Abstract
The resident prokaryotic microbiota of the metazoan gut elicits profound effects on the growth and development of the intestine. However, the molecular mechanisms of symbiotic prokaryotic–eukaryotic cross-talk in the gut are largely unknown. It is increasingly recognized that physiologically generated reactive oxygen species (ROS) function as signalling secondary messengers that influence cellular proliferation and differentiation in a variety of biological systems. Here, we report that commensal bacteria, particularly members of the genus Lactobacillus, can stimulate NADPH oxidase 1 (Nox1)-dependent ROS generation and consequent cellular proliferation in intestinal stem cells upon initial ingestion into the murine or Drosophila intestine. Our data identify and highlight a highly conserved mechanism that symbiotic microorganisms utilize in eukaryotic growth and development. Additionally, the work suggests that specific redox-mediated functions may be assigned to specific bacterial taxa and may contribute to the identification of microbes with probiotic potential.
Keywords: lactobacilli, microbiota, proliferation, ROS, symbiosis
Introduction
It is becoming increasingly evident that an optimal metazoan gut microbiota serves beneficial functions for the host that includes energy extraction, stimulation of immune development, and competitive exclusion of pathogenic microorganisms (Neish, 2009). In addition, experiments in germ-free animals have demonstrated a physiological role for the microbiota in regulation of epithelial homeostasis, as well as host immunity and metabolism (Hooper et al, 2012; Nicholson et al, 2012). Consistently, abnormalities (‘dysbiosis’) in the intestinal microbiota may be sufficient to provoke intestinal inflammation as seen in inflammatory bowel disease (IBD), and quantitative and/or qualitative abnormalities of the microbiota have been associated with other allergic, metabolic, systemic immune, and infectious disorders (Sartor, 2008). Indeed, therapeutic administration of exogeneous viable bacteria, termed probiotics, has also been reported to dampen inflammation, improve barrier function, and promote intestinal reparative responses in response to inflammatory and developmental disorders of the intestinal tract (Park and Floch, 2007).
Eukaryotes have evolved dedicated perception and signalling systems for monitoring potential pathogens, and necessarily, their own symbiotic microbiota. These pathways allow for recognition of microbes via pattern recognition receptors (PRRs) and activation of signal transduction cascades such as the MAPK and NF-κB pathways. While generally considered pro-inflammatory, basal low level PRR signalling has been implicated in normal homeostatic maintenance in the gut (Rakoff-Nahoum et al, 2004). Additionally, our research group has reported that the microbiota in the intestinal lumen modulates host redox biochemistry to limit pro-inflammatory signalling and activate reparative responses (Kumar et al, 2007, 2009; Wentworth et al, 2010; Swanson et al, 2011; Wentworth et al, 2011). Thus, there is increasing evidence that the gut microbiota is perceived by the host, and influences a wide range of physiological processes. However, little is known of how the microbiota mechanistically influences gut biology. Moreover, delineation of the molecular mechanisms that underlies host and microbe interactions would be instrumental in understanding this important symbiotic relationship, its role in heath and disease, and therapeutic the exploitation of beneficial bacteria.
The gut epithelium of both mammals and invertebrates is a highly adapted tissue that has evolved for both digestive and absorptive functions as well as providing a vital mechanical and immunological barrier against gut luminal contents. The epithelia of both are a two-dimensional, single cell sheet of enterocytes interspersed with pluripotent stem cell niches that serve as sources of cellular renewal (Micchelli and Perrimon, 2006; Ohlstein and Spradling, 2006; van der Flier and Clevers, 2009). In mammals, the stem cell compartment is at the base of the three-dimensional epithelial invaginations forming the crypt niche. The daughter progeny of mammalian stem cells further proliferate and migrate luminally defining the adjacent transient amplifying compartment prior to terminal differentiation into absorptive, mucus secreting, and neuroendocrine epithelial cells. It is estimated that the dynamic renewal of murine epithelia occurs within 4–5 days (van der Flier and Clevers, 2009). In Drosophila, the epithelial cells of the larval fly gut are initially supplied by dispersed single intestinal stem cells (ISCs) that proliferate into a multicellular niche over the course of larval life. The stem cells and progeny represent adult midgut precursors (AMPs) that serve as the primordia of the adult intestinal epithelium that forms during pupal metamorphosis. The adult Drosophila midgut comprises an enterocyte monolayer interspersed with hormone-producing enteroendocrine cells. The adult midgut enterocytes are continuously replenished by ISCs that adjoin the intestinal basement membrane (Micchelli and Perrimon, 2006; Ohlstein and Spradling, 2006). Together, these systems form an attractive and genetically tractable target system for the study of gut stem cell dynamics and the role of environmental influences in this process (Mathur et al, 2010).
Reactive oxygen species (ROS) are short-lived molecules derived from incomplete reduction of oxygen metabolites that at high levels have a microbicidal function in professional phagocytes. However, a lower ‘physiological’ level of ROS is increasingly recognized in the mediation of intracellular signalling events in a wide variety of cell types (Hernandez-Garcia et al, 2010). Importantly, ROS are induced in response to bacteria in virtually all forms of multicellular life ranging from plants and social amoebae to humans, thus representing a primordial form of microbial perception and control (Ha et al, 2005b; Kotchoni et al, 2006). Significantly, ROS are also increasingly recognized as mediators of cellular proliferation and differentiation in disparate biological systems such as plant root hair development (Tsukagoshi et al, 2010), and Drosophila haematopoiesis (Owusu-Ansah and Banerjee, 2009). Recently, we reported that some species of human gut bacteria can induce rapid, physiological generation of ROS that has potent regulatory effects on host immune function, intracellular signalling, and cytoskeletal dynamics (Kumar et al, 2007, 2009; Swanson et al, 2011; Wentworth et al, 2011). Cellular ROS are often produced via the catalytic action of NADPH oxidases. The archetypal member of this family, Nox2, was first identified in neutrophils, and was shown to play an important role in phagocyte microbicidal ROS generation in response to bacteria (oxidant burst). Subsequently, paralogues of Nox2 were identified in non-phagocytic tissues, including Nox1 and Duox2, which are strongly expressed in colonic intestinal epithelia of both flies and mice (Lambeth, 2004; Bedard and Krause, 2007; Ogier-Denis et al, 2008).
Herein, we show that the commensal Lactobacillus spp. are potent inducers of endogenous ROS generation, and of ROS-dependent cellular proliferation within intestines of two metazoan models, namely the fruitfly Drosophila melanogaster and the mouse. In addition, we show that Lactobacillus-induced ROS generation and cell proliferation is dependent on a functional Nox1 enzyme in intestinal epithelial cells. ROS production was absent in germ-free animals and was associated with suppressed epithelial growth. Together, these data indicate that bacteria-induced activation of an ROS generating enzyme in enterocytes influences cellular proliferation.
Results
Colonization of the Drosophila midgut by Lactobacillus plantarum induces cellular ROS generation
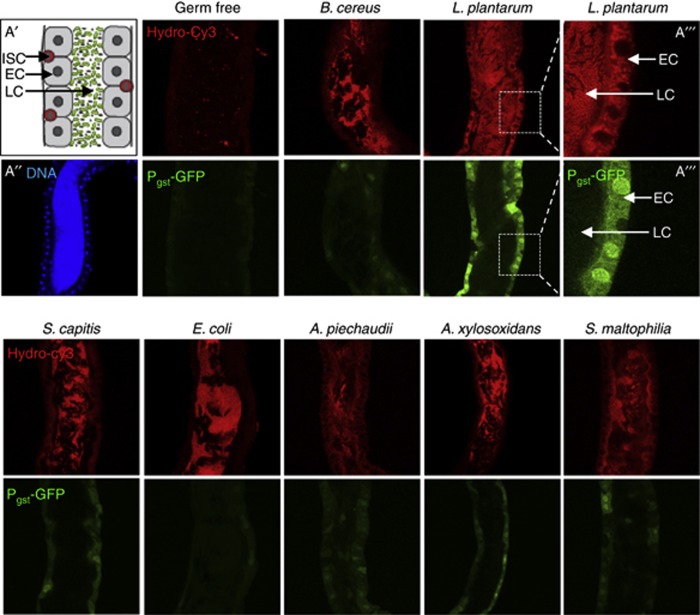
We previously demonstrated that contact with the human commensal (and commonly used probiotic) Lactobacillus rhamnosus GG with cultured epithelial cells induces the endogeneous generation of cellular ROS (Wentworth et al, 2011). Here, we assessed the ability of diverse strains of bacteria to elicit this response in viable Drosophila gut epithelia. To undertake these analyses, and as an improvement on previous techniques that used ROS detection dyes that were prone to auto-oxidation or photobleaching, we employed a new class of hydrocyanine dyes (hereafter referred to as hydro-Cy3) that exhibit far greater specificity and stability, enabling detection of ROS generation in tissue compartments in vivo (Kundu et al, 2009). Whereas the mammalian gut microbiota includes several hundred distinct species of bacteria, the Drosophila gut microbiota is markedly less complex (Wong et al, 2011). We isolated and cultured six distinct bacteria (three Gram negative and three Gram positive) from the luminal content of adult Drosophila (Supplementary Table S1). Pure cultures of the isolated bacteria were mixed with sterile media, and germ-free Drosophila embryos introduced into the vial. Thus, emerged first-instar larvae gnotobiotically ingested sterile media supplemented with pure cultures of the respective bacteria. Culture-based quantification revealed that each first-instar larvae typically ingested a total of about 103–104 CFUs (see Materials and methods). Fluorescent imaging revealed that gnotobiotic ingestion of L. plantarum potently induced the rapid generation of ROS in midgut enterocytes within 30 min, as detected by the oxidation of the hydro-Cy3 dye, and activation of the ROS-responsive gstD1-gfp reporter element (Sykiotis and Bohmann, 2008) (Figure 1). Ingestion of pure cultures of other members of the microbiota, particularly Gram-negative bacteria, elicited nearly undetectable levels of cellular ROS generation (Figure 1), and no ROS generation was detected when emerged germ-free larvae consumed sterilized food (Figure 1), although minor fragments of auto-fluorescent particles were visible. In addition to first-instar larvae, L. plantarum ingestion by germ-free third-instar larvae also induced the generation of cellular ROS, and ROS-responsive genetic elements within 30 min of feeding (Figures 2A and B). Importantly, ingestion of B. cereus isolated from the Drosophila gut, or the Drosophila pathogen Erwinia carotovora, did not induce ROS generation at up to 4 h post ingestion (Figures 2C and D). These results were recapitulated in adult Drosophila where ingestion of L. plantarum but not E. carotovora (detected in the figure by GFP activity) induced ROS generation after 4 h (Figure 2E).
Figure 1.
Ingestion of Lactobacillus plantarum by first-instar Drosophila larvae induces cellular ROS generation. Detection of ROS generation following the ingestion of indicated bacteria by germ-free newly emerged gstD1-gfp first-instar larvae for 30 min with the indicated Gram-positive or Gram-negative bacteria isolated from Drosophila midguts (Supplementary Table S1). Germ-free gstD1-gfp embryos were placed in a vial containing sterilized Drosophila growth media inoculated with 1 × 108 cfu of the indicated bacteria. ROS were detected by oxidation of the hydrocyanin ROS-sensitive dye (upper panels), that is present in the larval food. Larvae used also harbour an ROS inducible gstD1-gfp reporter gene (green lower panels). (A′) Cartoon of first-instar midgut. Enterocyte (EC), intestinal stem cell (ISC), luminal contents (LCs). (A″) Tissue orientation control by staining of first-instar midgut stained for DNA. (A′′′) Exploded view of the interface between the ECs and the LC in larvae fed L. plantarum. Numbers of bacteria ingested by larva were quantified and results are presented in Materials and methods.
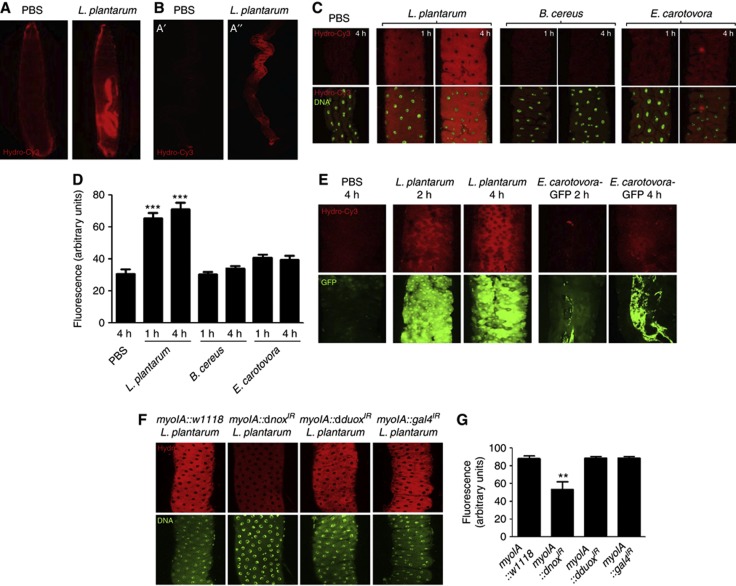
Figure 2.
Ingestion of Lactobacillus plantarum by Drosophila induces cellular ROS generation in the midgut. (A) ROS generation following the ingestion of L. plantarum by germ-free third-instar larvae over 1 h. ROS were detected by oxidation of the hydrocyanin ROS-sensitive dye also included in the media. (B) Microscopic analysis at × 4 magnification of larval midgut dissected from (A). (C) ROS generation in the third-instar midgut following the ingestion of L. plantarum, Bacillus cereus or Erwinia carotovora for up to 4 h. (D) Densitometric analysis of larval midguts described in (C). Results are an average for 5 dissected midguts from each assay. (E) ROS generation in the gstD1-gfp adult Drosophila midgut following the ingestion of L. plantarum, or Erwinia carotovora-GFP for up to 4 h. Note Hydro-Cy3 fluorescence and expression of GFP in enterocytes following the L. plantarum ingestion. Also note GFP fluorescence detected in the midgut following the ingestion of E. carotovora-GFP. (F) ROS generation following the ingestion of L. plantarum in adult Drosophila midguts of the indicated genotypes for 1 h. (G) Densitometric analysis of larval midguts described in (F). Results are an average for five dissected midguts from each assay. All histograms report densitometric analysis (arbitrary units) of hydro-Cy3 oxidation, using the ImageJ software. Ten identically sized areas within an image were measured. n=50. **P<0.01, ***P<0.0001.
As stated in the introduction, bacterial-induced ROS generation occurs in phagocytes via the enzymatic activity of the Nox2 NADPH oxidase. Indeed, it is well established that NADPH oxidases function in the anti-microbial response in mammalian phagocytes, and in epithelial cells of the Drosophila gut (Quinn and Gauss, 2004; Ha et al, 2005a). However, the function of ROS generated in response to colonization with commensal bacteria is not understood. We thus examined the extent to which the dNox or dDuox (the sole NADPH oxidases in Drosophila) function in L. plantarum-induced ROS generation within enterocytes. We expressed RNAi constructs against dNox and dDuox under the enterocyte-specific myoIA-GAL4 driver fly (Morgan et al, 1994). We confirm that the stock used significantly reduced the expression of dnox and dduox respectively compared to w1118 or gal4IR control flies (Supplementary Figure S1). Our analyses show that depletion in the levels of dNox but not dDuox markedly dampened induced ROS generation following the L. plantarum ingestion (Figure 2F and G). We conclude that initial ingestion of L. plantarum induces the rapid generation of ROS by a dNox-dependent mechanism.
Lactobacillus induces ROS-dependent cellular proliferation in the Drosophila intestine
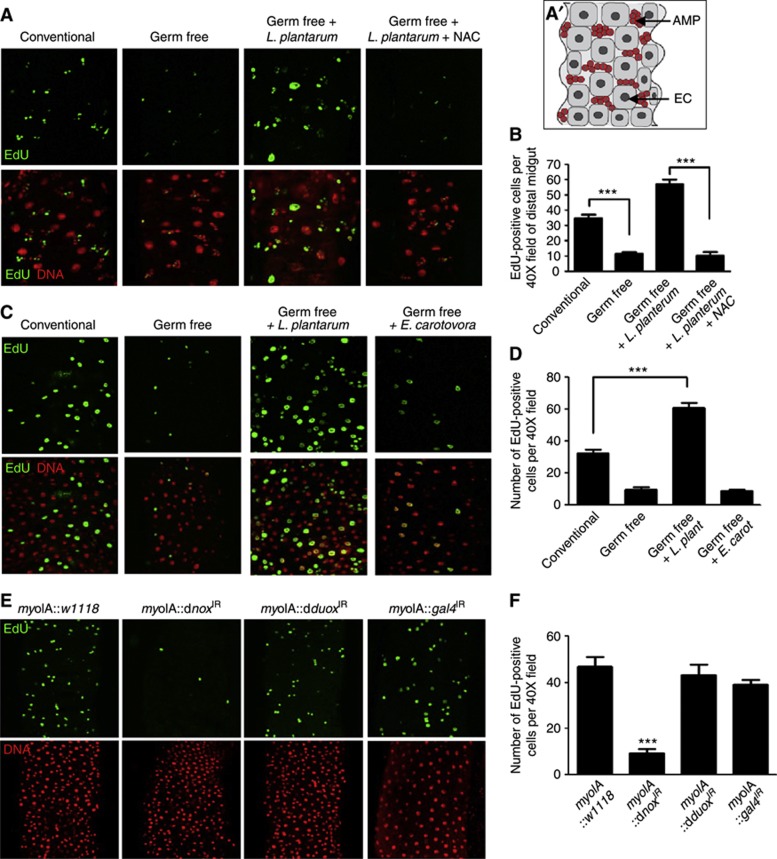
The Drosophila larvae midgut enterocytes are large polyploid cells that form the interface with the gut luminal contents. The stem cells are interspersed between enterocytes, and the stem cells expand over the larval life to form proliferative stem cell niches. These niches harbour adult midgut progenitors (AMPs), from which the adult intestinal epithelium is derived during pupal metamorphosis (Mathur et al, 2010), whereas the adult midgut enterocytes are continuously replenished by pluripotent ISCs (Micchelli and Perrimon, 2006; Ohlstein and Spradling, 2006). We assessed the number of proliferative niche cells in the midguts of conventionally raised and germ-free Drosophila larvae. Germ-free third-instar larvae had significantly fewer numbers of EdU-positive cells in the midgut stem cell niches, compared to conventionally raised larvae (Figure 3A and B), as well as fewer GFP-positive cells in germ-free esg-GAL4 UAS-gfp, a genetic marker of midgut niche stem cells and their progeny (Micchelli and Perrimon, 2006), compared to isogenic conventionally raised flies (Supplementary Figure S2). Strikingly, colonization of germ-free third instar Drosophila larvae with L. plantarum for 2 h significantly increased the number of proliferating niche cells in the midgut (Figure 3A and B). As stated in Introduction, ROS have been shown to function in cell proliferation within diverse tissues (Owusu-Ansah and Banerjee, 2009; Tsukagoshi et al, 2010). We corroborate these reports by showing that supplementing the N-acetylcysteine (NAC) (a glutathione precursor and direct antioxidant) into the fly media for 12 h before bacterial ingestion significantly decreased L. plantarum-induced cell proliferation (Figure 3A and B). We also detected similar responses in adult Drosophila. Germ-free adult Drosophila had significantly fewer EdU-positive ISCs and progeny than conventionally raised flies (Figure 3C and D). Similar to the response in larvae, L. plantarum ingestion for 12 h resulted in a significant increase in the number of EdU-positive ISCs and their progeny in the adult midgut while E. carotovora ingestion did not (Figure 3C and D).
Figure 3.
Ingestion of Lactobacillus plantarum induces ROS-dependent cellular proliferation in the Drosophila intestine. (A) EdU-positive cells in the midgut of w1118 germ-free third-instar larvae, or germ-free larvae fed with 1 × 108 cfu L. plantarum for 4 h. Where indicated, the media also contained 1 mM N-acetylcysteine (NAC). (A′) Cartoon of en face third-instar midgut. Enterocyte (EC) in grey, adult midgut progenitors (AMPs) in red. Note some large enterocytes are EdU positive due to endonuclear DNA replication in maturing larval. (B) Number of EdU-positive cells under conditions described in (A) n=20, ***P<0.001. (C) Detection of EdU-positive cells in the midgut of adult conventionally raised, germ-free, or germ-free adult Drosophila following the ingestion of L. plantarum or Erwinia carotovora for 12 h. (D) Number of EdU-positive cells under conditions described in (C) n=20, ***P<0.001. (E) Detection of EdU-positive cells in the midgut of adult where the levels of Nox or Duox are diminished under the enterocyte-specific myoIA-GAL4 driver. Full genotypes myoIA-GAL4;UAS-dnox-RNAi and myoIA-GAL4;UAS-dduox-RNAi, and myoIA-GAL4;UAS-gal4-RNAi. (F) Number of EdU-positive cells in (A). n=10, ***P<0.001.
As shown in Figure 2, the NADPH oxidase, dNox is required for L. plantarum-induced ROS generation in enterocytes. Here, we show that depletion of dNox, but not dDuox levels also significantly reduced the numbers of proliferating cells in conventionally raised larval midgut (Supplementary Figure S3), and in the 5-day-old adult midgut (Figure 3E and F). Consequentially, examination of the DNA counter stain in Figure 3E indicated changes in midgut histological architecture, and we detected significantly shorter lifespan in Drosophila expressing enterocyte-specific dnoxIR compared to control flies (Supplementary Figure S4). Examination of the adult midgut at 10, 20, and 30 days following the eclosure reveals only few detectable EdU-positive cells and marked observable changes in midgut histological architecture (Supplementary Figure S5). Similarly, using a GFP marker for enterocyte cells (Jiang et al, 2009), markedly altered enterocyte histological architecture was detected in dnoxIR compared to control flies (Supplementary Figure S6). Intriguingly, we also examined the influence of diminishing dNox levels in midgut ISCs using the ISC-specific escargot-GAL4 driver fly. We detected less influence on the numbers of EdU-positive cells in the midgut, although subtle changes in cell arrangement were seen (Supplementary Figure S7), suggesting that dNox-mediated pro-proliferative ROS production occurs predominantly in enterocytes. Together, these observations demonstrate that Lactobacillus-induced ROS generation promotes cell proliferation in the Drosophila intestine, by a mechanism dependent on dNox activity in enterocytes.
Contact of lactobacilli with cultured mammalian cells induces ROS generation
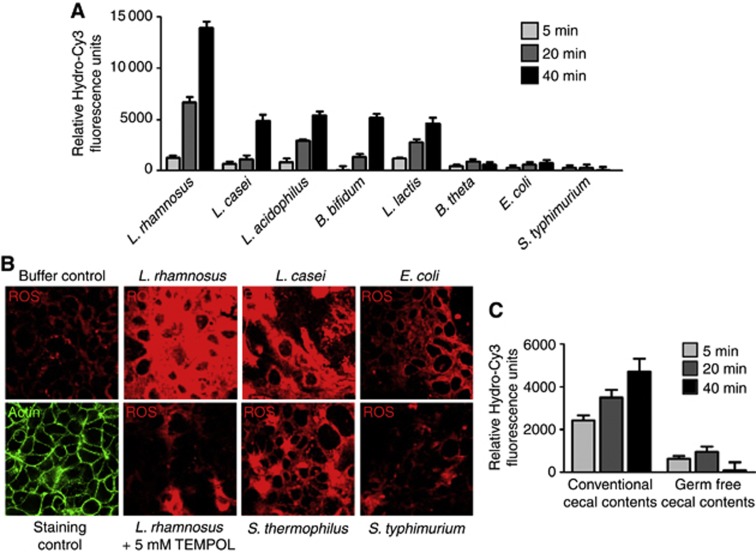
To determine whether the specific influence of ROS-inducing lactobacilli is conserved in mammalian systems, we assessed the ability of diverse strains of mammalian commensal bacteria to elicit this response in cultured Caco-2 cells. Cells were grown to confluency and pre-loaded with hydro-Cy3 for 30 min, before contact with 1 × 108 cfu of the candidate bacteria. Consistent with previous experiments, lactobacilli rapidly induced the generation of cellular ROS within minutes of contact (Figure 4A and B). Other Lactobacillus species tested, including L. acidophilus or L. casei, as well as the Gram-positive intestinal bacteria Bifidobacteria bifidum and Lactococcus lactis also induced the generation of cellular ROS, albeit to a lower extent than L. rhamnosus (Figure 4A and B). Importantly, L. rhamnosus-induced ROS generation was abolished in the presence of the superoxide dismutase (SOD) mimetic TEMPOL (Figure 4B). In contrast, Bacteroides thetaiotaomicron, a well-known enteric Gram-negative anaerobic microbe, as well as Gram-negative Escherichia coli or the mammalian pathogen Salmonella typhimurium could not induce cellular ROS generation. In addition, contact of cultured cells with the luminal contents isolated from conventionally raised mice also induced the generation of cellular ROS, whereas similar fecal preparations from germ-free mice could not (Figure 4C). These data confirm the conserved ability of members of the lactobacilli to induce the epithelial ROS generation across distant metazoan phyla.
Figure 4.
Contact of cultured cells with lactobacilli induces the generation of cellular reactive oxygen species (ROS). (A) Bacterial-induced ROS in cells contacted by the indicated bacteria for up to 40 min. Caco-2 cells seeded in a 96-well format were preloaded with 100 μM hydro-Cy3, then contacted with 3 × 108/100 μl viable bacteria for the indicated times. Cells were then washed three times with PBS before fluoromometric analysis at 575 nm. (B) Bacterial-induced ROS in cells treated as described in (A) detected by confocal microscopy at 585 nm. In some experiments, 5 mM TEMPOL (a membrane-permeable oxygen radical scavenger) was also included. (C) ROS generation in response to contact of Caco-2 cells as described in (A) by the cecal contents of a conventionally raised BL6, or of germ-free BL6 mice. ROS was detected by fluoromometric analysis at 575 nm.
Ingestion of Lactobacillus induces Nox1-dependent generation of cellular ROS in murine enterocytes
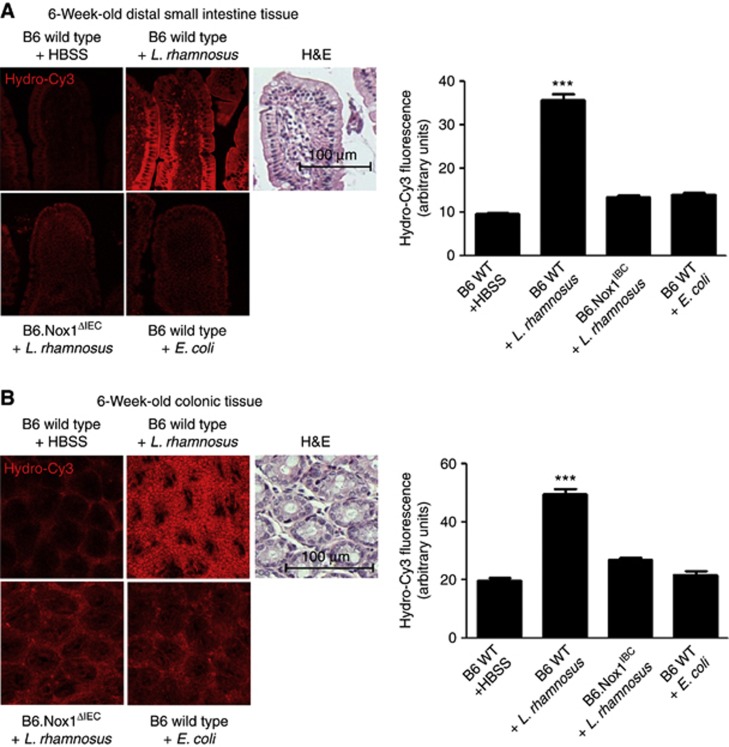
To examine the extent to which bacterially stimulated and Nox1-mediated ROS generation following the ingestion of lactobacilli can be detected in vivo in the murine model, we assessed the effects of feeding L. rhamnosus GG in 6-week-old and 2-day-old wild-type mice, and in intestinal epithelial cell-specific Nox1-deficient (B6.Nox1ΔIEC) animals. We recently described the generation of B6.Nox1ΔIEC mice and the intestinal epithelial expression pattern of Nox1 (Leoni et al, 2013). In addition, previous publications have reported strong expression of nox1 in colonic tissue (Suh et al, 1999; Geiszt et al, 2003). We confirm these findings, and also detect Nox1 activity in the distal small intestine (Supplementary Figure S8). To assess ROS generation, wild-type and B6.Nox1ΔIEC littermates were administered hydro-Cy3 by IP injection 30 min before oral gavage feeding with preparations of 1 × 109 cfu L. rhamnosus or E. coli. Preliminary experiments indicated the bacterial gavage contents reached the colon within 1 h. Examination of colonic tissues at 1 h post feeding revealed that L. rhamnosus, but not E. coli ingestion resulted in the generation of ROS within colonic and small intestine epithelial cells in 6-week-old mice (Figure 5A and B), and in 2-day-old mice (Supplementary Figure S9) by a Nox1-dependent mechanism. Furthermore, feeding germ-free 6-week-old mice L. rhamnosus potently induced ROS generation in the enterocytes of the small intestine (Supplementary Figure S10). Finally, pretreatment of murine subjects with the anti-oxidant NAC administered by IP at 1 h before oral gavage of L. rhamnosus GG completely abolished detectable ROS in the small intestine (Supplementary Figure S11). These data show that ingestion of L. rhamnosus GG results in the potent induction of Nox1-dependent ROS generation in enterocytes of the distal small intestine and the colon.
Figure 5.
Ingestion of Lactobacillus induces Nox1-dependent generation of cellular reactive oxygen species (ROS) in murine enterocytes. (A) Detection of ROS generation in the distal small intestine of B6.wild type or B6.Nox1ΔIEC 6-week-old mice at 1 h following the oral gavage feeding of L. rhamnosus GG or E. coli. Mice were administered 5 μl of 200 μM hydro-Cy3 at 15 min before bacterial feeding. (B) Detection of ROS generation in the colon of 6-week-old mice treated as described in (A). In each figure, Haematoxylin and Eosin (H&E) stain is included for tissue orientation and scale. Scale bar=100 μm. Histograms report densitometric analysis (arbitrary units) of hydro-Cy3 oxidation, using the ImageJ software. The densitometry of 10 identically sized areas within an image was measured. Results are an average for three mice for each treatment. n=30. ***P<0.001. Error bars indicate s.e.m.
Ingestion of Lactobacillus induces Nox1-dependent cell proliferation in the murine gut epithelium
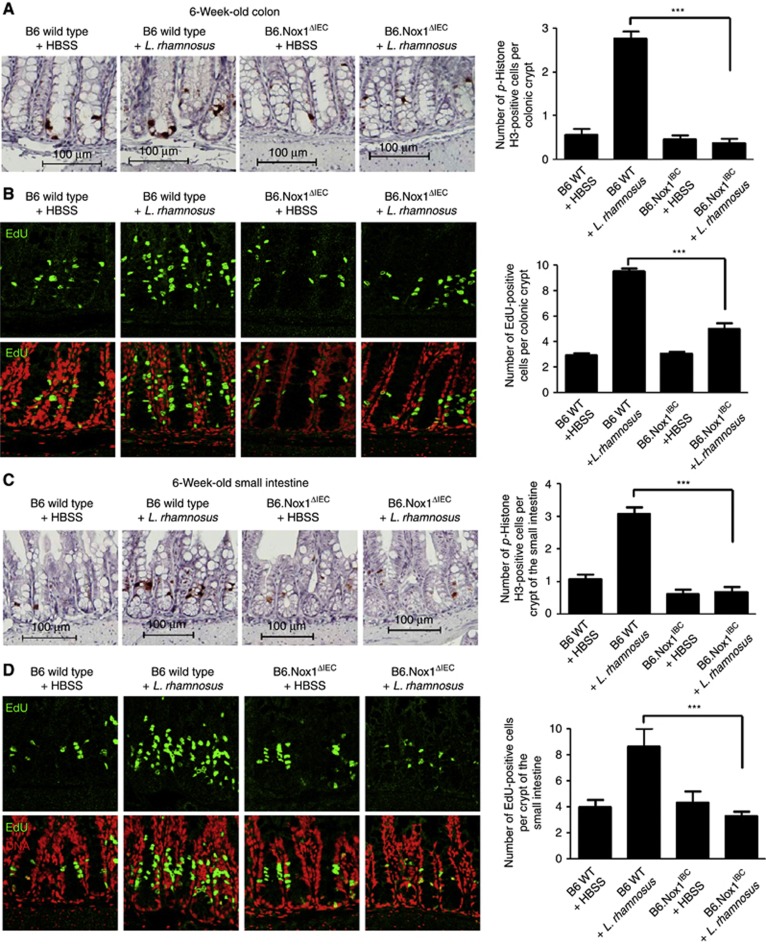
As stated previously, endogeneous and presumably enzymatic generation of ROS in cells have been implicated in the control of cellular proliferation in other biological systems (Owusu-Ansah and Banerjee, 2009; Tsukagoshi et al, 2010). We thus examined the extent to which L. rhamnosus-induced, and Nox1-dependent ROS generation influenced cellular proliferation in the murine intestine. Our experiments show that 6-week-old mice fed L. rhamnosus GG had significantly elevated levels of proliferating cells in the colon compared to controls at 5 h following the bacterial ingestion, as measured by the number of phospho-Histone H3- and EdU-positive cells (Figure 6A and B; Supplementary Figure S12), consistent with previous reports (Preidis et al, 2012). Importantly, we confirm that L. rhamnosus GG-induced cell proliferation in the colon is Nox1 dependent at 6 weeks and 2 days old (Figure 6A and B; Supplementary Figure S12). In addition, these data were recapitulated in the small intestines of or 6-week-old mice fed L. rhamnosus GG, again in a Nox1-dependent manner (Figure 6C and D), and upon initial colonization of 2-day-old mice (Supplementary Figure S13). Together, these data support the conclusion that L. rhamnosus is a potent inducer of Nox1-dependent ROS generation in enterocytes, and of ROS-dependent cellular proliferation in gut tissues (Supplementary Figure S14).
Figure 6.
Ingestion of Lactobacillus induces Nox1-dependent cell proliferation in the murine gut epithelium. (A) Detection and numeration of p-Histone H3 in cells within the colon of 6-week-old w.t. or B6.Nox1ΔIEC mice at 4 h following the oral gavage feeding of HBSS or L. rhamnosus GG. (B) Detection and numeration of EdU-positive cells within the colon of 6-week-old w.t. or B6.Nox1ΔIEC mice at 4 h following the oral gavage feeding of HBSS or L. rhamnosus GG. (C) Detection and numeration of p-Histone H3 in cells within the small intestine of 6-week-old w.t. or B6.Nox1ΔIEC mice at 4 h following the oral gavage feeding of HBSS or L. rhamnosus GG. (D) Detection and numeration of EdU-positive cells within the small intestine of 6-week-old w.t. or B6.Nox1ΔIEC mice at 4 h following the oral gavage feeding of 1 × 108 cfu of HBSS or L. rhamnosus GG. For each numeration, 40 × 20 fields were counted in three mice for each treatment. ***P<0.001.
Discussion
A growing body of evidence indicates that the gut microbiota beneficially affects intestinal homeostasis and, by extension, systemic organismal health. However, relatively little is known of how the host perceives non-pathogenic bacteria, or how the microbiota mechanistically influences gut biology. Here, we describe a highly conserved response (the generation of endogeneous cellular ROS via the action of NADPH oxidases) of host intestinal cells upon perception of bacteria that likely forms a fundamental component of the host–microbiota interaction.
Besides being by-products of metabolism, or active antimicrobial molecules, data are emerging that certain ROS species, especially non-radical forms such as H2O2, function as essential signalling molecules (Hernandez-Garcia et al, 2010). This notion became relevant with our recent discovery demonstrating that commensal gut bacteria induced rapid, generation of physiological levels of ROS from an unknown source within mammalian epithelial cells that had regulator effects (Kumar et al, 2007; Swanson et al, 2011; Wentworth et al, 2011). Cellular redox signalling requires ‘sensor’ proteins, which are generally regulatory enzymes whose activity can be modulated by ROS (Jones et al, 2012; Ray et al, 2012). These redox-sensitive proteins are modified by reversible H2O2-mediated oxidation of their active site cysteines, thus allowing for graded perception of intracellular H2O2 concentrations and control of critical steps in signal transduction pathways. As we and others have shown, such redox-sensitive thiolates are present in a limited but increasingly recognized subset of enzymes, including tyrosine phosphatases, MAP kinase phosphatases (MAPK-P or DUSPs) and regulatory enzymes of ubiquitin and ubiquitin-like proteins such as SUMO and Nedd8 (Jones et al, 2012). However, to date, little was known about the extent to which ROS modulates proteins that function to control cellular proliferation in the mammalian or insect intestine. Here, we propose a new paradigm where ROS generation stimulated by an exogeneous source—the gut microbiota—elicits the enzymatic generation of epithelial ROS, and subsequent modulatory effects on epithelial proliferation (Supplementary Figure S14). Identifying ROS-sensitive proteins that control cell proliferation and/or differentiation in the stem cell microenvironment will yield further insights into the molecular mechanisms that mediate symbiotic bacterial-induced promotion of epithelial homeostasis.
We show that candidate bacterial strains induce variable levels of ROS generation in contacted cells. Lactobacilli were found to be especially potent inducers of ROS generation in cultured cells and in vivo. On the basis of 16S ribosomal RNA sequencing, the candidate lactobacilli tested, L. rhamnosus, L. acidophilus and L. casei on mammalian cells, and L. plantarum in flies have been grouped into disparate phylogenic clades (O'Callaghan and O'Toole, 2013). Despite the divergence, each lactobacilli tested have seemingly evolved the ability to induce cellular ROS generation within its adapted host. Members of the Lactobacillus genus are well-known members of the human microbiota, and are prominently represented as candidate probiotic agents. These biological effects were highlighted in a recent report where Lactobacillus sp. stimulated intestinal epithelial growth (Preidis et al, 2012). Lactobacilli have been utilized in fermented food products for centuries. Their ability to catabolize milk, utilized by yogurt and cheese makers, may have also been exploited during evolution by the gut of neonatal mammals, which may explain the development of lactobacilli-mediated beneficial effects on mammals. Likewise, the role of lactobacilli in fruit spoilage may be relevant to the ability of Drosophila to use these organisms as a signal for growth. This notion is consistent with the recent reported that L. plantarum stably colonizes the Drosophila gut, is transmitted vertically, and promotes Drosophila systemic growth by modulating hormonal signals through TOR-dependent nutrient sensing (Storelli et al, 2011). This study concluded that the Drosophila microbiota sustains optimal larval development and that mono-ingestion of L. plantarum is sufficient to recapitulate the effects of the normal microbiota. Our investigations describe another role for L. plantarum in local epithelial development, that is, the stimulation of Nox-dependent epithelial ROS generation and the ROS-dependent promotion of gut proliferation under homeostatic conditions. In addition to ROS generation, we also detect activation of the Nrf2 pathway-responsive and ROS-sensitive anti-oxidant response element (ARE) (Figure 2E, lower panels). The Nrf2 pathway induces the upregulation of a battery of anti-oxidative genes that counterbalance ROS levels in the cytoplasm (Mitsuishi et al, 2012). Thus, ROS generation following the long-term colonization with lactobacilli is likely to be spatially and temporally variable, where ROS levels (and cell proliferation) are dynamically modulated. These events are the focus of current investigations within our research group.
We also identify the enzymatic basis of bacterial-induced ROS generation. Recently, Nox family members have been identified in many non-phagocytic cell types, with Nox1 and Duox2 strongly expressed in intestinal epithelia (Lambeth, 2004). Importantly, Nox1 has been shown to be important for migration of cultured colonic epithelial cells (Sadok et al, 2008), suggesting direct effects on intestinal epithelial related processes, including proliferation and/or migration. In this study, we confirm this function by showing that Nox1 is required for bacterial-induced intestinal cell proliferation in mice and Drosophila. In addition, depletion of Nox in Drosophila enterocytes reduced lifespan, although decreased viability was only detected following 30 days (Supplementary Figure S4). Depletion of dNox also resulted in markedly altered enterocyte histological architecture, with the degree of altered architecture correlating with age (Supplementary Figures S5 and S6). These observations strongly implicate dNox as functioning in a critical role during midgut development and/or homeostasis, consistent with previous reports (Coant et al, 2010). Further examination of signalling events in the ISC microenvironments where dNox levels are depleted will yield critical insights into the exact mechanisms by which dNox generated ROS controls and modulates pro-proliferative and differentiation events in the gut.
Orthologues of the Nox family mediate ROS generation throughout multicellular life, including Drosophila (Ha et al, 2005a). Interestingly, in the fly, ROS generation occurs in the epithelia, and is necessary for control of the luminal flora. In this case, the dDuox enzyme was shown to be responsible for antimicrobial ROS generation in response to pathogenic infection (Ha et al, 2005b; Chakrabarti et al, 2012). Furthermore, it was reported that the Drosophila intestine responds to E. carotovora infection and tissue damage by inducing ISC proliferation, through a mechanism regulated by the Imd and JAK-STAT pathways following 16 h infection (Buchon et al, 2009). Importantly, in our studies, E. carotovora did not induce detectable ROS generation using the hydro-Cy3 dye at the much shorter time points we examined, and moreover, we show that cell proliferation is also induced by a dNox-dependent response to symbiotic bacteria in conditions and time scale that do not involve tissue injury. It is important to contrast our results to recent studies that report the generation of HOCl in the gut following 2 h ingestion of E. carotovora by a Duox-dependent mechanism (Lee et al, 2013). HOCl generation is formed from hydrogen peroxide and chlorine by the enzyme myeloperoxide (MPO). In addition, the R19S dye used by Lee et al (2013) is sensitive only to HOCl (Chen et al, 2011), whereas the hydro-Cy3 dye used in our studies is sensitive to a broad range of ROS (Kundu et al, 2009). Thus, these data support the conclusion that Nox-catalysed ROS generation in response to lactobacilli modulates homeostatic cell signalling in the gut, whereas Duox and MPO-catalysed generation of HOCl is a response to pathogens.
In the mammalian gut, epithelial ROS generated in response to bacteria serves a signalling role and likely there are numerous ROS-sensitive enzymes that could be influenced by changes in cellular redox status. As has been discussed, reversible oxidative inactivation of a wide range of regulatory enzymes is an increasingly recognized mechanism of signal transduction (Ray et al, 2012). Delineating ROS-dependent outcomes on multiple cell signalling pathways in vivo will be the challenging future work. Alternatively (but not contradictorily), a role of bacterial-elicited ROS stimulating an epithelial antimicrobial response (as occurs in phagocytes and the Drosophila gut), especially in limited locations such as the intestinal crypt, is still to be resolved. Taken together, we suggest that ROS generation for signalling and microbicidal functions in the gut epithelia may represent the primordial ancestral response to bacteria, and that Nox1-dependent ROS generation in epithelial cells plays an important role in critical ROS-mediated intestinal homeostatic processes.
Materials and methods
Isolation of bacterial strains from the Drosophila intestine
w1118 third-instar larvae propagated on conventional Drosophila media were dissected and the gut homogenized in a 1.5-ml microcentrifuge tube in sterile PBS using a sterile plastic pestle. Serial dilutions of the homogenate were plated on three separate solid media: brain heart infusion (BD, Sparks, MD), tryptic soy (BD, Sparks, MD), and de-Man-Rogosa-Sharpe (BD, Sparks, MD) under both aerobic and anaerobic conditions. Anaerobiosis was achieved using a sealed chamber together with the Gas-Pak EZ system (BD, Sparks, MD). After incubation at 30°C for 24–48 h, individual, morphologically distinct colonies were inoculated into the corresponding liquid media and grown at 30°C for 24 h with or without aeration.
Purification of bacterial genomic DNA, 16S PCR and DNA sequencing, and quantitative PCR
Genomic DNA was purified from each bacterial isolate using the QIAamp DNA Stool Mini Kit (Qiagen, Valencia, CA). Short segments of 16S rDNA were amplified from each isolate by PCR using a combination of forward (16S63F, 5′-CAGGCCTAACACATGCAAGTC-3′; 16S338FV3, 5′-ACTCCTACGGGAGGCAGCAG-3′) and reverse (6S1387R, 5′-GGGCGGWGTGTACAAGGC-3′; 16S518RV3, 5′-ATTACCGCGGCTGCTGG-3′) primers. Amplified 16S rDNA products were purified using the Qiaquick PCR Purification Kit (Qiagen), and sequence analysis was performed at SeqWright DNA Technology Service (Houston, TX). The following bacterial strains were identified in the Drosophila gut: Bacillus cereus; Achromobacter xylosoxidans; Lactobacillus plantarum; Achromobacter piechaudii; Stenotrophomonas maltophilia; Staphylococcus capitis; Acetobacter pomorum (Supplementary Table S1). To determine the extent to which RNAi (IR) constructs obtained from the Vienna Drosophila RNAi Center (VDRC) silenced nox and duox expression, respectively, we dissected five adult Drosophila intestines and immerse them directly in TRIzol before RNA extraction. For each Drosophila line examined, RNA from three independent replicates (each containing five intestines) was extracted. Transcript levels were measured for each replicate in duplicate by qRT-PCR. nox transcript levels were amplified using primers Nox-RT-F 5′-ggctatctcctgcaagatcg-3′, and Nox-RT-R 5′-ccaactcaatcaggcggtat-3′, and duox transcripts amplified using primers Duox-RT-F, 5′-tggccaacgagatagtgatg-3′, and Duox-RT-R 5′-aaactgccatcaatccaagc-3′. nox and duox transcript levels were normalized against rp49 transcript levels measured using Rp49-F, 5′-tacaggcccaagatcgtgaa-3′ and Rp49-R, 5′-tctccttgcgcttcttgga-3′.
Fly culture and strains
Flies were maintained on standard media. The gstD-gfp reporter line was a gift from Dirk Bohmann (Sykiotis and Bohmann, 2008). MyoIA-GAL4 was a gift from Shigeo Takashima (Takashima et al, 2008), and Vienna Drosophila RNAi Center (VDRC) stocks 4913 (dnoxIR) and stock 2593 (dduoxIR) were used for depleting levels of NADPH oxidases in the midgut.
Detection of ROS generation and cell proliferation in the germ-free or colonized first and third instar Drosophila larvae
Drosophila embryos were collected and transferred to a cell strainer. Under a sterile hood, embryos were washed three times with sterile PBS, soaked in 50% bleach for 5 min, before washing again with sterile PBS. The mesh of the cell strainer was cut with a sterile blade, and transferred into a sterile Perti dish containing sterilize Drosophila food, and incubated for 24 h at 25°C. Then, first-instar larvae were transferred into another Petri dish containing 2 ml liquefied sterile Drosophila food containing a total of 1 × 106 cfu pure bacterial culture, and a final concentration of 100 μM of hydro-Cy3 ROS-sensitive reagent. After 30 min, the intestinal tract of first-instar larvae were dissected, fixed in 4% paraformaldehyde, and fluorescence detected by confocal microscopy. The procedure was similar for the detection of ROS generation in the intestine of third-instar larvae, save that embryos were incubated at 25°C for 4 days before transfer into liquefied Drosophila food. To determine that the first-instar larvae ingested similar numbers of bacterial monoculture, larval intestines were dissected into 1 ml sterile PBS, and cfu calculated by the plate count method. We detected 5.5 × 104 cfu L. plantarum per first-instar larval intestine (s.d.=2 × 104, n=5), 1.04 × 104 cfu B. cereus per intestine (s.d.=7.8 × 103, n=5), 1.16 × 104 cfu S. capitis per intestine (s.d.=3.4 × 103, n=5), 6.8 × 104 cfu E. coli per intestine (s.d.=1.51 × 104, n=5) 9.96 × 103 cfu A. piechaudii per intestine (s.d.=2.8 × 103, n=5), 3.3 × 104 cfu A. xylosoxidans per intestine (s.d.=1.1 × 104, n=5), and 6.2 × 104 cfu S. maltophilia per intestine (s.d.=1.6 × 104, n=5), respectively. For the detection of cellular proliferation in the intestine of third-instar larvae, EdU (final concentration of 0.4 mM) was added to liquefied sterile food (germ free) or to liquefied food containing 1 × 106 cfu pure bacterial culture. Larvae were incubated in the milieu for 4 h, the intestines dissected and EdU incorporation determined according to the manufacturer’s protocol (Roche Diagnostics GmbH, Germany).
Detection of ROS generation in cultured cells and murine models
ROS generation was measured using the Hydro-Cy3 ROS sensitive dye (Kundu et al, 2009). Briefly, cultured Caco-2 cells were grown to confluence on a 96-well plate. Then, 100 μM Hydro-Cy3 was added to the culture media (DMEM with 0.4% serum) incubated at 37°C and 5% CO2. After 1 h, media containing the Hydro-Cy3 dye was removed by washing with KRH (Krebs–Ringer–Hepes) buffer, before contacting cells with a total of 5 × 108 cfu bacteria suspended in KRH buffer. Fluorescence was measured using a fluorescence microplate reader (SpectraMax M2; Molecular Devices, Sunnyvale, CA, USA) after various time intervals with excitation at 544 nm and emission at 574 nm for Hydro-Cy3. For qualitative detection of ROS, cultured Caco-2 cells were grown to confluency in 8-well chamber slides, treated as described above, before detection of fluorescence by Confocal microscopy at 40 × objective. In some experiments, cells were incubated in the presence of 5 mM TEMPOL before bacterial contact. For the detection of ROS generation in the 6-week-old murine distal small intestine of 6-week-old mice, mice were fasted for 16 h before IP administration of 100 μl of 40 mM Hydro Cy-3 for 15 min. Then, 200 μl of 1 × 1010 cfu/ml of bacteria, or buffer control was fed to mice by oral gavage. After 1 h ingestion of bacteria, mice were sacrificed and tissues of the distal intestine sliced to small pieces, before visualization by confocal microscopy. All ROS fluorescence experiments repeated at least three times. The procedures for 2-day-old mice were identical to those for 6-week-old mice, save for no fasting period, 25 μl of 40 mM Hydro Cy-3 administered by IP for 15 min, and 100 μl of 1 × 109 cfu of bacteria fed by oral gavage and ingested for 30 min before sacrifice. For the detection of cell proliferation in murine tissues, 6-week-old mice were fed 200 μl of 5 × 109 cfu of bacteria for 2 h, and 2-day-old mice, were fed 100 μl of 1 × 109 cfu/ml of bacteria for 1 h before sacrifice.
Detection of EdU-positive cells and phospho-Histone H3-positive cells in the murine model
Proliferating cells in the intestine were identified by detecting EdU-positive or phospho-Histone H3-positive cells. For EdU analysis, mice were fed the 1 × 109 cfu of the indicated bacteria by oral gavage for 3 h. Then, mice were administered EdU by IP at a rate of 100 μg EdU per gram of mouse weight. Following a further 2 h, mice were euthanized and the intestinal epithelium was assessed for EdU-positive cells under confocal microscopy. For p-Histone H3 analysis, mice were fed 1 × 109 cfu of the indicated bacteria by oral gavage for 5 h. Frozen sections of the intestinal tissue were immunostained using the anti-phospho Histone H3 antibody (Cell Signaling, Danvers, MA), followed by (3,3′-diaminobenzidine) DAB detection. Numbers of p-Histone H3-positive cells were assessed by light microscopy. At least three mice were included in each experimental point, and 40 field views at × 20 magnification of each intestinal preparation were numerated for both EdU and p-Histone H3-positive cells.
Supplementary Material
Acknowledgments
RMJ is supported by NIH Grant K01DK081481, and ASN is supported, in part, by National Institutes of Health Grant R01DK071604 and RO1AI064462. PWD is supported by R01HD059122.
Author contributions: RMJ conceived most of the experiments, and performed some of the experiments for the manuscript. RMJ and LL performed all of the Drosophila-related experiments. JWM and CLG isolated and characterized bacteria from Drosophila luminal content. YMK performed all in vitro experiments for the detection of bacterial stimulated generation of ROS in cultured cells. CSA and PAS performed experiments on 6-week-old mice for the detection of bacterial-stimulated ROS. LL, AA, and HW performed experiments on 6-week-old mice for the detection of bacterial-stimulated cell proliferation. ANR performed all ROS detection and cell proliferation experiments on 2-day-old mice. JDL and PWD advised and gave conceptual support and advised on manuscript content. RMJ and ASN prepared final versions of the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bedard K, Krause KH (2007) The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol Rev 87: 245–313 [DOI] [PubMed] [Google Scholar]
- Buchon N, Broderick NA, Poidevin M, Pradervand S, Lemaitre B (2009) Drosophila intestinal response to bacterial infection: activation of host defense and stem cell proliferation. Cell Host Microbe 5: 200–211 [DOI] [PubMed] [Google Scholar]
- Chakrabarti S, Liehl P, Buchon N, Lemaitre B (2012) Infection-induced host translational blockage inhibits immune responses and epithelial renewal in the Drosophila gut. Cell Host Microbe 12: 60–70 [DOI] [PubMed] [Google Scholar]
- Chen X, Lee KA, Ha EM, Lee KM, Seo YY, Choi HK, Kim HN, Kim MJ, Cho CS, Lee SY, Lee WJ, Yoon J (2011) A specific and sensitive method for detection of hypochlorous acid for the imaging of microbe-induced HOCl production. Chem Commun (Camb) 47: 4373–4375 [DOI] [PubMed] [Google Scholar]
- Coant N, Ben Mkaddem S, Pedruzzi E, Guichard C, Treton X, Ducroc R, Freund JN, Cazals-Hatem D, Bouhnik Y, Woerther PL, Skurnik D, Grodet A, Fay M, Biard D, Lesuffleur T, Deffert C, Moreau R, Groyer A, Krause KH, Daniel F et al. (2010) NADPH oxidase 1 modulates WNT and NOTCH1 signaling to control the fate of proliferative progenitor cells in the colon. Mol Cell Biol 30: 2636–2650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiszt M, Lekstrom K, Brenner S, Hewitt SM, Dana R, Malech HL, Leto TL (2003) NAD(P)H oxidase 1, a product of differentiated colon epithelial cells, can partially replace glycoprotein 91phox in the regulated production of superoxide by phagocytes. J Immunol 171: 299–306 [DOI] [PubMed] [Google Scholar]
- Ha EM, Oh CT, Bae YS, Lee WJ (2005a) A direct role for dual oxidase in Drosophila gut immunity. Science 310: 847–850 [DOI] [PubMed] [Google Scholar]
- Ha EM, Oh CT, Ryu JH, Bae YS, Kang SW, Jang IH, Brey PT, Lee WJ (2005b) An antioxidant system required for host protection against gut infection in Drosophila. Dev Cell 8: 125–132 [DOI] [PubMed] [Google Scholar]
- Hernandez-Garcia D, Wood CD, Castro-Obregon S, Covarrubias L (2010) Reactive oxygen species: a radical role in development? Free Radic Biol Med 49: 130–143 [DOI] [PubMed] [Google Scholar]
- Hooper LV, Littman DR, Macpherson AJ (2012) Interactions between the microbiota and the immune system. Science 336: 1268–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Patel PH, Kohlmaier A, Grenley MO, McEwen DG, Edgar BA (2009) Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell 137: 1343–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RM, Mercante JW, Neish AS (2012) Reactive oxygen production induced by the gut microbiota: pharmacotherapeutic implications. Curr Med Chem 19: 1519–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotchoni SO, Kuhns C, Ditzer A, Kirch HH, Bartels D (2006) Over-expression of different aldehyde dehydrogenase genes in Arabidopsis thaliana confers tolerance to abiotic stress and protects plants against lipid peroxidation and oxidative stress. Plant Cell Environ 29: 1033–1048 [DOI] [PubMed] [Google Scholar]
- Kumar A, Wu H, Collier-Hyams LS, Hansen JM, Li T, Yamoah K, Pan Z-Q, Jones DP, Neish AS (2007) Commensal bacteria modulate cullin-dependent signaling via generation of reactive oxygen species. EMBO J 26: 4457–4466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Wu H, Collier-Hyams LS, Kwon YM, Hanson JM, Neish AS (2009) The bacterial fermentation product butyrate influences epithelial signaling via reactive oxygen species-mediated changes in cullin-1 neddylation. J Immunol 182: 538–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu K, Knight SF, Willett N, Lee S, Taylor WR, Murthy N (2009) Hydrocyanines: a class of fluorescent sensors that can image reactive oxygen species in cell culture, tissue, and in vivo. Angew Chem Int Ed Engl 48: 299–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambeth JD (2004) NOX enzymes and the biology of reactive oxygen. Nat Rev Immunol 4: 181–189 [DOI] [PubMed] [Google Scholar]
- Lee KA, Kim SH, Kim EK, Ha EM, You H, Kim B, Kim MJ, Kwon Y, Ryu JH, Lee WJ (2013) Bacterial-derived uracil as a modulator of mucosal immunity and gut-microbe homeostasis in Drosophila. Cell 153: 797–811 [DOI] [PubMed] [Google Scholar]
- Leoni G, Alam A, Neumann PA, Lambeth JD, Cheng G, McCoy J, Hilgarth RS, Kundu K, Murthy N, Kusters D, Reutelingsperger C, Perretti M, Parkos CA, Neish AS, Nusrat A (2013) Annexin A1, formyl peptide receptor, and NOX1 orchestrate epithelial repair. J Clin Invest 123: 443–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur D, Bost A, Driver I, Ohlstein B (2010) A transient niche regulates the specification of Drosophila intestinal stem cells. Science 327: 210–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micchelli CA, Perrimon N (2006) Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature 439: 475–479 [DOI] [PubMed] [Google Scholar]
- Mitsuishi Y, Motohashi H, Yamamoto M (2012) The Keap1-Nrf2 system in cancers: stress response and anabolic metabolism. Front Oncol 2: 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan NS, Skovronsky DM, Artavanis-Tsakonas S, Mooseker MS (1994) The molecular cloning and characterization of Drosophila melanogaster myosin-IA and myosin-IB. J Mol Biol 239: 347–356 [DOI] [PubMed] [Google Scholar]
- Neish AS (2009) Microbes in gastrointestinal health and disease. Gastroenterology 136: 65–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S (2012) Host-gut microbiota metabolic interactions. Science 336: 1262–1267 [DOI] [PubMed] [Google Scholar]
- O'Callaghan J, O'Toole PW (2013) Lactobacillus: host-microbe relationships. Curr Top Microbiol Immunol 358: 119–154 [DOI] [PubMed] [Google Scholar]
- Ogier-Denis E, Mkaddem SB, Vandewalle A (2008) NOX enzymes and Toll-like receptor signaling. Semin Immunopathol 30: 291–300 [DOI] [PubMed] [Google Scholar]
- Ohlstein B, Spradling A (2006) The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature 439: 470–474 [DOI] [PubMed] [Google Scholar]
- Owusu-Ansah E, Banerjee U (2009) Reactive oxygen species prime Drosophila haematopoietic progenitors for differentiation. Nature 461: 537–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Floch MH (2007) Prebiotics, probiotics, and dietary fiber in gastrointestinal disease. Gastroenterol Clin North Am 36: 47–63 [DOI] [PubMed] [Google Scholar]
- Preidis GA, Saulnier DM, Blutt SE, Mistretta TA, Riehle KP, Major AM, Venable SF, Finegold MJ, Petrosino JF, Conner ME, Versalovic J (2012) Probiotics stimulate enterocyte migration and microbial diversity in the neonatal mouse intestine. FASEB J 26: 1960–1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn MT, Gauss KA (2004) Structure and regulation of the neutrophil respiratory burst oxidase: comparison with nonphagocyte oxidases. J Leukoc Biol 76: 760–781 [DOI] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R (2004) Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 118: 229–241 [DOI] [PubMed] [Google Scholar]
- Ray PD, Huang BW, Tsuji Y (2012) Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell Signal 24: 981–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadok A, Bourgarel-Rey V, Gattacceca F, Penel C, Lehmann M, Kovacic H (2008) Nox1-dependent superoxide production controls colon adenocarcinoma cell migration. Biochim Biophys Acta 1783: 23–33 [DOI] [PubMed] [Google Scholar]
- Sartor RB (2008) Microbial influences in inflammatory bowel diseases. Gastroenterology 134: 577–594 [DOI] [PubMed] [Google Scholar]
- Storelli G, Defaye A, Erkosar B, Hols P, Royet J, Leulier F (2011) Lactobacillus plantarum promotes Drosophila systemic growth by modulating hormonal signals through TOR-dependent nutrient sensing. Cell Metab 14: 403–414 [DOI] [PubMed] [Google Scholar]
- Suh YA, Arnold RS, Lassegue B, Shi J, Xu X, Sorescu D, Chung AB, Griendling KK, Lambeth JD (1999) Cell transformation by the superoxide-generating oxidase Mox1. Nature 401: 79–82 [DOI] [PubMed] [Google Scholar]
- Swanson PA 2nd, Kumar A, Samarin S, Vijay-Kumar M, Kundu K, Murthy N, Hansen J, Nusrat A, Neish AS (2011) Enteric commensal bacteria potentiate epithelial restitution via reactive oxygen species-mediated inactivation of focal adhesion kinase phosphatases. Proc Natl Acad Sci USA 108: 8803–8808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykiotis GP, Bohmann D (2008) Keap1/Nrf2 signaling regulates oxidative stress tolerance and lifespan in Drosophila. Dev Cell 14: 76–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima S, Mkrtchyan M, Younossi-Hartenstein A, Merriam JR, Hartenstein V (2008) The behaviour of Drosophila adult hindgut stem cells is controlled by Wnt and Hh signalling. Nature 454: 651–655 [DOI] [PubMed] [Google Scholar]
- Tsukagoshi H, Busch W, Benfey PN (2010) Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell 143: 606–616 [DOI] [PubMed] [Google Scholar]
- van der Flier LG, Clevers H (2009) Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu Rev Physiol 71: 241–260 [DOI] [PubMed] [Google Scholar]
- Wentworth CC, Alam A, Jones RM, Nusrat A, Neish AS (2011) Enteric commensal bacteria induce ERK pathway signaling via formyl peptide receptor (FPR)-dependent redox modulation of Dual specific phosphatase 3 (DUSP3). J Biol Chem 286: 38448–38455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentworth CC, Jones RM, Kwon YM, Nusrat A, Neish AS (2010) Commensal-epithelial signaling mediated via formyl peptide receptors. Am J Pathol 177: 2782–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CN, Ng P, Douglas AE (2011) Low-diversity bacterial community in the gut of the fruitfly Drosophila melanogaster. Environ Microbiol 13: 1889–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.