Abstract
Listeria can escape host autophagy defense pathways through mechanisms that remain poorly understood. We show here that in epithelial cells, Listeriolysin (LLO)-dependent cytosolic escape of Listeria triggered a transient amino-acid starvation host response characterized by GCN2 phosphorylation, ATF3 induction and mTOR inhibition, the latter favouring a pro-autophagic cellular environment. Surprisingly, rapid recovery of mTOR signalling was neither sufficient nor necessary for Listeria avoidance of autophagic targeting. Instead, we observed that Listeria phospholipases PlcA and PlcB reduced autophagic flux and phosphatidylinositol 3-phosphate (PI3P) levels, causing pre-autophagosomal structure stalling and preventing efficient targeting of cytosolic bacteria. In co-infection experiments, wild-type Listeria protected PlcA/B-deficient bacteria from autophagy-mediated clearance. Thus, our results uncover a critical role for Listeria phospholipases C in the inhibition of autophagic flux, favouring bacterial escape from host autophagic defense.
Keywords: autophagy, innate immunity, Listeria, phospholipases C
Introduction
Listeria monocytogenes is a Gram-positive bacterial pathogen that invades host cells and rapidly lyses the entry vacuole to be free in the cytosol (Goebel and Kuhn, 2000). Once in the cytosol, Listeria uses actin-based motility to move intracellularly and spread from cell to cell. Bacterial escape from the entry vacuole requires expression of Listeriolysin O (LLO), a pore-forming toxin (Schnupf and Portnoy, 2007). Listeria also expresses two related membrane-digesting toxins, the phospholipases C (Plc) PlcA and PlcB, which are critical for Listeria escape from the double membrane surrounding the bacteria following cell-to-cell spread (Portnoy et al, 1992). PlcA/B also cooperate with LLO to mediate optimal rupture of the entry vacuole (Smith et al, 1995), although LLO is thought to play the predominant role at this step.
Autophagy is a critical cellular process through which cells degrade and recycle various intracellular cargos, such as organelles or multiprotein complexes (Klionsky, 2007). Autophagy is initiated at the level of pre-autophagosomal structures that are associated with the endoplasmic reticulum (ER), and the ER/mitochondria interface then provides the membranes necessary for the progressive extension of a double membrane that surrounds the cargo (Hamasaki et al, 2013), until full engulfment and formation of an autophagosome that is targeted to lysosomes for degradation of its content. While autophagy is inhibited by the checkpoint kinase mammalian target of rapamycin (mTOR) in metabolically replete cells, this process is strongly upregulated following mTOR inhibition in starved cells, allowing transient maintenance of metabolic supply through autophagic-mediated nutrient recycling (Wullschleger et al, 2006).
Although autophagy is emerging as a critical arm of the host defense against intracellular bacterial pathogens, the mechanism through which this process can be efficiently turned on in conditions of metabolic sufficiency remains poorly understood. We recently demonstrated that infection with Shigella and Salmonella causes a rapid induction of cytosolic AA starvation due to membrane damage (Tattoli et al, 2012a, 2012c), resulting in mTOR inhibition and autophagy induction. Because Listeria has been shown to escape bacterial autophagy through mechanisms that are incompletely understood, we aimed to characterize the interplay between AA starvation pathways, mTOR signalling and autophagy induction in Listeria-infected cells.
Results
Transient induction of amino-acid starvation pathways in Listeria-infected cells
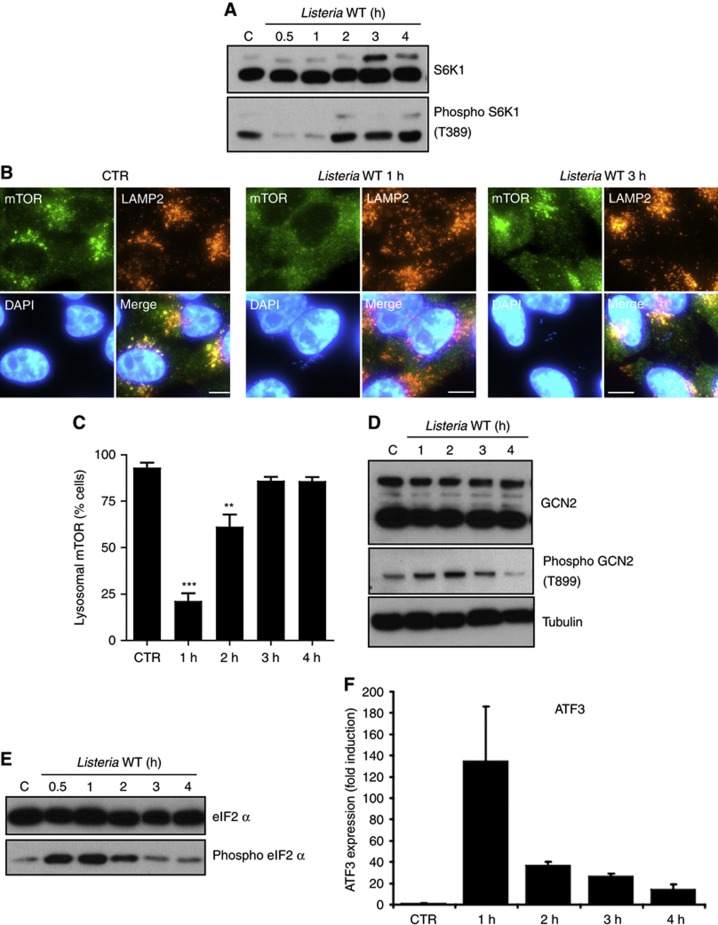
We infected human epithelial HeLa cells with Listeria and observed that S6K1, a major target of the kinase mTOR, was transiently dephosphorylated at 1 h post infection (p.i.), showing that the activity of mTOR was inhibited at this time (Figure 1A). In addition, mTOR localization to LAMP2-positive late endosomes and lysosomes was reduced at 1 h p.i. while recovering at later times (Figures 1B and C; Supplementary Figure S1A), suggesting that the transient inhibition of mTOR signalling at 1 h p.i. was due to the induction of a state of amino-acid (AA) starvation in Listeria-infected cells, as previously observed in the case of epithelial cells infected with Shigella and Salmonella (Tattoli et al, 2012a, 2012c). We noted that Listeria did not localize into LAMP2+ vesicles at either 1 h or 3 h p.i. (Figure 1B), in line with the well-characterized capacity of the bacterium to rapidly escape the invasion vacuole and reach the host cytosol. In support of an effect of Listeria on host AA starvation, we observed that the pathogen triggered the transient phosphorylation of the AA starvation kinase GCN2 (Figure 1D) and its target eIF2α (Figure 1E), as well as the transcriptional upregulation of the metabolic stress transcription factor ATF3 (Figure 1F), which are hallmarks of the integrated stress response (ISR) pathway induced by AA starvation. Thus, Listeria infection resulted in the transient upregulation of AA starvation response pathways, which peaked at ∼1 h p.i.
Figure 1.
Listeria induces a transient activation of host AA starvation pathways. (A) HeLa cells were infected with Listeria for 0.5–4 h, and lysates were analysed by blotting using indicated antibodies. (B) HeLa cells left unstimulated (CTR) or infected with Listeria for 1 or 3 h, analysed by IF using antibodies against mTOR and LAMP2. (C) Percentage of Listeria-infected cells displaying mTOR localization to LAMP2+ vesicles. Values are means±s.e.m. n=3. ***P<0.001 and **P<0.01 over uninfected. (D, E) HeLa cells were infected with Listeria for 0.5–4 h, and lysates were analysed by blotting using indicated antibodies. (F) qPCR analysis of ATF3 induction following infection of HeLa cells with Listeria for 1–4 h. Values are means±s.e.m. n=3. Scale bars: 10 μm.
Source data for this figure is available on the online supplementary information page.
LLO triggers AA starvation pathways in Listeria-infected cells
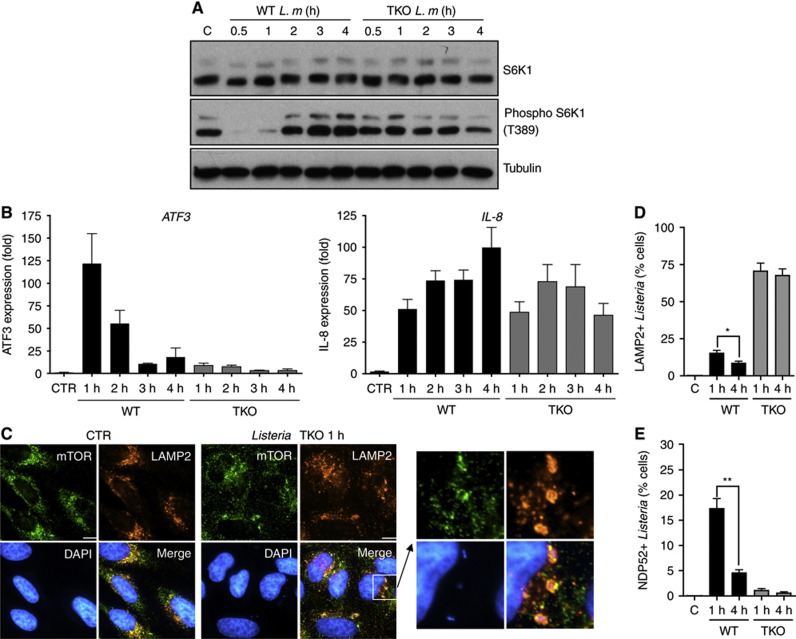
We next aimed to identify the bacterial determinants responsible for the early inhibition of mTOR signalling and ISR induction in Listeria-infected cells. Using a Listeria strain triple knockout (TKO) for the expression of the three major toxins, LLO, PlcA and PlcB, we observed that the transient downregulation of mTOR activity in Listeria-infected cells was toxin dependent (Figure 2A). ATF3 induction was also strongly blunted in cells infected with Listeria TKO compared to wild-type (WT) strain (Figure 2B, left), although both WT and TKO Listeria were detected by host cells similarly, as suggested by their comparable capacity to trigger IL-8 expression in infected cells (Figure 2B, right). Moreover, cytosolic dispersion of mTOR at 1 h p.i. was also not observed in cells infected with Listeria TKO (Figure 2C), thus implying that mTOR inhibition through AA starvation was not occurring with this mutant strain. Interestingly, both mTOR and LAMP2 were found to strongly accumulate at the surface of Listeria TKO-containing vacuoles (Figures 2C and D), as expected because Listeria TKO cannot escape the entry vacuole and is targeted to lysosomes for degradation. In contrast, WT but not TKO Listeria was found associated, although infrequently, with the membrane damage marker NDP52 at 1 h p.i. (Figure 2E; Supplementary Figure S1B), likely reflecting the fact that Listeria needs to damage the entry vacuole through the action of its toxins to escape to the cytosol. NDP52+ Listeria WT but not TKO vacuoles were also LAMP2+ and positive for the autophagy marker GFP-LC3 (Supplementary Figure S1C), indicating that damage to the Listeria vacuole is a signal for autophagy targeting, as previously proposed (Birmingham et al, 2007). The fact that Listeria targeting to NDP52+ autophagosomes remained infrequent at 1 h p.i. despite the concomitant inhibition of mTOR (see above) suggests that the bacterium can efficiently escape autophagy during the vacuole rupture phase, or that the vacuole escape is a very rapid event.
Figure 2.
Listeria toxins cause activation of host AA starvation pathways. (A) HeLa cells were infected with Listeria wild-type (WT) or the LLO/PlcA/PlcB triple knockout (TKO) mutant strain for 0.5–4 h, and lysates were analysed by blotting using indicated antibodies. (B) qPCR analysis of ATF3 and IL-8 induction following infection of HeLa cells with Listeria WT or TKO for 1–4 h. Values are means±s.e.m. n=3. (C) HeLa cells left unstimulated (CTR) or infected with Listeria TKO for 1 h, analysed by IF using antibodies against mTOR and LAMP2. (D, E) Percentage of cells infected with either WT or TKO Listeria strains displaying one or several LAMP2+ (D) or NDP52+ (E) Listeria vesicles. Values are means±s.e.m. n=3. *P<0.05; **P<0.01. Scale bars: 10 μm.
Source data for this figure is available on the online supplementary information page.
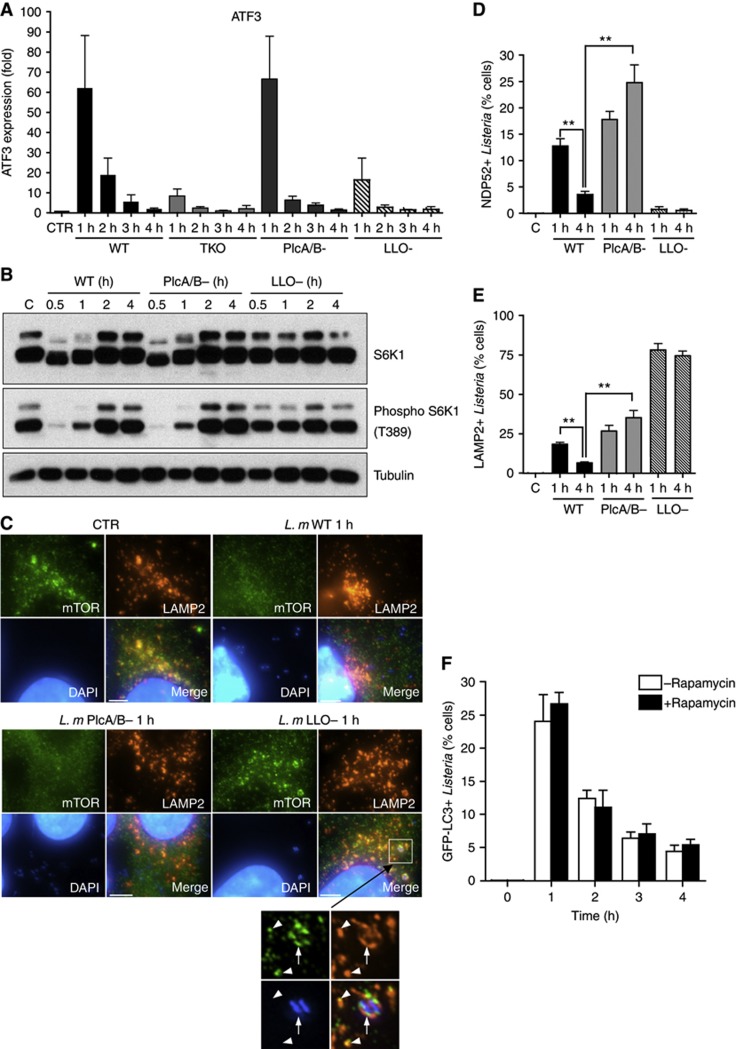
In order to identify the Listeria toxin(s) responsible for the induction of AA starvation pathways, we used strains deficient for either LLO expression (LLO−) or PlcA and PlcB expression (PlcA/B−). Similarly to the TKO strain, LLO− Listeria displayed blunted induction of ATF3 (Figure 3A) and unaltered levels of S6K1 phosphorylation at 1 h p.i. (Figure 3B), while PlcA/B− Listeria behaved like the WT strain (Figures 3A and B). Moreover, as for WT Listeria, PlcA/B− Listeria induced cytosolic dispersion of mTOR at 1 h p.i. (Figure 3C), while infection with LLO− Listeria did not affect mTOR localization to LAMP2+ late endosomes/lysosomes and resulted in accumulation of these proteins at the surface of LLO− Listeria-containing vacuoles (Figure 3C), similar to the results obtained with the TKO strain (see above). Together, these results demonstrate that Listeria phospholipases PlcA and PlcB do not contribute to the early induction of AA starvation pathways at 1 h p.i. and that this effect is mediated solely by the LLO pore-forming toxin.
Figure 3.
Distinct roles of LLO and PlcA/B in the modulation of host AA starvation pathways. (A) qPCR analysis of ATF3 induction following infection of HeLa cells with WT, TKO, PlcA/B− or LLO− Listeria strains for 1–4 h. Values are means±s.e.m. n=3. (B) HeLa cells were infected with WT, PlcA/B− or LLO− Listeria strains for 0.5–4 h, and lysates were analysed by blotting using indicated antibodies. (C) HeLa cells left unstimulated (CTR) or infected with WT, PlcA/B− or LLO− Listeria strains for 1 h, analysed by IF using antibodies against mTOR and LAMP2. The arrow shows a Listeria vacuole positive for mTOR and LAMP2, and arrowheads indicate mTOR- and LAMP2-positive late endosomes or lysosomes. (D, E) Percentage of cells infected with WT, PlcA/B− or LLO− Listeria strains displaying one or several LAMP2+ (D) or NDP52+ (E) Listeria vesicles. Values are means±s.e.m. n=3. **P<0.01. (F) Percentage of cells infected with WT Listeria, in the presence (black bars) or absence (white bars) of rapamycin, displaying one or several GFP-LC3+ Listeria autophagosomes. Values are means±s.e.m. n=2. Scale bars: 5 μm.
Source data for this figure is available on the online supplementary information page.
mTOR reactivation is neither necessary nor sufficient for Listeria autophagy escape
We analysed by immunofluorescence the recruitment of NDP52 and LAMP2 to the surface of LLO− and PlcA/B− Listeria vacuoles at 1–4 h p.i. NDP52 recruitment was LLO dependent but PlcA/B independent, thus showing that LLO is likely the only Listeria toxin responsible for host vacuole degradation at the cytosolic escape phase (Figure 3D). Concomitantly, a large number of LLO− Listeria vacuoles were LAMP2+ (Figures 3C–E) but GFP-LC3− (data not shown), suggesting that Listeria that remains trapped into their entry vacuole is routed to lysosomes through an endocytic rather than an autophagic mechanism. In the case of PlcA/B− Listeria, recruitment of NDP52 and LAMP2 to the vacuole occurred similarly to that of the WT at 1 h p.i. (Figures 3D and E), in line with the fact that this mutant strain displayed normal induction of AA starvation pathways at this early time point (see Figures 3A–C). However, we observed that significantly more PlcA/B− than WT Listeria were NDP52+ and LAMP2+ at 4 h p.i. (Figures 3D and E), and these bacteria were also GFP-LC3+ (Supplementary Figure S2), indicating that Listeria phospholipases C are required for efficient escape from autophagy at late times of infection, in agreement with previous observations (Birmingham et al, 2007; Py et al, 2007). Because induction of AA starvation pathways and mTOR inhibition were only transient in cells infected with the PlcA/B− strain (see Figures 3A–C), these results also imply that the normalization of host metabolic pathways cannot by itself account for Listeria autophagy escape at late times of infection. While mTOR signalling reactivation was not sufficient to support Listeria escape from autophagy, we next questioned whether, conversely, it was necessary in the case of the WT strain. To do so, cells were infected for 1–4 h with WT Listeria, in the presence or absence of rapamycin, a potent inhibitor of mTOR activity, and thus an inducer of autophagy. We observed that GFP-LC3 targeting of Listeria was similar in both conditions, and that it declined over time after an initial peak of autophagy targeting at 1 h p.i., in cells treated or not with rapamycin (Figure 3F). Thus, following an initial burst of metabolic stress pathway induction in Listeria-infected cells, mTOR reactivation appeared to be neither necessary nor sufficient to explain the capacity of the bacterium to escape autophagy at late time points of infection.
Listeria phospholipases C trigger the accumulation of NDP52+ cytosolic granules
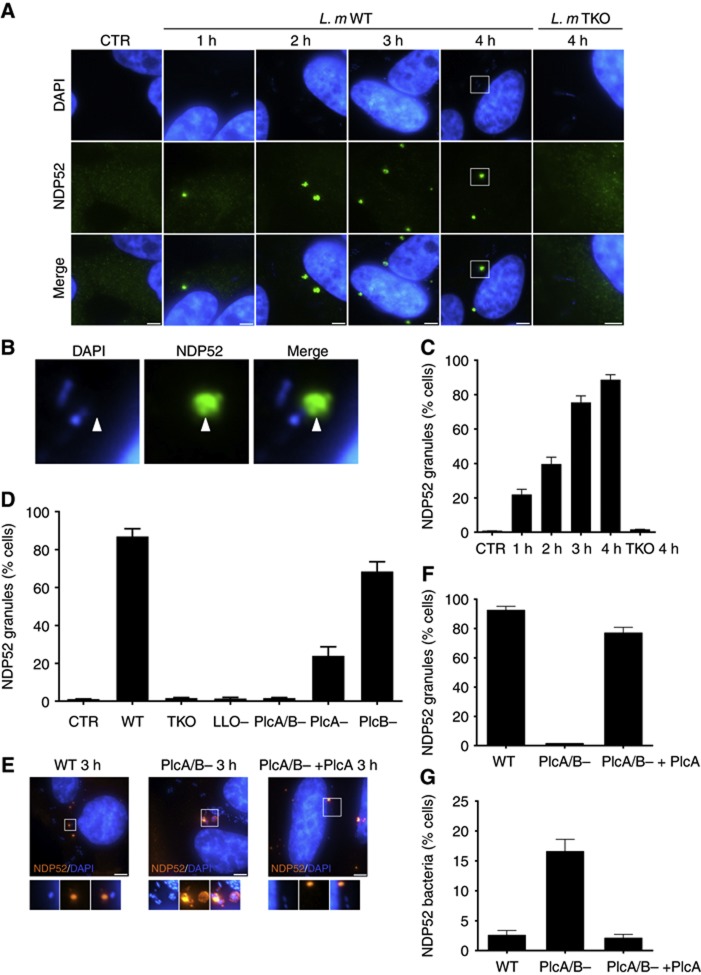
We next aimed to identify events occurring after 1 h p.i. that could account for the PlcA/B-dependent escape of Listeria from autophagic-mediated defense. While analysing the pattern of NDP52 immunofluorescence staining over time in Listeria-infected cells, we noticed that infected cells accumulated substantial amounts of NDP52+ granular structures (Figure 4A). These granules were frequently at the vicinity but generally not directly in contact with or surrounding intracellular bacteria (Figure 4B). There were few NDP52+ granules in cells infected for 1 h, but these structures progressively accumulated, and by 4 h p.i. most infected cells displayed one to a few (typically from 1 to 10 per cell) discrete granules scattered throughout the cytoplasm (Figure 4C). Importantly, the accumulation of these NDP52+ granules did not occur when cells were infected with the Listeria TKO strain, suggesting that their formation was either directly dependent upon the action of Listeria toxins, or required cytosolic exposure of Listeria (Figure 4A, right micrographs). Using Listeria strains deficient for individual toxins, we next showed that not only LLO but also surprisingly PlcA/B were required for the accumulation of NDP52+ granules (Figure 4D), suggesting that these structures were formed following exposure of PlcA/B to the host cytosol. Using Listeria mutants deleted for individual phospholipase C, we observed that both contributed to the formation of NDP52+ granules, although PlcA seemed to play the most critical role in this process (Figure 4D; Supplementary Figure S3A). Finally, we observed that ectopic expression of PlcA into PlcA/B− Listeria was sufficient to restore the capacity of the bacteria to trigger the formation of these granules (Figures 4E and F), while it dramatically reduced the targeting of bacteria deficient for the expression of endogeneous PlcA/B to NDP52+ autophagosomes (Figures 4E–G).
Figure 4.
PlcA/B-dependent accumulation of NDP52+ granules in Listeria-infected cells. (A) HeLa cells left unstimulated (CTR) or infected with Listeria WT for 1–4 h, or with Listeria TKO for 4 h and with analysed by IF using an antibody against NDP52. (B) Higher magnification view of the selected area from (A) showing that NDP52+ granules generally form at the vicinity of cytosolic Listeria. (C) Percentage of cells infected with Listeria WT for 1–4 h, or with Listeria TKO for 4 h, displaying one or several NDP52+ granules. Values are means±s.e.m. n=3. (D) Percentage of cells infected for 4 h with different Listeria mutants as indicated displaying one or several NDP52+ granules. Values are means±s.e.m. n=3. (E) HeLa cells infected with Listeria WT, PlcA/B− or PlcA/B− ectopically expressing PlcA for 3 h, analysed by IF using an antibody against NDP52. (F, G) Percentage of cells infected for 3 h with Listeria WT, PlcA/B− or PlcA/B− ectopically expressing PlcA displaying one or several NDP52+ granules (F) or bacteria entrapped in NDP52+ autophagosomes (G). Values are means±s.e.m. n=2. Scale bars: 5 μm.
PlcA/B-dependent NDP52+ granules are not associated with cell-to-cell spread
Because the most clearly identified function of Listeria phospholipases C is to degrade the double membrane vacuole formed after cell-to-cell spread, we questioned whether the accumulation of PlcA/B-dependent NDP52+ granules was associated with the event of cell-to-cell vacuole rupture. Similar accumulation of these granules was observed in cells infected with WT Listeria in the presence of cytochalasin D, an inhibitor of actin polymerization previously shown to block Listeria cell-to-cell spread (Tilney and Portnoy, 1989) (Supplementary Figure S3B). Next, we used ActA−Listeria, a strain that is deficient for actin-based motility and cannot spread from cell to cell. ActA−Listeria displayed transient induction of ATF3 expression (Supplementary Figure S3C), although ATF3 induction persisted longer in cells infected with ActA− than with WT Listeria. Moreover, ActA−Listeria triggered a transient inhibition of mTOR signalling (Supplementary Figure S3D) that was similar to that observed with the WT strain. ActA-Listeria was also indistinguishable from the WT strain with regard to the accumulation of NDP52+ granules (Supplementary Figure S3E), thus showing that the formation of these structures depends on PlcA/B but is independent from the role of these phospholipases C in the escape from the cell-to-cell spread vacuole.
NDP52+ granules contain damaged membrane markers and pre-autophagosomal structures
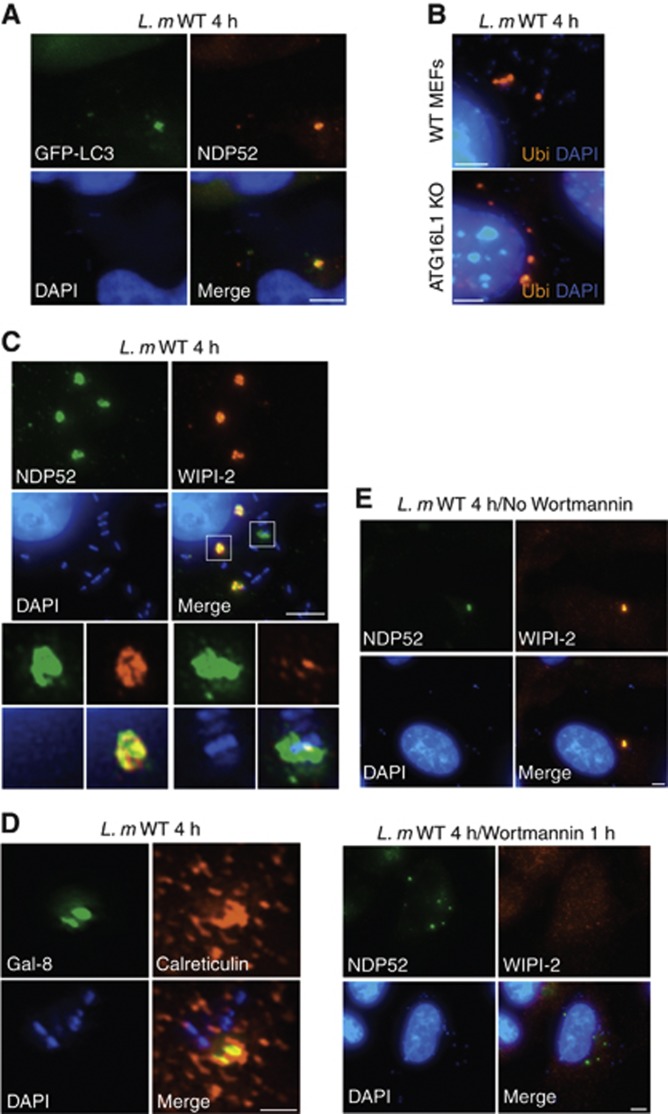
We next aimed to characterize the PlcA/B-dependent NDP52+ cytosolic granules. We first noted that these granules were also generally positive for ubiquitinated (Ubi) proteins (Supplementary Figure S4A), which represents another marker of bacteria-associated damaged membrane (Perrin et al, 2004), although NDP52+/Ubi− granules were also observed (Supplementary Figure S4A). Similarly to NDP52+ granules, we observed that Ubi+ granules strongly accumulated over time in Listeria-infected cells (Supplementary Figures S4B and C), and were absent in cells infected with the Listeria TKO strain (Supplementary Figure S4B). Galectin-8 has recently been identified as an NDP52-binding protein at the surface of Salmonella-containing vacuoles and other damaged membranes (Thurston et al, 2012), and we observed that Listeria-induced NDP52+ granules were also Galectin-8+ (Supplementary Figure S4D). However, the granules were LAMP2 negative, showing that these were distinct from late endosomes or lysosomes (Supplementary Figure S4E). Together, these results suggest that Listeria-induced granules contain damaged membranes, possibly representing membrane remnants of the degraded Listeria entry vacuole.
The role of damage membrane detection by the ubiquitin and NDP52/Galectin-8 in the recruitment of the autophagy protein LC3 is well documented in the case of the Salmonella-containing vacuole (Thurston et al, 2012). Interestingly, Listeria PlcA/B-dependent granules also contained GFP-LC3 (Figure 5A), as well as endogeneous LC3 (Supplementary Figure S5A), thus suggesting that Listeria-induced damaged membranes likely serve as a signal for the recruitment of autophagy proteins. The accumulation of ubiquitin in the granules was still observed in Listeria-infected atg16l1−/− mouse embryonic fibroblasts (MEFs) (Figure 5B), showing that membrane damage detection precedes and is independent from the recruitment of the LC3 conjugation machinery in the granules. In agreement, endogeneous ATG16L1 and the pre-autophagosomal structure marker DFCP-1 accumulated on bacterial autophagosomes at 1 h p.i. (Supplementary Figures S6–S7) as expected, and were both found in the accumulating granules at 4 h p.i. (Supplementary Figures S5B and S6).
Figure 5.
NDP52+ granules in Listeria-infected cells have characteristics of pre-autophagosomal structures. (A) HeLa cells transfected with GFP-LC3 and infected with Listeria WT for 4 h were analysed by IF using an antibody against NDP52. (B) Murine embryonic fibroblasts (MEFs) from WT or ATG16L1-deficient (ATG16L1 KO) mice infected with Listeria WT for 4 h were analysed by IF using an antibody against ubiquitinated proteins (Ubi). (C, D) HeLa cells infected with Listeria WT for 4 h analysed by IF using antibodies against NDP52 and WIPI-2 (C) or Galectin-8 (Gal-8) and calreticulin (D). (E) HeLa cells infected with Listeria WT for 4 h, in the absence or presence of Wortmannin added during the last hour of infection, analysed by IF using antibodies against NDP52 and WIPI-2. Scale bars: 5 μm for all panels except (D) (1.5 μm).
WIPI-2 is a phosphatidylinositol 3-phosphate (PI3P) binding protein that has been shown to localize to pre-autophagosomal structures or phagophores, but not to mature autophagosomes (Polson et al, 2010). The localization of WIPI-2 to the pre-autophagosomal structures is dependent on the activity of the Wortmannin-sensitive Vps34/Beclin-1 kinase complex, which generates PI3P from PI (Funderburk et al, 2010). Interestingly, we observed that PlcA/B-dependent Listeria granules were strongly WIPI-2+ (Figure 5C), thus implying that these structures contain maturing pre-autophagosomal structures, but not mature autophagosomes. Although autophagy-targeted Listeria was a rare event at 3–4 h p.i., we noted in this case that the bacteria were positive for NDP52 (Figure 5C) and LC3 (data not shown) but were WIPI-2 negative as expected since WIPI-2 does not localize to mature autophagosomes. In addition, we also observed that PlcA/B-dependent granules localized in close vicinity with the ER, which is the membrane source required for the maturing pre-autophagosomal structures (Figure 5D). Using Wortmannin, we next observed that WIPI-2 recruitment to the PlcA/B-dependent granules was Vps34/Beclin-1 dependent and thus likely required PI3P, as expected for maturing pre-autophagosomal structures (Figure 5E). Finally, we knocked down the expression of WIPI-2 using lentiviral-mediated transduction of a plasmid expression small hairpin RNA (shRNA) against WIPI-2 (Supplementary Figure S8A), and the functional knock-down was validated in AA-starved cells (Supplementary Figure S8B). Interestingly, silencing of WIPI-2 did not affect the accumulation of the membrane damage marker Galectin-8 to the granules, but inhibited the recruitment of LC3 (Supplementary Figure S9), showing that the PlcA/B-dependent granules are formed in a coordinated and sequential manner dependent on the activity of the autophagy protein WIPI-2.
Thus, these results demonstrate that WIPI-2+ PlcA/B-dependent granules that form away from bacteria are functionally distinct from mature autophagosomes that target Listeria, and represent a premature structure composed of damaged membranes and pre-autophagosomal structures.
PlcA/B-induced granules accumulate as a result of reduced autophagic flux and PI3P depletion
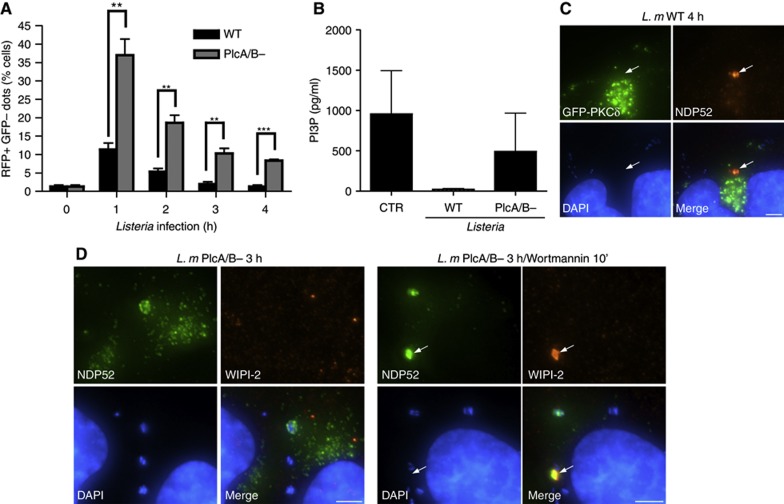
We next aimed to investigate the mechanism through which Listeria phospholipases C contribute to the accumulation of pre-autophagosomal structures. We first sought to determine whether these toxins had an impact on the autophagic flux in infected cells. To follow autophagic flux, cells were transfected with an expression vector encoding LC3 fused to a tandem of GFP and RFP. Because the fluorescence of GFP is more sensitive to pH than that of RFP, and is quenched at lysosomal pH, this construct can be used to quantify the rate of progression of autophagosomes (yellow fluorescence) to autolysosomes (red fluorescence) as a measure of the autophagic flux. Interestingly, while the autophagic flux appeared to be maximal at 1 h p.i. and declined sharply thereafter for cells infected with both Listeria WT and PlcA/B−, this flux was significantly stronger in the case of the PlcA/B− strain (Figure 6A), suggesting that Listeria phospholipases C inhibit the autophagic flux. In agreement, we also observed that NDP52+ granules in infected cells expressing GFP-RFP-LC3 were systematically positive for both GFP and RFP (Supplementary Figure S10), implying that these granules do not progress to autolysosomes.
Figure 6.
Accumulation of PlcA/B-induced granules as a result of reduced autophagic flux and PI3P depletion. (A) HeLa cells were transfected with GFP-RFP-LC3 and either were left uninfected (t=0) or were infected with Listeria WT or PlcA/B− for 1–4 h. GFP+ RFP+ and GFP− RFP+ puncta appear yellow and red, respectively. Percentage of cells infected with Listeria WT or PlcA/B− for 1–4 h displaying GFP− RFP+ red puncta were quantified. Values are means±s.e.m. n=3. **P<0.01; ***P<0.001. (B) HeLa cells left unstimulated (CTR) or infected with Listeria WT or PlcA/B− for 4 h were lysed and PI3P levels measured by competitive ELISA. (C) HeLa cells transfected with GFP-PKCδ and infected with Listeria WT for 4 h were analysed by IF using an antibody against NDP52. The arrow indicates an NDP52+ granule. (D) HeLa cells infected with Listeria PlcA/B− for 3 h, in the absence or presence of Wortmannin added during the last 10 min of infection, analysed by IF using antibodies against NDP52 and WIPI-2. The arrow indicates an NDP52+ granule. Scale bars: 5 μm.
PI3P plays a critical role in autophagic flux and autophagosome formation, and is generated from PI by the Vps34/Beclin-1 complex (Funderburk et al, 2010). Interestingly, bacterial phospholipases C are known to cleave PI into diacylglycerol (DAG) and myo-inositol very efficiently, although they cannot cleave PI3P (Heinz et al, 1998), raising the possibility that Listeria PlcA/B could affect PI3P levels indirectly by consuming PI. In line with this hypothesis, we observed that infection with WT but not with PlcA/B− Listeria resulted in a dramatic drop in the levels of cellular PI3P (Figure 6B). It is unlikely that Listeria PlcA/B directly hydrolysed PI3P at the maturing pre-autophagosomal structure, since DAG did not accumulate in the PlcA/B-dependent granules, as observed by the absence of targeting of the DAG-interacting protein PKCδ to these structures (Figure 6C). In order to demonstrate that Listeria PlcA/B triggered the formation of NDP52+ granules through the reduction in cellular levels of PI3P, we hypothesized that infection with PlcA/B− Listeria could result in granule formation if PI3P levels were lowered by Wortmannin-mediated inhibition of the Vps34/Beclin-1 complex. In agreement with this, cells infected with PlcA/B− Listeria for 3 h and incubated with Wortmannin during the last 10 min displayed NDP52+ granule accumulation while cells infected in the absence of Wortmannin did not (Figure 6D). Incubation with Wortmannin for longer periods resulted in the disappearance of the granules (data not shown), in agreement with the above findings (see Figure 5E), which demonstrate that long treatment (1 h) with Wortmannin inhibits NDP52+ granule accumulation.
Together, these results suggest that pre-autophagosomal structure stalling may occur when autophagic flux and cellular levels of PI3P fall under a threshold level. Moreover, because the PI3P-binding protein WIPI-2 still accumulated in PlcA/B-dependent granules, these observations suggest that pre-autophagosomal structure progression beyond the WIPI-2 step likely requires additional PI3P-dependent events with distinct sensitivities to PI3P depletion.
Inhibition of pre-autophagosomal maturation by Listeria PlcA/B protects against autophagy
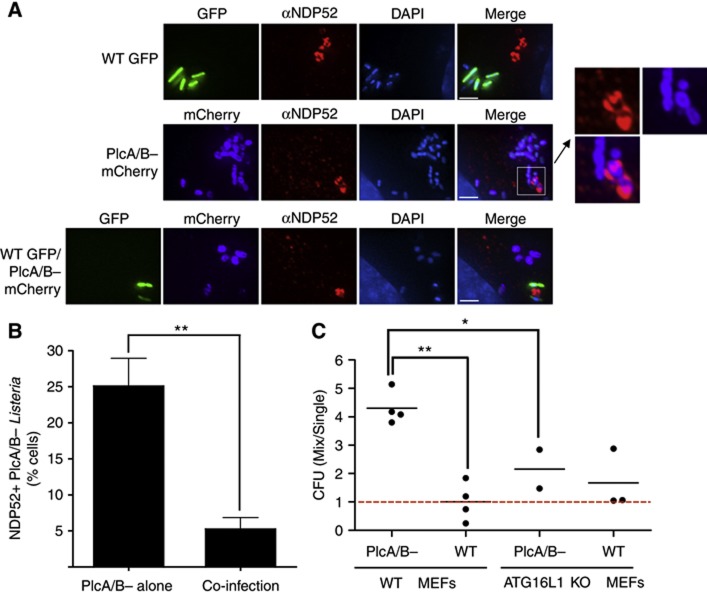
We next aimed to determine whether the capacity of Listeria PlcA/B to inhibit pre-autophagosomal elongation was solely responsible for PlcA/B-dependent escape from autophagy, or whether other mechanisms could account for this. In particular, it has been proposed that PlcA/B− Listeria could be more efficiently targeted by the autophagic machinery than WT Listeria because of a slower escape from the entry vacuole (Py et al, 2007). In order to test these two distinct mechanisms, we reasoned that if Listeria PlcA/B escaped autophagy at the step of the entry vacuole rupture, the protection conferred by the phospholipases C would be restricted to the bacteria producing these toxins (protection in cis). In contrast, in the case of overall autophagic flux inhibition and pre-autophagosomal structure stalling by cytosolic PlcA/B, inhibition of autophagic targeting could protect even bacteria that do not express the toxins (protection in trans). We thus used a WT Listeria strain expressing GFP (WT/GFP) and a PlcA/B− Listeria strain expressing mCherry (PlcA/B−/mCherry) in co-infection experiments. As controls, we observed that infection with WT/GFP alone for 4 h resulted in the formation of NDP52+ granules (Figure 7A), and that infection with PlcA/B−/mCherry alone did not result in NDP52+ granule formation while a number of bacteria were NDP52 decorated, indicative of autophagic targeting (Figure 7A). Interestingly, when cells were infected with both WT/GFP and PlcA/B−/mCherry, the number of NDP52-decorated PlcA/B−/mCherry was significantly reduced (Figures 7A and B), suggesting that phospholipases C produced by WT/GFP protected PlcA/B−/mCherry in trans against autophagic targeting. In line with these results, gentamycin protection assays revealed that ≈4x more PlcA/B− Listeria could be recovered from co-infection with WT Listeria than single infection in WT MEFs, while co-infection with PlcA/B− Listeria did not confer a growth advantage to WT Listeria (Figure 7C). Importantly, the protection conferred by WT Listeria on PlcA/B− Listeria growth in co-infection was significantly reduced in atg16l1−/− MEFs as compared to WT MEFs (Figure 7C), thus showing that WT Listeria protected PlcA/B− Listeria through its effect on autophagy.
Figure 7.
Inhibition of pre-autophagosomal maturation by Listeria PlcA/B protects Listeria against autophagy in trans. (A, B) HeLa cells infected for 4 h with Listeria WT expressing GFP (WT GFP), Listeria PlcA/B− expressing mCherry (PlcA/B− mCherry), or both Listeria strains at a 1:1 ratio, were analysed by IF using an antibody against NDP52 (A). (B) Quantification of the percentage of cells displaying NDP52+ PlcA/B− mCherry from the experiments described in (A), in cells infected with either PlcA/B− mCherry alone or WT GFP and PlcA/B−mCherry (co-infection). Values are means±s.e.m. n=3. **P<0.01. (C) Murine embryonic fibroblasts (MEFs) from WT or ATG16L1-deficient (ATG16L1 KO) mice infected for 4 h with WT GFP or Listeria PlcA/B− Listeria, either separately (Single) or in combination (Mix) were lysed and colony forming units (CFUs) determined on agar plates containing appropriate selection markers. Each point represents the value from individual independent experiments. **P<0.01; *P<0.05. Scale bars: 2.5 μm.
Together, these results support a model whereby the PlcA/B-dependent inhibition of pre-autophagosomal structure maturation, resulting in the formation of NDP52+ granules in the cytosol of infected cells, contributes to the protection of Listeria against autophagy-mediated clearance.
Discussion
Autophagy has emerged as a critical cellular process that mediates host defense against intracellular bacterial pathogens. Recent progress has been made towards the elucidation of the specific host molecules involved in the targeting of intracellular bacteria for autophagic clearance. Indeed, it was recently shown that molecules such as ubiquitinated proteins, NDP52, Galectin-8, p62 and DAG are part of molecular complexes that allow the recruitment of the autophagic machinery around bacteria (Shahnazari et al, 2010; Randow, 2011; Tattoli et al, 2012b). However, the mechanisms that govern the induction of this process in infected cells remain poorly understood. In particular, since autophagy is actively repressed in normally growing cells that do not experience metabolic starvation, understanding the mechanisms underlying metabolic transition in infected cells is of importance. We recently demonstrated that the intracellular bacterial pathogens Shigella and Salmonella rapidly inhibit mTOR signalling as a result of the induction of an AA starvation response in infected cells (Tattoli et al, 2012a, 2012c). Shigella-induced mTOR inhibition and AA starvation response is sustained while Salmonella only causes a transient metabolic stress response. Mechanistically, the induction of an AA starvation response in the infected cells results from host membrane damage likely caused by the insertion of the type III secretion system (TTSS) through the host entry vacuole (Tattoli et al, 2012a, 2012c). Antibacterial autophagic response is thus primed by the induction of a rapid AA starvation response in infected cells, highlighting how the modulation of host metabolic pathways impacts on cellular innate defense against bacterial pathogens.
We here demonstrate that, in addition to the Gram-negative bacterial pathogens Shigella and Salmonella, the Gram-positive bacterial pathogen Listeria also triggered a rapid induction of host AA starvation responses in epithelial cells, characterized by the inhibition of mTOR signalling and the upregulation of the GCN2-dependent ISR pathway, thus reinforcing the concept that AA starvation is a general host response to invasive bacterial pathogens. We also show that Listeria-induced AA starvation response was caused by LLO, a pore-forming toxin that inserts into and causes severe damage to host membranes, thus supporting our previous observations that bacteria-induced AA starvation was driven by host membrane damage (Tattoli et al, 2012a, 2012c). These results are also in agreement with recent reports demonstrating that several pore-forming toxins, including streptolysin O from Group A Streptococcus, a pore-forming toxin related to Listeria LLO, trigger GCN2 phosphorylation and promote autophagy through mTOR inhibition in mammalian cells (Kloft et al, 2010; von Hoven et al, 2012). Similarly, a pore-forming toxin from Pseudomonas entomophila has been shown to contribute to the inhibition of TOR and the activation of GCN2 in Drosophila, thus resulting in translation inhibition (Chakrabarti et al, 2012). It remains unclear how membrane damage results in AA starvation, but it has been recently proposed that bacterial pore-forming toxins, including LLO, could induce potassium efflux resulting in a decrease in cytoplasmic potassium (Kloft et al, 2010; Gonzalez et al, 2011), and this effect has been proposed to alter the activity of AA transporters (Kloft et al, 2010).
The analysis of mTOR and GCN2/ATF3 pathways revealed that LLO-mediated induction of AA starvation responses peaked at ∼1 h p.i. before a progressive normalization occurring at 3–4 h p.i. in Listeria-infected cells. These results are consistent with the kinetics of LLO-dependent entry vacuole escape and are also in line with the reported maximal targeting of Listeria to autophagosomes at ∼1 h p.i. (Birmingham et al, 2007; Py et al, 2007; Meyer-Morse et al, 2010). The transient nature of AA starvation responses in Listeria-infected cells is similar to the case of Salmonella-infected cells, which likely damage the Salmonella-containing vacuole by inserting the TTSS encoded by the Salmonella pathogenicity island 1 (Birmingham and Brumell, 2006; Tattoli et al, 2012a). However, a major difference between these two bacteria is that in the case of Salmonella, mTOR reactivation at the surface of the Salmonella-containing vacuole was critical for efficient escape from autophagy (Tattoli et al, 2012c), while Listeria appears to be capable of disarming autophagy independently from host metabolic pathways normalization.
Our study identified a key role for Listeria phospholipases C in bacterial escape from host autophagy by inhibiting autophagic flux and pre-autophagosomal structure maturation. It must be noted that other mechanisms of autophagy subversion have been proposed for this pathogen. Yoshikawa et al (2009) reported that Listeria efficiently disguises from autophagy targeting by recruiting the Arp2/3 complex and Ena/VASP in an ActA-dependent manner, and this action was independent from the known role for ActA in controlling actin-based motility. Similarly, Dortet et al (2011) showed that the bacterial cell wall-anchored protein InlK allowed Listeria to escape autophagy detection by recruiting the host major vault protein (MVP) (Dortet et al, 2011). Taken together, these results suggest that Listeria likely developed several complementary strategies to subvert host autophagy: (i) transient burst of LLO action, allowing rapid normalization of mTOR signalling; (ii) inhibition of cytosolic autophagy targeting using molecular coating complexes, such as InlK/MVP or ActA/Arp2/3; and (iii) stalling of pre-autophagosomal structures in a PlcA/B-dependent manner.
Listeria phospholipases C were identified over four decades ago and have been extensively studied. It was rapidly recognized that Listeria PlcA/B are essential for bacterial virulence in vivo (Camilli et al, 1991; Mengaud et al, 1991) and for cell-to-cell spread in vitro (Sun et al, 1990). Contrary to the entry vacuole formed after infection by extracellular Listeria (the primary vacuole), the vacuole formed after cell-to-cell spread (the secondary vacuole) is made of a double membrane, and PlcA/B have been shown to be essential for the cytosolic escape from the secondary vacuole through their capacity to degrade host phospholipids (Vazquez-Boland et al, 1992), thus allowing bacterial dissemination. It has been suggested that Listeria PlcA/B also contribute to accelerate LLO-dependent degradation of the primary vacuole (Smith et al, 1995), although PlcA/B− Listeria escape to the host cytosol efficiently. Our results report the first identification of a role for Listeria PlcA/B during the cytosolic phase of the infection, and suggest that, in addition to pre-autophagosomal structure stalling, other PI3P-dependent cellular processes such as vesicular trafficking could be affected by PlcA/B action.
Bacterial subversion of autophagy-mediated clearance has been characterized for several intracellular bacterial pathogens. While camouflaging from ongoing autophagy is a strategy used for instance by Shigella (Ogawa et al, 2005), several pathogens have evolved mechanisms to interfere with autophagic initiation or flux. Mycobacterium tuberculosis ESX-1 secretion system was found to be involved in the inhibition of autophagosome–lysosome fusion (Romagnoli et al, 2012) and Legionella pneumophila effector protein RazV was shown to inhibit autophagy by provoking an irreversible modification of Atg8, thus preventing its conjugation to phosphatidyl ethanolamine, a critical step in autophagosome maturation (Choy et al, 2012). The inhibitory effect of Listeria PlcA/B on pre-autophagosomal maturation is thus part of this second category of mechanisms evolved by bacterial pathogens to escape bacterial autophagy, which relies on the inhibition of the core autophagic machinery rather than on the camouflage from ongoing autophagy.
In sum, our results have characterized the interplay between Listeria and host metabolic stress pathways, which impact on autophagic defense programme in infected cells. While infected host cells were able to prime a pro-autophagic cellular environment through the induction of AA starvation pathways in response to LLO-mediated membrane damage, PlcA/B appeared to contribute to the bacterial escape from autophagy by lowering cellular levels of PI3P, likely causing inhibition of autophagic flux and premature stalling of maturing pre-autophagosomal structures. These results suggest that Listeria PlcA/B inhibit a yet uncharacterized PI3P-dependent step downstream of LC3 conjugation required for autophagosomal maturation in infected cells. Thus, these findings broaden our understanding of Listeria pathogenesis and provide new insights into the mechanisms that govern autophagy induction and maturation during infection with intracellular bacterial pathogens.
Materials and methods
Antibodies and reagents
Mouse anti-LAMP2 (ab25631), anti-GCN2 (ab70214), rabbit anti-Phospho-GCN2 (ab75836), rabbit anti-NDP52 (ab68588), mouse anti-Calreticulin (ab4), Abcam; mouse anti-tubulin clone DM1A (T9026), anti-ZFYVE-1 (DFCP-1; SAB2103609), Sigma; rabbit anti-mTOR (#2983, 7C10), rabbit anti-Phospho-p70S6 Kinase (Thr389) (#108D2), rabbit anti-p70S6 Kinase (#6198), anti-eIF2α (#9722), anti-phospho-eIF2α (#3597), anti-LC3 (#4108S), Cell Signaling Technology; mouse anti-Galectin 8, R&D Systems; mouse anti-ubiquitin (clone FK2), Millipore; anti-ATG16L1 (#PM040), MBL; mouse anti-WIPI-2 has been previously described (Polson et al, 2010); goat anti-rabbit IgG and Goat anti-mouse IgG peroxidase conjugated, Thermo Scientific USA; FITC-conjugated Goat anti-rabbit and Cy3-conjugated Goat anti-mouse, Jackson ImmunoResearch Laboratories, Canada; 4′,6-diamidio-2-phenylindole (DAPI), Vector Laboratories; Wortmannin, Sigma was used at 100 nM; Rapamycin, LKT Laboratories was used at 25 μg/ml. Cytochalasin D, Sigma was used at 1 μg/ml.
Bacterial strains and cell culture
Listeria monocytogenes (WT) 10403S and the isogenic Δhly (aka LLO−; DLP2116), plcA/B− (DLP1936), plcA− (DLP1552), plcB− (DLP1935) and actA− (DLP3078) strains were from Dr Dan Portnoy (U Berkeley) and grown in Brain Heart Infusion broth (BHI; Becton Dickinson). WT Listeria 10403S expressing GFP (WT/GFP) on a plasmid carrying resistance to chloramphenicol (NF-L1109) was a kind gift from Dr Nancy Freitag. When required, chloramphenicol and kanamycin were used at final concentrations of 10 and 30 μg/ml, respectively, for L. monocytogenes strains. A plasmid expressing Listeria PlcA (from Dr D Portnoy) was introduced into Listeria PlcA/B− strain by electroporation. The human epithelial cell line HeLa and MEFs WT and ATG16L1−/− (a kind gift from Dr Yoshimori, Osaka University) were cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS), 2 mM L-glutamine, 50 IU of penicillin and 50 μg/ml streptomycin. Cells were maintained in 95% air, 5% CO2 at 37°C. Endotoxin-free FCS and phosphate buffer saline (PBS) were from Wisent (St-Bruno, Quebec). FCS was used after inactivation at 56°C for 30 min.
Generation of mCherry-expressing Listeria ΔplcA/B (PlcA/B−)
The mCherry gene was amplified with the forward primer 5′-GGTACCATGGTGAGCAAG-3′ including a Kpn site (underlined) and with the reverse primer 5′-GTTAACTTACTTGTACAGCT-3′ including the EcoRI site (underlined) and cloned into pIMK2 expressing a kanamycin-resistance cassette, a gift from Dr Nancy E Freitag. The plasmid was electroporated (10 kV/cm, 400 Ω, and 25 μF) into PlcA/B− Listeria.
Bacterial infections
Cells cultured in antibiotic-free medium were infected with exponentially growing Listeria strains such that the multiplicity of infection was 50. Bacteria and cells were centrifuged at 2000 g for 10 min at 37°C, and incubated at 37°C/5% CO2 for 30 min. Cells were washed three times with PBS and treated with fresh gentamicin-containing complete medium (50 μg/ml). In co-infection experiments, WT Listeria 10403S chloramphenicol (Cm)-resistant strain (NF-L1109) WT/GFP was co-infected with ΔplcA/B kanamycin (Km)-resistant strain PlcA/B−/mCherry. Co-infected cells were lysed and serial dilution in 0.9% NaCl were plated onto BHI Agar containing either Cm or Km antibiotics to quantify the WT and mutant bacteria, respectively.
Immnunofluorescence microscopy
For immunofluorescence involving anti-LC3, anti-DFCP-1 and anti-ATG16L1, samples were fixed in 4% formaldehyde for 10 min at room temperature, rinsed 3 × in PBS saturated for 5 min, incubated in ice-cold 100% methanol for 10 min at −20°C. After washing three times in PBS, samples were saturated in blocking buffer (5% goat serum, 0.3% Triton X-100 in PBS) for 1 h, and following steps were as previously described (Tattoli et al, 2012c). For experiments involving other antibodies, immunofluorescence was performed as previously described (Tattoli et al, 2012c).
Expression vector and transfection
Expression vectors encoding for GFP-PKCδ and GFP-RFP-LC3 were from Drs Sergio Grinstein and Nicola Jones (both at HSC, Toronto), respectively, and GFP-LC3 was from Dr Yoshimori (Osaka University). Transfection was performed with PolyJet Transfection Reagent (FroggaBio) according to the manufacturer’s instruction.
PI3P ELISA
The total amount of PI3P was detected using the quantitative ELISA performed on lipid extracts from not infected and Listeria-infected cells according to the manufacturer’s recommendations (Echelon Biosciences). The samples were incubated in ice-cold 5% TCA in order to extract lipids. Extracted lipids were incubated with a PI3P detector protein, then added to a PI3P-precoated microplate. The secondary peroxidase-conjugated antibody against PI3P detector protein was added. PI3P levels were detected by measuring absorbance at 450 nm. Specific amounts were determined by comparison of values to a standard curve generated with simultaneous readings of known amounts of PI3P.
Western blot analysis
Western blotting was performed as previously described (Tattoli et al, 2012c).
Quantitative PCR
Quantitative PCR was performed as previously described (Benko et al, 2010) using primers previously described (Tattoli et al, 2012c).
Quantification of events observed in IF
The percentage of cells with colocalization of mTOR on LAMP2+ vesicles, as well as NDP52+, LAMP2+ or GFP-LC3+ Listeria was determined by IF. In each case, at least 100 cells from randomly selected fields were counted for each time point and condition, in at least three independent experiments. Cells that were poorly infected (<3 bacteria observed in DAPI channel) or very heavily infected (hundreds of bacteria and defects in the host cell nuclear morphology—a sign of ongoing cell death in these Listeria-infected cells) were excluded from the quantifications. For quantification of red and yellow puncta in cells transfected with GFP-RFP-LC3, cells were visualized using a dual excitation band fluorescence filter (FITC/Texas Red) and cells with three or more red puncta were considered as positive. Results are expressed as means±s.e.m. of data obtained in these independent experiments.
Statistical analysis
Results are expressed as means±s.e.m. of data obtained in independent experiments. Significant differences between mean values were evaluated using a one-sample or unpaired t-tests.
Supplementary Material
Acknowledgments
This work was supported by grants from the Burroughs Wellcome Fund (to SEG) and the CIHR (to SEG and DJP). CY acknowledges a fellowship from the Canadian Association of Gastroenterology (CAG). We thank Drs Dan Portnoy and Nancy Freitag for the Listeria strains used in this study.
Author contributions: IT, MTS and CY performed experiments. IT, SAT, DJP and SEG designed experiments and interpreted the results. SEG wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Benko S, Magalhaes JG, Philpott DJ, Girardin SE (2010) NLRC5 limits the activation of inflammatory pathways. J Immunol 185: 1681–1691 [DOI] [PubMed] [Google Scholar]
- Birmingham CL, Brumell JH (2006) Autophagy recognizes intracellular Salmonella enterica serovar Typhimurium in damaged vacuoles. Autophagy 2: 156–158 [DOI] [PubMed] [Google Scholar]
- Birmingham CL, Canadien V, Gouin E, Troy EB, Yoshimori T, Cossart P, Higgins DE, Brumell JH (2007) Listeria monocytogenes evades killing by autophagy during colonization of host cells. Autophagy 3: 442–451 [DOI] [PubMed] [Google Scholar]
- Camilli A, Goldfine H, Portnoy DA (1991) Listeria monocytogenes mutants lacking phosphatidylinositol-specific phospholipase C are avirulent. J Exp Med 173: 751–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti S, Liehl P, Buchon N, Lemaitre B (2012) Infection-induced host translational blockage inhibits immune responses and epithelial renewal in the Drosophila gut. Cell Host Microbe 12: 60–70 [DOI] [PubMed] [Google Scholar]
- Choy A, Dancourt J, Mugo B, O’Connor TJ, Isberg RR, Melia TJ, Roy CR (2012) The Legionella effector RavZ inhibits host autophagy through irreversible Atg8 deconjugation. Science 338: 1072–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dortet L, Mostowy S, Louaka AS, Gouin E, Nahori MA, Wiemer EA, Dussurget O, Cossart P (2011) Recruitment of the major vault protein by InlK: a Listeria monocytogenes strategy to avoid autophagy. PLoS Pathog 7: e1002168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funderburk SF, Wang QJ, Yue Z (2010) The Beclin 1-VPS34 complex--at the crossroads of autophagy and beyond. Trends Cell Biol 20: 355–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel W, Kuhn M (2000) Bacterial replication in the host cell cytosol. Curr Opin Microbiol 3: 49–53 [DOI] [PubMed] [Google Scholar]
- Gonzalez MR, Bischofberger M, Freche B, Ho S, Parton RG, van der Goot FG (2011) Pore-forming toxins induce multiple cellular responses promoting survival. Cell Microbiol 13: 1026–1043 [DOI] [PubMed] [Google Scholar]
- Hamasaki M, Furuta N, Matsuda A, Nezu A, Yamamoto A, Fujita N, Oomori H, Noda T, Haraguchi T, Hiraoka Y, Amano A, Yoshimori T (2013) Autophagosomes form at ER-mitochondria contact sites. Nature 495: 389–393 [DOI] [PubMed] [Google Scholar]
- Heinz DW, Essen LO, Williams RL (1998) Structural and mechanistic comparison of prokaryotic and eukaryotic phosphoinositide-specific phospholipases C. J Mol Biol 275: 635–650 [DOI] [PubMed] [Google Scholar]
- Klionsky DJ (2007) Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol 8: 931–937 [DOI] [PubMed] [Google Scholar]
- Kloft N, Neukirch C, Bobkiewicz W, Veerachato G, Busch T, von Hoven G, Boller K, Husmann M (2010) Pro-autophagic signal induction by bacterial pore-forming toxins. Med Microbiol Immunol 199: 299–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengaud J, Braun-Breton C, Cossart P (1991) Identification of phosphatidylinositol-specific phospholipase C activity in Listeria monocytogenes: a novel type of virulence factor? Mol Microbiol 5: 367–372 [DOI] [PubMed] [Google Scholar]
- Meyer-Morse N, Robbins JR, Rae CS, Mochegova SN, Swanson MS, Zhao Z, Virgin HW, Portnoy D (2010) Listeriolysin O is necessary and sufficient to induce autophagy during Listeria monocytogenes infection. PLoS One 5: e8610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Yoshimori T, Suzuki T, Sagara H, Mizushima N, Sasakawa C (2005) Escape of intracellular Shigella from autophagy. Science 307: 727–731 [DOI] [PubMed] [Google Scholar]
- Perrin AJ, Jiang X, Birmingham CL, So NS, Brumell JH (2004) Recognition of bacteria in the cytosol of Mammalian cells by the ubiquitin system. Curr Biol 14: 806–811 [DOI] [PubMed] [Google Scholar]
- Polson HE, De Lartigue J, Rigden DJ, Reedijk M, Urbe S, Clague MJ, Tooze SA (2010) Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy 6: 506–522 [DOI] [PubMed] [Google Scholar]
- Portnoy DA, Chakraborty T, Goebel W, Cossart P (1992) Molecular determinants of Listeria monocytogenes pathogenesis. Infect Immun 60: 1263–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Py BF, Lipinski MM, Yuan J (2007) Autophagy limits Listeria monocytogenes intracellular growth in the early phase of primary infection. Autophagy 3: 117–125 [DOI] [PubMed] [Google Scholar]
- Randow F (2011) How cells deploy ubiquitin and autophagy to defend their cytosol from bacterial invasion. Autophagy 7: 304–309 [DOI] [PubMed] [Google Scholar]
- Romagnoli A, Etna MP, Giacomini E, Pardini M, Remoli ME, Corazzari M, Falasca L, Goletti D, Gafa V, Simeone R, Delogu G, Piacentini M, Brosch R, Fimia GM, Coccia EM (2012) ESX-1 dependent impairment of autophagic flux by Mycobacterium tuberculosis in human dendritic cells. Autophagy 8: 1357–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnupf P, Portnoy DA (2007) Listeriolysin O: a phagosome-specific lysin. Microbes Infect 9: 1176–1187 [DOI] [PubMed] [Google Scholar]
- Shahnazari S, Yen WL, Birmingham CL, Shiu J, Namolovan A, Zheng YT, Nakayama K, Klionsky DJ, Brumell JH (2010) A diacylglycerol-dependent signaling pathway contributes to regulation of antibacterial autophagy. Cell Host Microbe 8: 137–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GA, Marquis H, Jones S, Johnston NC, Portnoy DA, Goldfine H (1995) The two distinct phospholipases C of Listeria monocytogenes have overlapping roles in escape from a vacuole and cell-to-cell spread. Infect Immun 63: 4231–4237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun AN, Camilli A, Portnoy DA (1990) Isolation of Listeria monocytogenes small-plaque mutants defective for intracellular growth and cell-to-cell spread. Infect Immun 58: 3770–3778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tattoli I, Philpott DJ, Girardin SE (2012a) The bacterial and cellular determinants controlling the recruitment of mTOR to the Salmonella-containing vacuole. Biol Open 1: 1215–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tattoli I, Sorbara MT, Philpott DJ, Girardin SE (2012b) Bacterial autophagy: The trigger, the target and the timing. Autophagy 8: 1848–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tattoli I, Sorbara MT, Vuckovic D, Ling A, Soares F, Carneiro LA, Yang C, Emili A, Philpott DJ, Girardin SE (2012c) Amino acid starvation induced by invasive bacterial pathogens triggers an innate host defense program. Cell Host Microbe 11: 563–575 [DOI] [PubMed] [Google Scholar]
- Thurston TL, Wandel MP, von Muhlinen N, Foeglein A, Randow F (2012) Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature 482: 414–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney LG, Portnoy DA (1989) Actin filaments and the growth, movement, and spread of the intracellular bacterial parasite, Listeria monocytogenes. J Cell Biol 109: 1597–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Boland JA, Kocks C, Dramsi S, Ohayon H, Geoffroy C, Mengaud J, Cossart P (1992) Nucleotide sequence of the lecithinase operon of Listeria monocytogenes and possible role of lecithinase in cell-to-cell spread. Infect Immun 60: 219–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hoven G, Kloft N, Neukirch C, Ebinger S, Bobkiewicz W, Weis S, Boller K, Janda KD, Husmann M (2012) Modulation of translation and induction of autophagy by bacterial exoproducts. Med Microbiol Immunol 201: 409–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN (2006) TOR signaling in growth and metabolism. Cell 124: 471–484 [DOI] [PubMed] [Google Scholar]
- Yoshikawa Y, Ogawa M, Hain T, Yoshida M, Fukumatsu M, Kim M, Mimuro H, Nakagawa I, Yanagawa T, Ishii T, Kakizuka A, Sztul E, Chakraborty T, Sasakawa C (2009) Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nat Cell Biol 11: 1233–1240 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.