Abstract
Malfunctioning of the protein α-synuclein is critically involved in the demise of dopaminergic neurons relevant to Parkinson’s disease. Nonetheless, the precise mechanisms explaining this pathogenic neuronal cell death remain elusive. Endonuclease G (EndoG) is a mitochondrially localized nuclease that triggers DNA degradation and cell death upon translocation from mitochondria to the nucleus. Here, we show that EndoG displays cytotoxic nuclear localization in dopaminergic neurons of human Parkinson-diseased patients, while EndoG depletion largely reduces α-synuclein-induced cell death in human neuroblastoma cells. Xenogenic expression of human α-synuclein in yeast cells triggers mitochondria-nuclear translocation of EndoG and EndoG-mediated DNA degradation through a mechanism that requires a functional kynurenine pathway and the permeability transition pore. In nematodes and flies, EndoG is essential for the α-synuclein-driven degeneration of dopaminergic neurons. Moreover, the locomotion and survival of α-synuclein-expressing flies is compromised, but reinstalled by parallel depletion of EndoG. In sum, we unravel a phylogenetically conserved pathway that involves EndoG as a critical downstream executor of α-synuclein cytotoxicity.
Keywords: α-synuclein, cell death, endonuclease G, mitochondria, Parkinson’s disease
Introduction
The pathogenesis of Parkinson’s disease (PD), one of the most prevalent neurodegenerative disorders, is influenced by a complex and largely elusive interplay between genetic and environmental factors. Death of dopaminergic neurons in the substantia nigra and accumulation of intracellular inclusions (Lewy bodies) constitute basic pathological features of PD. α-synuclein, a protein prominently expressed in the central nervous system, has been identified as the main component of the insoluble filaments forming Lewy bodies (Spillantini et al, 1997). In cultured human dopaminergic neurons, accumulation of α-synuclein may result in apoptosis mediated by reactive oxygen species (ROS) (Xu et al, 2002). Numerous studies using yeast, nematodes, flies, transgenic mice and cultured human cells indicate that α-synuclein functions in lipid metabolism and vesicular trafficking (Moore et al, 2005; Cooper et al, 2006). Additional evidence implicates mitochondrial (dys)function as a crucial factor in the pathogenesis of PD in general and α-synuclein toxicity in particular (Moore et al, 2005). In fact, several environmental toxins like rotenone or paraquat can accelerate the development of PD by interfering with mitochondrial function (Uversky, 2007). Furthermore, several proteins associated with familial PD, including parkin, DJ-1 and PINK1, are functionally linked to mitochondria (Moore et al, 2005; Abou-Sleiman et al, 2006). We previously reported that the death of yeast cells induced by expression of human α-synuclein strictly depends on functional, respiring mitochondria (Büttner et al, 2008). Here, we identify the mitochondrial pro-apoptotic nuclease endonuclease G (EndoG) as a crucial determinant of α-synuclein-inflicted cellular demise in yeast, nematodes, flies and neuroblastoma cells and show nuclear translocation of EndoG in dopaminergic neurons of PD-patient brain tissue samples.
Results
Yeast EndoG mediates α-synuclein-induced cell death
α-Synuclein has been demonstrated to physically interact with mitochondrial membranes, thereby interfering with mitochondrial function (Li et al, 2007; Cole et al, 2008; Chinta et al, 2010). These studies directly link the toxic consequences of α-synuclein to organelles that are pivotal determinants in cell death execution and constitute the major source of cellular ROS (Zamzami and Kroemer, 2001). Thus, we hypothesized that one or several mitochondrial factors may constitute the executor of α-synuclein-triggered neuronal cell death. To further explore the connection between α-synuclein and mitochondria, we took advantage of budding yeast (Saccharomyces cerevisiae), which is amenable to mitochondrial manipulation (Madeo et al, 1999). Consistent with data obtained in higher model systems, we found that a small portion of α-synuclein localized to purified mitochondria of yeast cells heterologously expressing human α-synuclein (Figure 1A). Immunoblotting using an antibody directed against the cytosolic 3-phosphoglycerat kinase Pgk1p excluded cytosolic contamination of the mitochondrial fractions (Supplementary Figure S1A). Mitochondrially located α-synuclein was lost upon proteinase K digest, indicating attachment to the outer mitochondrial membrane (Figure 1A). Automated quantification of ROS production was used to determine the precise contribution of known mitochondrial cell death mediators to α-synuclein cytotoxicity (Supplementary Figure S1B). Deletion of several genes involved in the regulation of mitochondrial dynamics, mitophagy and mitochondrial phospholipid metabolism (Supplementary Figure S1B) or deletion of the mitochondrial apoptosis-inducing factor AIF1 (Büttner et al, 2008) had no effect on α-synuclein-induced cell killing. Instead, deletion of yeast EndoG (NUC1) strongly suppressed α-synuclein-induced ROS overproduction and death (Figure 1B and C; Supplementary Figure S1B). Re-introduction of Nuc1p into Δnuc1 cells could re-install α-synuclein toxicity, while a point mutation within the active nuclease site of Nuc1p partly inhibited this complementation (Supplementary Figure S2). Although the pro-apoptotic mitochondrial nuclease EndoG has been associated with cellular degeneration in age-dependent muscle atrophy (Leeuwenburgh et al, 2005) and cerebral ischemia (Lee et al, 2005), thus far no links between EndoG and PD have been described.
Figure 1.
EndoG mediates yeast cell death upon α-synuclein expression. (A) Immunoblot analysis of mitochondria isolated from wild-type yeast cells expressing human α-synuclein (αSyn) or harbouring respective vector control (Ctrl.). Purified mitochondria were subjected to proteinase K (Prot. K) digest as indicated. Blots were probed with antibodies directed against FLAG epitope to detect FLAG-tagged αSyn, the NADH-cytochrome b5 reductase Mcr1p as a marker of the outer and inner mitochondrial membrane, and the mitochondrial matrix chaperone Mge1. (B, C) Clonogenic survival (B) and ROS production measured by assessing the ROS-driven conversion of dihydroethidium into ethidium, DHE→Eth (C) of wild-type (WT) and EndoG-deficient (Δnuc1) yeast cells upon galactose-induced expression of αSyn for 24 h (ROS production) or 36 h (survival). Means±s.e.m., n=8. ***P<0.001. (D) Nuclear translocation of Nuc1pFLAG in cells overexpressing Nuc1pFLAG with or without co-expression of αSyn under the control of a galactose promoter for 24 h quantified using immunoblot analysis. The Nuc1pFLAG signal was normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) in whole protein extracts and to histone H3 in isolated nuclei. The ratio of Nuc1pFLAG in cells expressing αSyn and cells harbouring the respective vector control was plotted. Representative blots are shown in Supplementary Figure S3. Means±s.e.m., n=6. *P<0.01. (E, F) Flow cytometric quantification of DNA fragmentation (E) and representative micrographs (F) using TUNEL staining of WT and Δnuc1 cells upon galactose-induced expression of αSyn for 36 h. Means±s.e.m., n=6. ***P<0.001. Scale bar represents 10 μm. (G) Immunoblot analysis of αSyn expression in wild-type and Δnuc1 cells using antibodies directed against FLAG epitope to detect FLAG-tagged α-synuclein and against GAPDH as a loading control. (H, I) Quantification of ROS production (DHE→Eth) (H) and clonogenic survival (I) of yeast cells expressing yeast EndoG (Nuc1p) under a galactose-inducible promoter or human α-synuclein under a methionine-repressible promoter or co-expressing both proteins. Of note, the growth conditions and expression vectors used here differ from those used in (B, C and E). Mid-exponential cells grown on glucose media were shifted to media containing 0.5% galactose and 1.5% glucose (instead of 1.5% galactose and 0.5% glucose) and subjected to determination of ROS production and survival after 24 or 36 h of growth, respectively. Expression vectors and growth conditions in which both proteins alone showed minor toxicity were applied (low galactose concentrations and no complete methionine depletion for low-level expression of Nuc1p and αSyn). Means±s.e.m., n=9–12. ***P<0.001.
Upon cell death induction, EndoG can translocate from mitochondria (its normal location) to the nucleus, where it mediates DNA fragmentation and eventually cell death (Li et al, 2001; Parrish et al, 2001, 2003; Büttner et al, 2007). Immunoblot analyses of subcellular fractions revealed that α-synuclein induced the mitochondria-nuclear translocation of the yeast orthologue of EndoG, Nuc1p (Figure 1D; Supplementary Figure S3A and B). Deletion of yeast EndoG significantly reduced nuclear DNA fragmentation induced by α-synuclein (Figure 1E and F). Expression levels of α-synuclein were unaffected by the absence of NUC1 (Figure 1G). Furthermore, enhanced levels of yeast EndoG due to overexpression exacerbated α-synuclein-driven ROS production and cell death (Figure 1H and I). Of note, in contrast to the experimental set-up used in Figure 1B and C, where high level expression of α-synuclein is driven by a galactose promoter, we applied expression vectors and growth conditions in which both proteins alone exhibited none or minor toxicity to visualize synergistic effects of Nuc1p and α-synuclein.
The pathway for α-synuclein-mediated cytotoxicity involves modulators of the permeability transition pore and the karyopherin Kap123p
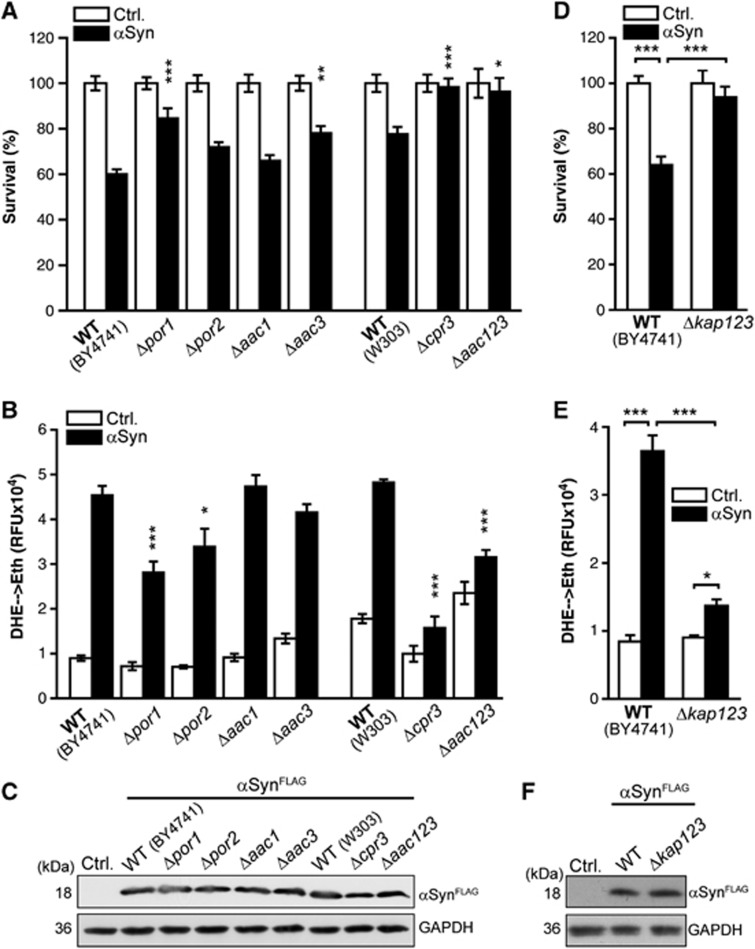
Yeast EndoG physically interacts with proteins that have been suggested to modulate the activity of the mitochondrial permeability transition pore (PTP) and depends on the adenine nucleotide translocator (ANT) to execute death (Büttner et al, 2007). Consistently, we identified regulators of the PTP as facilitators of α-synuclein cytotoxicity while analysing the contribution of various mitochondrial proteins to α-synuclein-mediated ROS production as described above (Supplementary Figure S1B). Yeast cells that lack the voltage-dependent anion channel (POR1), cyclophilin D (CPR3) or the three isoforms of the adenine-nucleotide translocator (AAC1/2/3, only viable in a W303 background) were all protected against α-synuclein-induced death and ROS production, although none of these yeast mutants affected the expression level of α-synuclein (Figure 2A–C; for unnormalized, absolute values of colony-forming units (CFUs), please see Supplementary Figure S4). In addition, yeast cells devoid of the karyopherin Kap123p, which functions in nuclear protein import and interacts with yeast EndoG, were not only protected against EndoG-mediated cell death (Büttner et al, 2007) but against α-synuclein cytotoxicity as well (Figure 2D and E). The absence of Kap123p largely inhibited α-synuclein-facilitated ROS overproduction (Figure 2E) and cell killing (Figure 2D), but did not compromise α-synuclein expression (Figure 2F).
Figure 2.
α-Synuclein cytotoxicity involves components of the permeability transition pore and the karyopherin Kap123p. (A, B) Clonogenic survival (A) and ROS production (DHE→Eth) (B) upon galactose-induced expression of αSyn in indicated deletion mutants and isogenic wild-type (WT) yeast cells for 24 h (DHE) and 36 h (survival). Means±s.e.m., n=8. ***P<0.001; **P<0.01; *P<0.05 compared to αSyn toxicity in the corresponding wild-type cells. Survival was normalized to isogenic vector control cells. For absolute values (colony-forming units), see Supplementary Figure S4. (C) Immunoblot analysis of αSyn expression in indicated deletion mutants and the corresponding wild-type cells using antibodies directed against FLAG epitope to detect FLAG-tagged αSyn and against glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as a loading control. (D, E) Survival determined by clonogenicity (D) and quantification of ROS production (DHE→Eth) (E) of wild-type (WT; BY4741) and Δkap123 cells upon galactose-induced expression of αSyn for 24 h (ROS) or 36 h (survival), and the corresponding vector control cells. Means±s.e.m., n=8. ***P<0.001; *P<0.05. (F) Immunoblot analysis of αSyn expression in wild-type and Δkap123 cells using antibodies directed against FLAG epitope to detect FLAG-tagged αSyn and against GAPDH as a loading control.
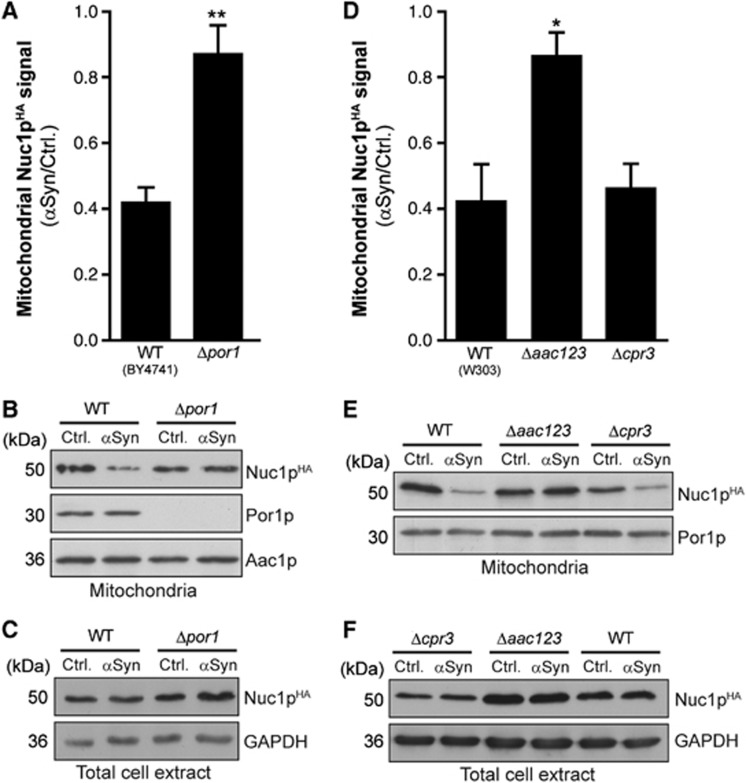
To test whether the amelioration of α-synuclein toxicity observed in deletion mutants of the PTP modulators occurs with a concomitant reduction in apoptotic EndoG release from mitochondria, we generated strains harbouring an endogeneously HA-tagged NUC1 version (Nuc1pHA). Isolation of mitochondria and subsequent immunoblot analysis of Nuc1pHA content demonstrated that the deletion of POR1 and of all three isoforms of AAC prominently inhibited α-synuclein-triggered mitochondrial release of EndoG (Figure 3A–F). While the overall expression of Nuc1pHA analysed by immunoblotting of total cell extracts seemed largely unaffected by α-synuclein, the deletion of the AAC genes (and to a lesser extent the deletion of POR1) provoked an increase in total cellular Nuc1pHA content (Figure 3C and F). Deletion of CPR3 did not affect the release of EndoG but instead caused a decrease in mitochondrial as well as total cellular Nuc1pHA levels per se (Figure 3D–F).
Figure 3.
α-Synuclein-induced mitochondrial release of EndoG requires regulators of the PTP. (A–C) Densitometric quantification (A) of the mitochondrial Nuc1pHA signal via immunoblot analysis and representative blots of isolated mitochondria (B) and total cell extracts (C) from wild-type cells and Δpor1 cells harbouring endogeneously HA-tagged NUC1 upon expression of αSyn for 36 h. Blots were probed with antibodies directed against HA epitope to detect Nuc1pHA, against Por1p, against the ADP/ATP translocator Aac1p as a mitochondrial loading control and against GAPDH. Mitochondrial Nuc1pHA signals were normalized to Aac1p, and this normalized mitochondrial Nuc1pHA content is shown as a ratio of αSyn and control cells. Means±s.e.m., n=8. **P<0.001. (D–F) Densitometric quantification (D) of the mitochondrial Nuc1pHA signal via immunoblot analysis of mitochondria isolated from W303 wild-type cells, Δcpr3 cells and cells deleted in all three isoforms of AAC (Δaac123) harbouring endogeneously HA-tagged NUC1. Representative blots of isolated mitochondria (E) and of total cell extract (F) are shown. Blots were probed with antibodies directed against HA epitope to detect Nuc1pHA, against Por1p as a mitochondrial loading control and against GAPDH. Means±s.e.m., n=4–8. *P<0.05.
Source data for this figure is available on the online supplementary information page.
α-Synuclein deregulates specific proteins at the mitochondrial outer membrane
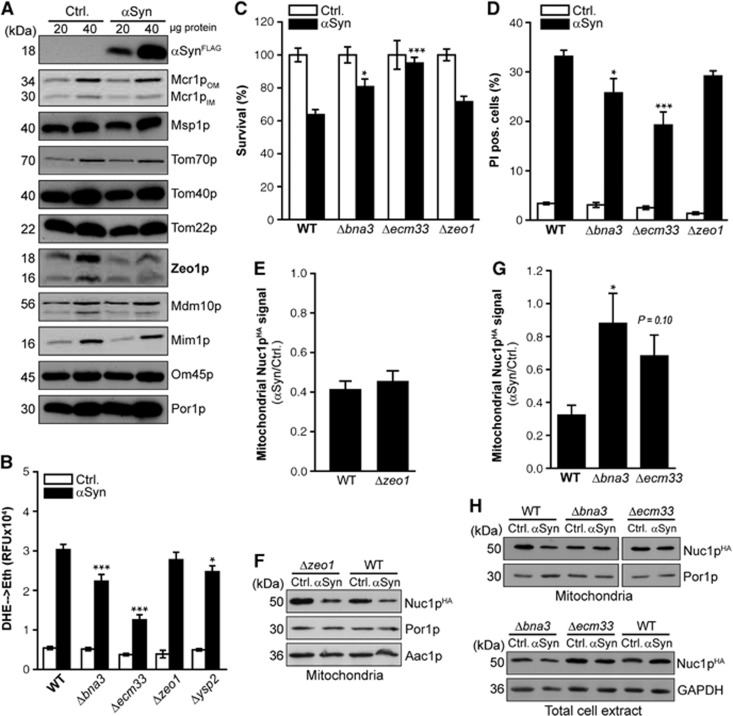
To further elucidate the mechanism underlying α-synuclein-induced mitochondrial cell death in general and the release of EndoG in particular, we searched for α-synuclein-triggered alterations within the mitochondrial protein composition using different approaches. Examining mitochondria isolated from cells expressing α-synuclein or harbouring respective vector control on SDS–PAGE followed by immunodecoration with antibodies directed against various mitochondrial proteins, we could not detect any changes in the levels of typical proteins located within the mitochondrial matrix, the intermembrane space, or the inner mitochondrial membrane (a selection of 30 proteins analysed via immunodecoration is shown in Supplementary Figure S5A). In addition, no alterations regarding the assembly of the supramolecular structures formed by respiratory complexes III and IV (Schägger and Pfeiffer, 2000) could by detected using blue native gel electrophoresis (BN-PAGE) (Supplementary Figure S5B). However, a protein found at the outer mitochondrial membrane, namely the cell integrity pathway protein Zeo1p, was specifically downregulated upon α-synuclein expression, while the levels of additional outer mitochondrial membrane proteins such as components of the TOM (translocase of the outer membrane) complex were unaffected (Figure 4A). Performing mass spectrometry-based quantitative proteomics using a ‘stable isotope labelling by amino acids in cell culture’ (SILAC) approach to analyse mitochondrial fractions for proteomic changes triggered by α-synuclein, we identified several proteins to be deregulated upon expression of α-synuclein (Supplementary Table S1), among them again Zeo1p. Interestingly, while all of the identified proteins have been demonstrated to co-purify with mitochondria, the majority is not exclusively mitochondrial but has been shown to display cytosolic and/or plasma membrane localization, as well (Huh et al, 2003; Sickmann et al, 2003; Brandina et al, 2006; Reinders et al, 2007). Thus, the expression of α-synuclein and its attachment to the outer mitochondrial membrane might influence the association of several proteins to mitochondria, thereby triggering alterations that subsequently allow PTP-dependent release of EndoG. To test whether one or several of the proteins identified to be deregulated upon α-synuclein expression is causally involved in its cytotoxicity, we quantified α-synuclein-driven ROS production in respective deletion mutants if viable (selected candidates are presented in Figure 4B; complete results are shown in Supplementary Table S1 and Supplementary Figure S6). While the absence of the yeast suicide protein Ysp2p, which has recently been shown to be involved in death following amiodarone treatment and intracellular acidification, thereby altering mitochondrial fragmentation (Pozniakovsky et al, 2005; Sokolov et al, 2006), slightly ameliorated ROS production driven by α-synuclein (Figure 4B), the deletion of ZEO1 did neither affect ROS production, loss of membrane integrity and clonogenic survival (Figure 4B–D) nor mitochondrial release of yeast EndoG induced by α-synuclein (Figure 4E and F). Interestingly, the most prominent cytoprotective effect was observed in cells devoid of Bna3p and Ecm33p, both of which have been found enriched in mitochondria of α-synuclein-expressing cells (Supplementary Table S1). Bna3p, a protein demonstrated to shuttle between the cytosol and mitochondria (Karniely et al, 2006), is essential for the formation of nicotinic acid from tryptophan, a biosynthetic route known as the kynurenine pathway. Interestingly, the kynurenine pathway has been repeatedly implicated in various human pathologies, including neurodegenerative disorders (Stone and Darlington, 2002). Several of its metabolites have been demonstrated to act either neuroprotective or neurotoxic, and sometimes even both in a concentration-dependent manner (Wu et al, 2000; Chiarugi et al, 2001; Schwarcz and Pellicciari, 2002; Smith et al, 2007; Tan et al, 2012). As Bna3p has been suggested to function either as arylformidase, catalysing the generation of kynurenine from N-formyl-kynurenine ((Panozzo et al, 2002), or more recently as kynurenine aminotransferase, thereby converting kynurenine into kynurenic acid (Wogulis et al, 2008), its exact influence on the different metabolites of this pathway remains to be clarified. Cytotoxicity of α-synuclein as indicated by accumulation of ROS, loss of plasma membrane integrity and cell death (Figure 4B–D) could be ameliorated via deletion of BNA3. Furthermore, the absence of Ecm33p, a plasma membrane protein with yet unknown function that has been found phosphorylated at mitochondria (Reinders et al, 2007), prominently inhibited the lethal consequences of α-synuclein expression (Figure 4B–D). The mitochondrial enrichment of both Bna3p and Ecm33p contributes to the release of yeast EndoG, as the absence of these proteins reduced the release of Nuc1p (Figure 4G and H). Thus, combining mitochondrial proteomics with yeast genetics and cell death assays enabled us to identify two causal regulatory proteins that promote EndoG release in the context of α-synuclein toxicity: Ecm33p and Bna3p.
Figure 4.
Cytotoxicity of α-synuclein involves Bna3p and Ecm33p. (A) Immunoblot analysis of mitochondria isolated from wild-type yeast cells expressing αSyn or harbouring respective vector control. Blots have been probed with indicated antibodies directed against proteins located at the outer mitochondrial membrane and against FLAG epitope to detect FLAG-tagged αSyn. For immunoblot analysis of proteins located at the inner mitochondrial membrane, the intermembrane space and the matrix, see Supplementary Figure S5. (B) Quantification of ROS production (DHE→Eth) upon galactose-induced expression of αSyn in indicated deletion mutants and isogenic wild-type (WT) yeast cells. Analysis of all deletion mutants corresponding to the mitochondrial proteins identified to be deregulated upon αSyn expression is depicted in Supplementary Figure S6. Means±s.e.m., n⩾8. ;**P<0.001; *P<0.05 compared to αSyn toxicity in wild-type cells. (C, D) Clonogenic survival (C) and loss of membrane integrity indicated by flow cytometric quantification of propidium iodide (PI) staining (D) of wild-type, Δbna3, Δecm33 and Δzeo1 cells expressing αSyn for 36 h or harbouring the empty vector control. Means±s.e.m., n=6–8. ***P<0.001; *P<0.05 compared to αSyn toxicity in wild-type cells. (E–H) Densitometric quantification (E, G) of the mitochondrial Nuc1pHA signal via immunoblot analysis and representative blots (F, H) of isolated mitochondria or whole protein extracts from wild-type, Δzeo1, Δbna3 and Δecm33 cells harbouring endogeneously HA-tagged NUC1 upon expression of αSyn for 36 h. Blots were probed with antibodies directed against HA epitope to detect Nuc1pHA and against Aac1p or Por1p as a mitochondrial loading control. For whole cell extracts, an antibody directed against glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as a loading control. Mitochondrial Nuc1pHA signals were normalized to Aac1p (E) or Por1p (G). The ratio of mitochondrial Nuc1pHA content in αSyn and control cells is shown. Means±s.e.m., n=4–8. *P<0.05.
Source data for this figure is available on the online supplementary information page.
EndoG is critical for α-synuclein neurotoxicity in nematodes and flies
Next, we examined the effects of EndoG depletion on α-synuclein-induced neurodegeneration in the nematode Caenorhabditis elegans and in the fruit fly Drosophila melanogaster (Auluck et al, 2002; Qiao et al, 2008). Nematodes expressing human α-synuclein were analysed for survival of specific dopaminergic neurons (also called cephalic sensilla, CEP), which were visualized via expression of GFP under the dopaminergic-specific dat-1 promoter (Figure 5A and B). The absence or presence of EndoG in the nematode genome did not affect the expression of α-synuclein (Figure 5C), yet had a dramatic effect on the neurotoxic action of α-synuclein. Expression of α-synuclein killed ∼50% of the dopaminergic neurons from wild-type nematodes but only ∼10% of these neurons in EndoG-deficient (cps-6(ok1718)) animals (Figure 5A and B).
Figure 5.
α-Synuclein-induced loss of dopaminergic neurons in C. elegans requires EndoG. (A, B) Representative micrographs (A) and quantification of survival (B) of dopaminergic neurons in wild-type (WT) or EndoG-deficient (cps-6(ok1718)) nematodes expressing Pdat-1GFP and Pdat-1α-Syn. The four CEP dopaminergic neurons in the head of the nematode visualized via Pdat-1GFP were scored as described previously (Qiao et al, 2008). Means±s.e.m., n=250. ***P<0.001. Arrowheads indicate neuronal cell bodies and arrows indicate intact neuronal processes. Scale bar represents 15 μm. (C) Immunoblot analysis of wild-type (WT) or EndoG-deficient (cps-6(ok1718)) nematodes expressing GFP-tagged α-synuclein in the body wall muscles (punc-54αSyn::GFP) using antibodies directed against α-synuclein or β-tubulin and respective secondary antibodies.
Flies expressing human α-synuclein under the control of the UAS-GAL4 system (Brand and Perrimon, 1993) only exhibited a modest decrease in viability, prompting us to combine this genetic manipulation with manganese administration, a known risk factor for PD (Powers et al, 2003). Pan-neuronal nsyb-GAL4-driven expression of α-synuclein significantly enhanced the manganese-induced death of male and female flies within days (Figure 6A and B) and caused an early impairment of climbing ability as indicated by a decline in negative geotaxis, which normally drives flies to walk upwards after being tapped to the bottom of a vial (Figure 6C). Depletion of EndoG by RNA interference (RNAi) (which resulted in ∼40% reduction in EndoG mRNA levels (Figure 6D) and did not affect the expression of α-synuclein, Supplementary Figure S7A and B) protected against the α-synuclein-induced decline in movement ability and later organismal death (Figure 6A–C), confirming that EndoG is pivotal for the neurotoxic activity of α-synuclein in flies. This neuroprotective effect of EndoG depletion was clearly specific for the α-synuclein-driven (as opposed to manganese-driven) neurodegeneration in female flies, while depletion of EndoG also had a minor beneficial effect on manganese toxicity in males (Figure 6B). Similar results were obtained when the neuroprotective effects of EndoG depletion were evaluated in flies expressing α-synuclein under the control of another pan-neuronal driver (elav-GAL4 instead of nsyb-GAL4) (Supplementary Figure S7C and D). Furthermore, these findings were confirmed using a second EndoG-RNAi line (Supplementary Figure S8A–D).
Figure 6.
EndoG is critical for α-synuclein neurotoxicity in flies. (A, B) Survival of female (A) and male (B) wild-type flies and of flies expressing either human αSyn or an RNAi specific for EndoG or both (driven by nsyb-GAL4 for pan-neuronal expression) upon supplementation of food (10% sucrose) with 20 mM Mn2+. Means±s.e.m., n=11–15 with 35–40 flies per experiment. ***P<0.001 and **P<0.01. (C) Climbing activity of female flies described in (A) after 36 h of Mn2+ treatment. Means±s.e.m., n=6 with 8 flies per experiment. **P<0.01. (D) Quantification of EndoG mRNA levels in brains (head extracts) of wild-type flies and of flies expressing either an RNAi depleting EndoG alone or in combination with human αSyn (driven by nsyb-GAL4) by Q-PCR normalized to mRNA levels of α-tubulin. Means±s.e.m., n=6–8. **P<0.01. (E) Survival of male wild-type flies and of flies expressing either αSyn or EndoG alone or co-expressing both proteins after supplementation of food with 20 mM Mn2+. Data represent means±s.e.m., n=8–12 with 35–40 flies per experiment. ***P<0.001, **P<0.01. (F, G) Total count of tyrosine hydroxylase (TH)-immunoreactive dopaminergic neurons (F) in the DM, PM and DL1 brain clusters of female flies expressing αSyn alone or in combination with an RNAi depleting EndoG after treatment with Mn2+ for 96 h. Representative confocal microscopy images of dissected brains immunostained for TH and for Bruchpilot (BRPNc82) to visualize brain structure are shown in (G). Neuronal counts were quantified by inspection of the individual planes of the z-stack. Means±s.e.m., n=5–8. ***P<0.001, *P<0.05. Scale bar represents 40 μm.
To evaluate whether high levels of EndoG can potentiate death inflicted by neuronal expression of α-synuclein, we generated flies simultaneously expressing both proteins under the control of the UAS-GAL4 system. Indeed, co-expression of EndoG did expedite and aggravate α-synuclein-induced organismal death (Figure 6E). α-Synuclein is known to provoke the selective loss of tyrosine hydroxylase-positive dopaminergic neurons in Drosophila PD models in defined clusters of the brain (DM, PM and DL1) (Auluck et al, 2002; Friggi-Grelin et al, 2003; Cooper et al, 2006; Du et al, 2010). Brains from flies expressing α-synuclein exhibited a significantly reduced number of dopaminergic neurons as compared to matched control flies, and this effect was largely revised by RNAi-mediated depletion of EndoG (Figure 6F and G).
EndoG mediates α-synuclein toxicity in SHSY5Y neuroblastoma cells and displays nuclear localization in dopaminergic neurons of Parkinson-diseased brain samples
We analysed the effect of EndoG depletion (Figure 7A) in a human neuronal cell culture model of synucleinopathy (Gerard et al, 2010). SHSY5Y neuroblastoma cells overexpressing α-synuclein subjected to oxidative stress inflicted by H2O2 and FeCl2 displayed prominent protein aggregation and apoptotic death, as determined by high-content microscopic analysis of inclusion body formation and apoptosis (Figure 7B and C; Supplementary Figure S9). Stable knockdown of EndoG mediated by lentiviral short-hairpin RNA (shRNA) largely inhibited α-synuclein-induced apoptosis in unstressed cells and in cells subjected to oxidative stress and also reduced the level of protein aggregation compared to cells transfected with a control shRNA (Figure 7B and C). Again, this suggests a pivotal role for EndoG in the α-synuclein-driven neuronal decay. Furthermore, performing immunocytochemistry, we could demonstrate that the overexpression of αSyn in neuroblastoma cells triggers the release of EndoG from mitochondria, leading to a diffuse localization throughout the cell (Figure 7D and E).
Figure 7.
EndoG mediates α-synuclein toxicity in SHSY5Y neuroblastoma cells and displays nuclear localization in dopaminergic neurons of Parkinson-diseased brain samples. (A) Quantification of EndoG mRNA levels in αSyn-overexpressing SHSY5Y cells either expressing a microRNA (miRNA)-based short-hairpin sequence against EndoG (miR-EndoG) or against mRFP as a control (miR-Control) by Q-PCR normalized to mRNA levels of β-actin. Means±s.e.m., n=3. *P<0.01. (B, C) High-content analysis of αSyn aggregation using Thioflavin S (ThioS) staining (B) and apoptosis indicated by nuclear condensation visualized via DAPI staining (C) in αSyn-overexpressing SHSY5Y cells transduced with a control miRNA (miR-Control) or with miR-EndoG for EndoG knockdown upon induction of oxidative stress using H2O2 and FeCl2 (Stress) or untreated (No stress). Means±s.e.m., n=16–30. ***P<0.001, **P<0.01. (D, E) Immunostaining to visualize EndoG localization in SHSY5Y control cells (Ctrl.) (D) and αSyn-overexpressing SHSY5Y cells (αSyn) (E). EndoG and αSyn were decorated with respective primary antibodies and visualized using secondary antibodies labelled with Alexa 488 and Alexa 555, respectively. Nuclei were stained using DAPI. Scale bar represents 10 μm. (F) Quantification of EndoG-immunoreactive nuclei in the substantia nigra in controls (Ctrl.) and individuals with Parkinson’s disease (PD). The percentage of EndoG-positive nuclei with either dense or granular immunoreactivity (IR) is depicted. Means±s.e.m., n=12–14. **P<0.01 comparing total counts of EndoG-positive nuclei (dense and granular) of control samples to PD-diseased samples. (G) Representative photomicrographs of immunostaining for EndoG of non-diseased control (Ctrl.) and Parkinson-diseased (PD) substantia nigra samples as quantified in (F). Scale bar represents 25 μm.
To examine whether our findings are relevant for human PD patients, we analysed brain tissue samples of the substantia nigra for the localization of EndoG via immunostaining. Sections from 12 PD patients (mean age at death±standard error: 78.92±2.29) and 14 non-diseased controls (77.36±2.48) were evaluated. Braak and Braak stages of neurofibrillary degeneration ranged between stages I and III. Braak stages of Lewy-related pathology in the PD cases were 4 (2 cases), 5 (7 cases) and 6 (3 cases). Immunostaining with an antibody directed against EndoG and subsequent quantification of EndoG-immunoreactive nuclei in the substantia nigra in controls and individuals with PD revealed a highly significant difference: While ∼40% of the nuclei in nigral neurons of PD patients displayed either dense or granular EndoG immunoreactivity, only ∼17% stained positive for EndoG in age-matched healthy controls (Figure 7F and G).
Discussion
In conclusion, our study identifies the pro-apoptotic nuclease EndoG as a major effector of α-synuclein cytotoxicity in yeast, nematodes, flies and human neurons. Notably, specific cell types are particularly susceptible to EndoG-triggered death: while overexpression of cytosolic Drosophila EndoG (lacking its mitochondrial targeting sequence) in neuronal tissues killed animals with 100% penetrance, its overexpression in epidermal tissues did not affect the vitality or morphology of flies (our unpublished observations). We conclude that mitochondrial EndoG triggers death in a highly tissue-specific manner and constitutes a cardinal downstream executor of α-synuclein-mediated neurotoxicity. In this line, post-mortem brain sections of PD patients display a ‘deadly’ nuclear localization of EndoG. Heterologous expression of human α-synuclein in yeast indicates a cell death mechanism that involves proteins suggested to modulate PTP as well as the karyopherin Kap123p. Given that yeast EndoG is physically and functionally connected to the PTP, it appears plausible that α-synuclein toxicity is exerted via opening of the PTP and subsequential translocation of mitochondrial EndoG to the nucleus, causing DNA fragmentation and death. Along this line, the mammalian ANT2, a regulator of the PTP, was found to be specifically upregulated in mesostriatal dopaminergic neurons, which preferentially degenerate in PD (Chung et al, 2005). Another putative PTP subunit, the peripheral benzodiazepine receptor homologue PBR, was found to be upregulated in a D. melanogaster Parkin mutant (Abou-Sleiman et al, 2006). Excessive cytosolic calcium rise is a hallmark of PD (Mattson, 2007; Surmeier et al, 2010) and also known to lead to PTP opening, which may be a conditio sine qua non for EndoG release. Cyclosporin A, an inhibitor of the PTP subunit cyclophilin D, protects primary neurons against cell death (El-Mir et al, 2008). In addition, cyclosporin A-mediated inhibition of PTP opening not only provides general protection in animal models of ischemia/reperfusion but also has been recently shown to reduce infarct size after reperfusion of coronary thrombosis in a clinical trial (Piot et al, 2008).
Accordingly, we demonstrate that mitochondrial attachment of α-synuclein coincides with proteomic changes at the outer mitochondrial membrane. We identify Bna3p and Ecm33p to be enriched in mitochondria of α-synuclein expressing cells and to gatekeep both EndoG release and cell death upon α-synuclein expression. These data advocate the view that Bna3p and Ecm33p are causally connected to α-synuclein-induced release of EndoG and subsequent cytotoxicity. While the function of Ecm33p is rather unknown, the involvement of Bna3p in the kynurenine pathway provides a previously established link to neurodegenerative demise, as various kynurenine metabolites have been demonstrated to be neuroactive (Stone and Darlington, 2002; Tan et al, 2012). Interestingly, inhibition of tryptophan degradation via deletion of the first enzyme of the kynurenine pathway as well as food supplementation with tryptophan abrogated α-synuclein cytotoxicity in a C. elegans PD model (Van der Goot et al, 2012). Whether the deletion of BNA3 protects cells from α-synuclein cytotoxicity due to an accumulation of tryptophan or rather due to changes in other metabolites of the kynurenine pathway remains unclear at this point.
Future will tell whether manipulation of the outer mitochondrial membrane permeability, for example by targeting components of the PTP or the kynurenine pathway, may prevent EndoG release and thus ameliorate PD pathology in humans.
Materials and methods
S. cerevisiae strains, genetics and cell death analysis
Experiments were carried out in BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) and respective null mutants (Euroscarf), or W303 (MATα ura3-52 trp1Δ2 leu2-3, 112 his3-11 ade2-1 can1-100) and respective mutants Δcpr3 and Δaac1/2/3 (gift from J Kolarov). All strains were grown on SC medium containing 0.17% yeast nitrogen base (Difco), 0.5% (NH4)2SO4 and 30 mg/l of all amino acids (except 80 mg/l histidine and 200 mg/l leucine), 30 mg/l adenine and 320 mg/l uracil with 2% glucose (SCD), or 1.5% galactose/0.5% glucose (SCG) for induction of expression of α-synuclein. Previously described α-synuclein constructs in pESC-His and pESC-Trp were deployed (Büttner et al, 2008). For co-expression of α-synuclein and yeast EndoG (Nuc1p), previously described NUC1 construct in pESC-His was used (Büttner et al, 2007), while α-synuclein was cloned into a modified version of pUG36. The EGFP from pUG36 was removed using XbaI and a C-terminal FLAG tag has been introduced using XhoI and the following primers: 5′-TCG AGA TGG ATT ACA AGG ATG ACG ACG ATA AGA TCT AA-3′ (forward) and 5′-TCG ATT AGA TCT TAT CGT CGT CAT CCT TGT AAT CCA TC-3′ (reverse). Human α-synuclein has been amplified using the primers 5′-ATC TAC TAG TAT GGA TGT ATT CAT GAA AGG ACT TTC-3′ (forward) and 5′-ATC TAT CGA TGG CTT CAG GTT CGT AGT CTT G-3′ (reverse) and cloned into the modified version of pUG36 with SpeI and ClaI.
To determine survival, oxidative stress and DNA fragmentation induced by α-synuclein, cells from overnight cultures were inoculated in SCD to OD600 0.1, grown to midlog phase and shifted to SCG for induction of α-synuclein expression. Aliquots were taken out to perform clonogenic survival plating at indicated time points as previously described (Madeo et al, 2002). Briefly, a CASY cell counter (Schärfe systems) was used to measure the cell counts, 500 cells were plated on full media (YEPD) agar plates and CFUs were quantified after 2 days of growth using a Scanalyzer Colony Counter (LemnaTec). To measure the level of cellular oxidative stress, cultures were subjected to dihydroethidium (DHE) staining followed by quantification using a fluorescence reader after 24 h of α-synuclein expression as previously described (Büttner et al, 2007). DNA fragmentation was quantified after 36 h of α-synuclein expression using TUNEL staining as previously described (Büttner et al, 2007). For quantifications using flow cytometry (BD FACSAria), 30 000 cells were evaluated and analysed with the BD FACSDiva software. Same cells were visualized via epifluorescence microscopy with the use of a FITC filter (Zeiss) on a Zeiss Axioskop microscope. Notably, at least four different clones were tested after plasmid transformation to rule out clonogenic variations.
S. cerevisiae subcellular fractionation and immunoblot analysis
Subcellular fractionation for purification of mitochondria and nuclei was accomplished as previously described (Wissing et al, 2004) using cells expressing both α-synucleinFLAG and Nuc1pFLAG from pESC plasmids (using His and Ura as a selection marker) under the control of a galactose promoter. Immunoblot analysis of mitochondrial and nuclear fractions as well as whole cell extracts was performed as described (Madeo et al, 2002). For quantification of mitochondria-nuclear translocation of Nuc1pFLAG, the nuclear FLAG signal was normalized to the histone H3 signal (as a nuclear loading control) and the FLAG signal in whole cell extracts was normalized to GAPDH for the overall expression level of Nuc1pFLAG. The ratio α-synuclein/vector control of Nuc1pFLAG signal for whole cell extracts and for nuclei was calculated. Blots were probed with monoclonal antibodies against FLAG epitope (Sigma), cytochrome c (Abcam), glyceraldehyd-3-phosphate dehydrogenase (GAPDH, Sigma), and histone H3 (ab1791, Abcam) and the respective peroxidase-conjugated affinity-purified secondary antibodies (Sigma). For quantification of mitochondrial release of endogeneously HA-tagged Nuc1p expressed under its own promoter, mitochondrial fractions were analysed using antibodies directed against HA epitope (Sigma), Por1p or Aac1p (gift from G Daum). The mitochondrial Nuc1pHA signal was normalized to either Por1p or Aac1p content as indicated.
For isolation of yeast mitochondria for analysis of steady-state protein levels and BN-PAGE, yeast cells were harvested after growth for 20 h and incubated for 45 min at 28°C in zymolyase buffer (1.2 M sorbitol, 20 mM potassium phosphate, pH 7.4) with 6 mg/g (wet weight yeast) zymolyase. The resulting spheroblasts were washed and disrupted by homogenization. Mitochondria were isolated by differential centrifugation as described before (Meisinger et al, 2006). Aliquots were stored in SEM buffer (250 mM sucrose, 1 mM EDTA, 10 mM MOPS-KOH, pH 7.2) at −80°C. For the proteinase K accessibility assay, 30 μg mitochondria were incubated in SEM buffer and increasing amounts of Proteinase K (2–50 μg/ml). After incubation on ice for 10 min, 4 mM PMSF (phenylmethylsulfonylfluoride in isopropanol) was added. Samples were washed in SEM and analysed by SDS–PAGE and immunodecoration. For BN-PAGE analysis, 50 μg mitochondria were solubilized in 1.0% digitonin buffer (20 mM Tris–HCl pH7.4, 0.1 mM EDTA, 50 mM NaCl, 10% (v/v) glycerol) and loaded on 4–13% gradient gels. Samples were further analysed by immunoblotting and immunodecoration with antibodies directed against Cox1p and Rip1p.
C. elegans strains, genetics, neurodegeneration analysis and immunoblot analysis
The following nematode strains were used: VC1253: cps-6(ok1718)I, BZ555: egIs1[pdat-1::GFP], UA44: baIn11[pdat-1::αSyn, pdat-1::GFP], UA49: baInl2[punc-54::αSyn::GFP, pRF4], cps-6(ok1718)I; baIn11[pdat-1::αSyn, pdat-1::GFP] and cps-6(ok1718)I; baInl2[punc-54::αSyn::GFP, pRF4]. Some strains used were provided by the C. elegans Gene Knockout Project at OMRF, which is a part of the International C. elegans Gene Knockout Consortium. The BZ555, UA44 and UA49 strains were provided by Guy Caldwell (University of Alabama). The VC1253 strain phenotype is superficially wild type. The ok1718 allele is a truncated form of cps-6 locus (676 bp deletion). The following set of primers was used to follow the mutant allele in genetic crosses: 5′-CGCGATAAGTGGAATGATTTGG-3′ (forward), 5′-AGCTGTTGCTGAGGAGAAAG-3′ (reverse). The PCR products have a length of 1117, bp and 441 bp for the wild-type and the mutant alleles, respectively. Seven-day old animals were used for αSyn-induced neurodegeneration quantification. The four CEP dopaminergic neurons in the head of the worm were scored as described previously (Qiao et al, 2008). Statistical analysis was performed using the GraphPad Prism software package (GraphPad Software Inc.). For immunoblotting, worms were sonicated in lysis buffer (50 mM Tris–HCl (pH 7.4), 1 mM EDTA, 1 mM PMSF, and protease inhibitor cocktail (Roche Diagnostics)) and centrifuged at 14 000 g for 20 min. Protein concentrations were determined by Bradford assay. Samples were boiled for 5 min in 3 × sample loading buffer, resolved on 12% SDS polyacrylamide gel and transferred onto nitrocellulose membrane. Blots were probed with primary mouse monoclonal antibody against α-synuclein (Abcam) or a rabbit polyclonal antibody against β-tubulin (Abcam) and the respective secondary antibodies.
D. melanogaster strains and genetics
The line UAS-α-synuclein was obtained from the Bloomington Stock Center (Indiana University, USA). The UAS-CG8862RNAi (EndoG homologue) lines (ID 38084 and 38085) were obtained from the Vienna Drosophila RNAi Center (VDRC, Austria). Lines overexpressing α-synuclein were crossed with these RNAi lines to create the following stable stocks of flies: UAS-CG8862RNAi/UAS-CG8862RNAi; UAS-αSyn/UAS-αSyn. Chromosome III-linked elav-GAL4 and nsyb-GAL4 enhancer trap lines were used to drive expression. To determine survival upon challenge with manganese, 1- to 3-day-old flies (both sexes, kept separately) were incubated at 29°C for 24 h and transferred into fresh vials with filter papers soaked with solution containing 10% sucrose and 20 mM MnCl2 as previously described (Büttner et al, 2012). Filters were kept wet at all times and numbers of dead flies were recorded at indicated time points. Each experiment was performed with 35–40 flies and repeated 8–15 times (as indicated in the figure legends). All experiments were performed using the EndoG-RNAi line referred to as ID 38085 at the Viena Drosophila RNAi center, and effects on survival were confirmed applying the second EndoG-RNAi line (ID 38084).
D. melanogaster determination of locomotive ability
To determine climbing ability upon supplementation of food with 20 mM Mn2+ ions for 36 h, 8–10 flies were placed into a vertical plastic tube with a diameter of 1.5 cm and gently tapped to the bottom of this vial. Flies reaching a specific mark (10 cm) within 10 s were counted. Experiments were conducted in the dark (red light). Six trials of climbing were performed for each set of 8–10 flies to determine the mean climbing activity per experiment, and at least five independent experiments were performed for each genotype.
D. melanogaster immunostaining and immunoblotting
Immunostaining was essentially performed as described before (Owald et al, 2010). Brains were dissected in HL3 on ice, fixed in cold 4% PBS for 20 min and washed four times for 15 min in 0.3% PBT. After 1 h in PBT with 10% NGS at RT, brains were incubated for 2 days in PBT with 5% NGS containing primary antibodies against tyrosine hydroxylase (TH, Millipore) to detect dopaminergic neurons and against Bruchpilot (BRPNc82) to visualize brain structure and then washed in PBT four times for 20 min. Then, brains were incubated in PBT with 5% NGS and the respective secondary antibodies labelled with FITC or Cy3 (Invitrogen) for 1 day. Finally, brains were washed four times in PBT and transferred onto slides in Vectashield (Vector laboratories). Image acquisition was performed with a confocal microscope (TCS SP5, Leica) using the LCS AF software (Leica). For immunoblot analysis, 20–30 fly heads were homogenized on ice in 50 μl 2% SDS with protease inhibitor cocktail (Roche Diagnostics). Equal volume of 2 × Lämmli was added, samples were incubated at 95°C for 5 min and then kept at RT for 5 min prior to centrifugation for 5 min at 13 000 g and subsequent SDS–PAGE analysis. Blots were probed with primary antibodies against α-tubulin (Abcam) and α-synuclein (Sigma) and respective secondary antibodies.
Cell culture and generation of stable EndoG knockdown cell lines
Human neuroblastoma α-synuclein-overexpressing SHSY5Y cells were grown in DMEM (Invitrogen) supplemented with 15% fetal calf serum (International Medical), 500 μg/ml gentamycin (Invitrogen) 1% non-essential amino acids (Invitrogen) and hygromycin B (200 μg/ml, Invitrogen) at 37°C and 5% CO2 in a humidified atmosphere. EndoG knockdown was achieved with lentiviral vector (LV)-based vectors encoding modified miRNA30-based sh sequence against human EndoG mRNA (5′-CTGATGGGAAATCCTA-3′) and a blasticidin-resistance cassette (Heeman et al, 2011). As a negative control, a miRNA30-based sh sequence against mRFP was designed. LV production was performed as described previously (Ibrahimi et al, 2009). SHSY5Y cells overexpressing α-synuclein were transduced with LV diluted in the cell culture medium. After 48 h, vector containing medium was replaced by medium with blasticidin (12 g/ml, Invitrogen) to select for transduced cells.
Reverse transcription quantitative PCR
To determine mRNA levels, total RNA was extracted from transduced cell lines (∼3 × 106 cells) using the Aurum™ RNA mini kit (Biorad). In all, 5 μg of total RNA was reverse-transcribed using the High Capacity cDNA Archive kit (Applied Biosystem) and cDNA was used in triplicate as a template for quantitative PCR amplification with SYBR Green-based detection for EndoG and TaqMan probe-based detection for the endogeneous housekeeping gene β-actin. The following primers and probe were used: human EndoG primers 5′-CTACCTGAGCAACGTGCG-3′ (forward) and 5′-TCCAGGTTGTTCCAGGATT-3′ (reverse); β-actin primers 5′-CACTGAGCGAGGCTACAGCTT-3′ (forward) and 5′-TTGATGTCGCGCACGATTT-3′ (reverse), and β-actin probe 5′-[HEX]ACCACCACGGCCGAGCGG[TAM]-3′. Cycling conditions were 3 min at 95°C, followed by 40 cycles of 10 s at 95°C and 30 s at 55°C. The obtained EndoG mRNA levels were normalized to the mRNA levels of the β-actin housekeeping gene present in the sample.
To determine mRNA levels in Drosophila brains, total RNA was extracted from respective strains using the Qiagen RNeasy kit (Qiagen) with ∼30 heads per extraction. Contaminating DNA was removed by DNase I digestion using Qiagen RNase-Free DNase Set and RNA was cleaned up according to the Qiagen RNA cleanup and concentration protocol. RNA concentrations were determined with a NanoDrop Spectrophotometer (NanoDrop Technologies) and 100 ng was used for the detection of EndoG mRNA and of α-tubulin mRNA (as an endogeneous housekeeping gene) via reverse transcription and quantitative PCR amplification using the SensiMix™ SYBR one-Step Kit (Bioline) and a Corbett Research RG6000 PCR machine. For each genotype, at least four independent RNA extractions have been subjected to reverse transcription quantitative PCR. The following primers were used: Drosophila EndoG primers 5′-CCACTGTACCTACCGCACAAGG-3′ (forward) and 5′-GATTGGGCATCACGTACGACTC-3′ (reverse), and α-tubulin primers 5′-TCATGGTCGACAACGAGGCTA-3′ (forward) and 5′-TACGTGGGTAGGGCACCAAGT-3′ (reverse). Cycling conditions were 10 min at 42°C and 10 min at 95°C, followed by 40 cycles of 15 s at 95°C, 15 s at 60°C and 15 s at 72°C. The obtained EndoG mRNA levels were normalized to the mRNA levels of the α-tubulin housekeeping gene within the same sample.
High-content analysis of α-synuclein aggregation and cell death
We used the cell culture model described by Gerard et al (2010) to induce α-synuclein aggregation (Gerard et al, 2010). Cells were plated in 96-well plates (1.5 × 104 cells/well). The next day, cells were treated for 72 h with 100 μM H2O2 and 5 mM freshly prepared FeCl2 in DMEM complete and filtered through a 0.20-μm filter (Corning Incorporated). After 3 days, cells were washed with PBS, fixed with 4% formaldehyde for 15 min and stored in PBS until analysis. Cells were incubated with 0.05% Thioflavin S (Sigma-Aldrich) for 20 min and washed twice with 70% ethanol for 2 min. Next, the cells in the wells were incubated with DAPI (1:10 000 in PBS). To quantify α-synuclein aggregation and apoptosis in our synucleinopathy cell culture model, the IN Cell Analyzer 1000 and IN Cell Investigator software (GE Healthcare) was used as described before (Gerard et al, 2010).
Immunocytochemistry of SHSY5Y cells
For immunocytochemistry, cells (SHSY5Y and SHSY5Y α-SYN) were grown in an incubation chamber (2 × 104 cells/well) for 3 days. Cells were fixed with 4% formaldehyde for 15 min after two wash steps with PBS. For permeabilization, they were incubated for 10 min with PBS-0.1% Triton X-100. After a blocking step with 10% goat serum for 20 min, cells were incubated overnight with the primary antibodies (rabbit anti-endo G, 1:500, Abcam an9467, mouse anti-α-SYN211, 1:500, Sigma S5566) in PBS with 0.1% Triton X-100. After washing three times with PBS for 5 min, the cells were incubated for 1 h with secondary antibodies (Alexafluor 488 or 555-conjugated antibodies, 1:500, Invitrogen). After washing three times in PBS for 5 min, the cells were mounted with Mowiol solution containing DAPI (1:1000, Sigma). Fluorescent double staining was visualized by confocal microscopy with an LSM 510 unit (Zeiss, Belgium). Images were further analysed using the Zeiss co-localization software.
Immunostaining of substantia nigra samples
Tissue samples for the study on individuals affected by PD and without neurodegenerative disease originating from the Institute of Neurology, Medical University of Vienna, were collected following local regulations for diagnostic purposes. Anonymized tissue samples remaining after the diagnostic evaluation were used in this project in the frame of a study (‘Molecular neuropathologic investigation of neurodegenerative diseases’; Collection named as KIN-Biobank where KIN: ‘Klinisches Institut für Neurologie’) approved by the Ethical Committee of the Medical University of Vienna (Nr. 396/2011) and following the principles of the Helsinki declaration. All cases underwent detailed neuropathological evaluation following established protocols (Kovacs and Budka, 2010). Formalin-fixed, paraffin-embedded tissue section of the substantia nigra was evaluated using immunostaining for anti-Endo G (rabbit polyclonal ab9647, corresponding to aa 55–70 of human Endo G; 1:100 dilution; 10 min epitope retrieval with citrate buffer, pH 6; Abcam, Cambridge, UK). Five high-power fields (× 400 magnification) were evaluated: the total number of neurons with visible nucleus and the number of neurons showing granular or dense nuclear immunoreactivity were evaluated. The presence of neuromelanin pigment in the cytoplasm of the cell and large nucleus with a visible nucleolus clearly defines neurons of the substantia nigra and is distinguishable from glial cells, which have smaller nucleus and lack of neuromelanin in their thin cytoplasm. The proportion for each type of immunoreactivity was calculated by dividing the number of positive cells to the total number of neurons visible.
Statistical analysis
For Drosophila survival experiments, a two-way ANOVA with time and strain as independent factors was used. For comparison of only two groups, a Mann–Whitney test was used. For all other experiments, significances have been calculated using a one-way ANOVA followed by a Tukey post-hoc test.
Sample preparation for mass spectrometry
For stable isotope labelling of cultures, a BY4741 strain with additional deficiencies in ARG4 and LYS2 was transfected with α-synuclein or respective vector control. Cells expressing α-synuclein were grown in media supplemented with Arg10 and Lys4 (Sigma) and vector control cells on regular media and vice versa for a biological replicate. Mitochondria were isolated after 20 h of growth as described above. Samples were reduced with 1 mM DTT (Sigma) for 5 min at 95°C and alkylated using 5.5 mM iodoacetamide (Sigma) for 30 min at 25°C. Protein mixtures were separated by SDS–PAGE using 4–12% Bis-Tris mini gradient gels (NuPAGE, Invitrogen). The gel lanes were cut into 10 equal slices, which were in-gel digested with Trypsin (Promega) (Shevchenko et al, 2006), and the resulting peptide mixtures were processed on STAGE tips as described (Rappsilber et al, 2007).
Mass spectrometry
Mass spectrometric measurements were performed on an LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific) coupled to an Agilent 1200 nanoflow-HPLC (Agilent Technologies GmbH). HPLC-column tips (fused silica) with 75 μm inner diameter (New Objective) were self packed with Reprosil-Pur 120 ODS-3 (Dr Maisch, Ammerbuch, Germany) to a length of 20 cm. Samples were applied directly onto the column without pre-column. A gradient of A (0.5% acetic acid (high purity, LGC Promochem) in water (HPLC gradient grade, Mallinckrodt Baker B.V)) and B (0.5% acetic acid in 80% ACN (LC-MS grade, Wako) in water) with increasing organic proportion was used for peptide separation (loading of sample with 2% B; separation ramp: from 10 to 30% B within 80 min). The flow rate was 250 nl/min and for sample application 500 nl/min. The mass spectrometer was operated in the data-dependent mode and switched automatically between MS (max. of 1 × 106 ions) and MS/MS. Each MS scan was followed by a maximum of five MS/MS scans in the linear ion trap using normalized collision energy of 35% and a target value of 5000. Parent ions with a charge state from z=1 and unassigned charge states were excluded for fragmentation. The mass range for MS was m/z=370–2000. The resolution was set to 60 000. Mass-spectrometric parameters were as follows: spray voltage 2.3 kV; no sheath and auxiliary gas flow; ion-transfer tube temperature 125°C.
Identification of proteins and protein ratio assignment using MaxQuant
The MS raw data files were uploaded into the MaxQuant software version 1.3.0.5 (Cox and Mann, 2008) and searched with the Andromeda search engine (Cox et al, 2011) against the yeast UniProt database (release 2012_11). Carbamidomethyl cysteine was set as fixed modification; methionine oxidation and protein amino-terminal acetylation were set as variable modifications. Triple SILAC was chosen as a quantitation mode. Two miscleavages were allowed, enzyme specificity was Trypsin/P+DP, and the MS/MS tolerance was set to 0.5 Da. The average mass precision of identified peptides was in general <1 p.p.m. after recalibration. Peptide lists were further used by MaxQuant to identify and relatively quantify proteins using the following parameters: peptide, and protein false discovery rates (FDRs) were set to 0.01, maximum peptide posterior error probability (PEP) was set to 1, minimum peptide length was set to 6, minimum number of peptides for identification and quantitation of proteins was set to two unique peptides, minimum ratio count was set to two, and identified proteins have been re-quantified. The ‘match-between-run’ option (2 min) was used.
Data analysis
The obtained protein list was filtered for mitochondrial GO terms by using the freely available GProX software (Rigbolt et al, 2011) and further processed in Perseus (Cox et al, 2011). Only proteins identified in both biological replicates were considered and combined for analysis. Significance A with Benjamini–Hochberg FDR set to 0.05 was used to identify significantly upregulated and downregulated proteins between α-synuclein and vector control cells.
Supplementary Material
Acknowledgments
This work was supported by the Austrian Science Fund FWF (grants T414-B09 and V235-B09 to SB, S-9304-B05 to FM and DC-G, LIPOTOX, P24381 and P23490 to FM and DK-MCD to LH and FM), the European Research Council (ERC, to NT), the European Commission (Apo-Sys to FM and TE), the research council of the KULeuven (DBOF fellowship to FM, OT 08-052A), IWT-Vlaanderen (SBO NeuroTARGET to JW and VB), and the FWF and the Deutsche Forschungsgemeinschaft DFG for grant I2000 to FM and the DFG, Excellence Initiative of the German Federal and State Governments (EXC 294 BIOSS; GSC-4 Spemann Graduate School to AAT). TE is recipient of an APART fellowship of the Austrian Academy of Sciences at the Institute of Molecular Biosciences, University of Graz. In addition, this work is supported by grants to GK from the Ligue Nationale contre le Cancer (Equipes labellisée), Agence Nationale pour la Recherche (ANR), the Longevity Research Chair of the AXA Foundation, Association pour la Recherche sur le Cancer, European Commission (ArtForce), European Research Council (Advanced Investigator Award), Fondation pour la Recherche Médicale, Institut National du Cancer, Cancéropôle Ile-de-France, Fondation Bettencourt-Schueller, the LabEx Onco-Immunology, and the Paris Alliance of Cancer Research Institutes.
Author contributions: SB, FM, GK and SJS planned the project and wrote the manuscript. SB, FM, LH, FB, FNV, VB, NT, GGK, JD, CM and SJS analysed the data. SB, LH, FB, DR, FNV, MV, FrM, VK, DC-G, TE, JR, MM, AAT, StB, CR, RB, CVdH, TB, AvdP and GGK performed the experiments. SB, FM, K-UF, JW, VB, NT, CM and SJS conceived and designed the experiments.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abou-Sleiman PM, Muqit MM, Wood NW (2006) Expanding insights of mitochondrial dysfunction in Parkinson’s disease. Nat Rev Neurosci 7: 207–219 [DOI] [PubMed] [Google Scholar]
- Auluck PK, Chan HYE, Trojanowski JQ, Lee VMY, Bonini NM (2002) Chaperone suppression of alpha-synuclein toxicity in a Drosophila model for Parkinson’s disease. Science 295: 865–868 [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415 [DOI] [PubMed] [Google Scholar]
- Brandina I, Graham J, Lemaitre-Guillier C, Entelis N, Krasheninnikov I, Sweetlove L, Tarassov I, Martin RP (2006) Enolase takes part in a macromolecular complex associated to mitochondria in yeast. Biochim Biophys Acta 1757: 1217–1228 [DOI] [PubMed] [Google Scholar]
- Büttner S, Bitto A, Ring J, Augsten M, Zabrocki P, Eisenberg T, Jungwirth H, Hutter S, Carmona-Gutierrez D, Kroemer G, Winderickx J, Madeo F (2008) Functional mitochondria are required for alpha-synuclein toxicity in aging yeast. J Biol Chem 283: 7554–7560 [DOI] [PubMed] [Google Scholar]
- Büttner S, Eisenberg T, Carmona-Gutierrez D, Ruli D, Knauer H, Ruckenstuhl C, Sigrist C, Wissing S, Kollroser M, Frohlich KU, Sigrist S, Madeo F (2007) Endonuclease G regulates budding yeast life and death. Mol Cell 25: 233–246 [DOI] [PubMed] [Google Scholar]
- Büttner S, Faes L, Reichelt WN, Broeskamp F, Habernig L, Benke S, Kourtis N, Ruli D, Carmona-Gutierrez D, Eisenberg T, D'hooge P, Ghillebert R, Franssens V, Harger A, Pieber TR, Freudenberger P, Kroemer G, Sigrist SJ, Winderickx J, Callewaert G et al. (2012) The Ca(2+)/Mn(2+) ion-pump PMR1 links elevation of cytosolic Ca(2+) levels to α-synuclein toxicity in Parkinson’s disease models. Cell Death Differ 20: 465–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiarugi A, Meli E, Moroni F (2001) Similarities and differences in the neuronal death processes activated by 3OH-kynurenine and quinolinic acid. J Neurochem 77: 1310–1318 [DOI] [PubMed] [Google Scholar]
- Chinta SJ, Mallajosyula JK, Rane A, Andersen JK (2010) Mitochondrial α-synuclein accumulation impairs complex I function in dopaminergic neurons and results in increased mitophagy in vivo. Neurosci Lett 486: 235–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung CY, Seo H, Sonntag KC, Brooks A, Lin L, Isacson O (2005) Cell type-specific gene expression of midbrain dopaminergic neurons reveals molecules involved in their vulnerability and protection. Hum Mol Genet 14: 1709–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole NB, DiEuliis D, Leo P, Mitchell DC, Nussbaum RL (2008) Mitochondrial translocation of α-synuclein is promoted by intracellular acidification. Exp Cell Res 314: 2076–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper AA, Gitler AD, Cashikar A, Haynes CM, Hill KJ, Bhullar B, Liu K, Xu K, Strathearn KE, Liu F, Cao S, Caldwell KA, Caldwell GA, Marsischky G, Kolodner RD, Labaer J, Rochet JC, Bonini NM, Lindquist S (2006) Alpha-synuclein blocks ER-Golgi traffic and Rab1 rescues neuron loss in Parkinson’s models. Science 313: 324–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26: 1367–1372 [DOI] [PubMed] [Google Scholar]
- Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res 10: 1794–1805 [DOI] [PubMed] [Google Scholar]
- Du G, Liu X, Chen X, Song M, Yan Y, Jiao R, Wang C-C (2010) Drosophila histone deacetylase 6 protects dopaminergic neurons against {alpha}-synuclein toxicity by promoting inclusion formation. Mol Biol Cell 21: 2128–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Mir M-Y, Detaille D, R-Villanueva G, Delgado-Esteban M, Guigas B, Attia S, Fontaine E, Almeida A, Leverve X (2008) Neuroprotective role of antidiabetic drug metformin against apoptotic cell death in primary cortical neurons. J Mol Neurosci 34: 77–87 [DOI] [PubMed] [Google Scholar]
- Friggi-Grelin F, Coulom H, Meller M, Gomez D, Hirsh J, Birman S (2003) Targeted gene expression in Drosophila dopaminergic cells using regulatory sequences from tyrosine hydroxylase. J Neurobiol 54: 618–627 [DOI] [PubMed] [Google Scholar]
- Gerard M, Deleersnijder A, Daniëls V, Schreurs S, Munck S, Reumers V, Pottel H, Engelborghs Y, Van den Haute C, Taymans J-M, Debyser Z, Baekelandt V (2010) Inhibition of FK506 binding proteins reduces alpha-synuclein aggregation and Parkinson’s disease-like pathology. J Neurosci 30: 2454–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heeman B, Van den Haute C, Aelvoet S-A, Valsecchi F, Rodenburg RJ, Reumers V, Debyser Z, Callewaert G, Koopman WJH, Willems PHGM, Baekelandt V (2011) Depletion of PINK1 affects mitochondrial metabolism, calcium homeostasis and energy maintenance. J Cell Sci 124: 1115–1125 [DOI] [PubMed] [Google Scholar]
- Huh W-K, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK (2003) Global analysis of protein localization in budding yeast. Nature 425: 686–691 [DOI] [PubMed] [Google Scholar]
- Ibrahimi A, Vande Velde G, Reumers V, Toelen J, Thiry I, Vandeputte C, Vets S, Deroose C, Bormans G, Baekelandt V, Debyser Z, Gijsbers R (2009) Highly efficient multicistronic lentiviral vectors with peptide 2A sequences. Hum Gene Ther 20: 845–860 [DOI] [PubMed] [Google Scholar]
- Karniely S, Rayzner A, Sass E, Pines O (2006) Alpha-complementation as a probe for dual localization of mitochondrial proteins. Exp Cell Res 312: 3835–3846 [DOI] [PubMed] [Google Scholar]
- Kovacs GG, Budka H (2010) Current concepts of neuropathological diagnostics in practice: neurodegenerative diseases. Clin Neuropathol 29: 271–288 [DOI] [PubMed] [Google Scholar]
- Lee BI, Lee DJ, Cho KJ, Kim GW (2005) Early nuclear translocation of endonuclease G and subsequent DNA fragmentation after transient focal cerebral ischemia in mice. Neurosci Lett 386: 23–27 [DOI] [PubMed] [Google Scholar]
- Leeuwenburgh C, Gurley CM, Strotman BA, Dupont-Versteegden EE (2005) Age-related differences in apoptosis with disuse atrophy in soleus muscle. Am J Physiol Regul Integr Comp Physiol 288: R1288–R1296 [DOI] [PubMed] [Google Scholar]
- Li LY, Luo X, Wang X (2001) Endonuclease G is an apoptotic DNase when released from mitochondria. Nature 412: 95–99 [DOI] [PubMed] [Google Scholar]
- Li W-W, Yang R, Guo J-C, Ren H-M, Zha X-L, Cheng J-S, Cai D-F (2007) Localization of alpha-synuclein to mitochondria within midbrain of mice. Neuroreport 18: 1543–1546 [DOI] [PubMed] [Google Scholar]
- Madeo F, Frohlich E, Ligr M, Grey M, Sigrist SJ, Wolf DH, Frohlich KU (1999) Oxygen stress: a regulator of apoptosis in yeast. J Cell Biol 145: 757–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo F, Herker E, Maldener C, Wissing S, Lachelt S, Herlan M, Fehr M, Lauber K, Sigrist SJ, Wesselborg S, Fröhlich KU (2002) A caspase-related protease regulates apoptosis in yeast. Mol Cell 9: 911–917 [DOI] [PubMed] [Google Scholar]
- Mattson MP (2007) Calcium and neurodegeneration. Aging Cell 6: 337–350 [DOI] [PubMed] [Google Scholar]
- Meisinger C, Pfanner N, Truscott KN (2006) Isolation of yeast mitochondria. Methods Mol Biol 313: 33–39 [DOI] [PubMed] [Google Scholar]
- Moore DJ, West AB, Dawson VL, Dawson TM (2005) Molecular pathophysiology of Parkinson’s disease. Annu Rev Neurosci 28: 57–87 [DOI] [PubMed] [Google Scholar]
- Owald D, Fouquet W, Schmidt M, Wichmann C, Mertel S, Depner H, Christiansen F, Zube C, Quentin C, Körner J, Urlaub H, Mechtler K, Sigrist SJ (2010) A Syd-1 homologue regulates pre- and postsynaptic maturation in Drosophila. J Cell Biol 188: 565–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panozzo C, Nawara M, Suski C, Kucharczyka R, Skoneczny M, Bécam AM, Rytka J, Herbert CJ (2002) Aerobic and anaerobic NAD+ metabolism in Saccharomyces cerevisiae. FEBS Lett 517: 97–102 [DOI] [PubMed] [Google Scholar]
- Parrish J, Li L, Klotz K, Ledwich D, Wang X, Xue D (2001) Mitochondrial endonuclease G is important for apoptosis in C. elegans. Nature 412: 90–94 [DOI] [PubMed] [Google Scholar]
- Parrish JZ, Yang C, Shen B, Xue D (2003) CRN-1, a Caenorhabditis elegans FEN-1 homologue, cooperates with CPS-6/EndoG to promote apoptotic DNA degradation. EMBO J 22: 3451–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piot C, Croisille P, Staat P, Thibault H, Rioufol G, Mewton N, Elbelghiti R, Cung TT, Bonnefoy E, Angoulvant D, Macia C, Raczka F, Sportouch C, Gahide G, Finet G, André-Fouët X, Revel D, Kirkorian G, Monassier JP, Derumeaux G et al. (2008) Effect of cyclosporine on reperfusion injury in acute myocardial infarction. N Engl J Med 359: 473–481 [DOI] [PubMed] [Google Scholar]
- Powers KM, Smith-Weller T, Franklin GM, Longstreth WT Jr, Swanson PD, Checkoway H (2003) Parkinson’s disease risks associated with dietary iron, manganese, and other nutrient intakes. Neurology 60: 1761–1766 [DOI] [PubMed] [Google Scholar]
- Pozniakovsky AI, Knorre DA, Markova OV, Hyman AA, Skulachev VP, Severin FF (2005) Role of mitochondria in the pheromone- and amiodarone-induced programmed death of yeast. J Cell Biol 168: 257–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao L, Hamamichi S, Caldwell KA, Caldwell GA, Yacoubian TA, Wilson S, Xie ZL, Speake LD, Parks R, Crabtree D, Liang Q, Crimmins S, Schneider L, Uchiyama Y, Iwatsubo T, Zhou Y, Peng L, Lu Y, Standaert DG, Walls KC et al. (2008) Lysosomal enzyme cathepsin D protects against alpha-synuclein aggregation and toxicity. Mol Brain 1: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Mann M, Ishihama Y (2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc 2: 1896–1906 [DOI] [PubMed] [Google Scholar]
- Reinders J, Wagner K, Zahedi RP, Stojanovski D, Eyrich B, Laan M, van der, Rehling P, Sickmann A, Pfanner N, Meisinger C (2007) Profiling phosphoproteins of yeast mitochondria reveals a role of phosphorylation in assembly of the ATP synthase. Mol Cell Proteomics 6: 1896–1906 [DOI] [PubMed] [Google Scholar]
- Rigbolt KTG, Vanselow JT, Blagoev B (2011) GProX, a user-friendly platform for bioinformatics analysis and visualization of quantitative proteomics data. Mol Cell Proteomics 10: O110.007450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarcz R, Pellicciari R (2002) Manipulation of brain kynurenines: glial targets, neuronal effects, and clinical opportunities. J Pharmacol Exp Ther 303: 1–10 [DOI] [PubMed] [Google Scholar]
- Schägger H, Pfeiffer K (2000) Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J 19: 1777–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1: 2856–2860 [DOI] [PubMed] [Google Scholar]
- Sickmann A, Reinders J, Wagner Y, Joppich C, Zahedi R, Meyer HE, Schonfisch B, Perschil I, Chacinska A, Guiard B, Rehling P, Pfanner N, Meisinger C (2003) The proteome of Saccharomyces cerevisiae mitochondria. Proc Natl Acad Sci USA 100: 13207–13212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AJ, Stone TW, Smith RA (2007) Neurotoxicity of tryptophan metabolites. Biochem Soc Trans 35: 1287–1289 [DOI] [PubMed] [Google Scholar]
- Sokolov S, Knorre D, Smirnova E, Markova O, Pozniakovsky A, Skulachev V, Severin F (2006) Ysp2 mediates death of yeast induced by amiodarone or intracellular acidification. Biochim Biophys Acta 1757: 1366–1370 [DOI] [PubMed] [Google Scholar]
- Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, Goedert M (1997) Alpha-synuclein in Lewy bodies. Nature 388: 839–840 [DOI] [PubMed] [Google Scholar]
- Stone TW, Darlington LG (2002) Endogenous kynurenines as targets for drug discovery and development. Nat Rev Drug Discov 1: 609. [DOI] [PubMed] [Google Scholar]
- Surmeier DJ, Guzman JN, Sanchez-Padilla J (2010) Calcium, cellular aging, and selective neuronal vulnerability in Parkinson’s disease. Cell Calcium 47: 175–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Yu J-T, Tan L (2012) The kynurenine pathway in neurodegenerative diseases: mechanistic and therapeutic considerations. J Neurol Sci 323: 1–8 [DOI] [PubMed] [Google Scholar]
- Uversky VN (2007) Neuropathology, biochemistry, and biophysics of alpha-synuclein aggregation. J Neurochem 103: 17–37 [DOI] [PubMed] [Google Scholar]
- Van der Goot AT, Zhu W, Vázquez-Manrique RP, Seinstra RI, Dettmer K, Michels H, Farina F, Krijnen J, Melki R, Buijsman RC, Ruiz Silva M, Thijssen KL, Kema IP, Neri C, Oefner PJ, Nollen EA (2012) Delaying aging and the aging-associated decline in protein homeostasis by inhibition of tryptophan degradation. Proc Natl Acad Sci USA 109: 14912–14917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissing S, Ludovico P, Herker E, Büttner S, Engelhardt SM, Decker T, Link A, Proksch A, Rodrigues F, Corte-Real M, Fröhlich KU, Manns J, Candé C, Sigrist SJ, Kroemer G, Madeo F (2004) An AIF orthologue regulates apoptosis in yeast. J Cell Biol 166: 969–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wogulis M, Chew ER, Donohoue PD, Wilson DK (2008) Identification of Formyl Kynurenine Formamidase and Kynurenine Aminotransferase from Saccharomyces cerevisiae Using Crystallographic, Bioinformatic and Biochemical Evidence‡. Biochemistry 47: 1608–1621 [DOI] [PubMed] [Google Scholar]
- Wu HQ, Guidetti P, Goodman JH, Varasi M, Ceresoli-Borroni G, Speciale C, Scharfman HE, Schwarcz R (2000) Kynurenergic manipulations influence excitatory synaptic function and excitotoxic vulnerability in the rat hippocampus in vivo. Neuroscience 97: 243–251 [DOI] [PubMed] [Google Scholar]
- Xu J, Kao SY, Lee FJ, Song W., Jin LW, Yankner BA (2002) Dopamine-dependent neurotoxicity of alpha-synuclein: a mechanism for selective neurodegeneration in Parkinson disease. Nat Med 8: 600–606 [DOI] [PubMed] [Google Scholar]
- Zamzami N, Kroemer G (2001) The mitochondrion in apoptosis: how Pandora’s box opens. Nat Rev Mol Cell Biol 2: 67–71 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.