated with distinct clinicopathological features including differences in age distribution (higher rates in older patients), localisation (higher rates in tumours localised in the head and neck) and a worse distant metastasis-free survival (Menzies et al, 2012). However, this mutation type might be missed with some methods (Anderson et al, 2012; Heinzerling et al, 2013) and consequently these patients would be excluded from clinical trials with BRAF inhibitors or regular treatment with vemurafenib or dabrafenib (Flaherty et al, 2010). In previous studies ∼6–30% of all BRAF mutations represented variant mutations distinct from the more common V600E genotype (Rubinstein et al, 2010; Beadling et al, 2011; Long et al, 2011; Lovly et al, 2012). In fact, among BRAF V600 mutations, 79%, 12%, 5%, and 4% were V600E, V600K, V600R, and V600M, respectively (Lovly et al, 2012). Interestingly, in vitro and in vivo data indicate that BRAF inhibitors could be similarly effective in these patients (Rubinstein et al, 2010; Chapman et al, 2011).
As treatment of BRAF mutation-positive patients with BRAF inhibitors has a profound impact on disease and overall survival, with some evidence for tumour regression in up to 90% of patients (Long et al, 2011), the correct identification of the mutation status is crucial. Although there seems to exist a certain consistency in BRAF mutation status of multiple metastases within the same patient, variation of mutation findings between distant metastases, lymph node metastases or the primary tumours has been observed with, for example, higher mutation rates of 41–55% in metastases compared with 33–47% in primary tumours (Long et al, 2011). Interestingly, there is also variation dependent on the site of the tumour with mutations being detected more frequently in skin metastases compared with visceral lesions (Colombino et al, 2012). Branched evolution in metastatic disease has been shown to create a remarkable genetic heterogeneity among different metastases of one patient (Gerlinger et al, 2012; Yancovitz et al, 2012) and within single metastases (Lin et al, 2011).
This study investigates the frequency, type and intraindividual concordance of rare V600 BRAF mutations in primary tumours and different metastases of melanoma patients, compares different detection methods, and correlates the BRAF genotype with clinical characteristics and survival.
Materials and Methods
Patients
A total of 300 tumour samples from 187 consecutive patients with metastatic stage IV melanoma consulting the University Hospital Erlangen were analysed within this study excluding patients with uvea melanoma. For patients with multiple tumour samples data on tumour type, treatment and course of disease were gathered from patient files. Primary tumour tissue was requested for all patients with multiple samples. However, for some of the patients tissue was no longer available. Survival data were obtained at the tumour registry Nürnberg–Erlangen if not accessible from the clinic. Patients had a median age of 60 years, 39% being female.
The investigations were approved by the local ethics committee of the University Erlangen.
DNA extraction and mutation analysis
Genomic DNA was extracted from 2–3.5 μm sections of formalin-fixed paraffin-embedded tissue blocks. The relevant tumour area was marked by a pathologist (AH, AA) and in some cases by the dermatopathologist. After deparaffinisation, DNA was prepared as described recently (Heinzerling et al, 2013) using the NucleoSpin Tissue kit according to the manufacturer's instructions (Daniels et al, 2011).
DNA was amplified using the multiplex PCR-kit according to the instructions of the manufacturer (Qiagen, Hilden, Germany) using the following primers: forward: 5′-TGA AGA CCT CAC AGT AAA AAT AGG-3′, and reverse: 5′-Biotin AAA ATG GAT CCA GAC AAC TGT TC-3′. The cycling was performed as follows: a single cycle of denaturation at 95 °C for 15 min, 42 cycles of 95 °C for 20 s, 61 °C for 30 s, and 72 °C for 5 min, and a final 5 min extension at 72 °C. For pyrosequencing (PyroMark Q24, Qiagen) single-stranded DNA was prepared from 40 μl biotinylated PCR product with streptavidin-coated sepharose and 0.5 μM of the sequencing primer: 5′-GGTGATTTTGGTCTAGC-3′ using the PSQ Vacuum Prep Tool (Qiagen). The set up for the pyrosequencing assay was selected with the following sequence in ‘sequence to analyse': TACAGA/TGAAA. The underlined A/T describes the hot spot mutation site at codon 600 and primarily describes the V600E with a substitution of GTG (valine) by GAG (glutamic acid). The following dispensation order was used: GTACACGATG. The underlined ‘C' was included as an internal control.
Statistical analyses
For analysing differences of survival times in the different groups the Mann–Whitney U-test was applied.
Results
A total of 300 melanoma tissue samples from 187 patients were analysed. Tissue samples included primary tumours (n=44), skin (n=137), lymph node (n=20), distant metastases (liver, lung, gastric, pancreas, brain, intestinal and soft tissue; n=37) and tumour tissue of unknown origin (n=62).
Variation in the mutation status detectable in a subset of patients
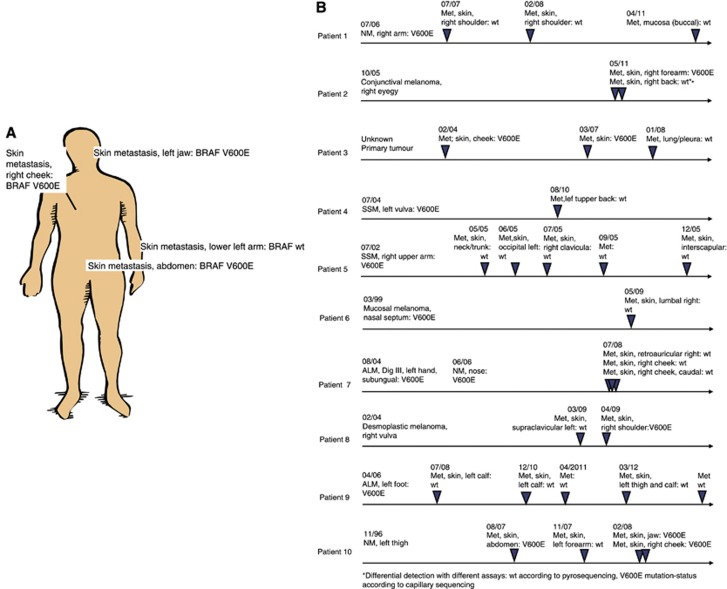
In 53 out of 187 patients multiple tumour samples (2–13 biopsies) were available for analysis. Tumour samples were obtained from primary tumours and skin, lymph node, soft tissue, lung, visceral organ and brain metastases. In a total of 10 out of these 53 patients (18.9%) both wild-type and mutation-positive metastases were found. There was one patient (patient #2; Table 1C), who showed different results in one tumour probe depending on the assay (wild type in pyrosequencing, V600E mutation-positive in capillary sequencing) and was regarded as wild-type for further analyses. Two patients showed both, tumours with V600E as well as rare BRAF mutations (patient #1 and #7; Table 1B). In 12 patients with multiple tumour probes only rare BRAF mutations were seen. From the remaining 41 patients 31 (75.6%) were concordant with respect to BRAF mutation status (Table 1A, B), whereas 10 patients showed differences in mutation status in different tumour samples, that is, with some tumours that were BRAF wild-type and some tumours that showed the BRAF mutation (Table 1C). In these discordant patients no clear association of mutation status with duration of disease was seen. Indeed when analysing the time of occurrence of mutation in metastases of patients with multiple metastases there was no accumulation of mutations over time (Figure 1B). For example, one patient (patient #1; Table 1C) showed a mutated primary tumour but multiple subsequent metastases revealed a wild-type genotype.
Table 1A. Patients with concordant BRAF wt results in analyses of multiple tumour samples and clinical characteristics.
| Patient-ID | Gender | Age | Primary melanoma (ALM, NM, SSM) | Localisation of primary melanoma | Region of primary melanoma | BRAF status | Metastases examined | Localisation of metastases | Time of occurrence | Survival in stage IV (months) | Number of probes analysed |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
M |
55 |
Desmoplastic melanoma |
Right cheek |
Head and neck |
No data |
|
|
08/2007 |
30 |
2 |
| |
|
|
|
|
|
wt |
No data |
No data |
No data |
|
|
| |
|
|
|
|
|
wt |
Liver |
Liver parts ii, iii, iv |
08/2010 |
|
|
| 2 |
M |
81 |
SSM |
Right heel |
Extremities |
wt |
|
|
06/2004 |
20 |
3 |
| |
|
|
|
|
|
wt |
Skin |
Right lower leg, medial |
05/2005 |
|
|
| |
|
|
|
|
|
wt |
Skin |
Right heel |
12/2008 |
|
|
| 3 |
M |
54 |
Unknown primary |
— |
— |
— |
|
|
|
87 |
2 |
| |
|
|
|
|
|
wt |
Skin |
Axilla |
01/2007 |
|
|
| |
|
|
|
|
|
wt |
Lymph node |
Cervical lymph node |
06/2010 |
|
|
| 4 |
F |
>82 |
ALM |
Plantar of the left foot |
Extremities |
no data |
|
|
11/2007 |
>11 |
2 |
| |
|
|
|
|
|
wt |
Lymph node |
Inguinal, left |
04/2009 |
|
|
| |
|
|
|
|
|
wt |
No data |
No data |
02/2013 |
|
|
| 5 |
M |
43 |
NM |
Right thigh |
Extremities |
no data |
|
|
12/2007 |
10 |
2 |
| |
|
|
|
|
|
wt |
Skin |
Right thigh |
09/2008 |
|
|
| |
|
|
|
|
|
wt |
Skin |
Right thigh |
07/2009 |
|
|
| 6 |
M |
70 |
Ulcerated ALM |
Right dorsum of the foot, interdigital |
Extremities |
wt |
|
|
03/2007 |
5 |
4 |
| |
|
|
|
|
|
wt |
Skin |
Left thigh |
01/2009 |
|
|
| |
|
|
|
|
|
wt |
Skin |
Right lower leg, lateral |
02/2009 |
|
|
| |
|
|
|
|
|
wt |
No data |
No data |
no data |
|
|
| 7 |
F |
70 |
Nodular SSM |
Left upper arm |
Extremities |
wt |
|
|
07/2008 |
6 |
2 |
| |
|
|
|
|
|
wt |
Skin |
Abdomen |
07/2009 |
|
|
| 8 |
M |
29 |
Unknown primary |
— |
— |
— |
|
|
— |
51 |
2 |
| |
|
|
|
|
|
wt |
No data |
No data |
05/2011 |
|
|
| |
|
|
|
|
|
wt |
Skin |
Left upper arm, lateral |
10/2011 |
|
|
| 9 |
M |
48 |
Unknown primary |
— |
— |
— |
|
|
— |
59 |
5 |
| |
|
|
|
|
|
wt |
Lymph node |
Right axilla or cervical, right |
11/2003 |
|
|
| |
|
|
|
|
|
wt |
No data |
No data |
11/2003 |
|
|
| |
|
|
|
|
|
wt |
Skin |
Sternal |
08/2005 |
|
|
| |
|
|
|
|
|
wt |
Skin and soft tissue |
Right breast |
12/2005 |
|
|
| |
|
|
|
|
|
wt |
Skin |
Pectoral, right |
05/2007 |
|
|
| 10 |
F |
>46 |
Ulcerated NM |
Bottom lip |
Head and neck |
No data |
|
|
05/2011 |
>12 |
2 |
| |
|
|
|
|
|
wt |
No data |
No data |
no data |
|
|
| |
|
|
|
|
|
wt |
Lung |
Left inferior lobe |
06/2012 |
|
|
| 11 |
M |
60 |
Mucosal melanoma |
Penis |
Mucosa |
wt |
|
|
07/2009 |
15 |
2 |
| |
|
|
|
|
|
wt |
Skin |
Right groin |
04/2010 |
|
|
| 12 |
M |
53 |
NM |
Back |
Trunk |
wt |
|
|
02/2006 |
8 |
2 |
| |
|
|
|
|
|
wt |
Skin |
Left breast |
02/2010 |
|
|
| 13 |
F |
69 |
Unknown primary |
— |
— |
— |
|
|
— |
55 |
4 |
| |
|
|
|
|
|
wt |
Skin |
Back, left |
10/2006 |
|
|
| |
|
|
|
|
|
wt |
Skin |
Left upper arm |
12/2006 |
|
|
| |
|
|
|
|
|
wt |
Skin |
Right scapula |
01/2007 |
|
|
| |
|
|
|
|
|
wt |
skin |
Left upper arm, dorsal |
01/2007 |
|
|
| 14 |
M |
40 |
Nodular SSM |
Left shoulder |
Trunk |
no data |
|
|
07/2002 |
12 |
2 |
| |
|
|
|
|
|
wt |
Skin |
Left shoulder |
08/2008 |
|
|
| |
|
|
|
|
|
wt |
Skin |
Trunk, left, proximal |
12/2008 |
|
|
| 15 |
M |
>85 |
Ulcerated NM |
Left calf |
Extremities |
no data |
|
|
05/2002 |
>72 |
2 |
| |
|
|
|
|
|
wt |
Soft tissue |
Abdomen |
06/2007 |
|
|
| |
|
|
|
|
|
wt |
Soft tissue |
Abdomen |
06/2012 |
|
|
| 16 |
M |
68 |
Naevoid malignant melanoma |
Back, left lumbal |
Trunk |
no data |
|
|
06/2001 |
59 |
2 |
| |
|
|
SSM |
Left elbow |
Extremities |
no data |
|
|
04/2004 |
|
|
| |
|
|
|
|
|
wt |
Skin |
Back |
08/2005 |
|
|
| |
|
|
|
|
|
wt |
Lymph node |
Right axilla |
11/2007 |
|
|
| 17 |
M |
61 |
Mucosal melanoma |
Oral cavity |
Mucosa |
No data |
|
|
09/2006 |
3 |
3 |
| |
|
|
|
|
|
wt |
Skin |
Cervical, left |
07/2007 |
|
|
| |
|
|
|
|
|
wt |
Skin |
Cappilitum |
07/2007 |
|
|
| |
|
|
|
|
|
wt |
Skin |
Cervical, left |
09/2009 |
|
|
| 18 |
M |
64 |
Unknown primary |
— |
— |
— |
|
|
— |
52 |
4 |
| |
|
|
|
|
|
wt |
Skin |
Left axilla |
05/2008 |
|
|
| |
|
|
|
|
|
wt |
Skin |
Right thorax |
04/2009 |
|
|
| |
|
|
|
|
|
wt |
Lymph node |
Right axilla |
02/2010 |
|
|
| |
|
|
|
|
|
wt |
Lymph node |
No data |
01/2011 |
|
|
| 19 |
F |
53 |
NM |
Above right popliteal fossa |
Extremities |
No data |
|
|
03/2006 |
5 |
2 |
| |
|
|
|
|
|
wt |
Skin |
Right thigh, medial |
03/2007 |
|
|
| |
|
|
|
|
|
wt |
Skin |
Right thigh, medial |
03/2007 |
|
|
| 20 |
M |
43 |
Partly NM, partly SSM |
Right heel |
Extremities |
No data |
|
|
|
12 |
2 |
| |
|
|
|
|
|
wt |
Skin |
Right thigh |
11/2010 |
|
|
| wt | Skin | Thigh | 05/2011 |
Abbreviations: ALM=acrolentiginous melanoma; F=female; M=male; NM=nodular melanoma; SSM=superficial spreading melanoma; wt=wild-type.
Table 1B. Patients with concordant V600-mutated results in analyses of multiple tumour samples and clinical characteristics.
| Pat-ID | Gender | Age | Primary melanoma (SSM, NM) | Localisation of primary melanoma | Region of primary melanoma | BRAF status | Metastases examined | Localisation of metastases | Time of occurrence | Survival in stage IV (months) | Number of probes analysed |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1a |
F |
50 |
SSM |
Left shoulder |
Trunk |
V600K |
|
|
11/1992 |
101 |
4 |
| |
|
|
SSM |
Retroauricular, right |
Head and neck |
V600K |
|
|
04/1999 |
|
|
| |
|
|
|
|
|
V600E |
Skin |
Supraumbilical |
11/2009 |
|
|
| |
|
|
|
|
|
V600E |
Skin |
No data |
12/2009 |
|
|
| 2 |
M |
51 |
SSM |
Thoracal, left |
Trunk |
No data |
|
|
07/2002 |
22 |
5 |
| |
|
|
|
|
|
V600E |
Skin |
Left axilla |
07/2005 |
|
|
| |
|
|
|
|
|
V600E |
Skin |
Periumbilical, left |
06/2006 |
|
|
| |
|
|
|
|
|
V600E |
Skin |
Left lower abdomen |
11/2006 |
|
|
| |
|
|
|
|
|
V600E |
Skin |
Inguinal, right |
11/2006 |
|
|
| |
|
|
|
|
|
V600E |
Skin |
cervical |
02/2007 |
|
|
| 3 |
M |
26 |
NM |
Perineal |
Trunk |
V600E |
|
|
08/2006 |
12 |
3 |
| |
|
|
|
|
|
V600E |
Skin |
Capillitum, centre |
01/2008 |
|
|
| |
|
|
|
|
|
V600E |
Skin |
Capillitum |
02/2008 |
|
|
| 4 |
F |
70 |
Ulcerated NM |
Right upper arm |
Extremities |
V600E |
|
|
03/2007 |
11 |
3 |
| |
|
|
|
|
|
V600E |
Skin |
Axillary line |
10/2007 |
|
|
| |
|
|
|
|
|
V600E |
Skin |
back |
02/2008 |
|
|
| 5 |
F |
86 |
MM |
Right thigh |
Extremities |
No data |
|
|
07/1965 |
12 |
2 |
| |
|
|
|
|
|
V600E |
Skin |
Right thigh |
01/2006 |
|
|
| |
|
|
|
|
|
V600E |
Skin |
Lumal |
01/2007 |
|
|
| 6 |
M |
71 |
Polypoid melanoma |
Paravertebral, right |
Trunk |
V600E |
|
|
11/2004 |
10 |
3 |
| |
|
|
|
|
|
V600E |
Skin |
Right abdomen |
04/2008 |
|
|
| |
|
|
|
|
|
V600E |
Skin |
Right breast |
04/2008 |
|
|
| 7a |
F |
31 |
SSM |
Capillitum, parietal, left |
Head and neck |
V600K |
|
|
07/2006 |
14 |
4 |
| |
|
|
|
|
|
V600E |
Skin |
Left axilla |
05/2010 |
|
|
| |
|
|
|
|
|
V600E |
No data |
No data |
06/2010 |
|
|
| |
|
|
|
|
|
V600E |
Skin |
Collar, right |
06/2010 |
|
|
| 8 |
M |
54 |
NM |
Paravertebral, right |
Trunk |
— |
|
|
07/2002 |
18 |
|
| |
|
|
|
|
|
V600E |
Skin |
Right axilla, dorsal |
09/2008 |
|
|
| |
|
|
|
|
|
V600E |
Skin |
Back |
02/2010 |
|
|
| 9 |
M |
62 |
Ulcerated NM |
Right epigastrium |
Trunk |
V600E |
|
|
04/2004 |
14 |
2 |
| |
|
|
|
|
|
V600E |
Skin |
Thorax, right |
04/2007 |
|
|
| 10 |
F |
37 |
Unknown primary |
— |
— |
— |
|
|
— |
27 |
5 |
| |
|
|
|
|
|
V600E |
Skin |
Pectoral, left |
11/2005 |
|
|
| |
|
|
|
|
|
V600E |
Skin |
Left axilla |
12/2005 |
|
|
| |
|
|
|
|
|
V600E |
Skin and intramuscular |
Left cheek |
06/2007 |
|
|
| |
|
|
|
|
|
V600E |
Skin |
Neck |
06/2007 |
|
|
| |
|
|
|
|
|
V600E |
Skin |
Frontal |
06/2007 |
|
|
| 11 |
F |
44 |
Unknown primary |
— |
— |
— |
|
|
— |
6 |
2 |
| |
|
|
|
|
|
V600E |
Skin |
Neck, left |
06/2009 |
|
|
| V600E | Skin | Mons pubis | 07/2009 |
Abbreviations: F=female; M=male; NM=nodular melanoma; SSM=superficial spreading melanoma; wt=wild-type.
These patients showed different mutations (V600E and V600K).
Table 1C. Patients with discordant results with respect to BRAF mutation status in analyses of multiple tumour samples and clinical characteristics.
| Patient-ID | Gender | Age | Primary melanoma (SSM, NM) | Localisation of primary melanoma | Region of primary melanoma | BRAF status | Metastases examined | Localisation of metastasis | Time of occurrence | Survival in stage IV (months) | Number of probes analysed |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
F |
74 |
NM |
Right upper arm, proximal |
Extremities |
V600E |
|
|
07/2006 |
46 |
4 |
| |
|
|
|
|
|
wt |
Skin |
Right shoulder |
07/2007 |
|
|
| |
|
|
|
|
|
wt |
Skin |
Right shoulder |
02/2008 |
|
|
| |
|
|
|
|
|
wt |
Mucosa |
Cheek |
04/2011 |
|
|
| 2 |
M |
58 |
Conjunctival melanoma |
Right eye |
Mucosa |
No data |
|
|
10/2005 |
6 |
2 |
| |
|
|
|
|
|
wta |
Skin |
Back, right |
05/2011 |
|
|
| |
|
|
|
|
|
V600E |
Skin |
Right forearm |
05/2011 |
|
|
| 3 |
M |
>31 |
Unknown primary |
— |
— |
— |
|
|
|
>73 |
3 |
| |
|
|
|
|
|
V600E |
Skin |
Cheek |
02/2004 |
|
|
| |
|
|
|
|
|
V600E |
Skin |
No data |
03/2007 |
|
|
| |
|
|
|
|
|
wt |
|
Lung/pericard/pleura |
01 o. 02/2008 |
|
|
| 4 |
F |
71 |
SSM |
Left vulva |
Trunk |
V600E |
|
|
07/2004 |
31 |
2 |
| |
|
|
|
|
|
wt |
Skin |
Left, upper back |
08/2010 |
|
|
| 5 |
F |
35 |
SSM |
Right upper arm |
Extremities |
V600E |
|
|
07/2002 |
12 |
6 |
| |
|
|
|
|
|
wt |
Skin |
No data |
05/2005 |
|
|
| |
|
|
|
|
|
wt |
Skin |
Occipital, left |
06/2005 |
|
|
| |
|
|
|
|
|
wt |
Skin |
Right clavicula |
07/2005 |
|
|
| |
|
|
|
|
|
wt |
No data |
No data |
09/2005 |
|
|
| |
|
|
|
|
|
wt |
Skin |
Interscapular |
12/2005 |
|
|
| 6 |
F |
54 |
Mucosal melanoma |
Nasal septum |
Mucosa |
No data |
|
|
03/1999 |
40 |
3 |
| |
|
|
|
|
|
wt |
No data |
No data |
12/2010 |
|
|
| |
|
|
|
|
|
V600E |
Relapse of primary tumour |
Frontal sinus |
07/2008 |
|
|
| |
|
|
|
|
|
wt |
Skin |
Lumbal, right |
05/2009 |
|
|
| 7 |
M |
74 |
ALM |
Left middle finger, subungual |
Extremities |
V600E |
|
|
08/2004 |
13 |
5 |
| |
|
|
NM |
Tip of the nose |
Head and neck |
V600E |
|
|
06/2006 |
|
|
| |
|
|
|
|
|
wt |
Skin |
Right cheek |
07/2008 |
|
|
| |
|
|
|
|
|
wt |
Skin |
Retroauricular, right |
07/2008 |
|
|
| |
|
|
|
|
|
wt |
Skin |
Right cheek |
07/2008 |
|
|
| 8 |
F |
76 |
Desmoplatisches MM |
Right vulva |
Trunk |
No data |
|
|
02/2004 |
44 |
2 |
| |
|
|
|
|
|
wt |
Skin |
Supraclavicular, left |
03/2009 |
|
|
| |
|
|
|
|
|
V600E |
Skin |
Right shoulder |
04/2009 |
|
|
| 9 |
M |
75 |
Ulcerated ALM |
Left foot, lateral |
Extremitis |
V600E |
|
|
04/2006 |
42 |
5 |
| |
|
|
|
|
|
wt |
Skin |
Left calf |
07/2008 |
|
|
| |
|
|
|
|
|
wt |
Skin |
Left calf |
12/2010 |
|
|
| |
|
|
|
|
|
wt |
No data |
No data |
04/2011 |
|
|
| |
|
|
|
|
|
wt |
No data |
No data |
No data |
|
|
| 10 |
M |
50 |
NM |
Left thigh |
Extremities |
No data |
|
|
11/1996 |
20 |
4 |
| |
|
|
|
|
|
V600E |
Skin |
Abdomen |
08/2007 |
|
|
| |
|
|
|
|
|
wt |
Skin |
Left forearm |
11/2007 |
|
|
| |
|
|
|
|
|
V600E |
Skin |
Left jaw |
02/2008 |
|
|
| V600E | Skin | Right cheek | 02/2008 |
Abbreviations: F=female; M=male; NM=nodular melanoma; SSM=superficial spreading melanoma; wt=wild-type.
Differential detection with different assays (wild type according to pyrosequencing; V600E according to capillary sequencing).
Figure 1.
Distribution of tumours evaluated as BRAF-mutated and non-mutated in discordant patients. (A) Example of different localisations in patient #10 (B) Mutations appear over the course of disease (patients #1–10).
Overall, wild-type BRAF was present in 60.4% of patients (113 patients) and 52.0% of samples (156 samples) and mutant BRAF in 44.9% of patients (84 patients) and 48.0% of samples (144 samples). In primary tumours, the BRAF mutation rate was 45.5%, whereas in metastases mutations were detected in 51.6%. Out of the BRAF-mutated patients 69 (82.1%) were V600E in a total of 94 tumour samples, whereas rare BRAF mutations were found in 17 patients (20%) in a total of 50 tumour samples.
There was no association of mutation status with localisation of primary tumour or metastases
In the patients with multiple analysed tumours, BRAF was detected as mutated in 100% of primary tumours on head/neck (3 out of 3), 83.3% on the trunk (5 out of 6) and 62.5% on the extremities (5 out of 8). For metastases these numbers were 42.5% for skin (34 out of 46), 0% for lymph node (0 out of 6) and 0% for visceral metastases (0 out of 6), respectively (Table 1A, B, C).
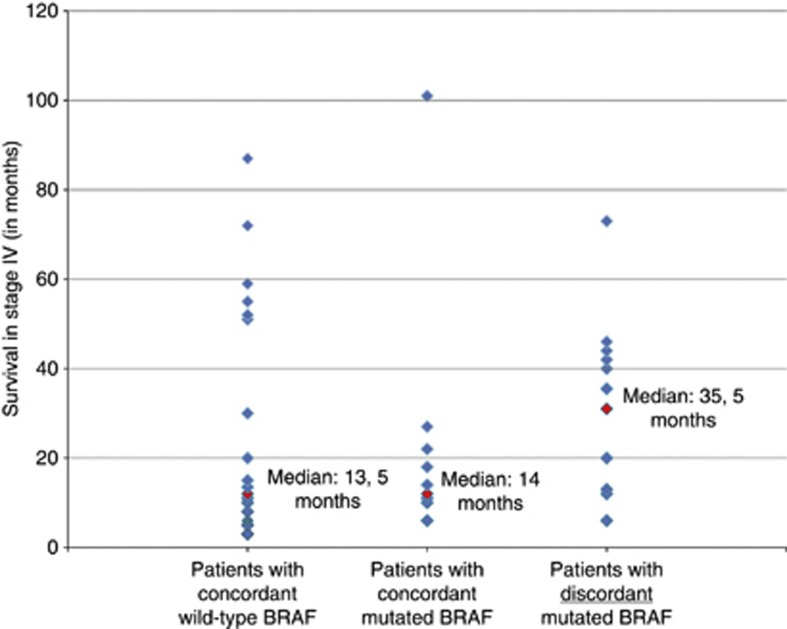
Patients with discordant mutation status showed a tendency towards better median survival when compared to concordant BRAF-mutated patients
Patients with discordant mutation status showed a median survival of 35.5 months (Figure 2) as compared with 14 months in patients with BRAF mutations in all tumour probes. Though this difference was not statistically significant (P=0.112), it shows a tendency towards better median survival of patients with discordant mutation status. Interestingly, two patients showed a rare mutation (V600K) in their primary tumours and the more common V600E mutation in their metastases (patient #1 and #7; Table 1B). Patients with a BRAF V600E mutation (discordant and concordant mutated; n=21) showed a median survival of 18 months (Table 1B, C) as compared with 13.5 months in patients with wild-type BRAF (n=20; Table 1A). This difference was not statistically significant (P=0.695). These comparable survival times result from the relatively long median survival of patients with discordant BRAF mutation status and the relatively short survival time of patients with a concordant mutation status.
Figure 2.
Survival of different patient groups. Each patient's survival is represented by a blue rhomb, median survival is marked with red rhomb.
There was no accumulation of mutations in the course of disease
Occurrence of mutations did not correlate with time from diagnosis. Also all patterns of occurrence of wild-type and mutated tumours were observed. See representatively Figure 1A (patient #10; Table 1C), whose metastases are distributed on head (V600E), upper extremity (wild-type) and trunk (V600E). Indeed, we found four patients with V600E-mutated primaries that did not show the mutation in any of the analyses of subsequently developed metastases (Table 1C; patients #1, #4, #5, #7 and #9) and simultaneously developed metastases with different mutation status (Figure 1A; patient #8).
Response to therapy with BRAF inhibitors
There was only one patient treated with a BRAF inhibitor since at the time of sample collection BRAF inhibitors were not yet available. This patient with discordant mutation status experienced a partial remission under treatment with vermurafenib (patient #3). Interestingly, in this patient tumour nodes that were evaluated as wild-type also regressed under treatment with vemurafenib.
Immunohistochemistry showed intratumoural heterogeneity in a subset of patients
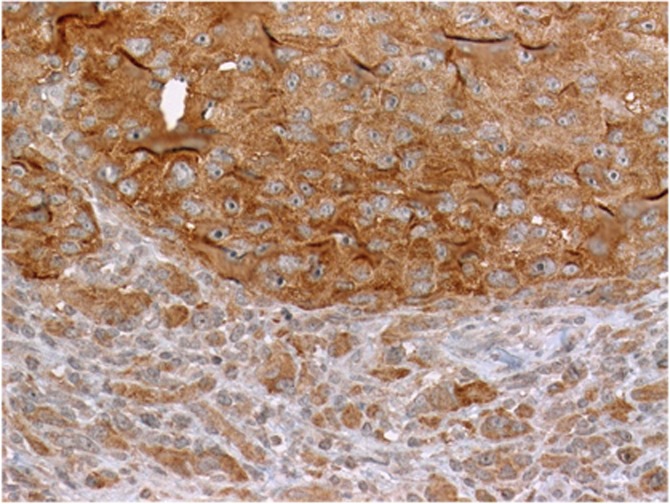
The V600E protein was stained by immunohistochemistry using the V600E mutation-specific antibody as previously reported (Heinzerling et al, 2013). Interestingly, some of the analysed tissue samples showed intratumoural heterogeneity (Figure 3).
Figure 3.
Intratumoural heterogeneity of BRAF V600E expression within melanoma metastasis. Example of a mutation-positive lymph node metastasis showed strong staining of a tumour clone (upper field) juxtaposed to nonstaining tumour tissue (lower field).
Discussion
This study characterises for the first time intraindividual heterogeneity of BRAF mutation findings in a larger melanoma patient population with up to 13 analysed tumours per patient. It analyses clinical data and survival of melanoma patients with respect to BRAF V600 mutation status.
In clinical practice, mutation analyses are performed from a tissue sample available, preferably from a recently detected biopsied or resected metastasis. However, heterogeneity in BRAF mutation findings has been documented between primary tumour and metastases (Houben et al, 2004) and between different metastases (Lin et al, 2011; Yancovitz et al, 2012). For example, discordant BRAF status among metastases was detected in 26% of patients (with 2 metastases each) and between primary tumour and one metastases in 33% (6/18 patients, Yancovitz et al, 2012). Though analysing a higher number of samples per patient (up to 13 tumour samples) our discordance rate was similar with 18.9% of patients showing discordant results (10 out of 53 patients). Discordance rate between primary tumours and metastases on the other hand was higher in our series with 44%. Analysis of the intratumoural mutation spectrum shows discordance in some instances (Yancovitz et al, 2012), so this observed heterogeneity could be a result of sampling or of the detection method used. However, it could also be associated with biological differences. The trend towards longer survival in the discordant group as well as the absence of these high discordance rate in patients with rare mutations of V600 (Richtig et al, 2012) would hint to the latter explanation.
In contrast to previous reports on three patients (Lin et al, 2011), we showed no correlation of mutation with progression of disease. This has profound implications for testing as the determination of the BRAF mutation is a prerequisite for treatment with the BRAF inhibitor. Selective BRAF inhibitors consistently produce an objective response rate of ∼50%. Furthermore, around 90% of treated patients show some evidence of tumour regression. Thus, it is especially alarming that according to our data potentially 18.9% of patients could be excluded from BRAF inhibitor therapy despite the presence of mutated metastases. Furthermore, a small subset of patients experience disease progression early during BRAF-targeted therapy. Although secondary resistance can be due to alternative mechanisms (pathways) and secondary mutations including activating NRAS mutations, BRAF gene amplification, overexpression of MAP3K8/COT, a kinase that directly activates MEK and ERK, and alternative splicing of BRAF mRNA (Romano et al, 2013; Sullivan and Flaherty, 2013) molecular heterogeneity with some wild-type metastases could be responsible for this early tumour progression under BRAF inhibition. However, data on this aspect are currently lacking as mutation testing in clinical practice usually is limited to one sample and sampling all metastatic lesions in one patient is not feasible. Thus, sampling a unifocal progressive disease would be of clinical relevance for patient treatment if new alternative drugs targeting resistant (BRAF wild-type) tumour clones are becoming available, particularly within clinical trials.
Our results demonstrate that single testing is not sufficient to identify all carriers of BRAF mutations, which could potentially benefit from the therapy.
Whether BRAF inhibitors have the same effectiveness in patients with concordant and discordant BRAF mutation findings has to be evaluated.
Acknowledgments
We thank Waltraud Leisgang for continuous support in the processing of tissue samples for DNA extraction. We are also grateful that the following Departments of Pathology/Dermatohistology sent us tumour tissue: Dermatology Dr Maciejewski (Munich), Pathology Nuremberg (Professor Dr Papadopoulos), Dermatohistology PD Dr Nilles (Gießen), Pathology Dr Vieth (Bayreuth), University of Baltimore (J Brooks Jackson, MD, MBA), Pathology Professor Dr Stömmer, Dr Langer (Augsburg), Dermatopathology Dres Kutzner, Rütten, Mentzel, Hantschke, Paredes, Schärer (Friedrichshafen), Pathology, University Regensburg (Professor Dr Hofstädter). The work was supported by the Staedtler-Stiftung (Germany) to LH and RSS.
Footnotes
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
References
- Anderson S, Bloom KJ, Vallera DU, Rueschoff J, Meldrum C, Schilling R, Kovach B, Lee JR, Ochoa P, Langland R, Halait H, Lawrence HJ, Dugan MC. Multisite analytic performance studies of a real-time polymerase chain reaction assay for the detection of BRAF V600E mutations in formalin-fixed paraffin-embedded tissue specimens of malignant melanoma. Arch Pathol Lab Med. 2012;136 (11:1385–1391. doi: 10.5858/arpa.2011-0505-OA. [DOI] [PubMed] [Google Scholar]
- Beadling C, Heinrich MC, Warrick A, Forbes EM, Nelson D, Justusson E, Levine J, Neff TL, Patterson J, Presnell A, McKinley A, Winter LJ, Dewey C, Harlow A, Barney O, Druker BJ, Schuff KG, Corless CL. Multiplex mutation screening by mass spectrometry evaluation of 820 cases from a personalized cancer medicine registry. J Mol Diagn. 2011;13 (5:504–513. doi: 10.1016/j.jmoldx.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M, Hogg D, Lorigan P, Lebbe C, Jouary T, Schadendorf D, Ribas A, O'Day SJ, Sosman JA, Kirkwood JM, Eggermont AM, Dreno B, Nolop K, Li J, Nelson B, Hou J, Lee RJ, Flaherty KT, McArthur GA. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364 (26:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombino M, Capone M, Lissia A, Cossu A, Rubino C, De G,V, Massi D, Fonsatti E, Staibano S, Nappi O, Pagani E, Casula M, Manca A, Sini M, Franco R, Botti G, Caraco C, Mozzillo N, Ascierto PA, Palmieri G. BRAF/NRAS mutation frequencies among primary tumours and metastases in patients with melanoma. J Clin Oncol. 2012;30 (20:2522–2529. doi: 10.1200/JCO.2011.41.2452. [DOI] [PubMed] [Google Scholar]
- Curtin JA, Fridlyand J, Kageshita T, Patel HN, Busam KJ, Kutzner H, Cho KH, Aiba S, Brocker EB, LeBoit PE, Pinkel D, Bastian BC. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;353 (20:2135–2147. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- Daniels M, Lurkin I, Pauli R, Erbstosser E, Hildebrandt U, Hellwig K, Zschille U, Luders P, Kruger G, Knolle J, Stengel B, Prall F, Hertel K, Lobeck H, Popp B, Theissig F, Wunsch P, Zwarthoff E, Agaimy A, Schneider-Stock R. Spectrum of KIT/PDGFRA/BRAF mutations and phosphatidylinositol-3-kinase pathway gene alterations in gastrointestinal stromal tumors (GIST) Cancer Lett. 2011;312 (1:43–54. doi: 10.1016/j.canlet.2011.07.029. [DOI] [PubMed] [Google Scholar]
- Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, O'Dwyer PJ, Lee RJ, Grippo JF, Nolop K, Chapman PB. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363 (9:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, Varela I, Phillimore B, Begum S, McDonald NQ, Butler A, Jones D, Raine K, Latimer C, Santos CR, Nohadani M, Eklund AC, Spencer-Dene B, Clark G, Pickering L, Stamp G, Gore M, Szallasi Z, Downward J, Futreal PA, Swanton C. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366 (10:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauschild A, Grob JJ, Demidov LV, Jouary T, Gutzmer R, Millward M, Rutkowski P, Blank CU, Miller WH, Jr., Kaempgen E, Martin-Algarra S, Karaszewska B, Mauch C, Chiarion-Sileni V, Martin AM, Swann S, Haney P, Mirakhur B, Guckert ME, Goodman V, Chapman PB. Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet. 2012;380 (9839:358–365. doi: 10.1016/S0140-6736(12)60868-X. [DOI] [PubMed] [Google Scholar]
- Heinzerling L, Kuhnapfel S, Meckbach D, Baiter M, Kaempgen E, Keikavoussi P, Schuler G, Agaimy A, Bauer J, Hartmann A, Kiesewetter F, Schneider-Stock R. Rare BRAF mutations in melanoma patients: implications for molecular testing in clinical practice. Br J Cancer. 2013;108 (10:2164–2171. doi: 10.1038/bjc.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houben R, Becker JC, Kappel A, Terheyden P, Brocker EB, Goetz R, Rapp UR. Constitutive activation of the Ras-Raf signaling pathway in metastatic melanoma is associated with poor prognosis. J Carcinog. 2004;3 (1:6. doi: 10.1186/1477-3163-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Goto Y, Murata H, Sakaizawa K, Uchiyama A, Saida T, Takata M. Polyclonality of BRAF mutations in primary melanoma and the selection of mutant alleles during progression. Br J Cancer. 2011;104 (3:464–468. doi: 10.1038/sj.bjc.6606072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long GV, Menzies AM, Nagrial AM, Haydu LE, Hamilton AL, Mann GJ, Hughes TM, Thompson JF, Scolyer RA, Kefford RF. Prognostic and clinicopathologic associations of oncogenic BRAF in metastatic melanoma. J Clin Oncol. 2011;29 (10:1239–1246. doi: 10.1200/JCO.2010.32.4327. [DOI] [PubMed] [Google Scholar]
- Lovly CM, Dahlman KB, Fohn LE, Su Z, Dias-Santagata D, Hicks DJ, Hucks D, Berry E, Terry C, Duke M, Su Y, Sobolik-Delmaire T, Richmond A, Kelley MC, Vnencak-Jones CL, Iafrate AJ, Sosman J, Pao W. Routine multiplex mutational profiling of melanomas enables enrollment in genotype-driven therapeutic trials. PLoS One. 2012;7 (4:e35309. doi: 10.1371/journal.pone.0035309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzies AM, Haydu LE, Visintin L, Carlino MS, Howle JR, Thompson JF, Kefford RF, Scolyer RA, Long GV. Distinguishing Clinicopathologic features of patients with V600E and V600K BRAF-mutant metastatic melanoma. Clin Cancer Res. 2012;18 (12:3242–3249. doi: 10.1158/1078-0432.CCR-12-0052. [DOI] [PubMed] [Google Scholar]
- Richtig E, Schrama D, Ugurel S, Fried I, Niederkorn A, Massone C, Becker JC. BRAF mutation analysis of only one metastatic lesion can restrict the treatment of melanoma: a case report. Br J Dermatol. 2012;168 (2:428–430. doi: 10.1111/j.1365-2133.2012.11121.x. [DOI] [PubMed] [Google Scholar]
- Romano E, Pradervand S, Paillusson A, Weber J, Harshman K, Muehlethaler K, Speiser DE, Peters S, Rimoldi D, Michielin O. Identification of multiple mechanisms of resistance to vemurafenib in a patient with BRAFV600E-mutated cutaneous melanoma successfully rechallenged after progression. Clin Cancer Res. 2013;19 (20:5749–5757. doi: 10.1158/1078-0432.CCR-13-0661. [DOI] [PubMed] [Google Scholar]
- Rubinstein JC, Sznol M, Pavlick AC, Ariyan S, Cheng E, Bacchiocchi A, Kluger HM, Narayan D, Halaban R. Incidence of the V600K mutation among melanoma patients with BRAF mutations, and potential therapeutic response to the specific BRAF inhibitor PLX4032. J Transl Med. 2010;8:67. doi: 10.1186/1479-5876-8-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan RJ, Flaherty KT. Resistance to BRAF-targeted therapy in melanoma. Eur J Cancer. 2013;49 (6:1297–1304. doi: 10.1016/j.ejca.2012.11.019. [DOI] [PubMed] [Google Scholar]
- Yancovitz M, Litterman A, Yoon J, Ng E, Shapiro RL, Berman RS, Pavlick AC, Darvishian F, Christos P, Mazumdar M, Osman I, Polsky D. Intra- and inter-tumour heterogeneity of BRAF (V600E) mutations in primary and metastatic melanoma. PLoS One. 2012;7 (1:e29336. doi: 10.1371/journal.pone.0029336. [DOI] [PMC free article] [PubMed] [Google Scholar]