Abstract
The body is composed of various tissue microenvironments with finely tuned local immunosurveillance systems, many of which are in close apposition with distinct commensal niches. Mammals have formed an evolutionary partnership with the microbiota that is critical for metabolism, tissue development and host defense. Despite our growing understanding of the impact of this host-microbe alliance on immunity in the gastrointestinal tract, the extent to which individual microenvironments are controlled by resident microbiota remains unclear. In this Perspective we discuss how resident commensals outside the gastrointestinal tract can control unique physiological niches and the potential implications of the dialog between these commensals and the host for the establishment of immune homeostasis, protective responses and tissue pathology.
Maintenance of tolerance and restoration of host homeostasis after insults or exposure to pathogens relies on complex and coordinated innate and adaptive responses. To this end, specialized populations of cells have to integrate local cues, such as defined metabolites, cytokines or hormones, to induce responses in a way that preserves the physiological and functional requirements of each tissue. To insure these distinct responses, unique subsets of antigen-presenting cells1-4, innate lymphoid cells5 and stromal cells6,7 seed and are locally conditioned by each microenvironment8. These tissue-tailored immunological networks are essential for maintaining tissue or exogenous tolerance and the development of appropriate protective and controlled immune responses.
Tissue-specific responses have been particularly explored at barrier tissues such as the lung, skin and the gastrointestinal tract—sites that are constitutively colonized by highly diverse and site-specific flora. Mammals have an evolutionary partnership with the microbiota that is critical for metabolism, tissue development and host defense. In the gastrointestinal tract, part of the local immune response is aimed at maintaining a peaceful coexistence with the resident microbiota. These microbes can in turn control many aspects of both innate and adaptive responses9,10. As such, dysbiosis of the gut microbiota has been associated with severe pathologies ranging from inflammatory bowel diseases to malnutrition11,12. Despite our growing understanding of the complexity and diversity of commensal populations at all barrier sites, such as the skin, the oral cavity and the airways, how tissue-resident microbes outside of the gastrointestinal tract control local and systemic responses remains poorly explored.
Each barrier tissue is a complex and in some cases unstable composite of microbes and host structural, hormonal, nervous and immunological networks, with each of these systems potentially controlled by resident microbiota. Thus, the delicate balance between a healthy or disease state may hinge on discrete interactions that occur between commensals and many host components in a given environment. Based on our understanding of tissue specialization, we could postulate that these unique microbial communities feed into tissue complexity and have coevolved with their host to finely tune the unique requirement of each site. In this Perspective, we will discuss recent evidence linking tissue-resident microbiota and microbe-derived metabolites in the control of local and systemic immune responses and discuss the current gaps in our understanding of these responses.
Distinct anatomical sites have unique commensal communities
Epithelial surfaces sustain diverse communities of commensals that include bacteria, archaea, fungi, protozoa and viruses13-17. With an estimated composition of 100 trillion cells, commensals outnumber host cells by at least a factor of ten and encode at least 100-fold more unique genes than their host’s genome18. An appreciation for this complexity was precipitated by recent advances in high-throughput sequencing approaches that uncovered the remarkable diversity of the human microbiota19-21. These surveys also unveiled the spatial patterning of the commensal communities in the human body. The gastrointestinal tract is home to the most abundant commensal niche and was the first and most thoroughly examined commensal community in the body22. These studies indicated that Firmicutes and Bacteriodetes are the dominant bacterial phyla of a healthy gut23. Commensal communities in the intestine are highly sensitive to environmental perturbations caused by host metabolic and inflammatory stress, and dysbiosis in these communities is associated with various disease states24-26.
Survey of the flora of 27 different body sites, including the skin, nostril, hair and oral cavity revealed that distinct anatomical sites house unique communities of bacteria and that community structure composition is determined by the ecology of each body site21. Similarly, a comparative analysis of the oral and respiratory viromes highlighted the importance of habitat in determining viral composition27. Analysis of the skin, oral cavity, airways, gastrointestinal tract and vagina revealed that each body habitat has distinct dominant bacterial taxa; signature clades identified in each anatomical region of the body were as follows: Actinobacteria, Firmicutes and Proteobacteria dominantly colonize the skin, Lactobacillus spp. predominates in the vaginal mucosa, and Bacteroidetes, Firmicutes, Fusobacteria and Proteobacteria prevail in the oral cavity28,29.
Global surveys of the composition of flora highlighted the importance of tissue-specific physical, metabolic and immunological factors in shaping the composition of commensals28,29. The skin, for instance, is a topographically heterogeneous organ that has an abundance of folds and invaginations, such as hair follicles, sebaceous glands and sweat glands, capable of sustaining unique communities of microbes30 (Fig. 1). Characterization of flora from distinct skin sites uncovered niche specialization of the skin microbiome31. For instance, Propinobactium spp. can metabolize lipids in sebum and therefore flourish in skin sites enriched in sebaceous glands, Corynebacterium spp inhabit moist sites, and Proteobacteria spp. dominated in dry sites31. Analysis of microbes from the oral cavity revealed that whereas Streptococcus spp. are prevalent in most oral habitats, Haemophilus spp. abundantly colonize the buccal mucosa, Actinomyces spp. the supragingival plaque and Prevotella spp. the subgingival plaque29,32. The airways can also be segregated into distinct ecological regions33. Although these sites have a homogenous microbial composition, the upper airways have a two- to fourfold greater biomass than the lung and lower respiratory tract 33,34 (Fig. 1). Although the composition of commensals is likely a major determinant in the control of each tissue, differences in biomass may also have profound implications in the physiology of the colonized site.
Figure 1.
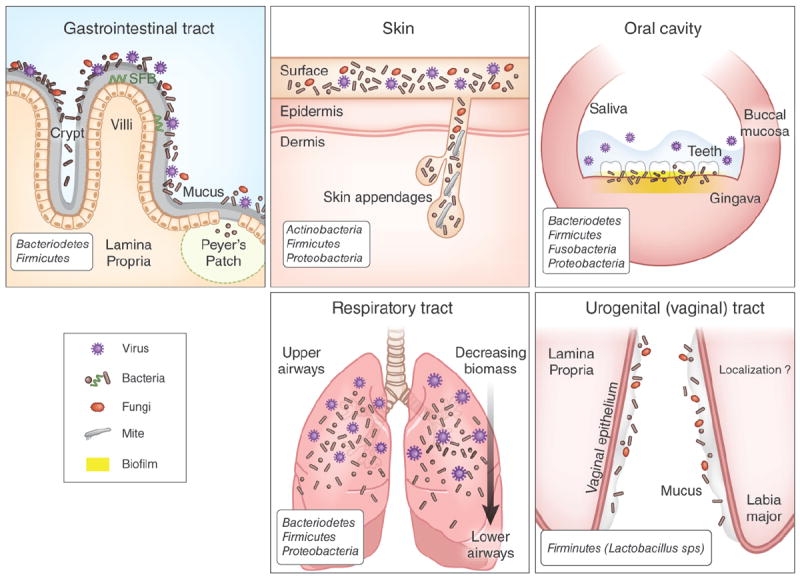
Tissue-specific modes of host-commensal interactions at distinct barrier sites. The gastrointestinal tract has the most abundant commensal niches in the body. A thick mucus layer separates the intestinal epithelium from resident microbes. Certain commensal species such as segmented filamentous bacteria (SFB) can penetrate the intestinal mucosal layer and reside in intimate contact with epithelial cells and in Peyer’s patches. By virtue of their localization, these species are uniquely poised to influence immune functions. Commensal microorganisms reside on the surface of the skin and appendages, such as hair follicles, sebaceous glands and sweat glands. These appendages may be critical sites of interactions between immune cells and commensals in the skin. The oral cavity contains several microenvironments that house commensal microbes including buccal mucosa, saliva, teeth and gingiva. Individual teeth house bacteria both above and below the gumline, that have been shown to modulate immunity in the surrounding gingiva; additionally, commensal bacteria constitutively form biofilm at this tissue site. In the respiratory tract, the composition of commensals is conserved across different geographical locations but the density of commensals is greatest in the upper airways and is less in the lower airways. The vaginal mucosa is dominantly colonized by Lactobacillus spp., but little is known about the precise localization of commensals in this niche and how fluctuations associated with sexual activity, menstrual cycle and pregnancy impact the microbiota in this site.
Whereas sequencing of the 16S DNA has allowed for a comprehensive view of human microbial communities, whole-genome shotgun metagenomic sequencing is beginning to inform us of the function of these communities. Metagenomic sequencing has revealed the presence of ‘core’ pathways that are essential for microbial survival in any environment, such as ribosome and translational machinery, nucleotide charging, and ATP synthesis and glycolysis29,35. Notably, commensal communities are enriched for metabolic capabilities based on the carbohydrate signature of their local microenvironment36. For instance, microbial communities in the oral cavity catabolize simple sugars, intestinal commensals degrade complex polysaccharides and glycogen, and communities in the vagina are enriched in the capacity to degrade peptidoglycan36. As we discuss bellow, byproducts of these tissue-specific microbial metabolic processes can have a central role in conditioning defined microenvironments.
Bystander control of peripheral tissues by gut commensals
Abundant experimental and clinical data support the idea that commensals residing in the gastrointestinal tract can calibrate both innate and adaptive responses10. Unique groups of commensals as well as defined byproducts or metabolites of commensals also can have key roles in the control of mucosal responses10. Additionally, despite being contained by the mucosal firewall9, the gut microbiota can also set the tone of immune responses at distal sites in the steady state and during inflammation (Fig. 2). Notably, a decrease in the number of gut commensals via treatment with a broad-spectrum antibiotic resulted in blunted T cell and B cell response to an intranasal infection with the A/PR8 strain of influenza37. This effect of the microbiota is linked to a capacity of microbiota to promote inflammasome-mediated induction of the secretion of interleukin 1β (IL-1β) and IL-1837. In this setting, rectal administration of Toll-like receptor (TLR) agonists restored the immune response in antibiotic-treated mice, indicating that either the microbial products can diffuse systemically or that activation of the inflammasome does not need to occur at the site of infection37. Treatment with an antibiotic also impaired adaptive and innate antiviral responses after exposure to systemic lymphocytic choriomeningitis virus and musosal influenza virus38. Genome-wide transcriptional profiling of macrophages revealed a broad decrease in the expression of genes associated with antiviral immunity38.
Figure 2.
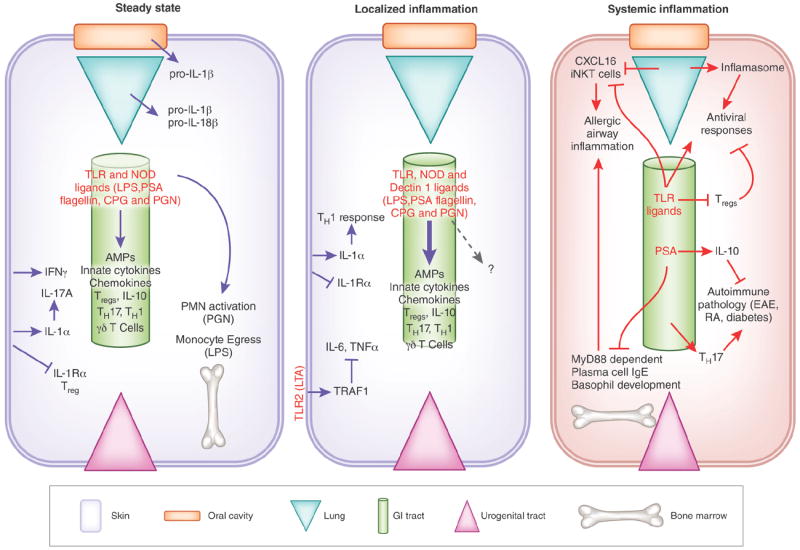
Localized and systemic regulation of the immune system by distinct commensal niches. (a) Commensal bacteria in the skin dynamically regulate the cutaneous effector and Treg cells by amplifying inflammatory signals (IL-1). Similarly, commensal signals in the oral mucosa and respiratory tract promote the production of IL-1, and of IL-1 and IL-18, respectively. In the gastrointestinal tract, commensal ligands (TLR, NLR and Nod ligands) and commensal metabolites (short-chain fatty acids) instruct immunity locally. Trace amounts of commensal byproducts enter blood circulation and localize to the bone marrow where they control immune cell development and function. (b) Skin commensals control protective TH1 responses during a dermal infection in an IL-1–dependent manner. A TLR2 ligand, lipoteichoic acid (LTA) specifically derived from S. epidermidis ameliorates exuberant production of TNF and IL-6 during skin inflammation. The signals involved in regulation of intestinal immunity during localized inflammatory responses are similar to those involved in controlling immune homeostasis, but these commensal derived signals are greatly amplified during an inflammatory response. Signals from gut microbiota may diffuse more readily into systemic circulation during gut inflammation. (c) Intestinal microbiota has been identified as a key modulator of systemic immunity. Gut-dwelling commensals can promote pathology in various mouse models of autoimmunity (experimental autoimmune encephalomyelitis (EAE), diabetes and arthritis). The intestinal microbiota also regulates viral immunity in the lung by controlling macrophage responses and inflammasome activation. Allergic airway inflammation is negatively regulated by signals from the flora that down-modulate responses of immune effectors, including production of IgE by plasma cells, development of basophils and accumulation of iNKT in the gut and lung. innate Natural Killer T cells (iNKT), experimental autoimmune encephalomyelitis (EAE), Rheumatoid arthritis (RA), polysaccharide A(PSA), polymorphonuclear neutrophils (PMN), Peptidoglycan (PGN), Antimicrobial peptides (AMPs).
In addition to their capacity to promote protective immunity, gut commensals can also influence autoimmune and allergic conditions. Mice lacking intestinal microbiota develop less severe disease in models of arthritis and experimental autoimmune encephalomyelitis39,40. In contrast, colonization with a group of gut commensals, segmented filamentous bacteria, promotes autoimmune arthritis through the induction of antigen-specific interleukin 17–producing T helper cells, which promote production of auto-antibodies via expansion of B cells in germinal centers. Furthermore, recruitment and activation of autoantibody-producing B cells from the endogenous immune repertoire depends on the availability of the target autoantigen and commensal microbiota41. The commensal microbiota can also decrease inflammation, as colonization of mice with Bacteroides fragilis results in the expansion of IL-10–producing regulatory T (Treg) cells, which limit the proinflammatory mechanisms of experimental autoimmune encephalomyelitis in a TLR2-dependent manner39. Additionally, induction of diabetes in the nonobese diabetic mouse model has been linked to the presence of defined intestinal microbes42,43, and signals derived from commensal bacteria can regulate basophil hematopoiesis and consequently allergic inflammation44.
This bystander control of peripheral responses can be, at least in part, explained by the unique requirement of the gastrointestinal tract for absorption, resulting in the constant diffusion of a low level of microbial products or metabolites into the bloodstream. For instance, peptidoglycan from radiolabeled Escherichia coli can be found in the serum and can improve the killing of Streptoccocus pneumoniae and Staphylococcus aureus by bone marrow–derived neutrophils in a Nod1-dependent manner45. Experimental evidence suggests that the diffusion of low amounts of commensal products into the bloodstream can contribute to monocyte exit from the bone marrow at steady-state and alter hematopoiesis46. Another example of such communication is the metabolism of dietary fiber by commensal bacteria of the Bacteroidetes phylum into immunomodulatory short-chain fatty acids, such as butyrate and acetate47. Binding of these fatty acids to the GPR43 receptor on neutrophils restrains their activation47. Consequently, mice lacking GPR43 have augmented inflammatory responses, and exogenous administration of short-chain fatty acids to wild-type mice is clinically beneficial47. These data suggest that the constant exposure to low amounts of commensal products and metabolites is critical to functionally tune the peripheral immune system and that subtle changes in this conditioning are likely to have profound consequences on tissue physiology. Nevertheless, although these observations collectively suggest that gut commensals can control the systemic threshold of activation of innate and adaptive cells, these studies cannot exclude a direct role of commensals residing in the lung, skin or other barrier sites in the control of local immunity (Fig. 2). Inflammatory conditions at all barrier sites have been associated with regional perturbations in the indigenous microflora48-52, emphasizing the local role of commensal flora. Below we discuss experimental evidence that supports a role for tissue-resident commensals in the calibration of systemic immunity, regional immunity and inflammation, and highlight the gaps in our current understanding of this control.
Commensals in liver immune function
The liver, although not directly colonized by commensals, is a unique environment in regard to its relationship with the microbiota. This vital organ with a wide range of metabolic functions, including detoxification and production of factors necessary for digestion, receives 80% of its blood from the portal vein, the draining tributary of the intestines, spleen and pancreas. In humans, the entire blood volume circulates through the liver about 360 times per day, and a third of our blood volume passes through the liver each minute53. Consequently, although the liver is not in direct contact with live commensals, its immune function is likely conditioned by its constant exposure to microbe-derived ligands or metabolites54. For example, exposure of liver sinusoidal endothelial cells to a low level of lipopolysaccharide results in the loss of TLR4 expression and lipopolysaccharide insensitivity55. Although not thoroughly characterized, ligands for TLR9, TLR2 and TLR5 are among the most represented microbial ligands in the portal blood56. As such, gut-derived bacterial products control liver-resident dendritic cells by stimulating hepatic IL-6–STAT3 signaling56. This signaling inhibits activation and/or maturation of hepatic dendritic cells, elevating the threshold needed for the induction of effector responses56. Kupffer cells, the most abundant of all tissue macrophages, reside alongside the sinusoidal endothelium and are the first to encounter commensal products or metabolites. As such, the numbers, functional activity and maturation status of Kupffer cell are directly controlled by the presence of gut flora54. In germ-free mice, there are substantially fewer adhesion molecules such as ICAM-1 on liver sinusoidal endothelial cells compared to conventionally raised mice54 suggesting that seeding by precursors of Kupffer cells and/or recruitment of other cells may be constitutively controlled by commensal products. Considering our growing understanding of the diversity and abundance of commensal-derived metabolites in the bloodstream, we speculate that the liver is optimally equipped to be conditioned by many microbial signals to maintain its fundamental metabolic function. Further, we propose that the liver may readily sense subtle alterations in this microbial fingerprinting and translate these changes into alarm responses for the immune system.
Ex vivo analysis of the activation of antigen-presenting cells (APCs) revealed a similar pattern of activation between germ-free and conventionally raised mice in secondary lymphoid organs, including mesenteric lymph nodes57,58, suggesting that under steady-state conditions, the control of activation of APCs may be tissue-specific. However, it is worth noting that the diet of germ-free mice contains microbial ligands that can provide surrogate signals to the ones normally provided by the flora59. Coupled with activation induced by tissue dissociation, this is likely to blunt any potential differences resulting from the absence of commensals. A better characterization of the effect of commensals’ metabolites and products on resident APCs in distinct tissue is clearly needed.
Skin microbiota in immune responses
The skin, the largest organ of the body, is the primary interface between the host and the environment. Microbial profiling has revealed the presence of highly diverse and specific commensal niches along distinct topographical sites of the skin21,31. Although the skin is a rather inhospitable environment, poor in nutrients, one billion bacteria inhabit a typical square centimeter of human skin, covering the surface and extending down into the appendages such as sebaceous glands and hair follicles60. Notably, the hair follicle is a very unique structure, with limited contact with the immune system61 and capable of orchestrating the organization of the skin immune system in inflammatory settings62.
Recent evidence supports the idea that skin microbiota has a fundamental and complex role in the control of skin physiology. Notably, Staphylococcus epidermidis, the dominant commensal bacterium found in the skin microflora, produces several antimicrobial proteins and proteases that can limit biofilm formation of pathogenic species63. Gram positive bacteria such as Lactococcus, Streptococcus and Streptomyces spp. also produce factors known as bacteriocins that inhibit the growth of other bacterial strains64. In the gastrointestinal tract, commensals have been involved in the induction of antimicrobial proteins such as RegIIIγ65. How skin-resident commensals control the host production of antimicrobial proteins remains an open question.
On the one hand, in contrast to the known role of the gut microbiota in promoting the gut associated lymphoid tissue (GALT) development, skin commensals are not required for the seeding of immune cells and overall development of the tissue66. On the other hand, the skin-resident bacteria can control fundamental aspects of local immunity and tissue repair. The skin is colonized by various species of Staphylococcus, including S. epidermidis, that normally reside in contact with keratinocytes. In a setting of skin injury in which pathology is dependent on TLR3, a defined product of this ubiquitous group of bacteria, lipoteichoic acid, can mitigate inflammation in a TLR2-dependent manner67. More specifically, lipoteichoic acid suppresses local production of various inflammatory mediators such as IL-6 and tumor necrosis factor (TNF)67. Thus, via their capacity to constrain inflammatory responses after tissue injury, defined skin commensal products can limit detrimental inflammatory responses and contribute to wound healing. In contrast, aberrant composition of skin flora or an increase in bacterial load may contribute to failure of tissue repair. Notably, most chronic wounds including diabetic, venous and pressure wounds are characterized by sustained inflammatory responses68 and in a mouse model of type 2 diabetes, nonhealing wounds were associated with increased abundance of Staphylococcus spp.69, highlighting a possible link between bacterial load and local pathologies. Thus, in the context of metabolic syndromes, altered nutrient availability and sustained inflammatory states could contribute to the emergence and dominance of bacteria that either qualitatively or quantitatively alter the local inflammatory milieu and promote local pathologies.
Skin microbiota also has a nonredundant role in controlling regional immunity (Fig. 2). Skin microbiota directly controls activation of skin-resident lymphocytes at steady state, and in the absence of skin commensals the frequency of Foxp3+ Treg cells is dramatically increased66. Skin commensals can promote protective immunity to a dermal pathogen, Leishmania major37. Thus, commensals in the skin, and potentially at other barrier sites such as the lung or the oral cavity, could act as local adjuvants required to finely tune the activation of effector cells at the site of the response. The action of skin commensals is discrete and specific. As such, skin commensals do not affect the capacity of T cells to be primed or to migrate to the skin but modulate function of dermal T cells by tuning the cutaneous inflammatory milieu. Particularly, skin-resident commensals promote, via mechanisms that remain to be addressed, the production of IL-1α that in turn directly controls the capacity of dermal resident T cells to produce inflammatory cytokines such as interferon γ (IFN-γ) and IL-17A37. Of importance, the skin flora controls immune homeostasis and responses to infection in an autonomous manner and independently of the gut flora37. Moreover, the IL-1 pathway is not necessary for the production of inflammatory cytokines by gut-resident T cells70. Thus, under steady-state conditions, or in the context of local inflammation, each barrier site is likely to be controlled independently of other commensal niches, and commensals may use unique mechanisms to control each tissue (Fig. 2). This compartmentalization of responses may have evolved as a mechanism to constrain the adjuvant properties of commensals and the unwanted consequences associated with systemic increases in inflammatory responses.
As a corollary of the capacity of the flora to promote local inflammation, clinical reports have implicated skin microbiota in the etiology of cutaneous inflammatory conditions. Disorders such as psoriasis, atopic dermatitis and rosacea are associated with dysbiosis of the skin flora71. In atopic dermatitis (eczema), a chronic relapsing inflammatory skin disorder, microbial community structures at sites of disease were dramatically altered compared to controls72. In patients with atopic dermatitis, the proportion of Staphylococcus spp. sequences, and in particular those of S. aureus, is greater during disease ‘flares’ and correlated with increased disease severity72. Similarly, patients with psoriasis, a chronic T cell–mediated disease affecting 2–3% of the population in the United States and 0.6–1.5% of the population in Europe exhibit a decrease in skin bacterial diversity. Notably, lesional skin from patients with plaque psoriasis is enriched in Streptococcus spp. and harbor less P. acnes compared to controls73. Mechanistically, skin commensals could contribute to the initiation or amplification of skin pathologies via several mechanisms. Local expansion of unique commensals with enhanced inflammatory potential and/or enrichment in commensal load may trigger aberrant production of antimicrobial peptides, contribute to aberrant keratinocyte proliferation or, as previously discussed, promote the local production of inflammatory mediators37. The IL-1 pathway that is promoted by skin-resident commensals is linked to many chronic inflammatory disorders such as arthritis and asthma, as well as psoriasis and other cutaneous disorders74. Psoriatic plaques are also characterized by marked infiltration of activated T cells producing inflammatory cytokines, in particular cytokines of the IL-17 family, such as IL-17A75-77, that have been associated with the pathogenesis of the disease78,79. Some of the pathogenic role of IL-17A results from its capacity to amplify various inflammatory pathways in the skin, leading to hyperproliferation of keratinocytes and formation of lesions in psoriasis80,81. In the context of inflammation, changes in barrier permeability and enhanced contact with commensals could also promote the local inflammatory process. Thus, alterations in the composition or sensing of commensals—via the capacity of commensals to directly and/or indirectly set the threshold of activation of skin-resident T cells, keratinocytes and APCs—are likely the primary drivers and amplifiers of skin pathologies.
In addition to organ-specific microbiota, dramatic differences in microbial composition exist in each tissue. In humans, skin microbial diversity varies dramatically across various ecological niches60. These differences in bacterial composition and/or density may explain why some skin sites are more prone to inflammatory disorders and why severe skin responses in patients with graft versus host disease occur at sites enriched in commensals niches. How indigenous commensal communities control immune cells in distinct ecological niches in a single organ is poorly understood.
Oral microbiota in local immunity
The oral cavity harbors a diverse and complex microbial community that accumulates on both the hard and soft oral tissues in sessile biofilms82. Whereas biofilms are extracellular, the oral microbiota also includes communities of intracellular bacteria that invade the gingival and buccal epithelial cells of the mouth. A prominent member of this community is the opportunistic intracellular species, Porphyromonas gingivalis, a pathobiont that can behave either as a commensal or pathogen83. Recent evidence suggests that the oral microbiome can control innate host responses of the periodontal tissue and in some settings, contribute to periodontal pathologies84. In particular, oral commensals promote the expression of IL-1β protein in the oral mucosa84. In contrast, that amount of mRNA encoding IL-1β was not decreased in gingival samples from germ-free mice, suggesting that in the oral cavity commensals may promote inflammasome activity84. These results are consistent with the notion that commensal bacteria, in most tissues, can establish a threshold of activation required for immune fitness. As proposed for other barrier sites, commensals can also contribute to tissue pathogenesis. The pathology associated with infection by P. gingivalis, a low-abundance oral anaerobic pathogen85, relies on the presence of oral microbiota86. This species of bacteria, even at very low colonization levels, triggers shifts in the composition of oral microbiota and promotes a substantial increase in the overall abundance of commensal density86. During infection by P. gingivalis, activation of both the microbiota and the complement cascade are required for bone loss86. The mechanism by which oral commensals contribute to this pathology remains unclear, but alteration of commensal populations and increased commensal density have been reported in various settings of barrier inflammation. Acute gastrointestinal infections are often characterized by substantial shifts in the microbiota and dominance of bacteria with enhanced invasive and inflammatory properties87-90. Understanding the key metabolic shift responsible for the emergence of commensals with pathogenic potential in each microenvironment will be central to our understanding of tissue-specific pathology.
Vaginal commensals in immune responses
The human vaginal mucosa, composed of a non-keratinized stratified squamous epithelium, is home to an abundant microflora91. This highly specialized environment is exposed to foreign substances including spermatozoa and a wide array of pathogens. Adding to this complexity, various parts of the female genital tract are influenced by sex hormones during the menstrual cycle. As such, the vaginal microbial ecosystem undergoes substantial structural changes at various stages in a woman’s life91. In addition, the structure and composition of vaginal microbial communities in healthy women are influenced by natural changes, such as aging, time in the menstrual cycle, menstruation, pregnancy and stress92. This implies that the vaginal mucosal innate and adaptive immune system is endowed with the complex task of maintaining the delicate balance between commensal and pathogen across various stages of a woman’s life, and induce tolerance required for successful pregnancy. The commensal microflora is an important component of the vaginal mucosal defense against infections. A key feature of the predominant colonization of the vaginal tract by Lactobacillus spp is a relatively low pH, owing to their production of large amounts of lactic acid93. Lactobacilli are also thought to contribute to the health of the vagina via the production of H2O2, the induction of mucus as well as the direct production of antimicrobial factors94. In vitro colonization of vaginal epithelial cell multilayers by common vaginal commensals has revealed that these bacteria do not trigger epithelial cells to produce cytokines95. Moreover, various isolates of Lactobacilli suppress the capacity of epithelial cells to respond to various TLR ligands95. Lactic acid is a major product of Lactobacilli metabolism96. Exposure of peripheral blood mononuclear cells to this metabolite promotes production of IL-23 in response to lipopolysaccharide97. In support of a link between vaginal commensals and the local immune responses, vaginal concentrations of IL-1 receptor agonist are elevated in the presence of anaerobic gram negative rods and/or Gardnerella vaginalis98. It is important to address the role of Lactobacilli on the innate and adaptive response of the vaginal environment in vivo and the interplay of this control with cyclic hormonal changes.
Airway microbiome in immune responses
The airways can be segregated into distinct ecological regions, which include nasopharynx, oropharynx, upper respiratory tract and lower respiratory tract33. The epithelial surfaces of the airway contain cilia and are covered by a mucus layer. A feature of the airway is the difference in bacterial density across its length, with the upper airways colonized by the highest load of bacteria33,34. Although the role of the endogenous airway microbiota in the control of local immunity has not been addressed, evidence suggests that local manipulation of the composition of the flora may have profound consequences on the capacity of the host to mount protective responses. In particular, intranasal priming of mice with live or heat-inactivated Lactobacillus spp. protects against lethal sequelae infection with the virulent pathogen pneumonia virus of mice. This protective response is associated with diminished expression of proinflammatory cytokines and recovery of less virus99,100. Priming with live Lactobacilli results in lower granulocyte recruitment, diminished expression of multiple proinflammatory cytokines (CXCL10, CXCL1, CCL2 and TNF) and recovery of less virus. Notably, protection is not unique to Lactobacillus spp. and also has been observed in response to priming with nonpathogenic gram-positive Listeria spp., suggesting that microbial load, rather than specific groups of bacteria may control this adjuvant effect99,100. Of note, this protective effect was independent of TLR signaling, and individual microbial ligands, such as peptidoglycan or bacterial DNA, did not confer protection100. Along with the studies mentioned above, this highlights our limited understanding of the mechanisms by which commensals control immune functions of tissue.
In in germ-free mice, the number of infiltrating TH2 lymphocytes and eosinophils was higher compared conventionally raised mice, and the composition and status of activation of lung dendritic cells was altered during airway inflammation101. This effect could be reversed by colonization of the mice with the complex commensal flora of conventionally raised mice101. The relative contribution of gut versus lung microbiota to this effect remains to be addressed. A recent comparative microbiome profiling of the sinus of patients and healthy controls revealed lower bacterial diversity in the former, with specific depletion of lactic bacteria and a relative increase in a single species, Corynebacterium tuberculostearicum49. Experimental approaches in mice have demonstrated that the mucosal microbiota is required to protect against the pathogenic role of C. tuberculostearicum49. Moreover, Lactobacillus sakei, identified in a human cohort of protected patients, antagonized C. tuberculostearicum sinus infection, even in the absence of a complete microbiota. These studies suggest that sinus mucosal immune defense is highly dependent on the composition of the resident microbiota, albeit the mechanism of such control remains to be addressed.
Tissue ‘memory’: long-term consequences of tissue insults
After pathogenic or structural insults, tissues maintain traces or memory that extend beyond adaptive responses of T cells and B cells. These changes include profound structural remodeling, such as scars imposed by excessive fibrotic responses, epigenetic alteration of innate cells and stromal cells as well as modification of the APC network. Additionally, tissue ‘memory’ can also result from long-term alteration of its resident microbiota. A long-term consequence of tissue damage and infection could be the induction of immune responses to commensals. Indeed, during gastrointestinal infections, tolerance to commensals can be lost and microbiota-specific T cells become activated102. In contrast to steady-state responses to commensal bacteria, during infection commensal-specific T cells, much like pathogen-specific T cells, differentiate to an inflammatory phenotype. In the gastrointestinal tract, these commensal-specific T cells form memory cells that are phenotypically and functionally indistinguishable from pathogen-specific T cells102. All barrier environments are also primary sites of exposure to pathogens, and sites such as the skin are often subject to barrier breach because of physical damage or infection. These events could lead to enhanced exposure to the microbiota and to immunity to resident commensals at diverse barrier sites. Because of the extraordinary amount of potential antigens expressed by the host microbiota at all body surfaces, this would imply that a substantial fraction of memory cells are expected to be commensal-specific. Such memory cells will develop over time in response to successive infections and/or various barrier breaches. In support of this hypothesis, healthy human serum contains antibodies to skin and intestinal microbiota103. Thus, primary exposure to a pathogen in the skin, lung and gastrointestinal tract is likely to occur in the context of a much broader recall response against commensal bacteria. Additional exploration of the memory-cell compartment and the antigen specificity of lymphocytes residing at all barrier sites would inform of the potential impact of these complex responses on tissue physiology and subsequent pathologies. It would be of particular interest to address how responses to conserved bacterial antigens across barrier sites impact local and systemic tissue responses.
Conclusion
Although we could propose that under steady-state conditions or during local inflammatory responses commensals control each tissue in a highly specialized and compartmentalized manner, this regional segregation is likely to be overruled by systemic inflammation (Fig. 2). Low-level diffusion of microbial metabolites to the blood stream and alterations of these signals during pathological states add an additional layer of complexity to our understanding of tissue regulation. The challenge of the next few years will be to devise approaches to explore the complex interplay between these various commensal niches under steady-state conditions and disease states. This will require a better understanding of the keystone bacterial species that control the immune responses at individual sites, and exploration of the mechanisms and cellular target of this control, and an in-depth exploration of the precise mode of interaction between commensals and host cells in their specific ecological niches.
Acknowledgments
This work was supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, US National Institutes of Health. We thank members of the Belkaid laboratory for helpful discussions, particularly S. Spencer and J. Grainger for critical reading of the manuscript.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Shklovskaya E, et al. Langerhans cells are precommitted to immune tolerance induction. Proc Natl Acad Sci USA. 2011;108:18049–18054. doi: 10.1073/pnas.1110076108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chu CC, et al. Resident CD141 (BDCA3)+ dendritic cells in human skin produce IL-10 and induce regulatory T cells that suppress skin inflammation. J Exp Med. 2012;209:935–945. doi: 10.1084/jem.20112583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Igyarto BZ, et al. Skin-resident murine dendritic cell subsets promote distinct and opposing antigen-specific T helper cell responses. Immunity. 2011;35:260–272. doi: 10.1016/j.immuni.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scott CL, Aumeunier AM, Mowat AM. Intestinal CD103+ dendritic cells: master regulators of tolerance? Trends Immunol. 2011;32:412–419. doi: 10.1016/j.it.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 5.Spits H, Cupedo T. Innate lymphoid cells: emerging insights in development, lineage relationships, and function. Annu Rev Immunol. 2012;30:647–675. doi: 10.1146/annurev-immunol-020711-075053. [DOI] [PubMed] [Google Scholar]
- 6.Owens BM, Simmons A. Intestinal stromal cells in mucosal immunity and homeostasis. Mucosal Immunol. 2013;6:224–234. doi: 10.1038/mi.2012.125. [DOI] [PubMed] [Google Scholar]
- 7.Malhotra D, Fletcher AL, Turley SJ. Stromal and hematopoietic cells in secondary lymphoid organs: partners in immunity. Immunol Rev. 2013;251:160–176. doi: 10.1111/imr.12023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matzinger P, Kamala T. Tissue-based class control: the other side of tolerance. Nat Rev Immunol. 2011;11:221–230. doi: 10.1038/nri2940. [DOI] [PubMed] [Google Scholar]
- 9.Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol. 2010;10:159–169. doi: 10.1038/nri2710. [DOI] [PubMed] [Google Scholar]
- 10.Molloy MJ, Bouladoux N, Belkaid Y. Intestinal microbiota: shaping local and systemic immune responses. Semin Immunol. 2012;24:58–66. doi: 10.1016/j.smim.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blumberg R, Powrie F. Microbiota, disease, and back to health: a metastable journey. Sci Transl Med. 2012;4:137rv137. doi: 10.1126/scitranslmed.3004184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith MI, et al. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science. 2013;339:548–554. doi: 10.1126/science.1229000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foulongne V, et al. Human skin microbiota: high diversity of DNA viruses identified on the human skin by high throughput sequencing. PLoS ONE. 2012;7:e38499. doi: 10.1371/journal.pone.0038499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iliev ID, et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science. 2012;336:1314–1317. doi: 10.1126/science.1221789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reyes A, et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466:334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eckburg PB, Lepp PW, Relman DA. Archaea and their potential role in human disease. Infect Immun. 2003;71:591–596. doi: 10.1128/IAI.71.2.591-596.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grice EA, Segre JA. The human microbiome: our second genome. Annu Rev Genomics Hum Genet. 2012;13:151–170. doi: 10.1146/annurev-genom-090711-163814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 19.Medini D, et al. Microbiology in the post-genomic era. Nat Rev Microbiol. 2008;6:419–430. doi: 10.1038/nrmicro1901. [DOI] [PubMed] [Google Scholar]
- 20.Kuczynski J, et al. Experimental and analytical tools for studying the human microbiome. Nat Rev Genet. 2012;13:47–58. doi: 10.1038/nrg3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Costello EK, et al. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eckburg PB, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tilg H, Kaser A. Gut microbiome, obesity, and metabolic dysfunction. J Clin Invest. 2011;121:2126–2132. doi: 10.1172/JCI58109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tlaskalova-Hogenova H, et al. The role of gut microbiota (commensal bacteria) and the mucosal barrier in the pathogenesis of inflammatory and autoimmune diseases and cancer: contribution of germ-free and gnotobiotic animal models of human diseases. Cell Mol Immunol. 2011;8:110–120. doi: 10.1038/cmi.2010.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pride DT, et al. Evidence of a robust resident bacteriophage population revealed through analysis of the human salivary virome. ISME J. 2012;6:915–926. doi: 10.1038/ismej.2011.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Human Microbiome Project Consortium. A framework for human microbiome research. Nature. 2012;486:215–221. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol. 2011;9:244–253. doi: 10.1038/nrmicro2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grice EA, et al. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324:1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Segata N, et al. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 2012;13:R42. doi: 10.1186/gb-2012-13-6-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beck JM, Young VB, Huffnagle GB. The microbiome of the lung. Transl Res. 2012;160:258–266. doi: 10.1016/j.trsl.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Charlson ES, et al. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med. 2011;184:957–963. doi: 10.1164/rccm.201104-0655OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abubucker S, et al. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput Biol. 2012;8:e1002358. doi: 10.1371/journal.pcbi.1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cantarel BL, Lombard V, Henrissat B. Complex carbohydrate utilization by the healthy human microbiome. PLoS ONE. 2012;7:e28742. doi: 10.1371/journal.pone.0028742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ichinohe T, et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci USA. 2011;108:5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abt MC, et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity. 2012;37:158–170. doi: 10.1016/j.immuni.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ochoa-Reparaz J, et al. Central nervous system demyelinating disease protection by the human commensal Bacteroides fragilis depends on polysaccharide A expression. J Immunol. 2010;185:4101–4108. doi: 10.4049/jimmunol.1001443. [DOI] [PubMed] [Google Scholar]
- 40.Wu HJ, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berer K, et al. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature. 2011;479:538–541. doi: 10.1038/nature10554. [DOI] [PubMed] [Google Scholar]
- 42.Wen L, et al. Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kriegel MA, et al. Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. Proc Natl Acad Sci USA. 2011;108:11548–11553. doi: 10.1073/pnas.1108924108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hill DA, et al. Commensal bacteria-derived signals regulate basophil hematopoiesis and allergic inflammation. Nat Med. 2012;18:538–546. doi: 10.1038/nm.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clarke TB, et al. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med. 2010;16:228–231. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi C, et al. Bone marrow mesenchymal stem and progenitor cells induce monocyte emigration in response to circulating toll-like receptor ligands. Immunity. 2011;34:590–601. doi: 10.1016/j.immuni.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maslowski KM, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kong HH, et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 2012;22:850–859. doi: 10.1101/gr.131029.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abreu NA, et al. Sinus microbiome diversity depletion and Corynebacterium tuberculostearicum enrichment mediates rhinosinusitis. Sci Transl Med. 2012;4:151ra124. doi: 10.1126/scitranslmed.3003783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hilty M, et al. Disordered microbial communities in asthmatic airways. PLoS ONE. 2010;5:e8578. doi: 10.1371/journal.pone.0008578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Belda-Ferre P, et al. The oral metagenome in health and disease. ISME J. 2012;6:46–56. doi: 10.1038/ismej.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Srinivasan S, et al. Temporal variability of human vaginal bacteria and relationship with bacterial vaginosis. PLoS ONE. 2010;5:e10197. doi: 10.1371/journal.pone.0010197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Crispe IN. The liver as a lymphoid organ. Annu Rev Immunol. 2009;27:147–163. doi: 10.1146/annurev.immunol.021908.132629. [DOI] [PubMed] [Google Scholar]
- 54.Corbitt N, et al. Gut bacteria drive Kupffer cell expansion via MAMP-mediated ICAM-1 induction on sinusoidal endothelium and influence preservation-reperfusion injury after orthotopic liver transplantation. Am J Pathol. 2013;182:180–191. doi: 10.1016/j.ajpath.2012.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bigorgne AE, Crispe IN. TLRs in hepatic cellular crosstalk. Gastroenterol Res Pract. 2010;2010:618260. doi: 10.1155/2010/618260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lunz JG, III, Specht SM, Murase N, Isse K, Demetris AJ. Gut-derived commensal bacterial products inhibit liver dendritic cell maturation by stimulating hepatic interleukin-6/signal transducer and activator of transcription 3 activity. Hepatology. 2007;46:1946–1959. doi: 10.1002/hep.21906. [DOI] [PubMed] [Google Scholar]
- 57.Wilson NS, et al. Normal proportion and expression of maturation markers in migratory dendritic cells in the absence of germs or Toll-like receptor signaling. Immunol Cell Biol. 2008;86:200–205. doi: 10.1038/sj.icb.7100125. [DOI] [PubMed] [Google Scholar]
- 58.Walton KL, He J, Kelsall BL, Sartor RB, Fisher NC. Dendritic cells in germ-free and specific pathogen-free mice have similar phenotypes and in vitro antigen presenting function. Immunol Lett. 2006;102:16–24. doi: 10.1016/j.imlet.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 59.Hill DA, et al. Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunol. 2009;3:148–158. doi: 10.1038/mi.2009.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Grice EA, et al. A diversity profile of the human skin microbiota. Genome Res. 2008;18:1043–1050. doi: 10.1101/gr.075549.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Christoph T, et al. The human hair follicle immune system: cellular composition and immune privilege. Br J Dermatol. 2000;142:862–873. doi: 10.1046/j.1365-2133.2000.03464.x. [DOI] [PubMed] [Google Scholar]
- 62.Nagao K, et al. Stress-induced production of chemokines by hair follicles regulates the trafficking of dendritic cells in skin. Nat Immunol. 2012;13:744–752. doi: 10.1038/ni.2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Iwase T, et al. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature. 2010;465:346–349. doi: 10.1038/nature09074. [DOI] [PubMed] [Google Scholar]
- 64.Gallo RL, Hooper LV. Epithelial antimicrobial defence of the skin and intestine. Nat Rev Immunol. 2012;12:503–516. doi: 10.1038/nri3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Naik S, et al. Compartmentalized control of skin immunity by resident commensals. Science. 2012;337:1115–1119. doi: 10.1126/science.1225152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lai Y, et al. Commensal bacteria regulate Toll-like receptor 3–dependent inflammation after skin injury. Nat Med. 2009;15:1377–1382. doi: 10.1038/nm.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Loots MA, et al. Differences in cellular infiltrate and extracellular matrix of chronic diabetic and venous ulcers versus acute wounds. J Invest Dermatol. 1998;111:850–857. doi: 10.1046/j.1523-1747.1998.00381.x. [DOI] [PubMed] [Google Scholar]
- 69.Grice EA, et al. Longitudinal shift in diabetic wound microbiota correlates with prolonged skin defense response. Proc Natl Acad Sci USA. 2010;107:14799–14804. doi: 10.1073/pnas.1004204107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ivanov II, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gallo RL, Nakatsuji T. Microbial symbiosis with the innate immune defense system of the skin. J Invest Dermatol. 2011;131:1974–1980. doi: 10.1038/jid.2011.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kong HH, et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 2012;22:850–859. doi: 10.1101/gr.131029.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gao Z, Tseng CH, Strober BE, Pei Z, Blaser MJ. Substantial alterations of the cutaneous bacterial biota in psoriatic lesions. PLoS ONE. 2008;3:e2719. doi: 10.1371/journal.pone.0002719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sims JE, Smith DE. The IL-1 family: regulators of immunity. Nat Rev Immunol. 2010;10:89–102. doi: 10.1038/nri2691. [DOI] [PubMed] [Google Scholar]
- 75.Nestle FO, Kaplan DH, Barker J. Psoriasis. N Engl J Med. 2009;361:496–509. doi: 10.1056/NEJMra0804595. [DOI] [PubMed] [Google Scholar]
- 76.Ortega C, et al. IL-17-producing CD8+ T lymphocytes from psoriasis skin plaques are cytotoxic effector cells that secrete TH17-related cytokines. J Leukoc Biol. 2009;86:435–443. doi: 10.1189/JLB.0109046. [DOI] [PubMed] [Google Scholar]
- 77.Eyerich S, et al. Th22 cells represent a distinct human T cell subset involved in epidermal immunity and remodeling. J Clin Invest. 2009;119:3573–3585. doi: 10.1172/JCI40202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Leonardi C, et al. Anti-interleukin-17 monoclonal antibody ixekizumab in chronic plaque psoriasis. N Engl J Med. 2012;366:1190–1199. doi: 10.1056/NEJMoa1109997. [DOI] [PubMed] [Google Scholar]
- 79.Papp KA, et al. Brodalumab, an anti-interleukin-17-receptor antibody for psoriasis. N Engl J Med. 2012;366:1181–1189. doi: 10.1056/NEJMoa1109017. [DOI] [PubMed] [Google Scholar]
- 80.Zheng Y, et al. Interleukin-22, a TH17 cytokine, mediates IL-23–induced dermal inflammation and acanthosis. Nature. 2007;445:648–651. doi: 10.1038/nature05505. [DOI] [PubMed] [Google Scholar]
- 81.Lai Y, et al. The antimicrobial protein REG3A regulates keratinocyte proliferation and differentiation after skin injury. Immunity. 2012;37:74–84. doi: 10.1016/j.immuni.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Avila M, Ojcius DM, Yilmaz O. The oral microbiota: living with a permanent guest. DNA Cell Biol. 2009;28:405–411. doi: 10.1089/dna.2009.0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yilmaz O. The chronicles of Porphyromonas gingivalis: the microbium, the human oral epithelium and their interplay. Microbiology. 2008;154:2897–2903. doi: 10.1099/mic.0.2008/021220-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dixon DR, Reife RA, Cebra JJ, Darveau RP. Commensal bacteria influence innate status within gingival tissues: a pilot study. J Periodontol. 2004;75:1486–1492. doi: 10.1902/jop.2004.75.11.1486. [DOI] [PubMed] [Google Scholar]
- 85.Desvarieux M, et al. Periodontal microbiota and carotid intima-media thickness: the oral infections and vascular disease epidemiology study (INVEST) Circulation. 2005;111:576–582. doi: 10.1161/01.CIR.0000154582.37101.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hajishengallis G, et al. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe. 2011;10:497–506. doi: 10.1016/j.chom.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Egan CE, Cohen SB, Denkers EY. Insights into inflammatory bowel disease using Toxoplasma gondii as an infectious trigger. Immunol Cell Biol. 2012;90:668–675. doi: 10.1038/icb.2011.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Heimesaat MM, et al. Gram-negative bacteria aggravate murine small intestinal Th1-type immunopathology following oral infection with Toxoplasma gondii. J Immunol. 2006;177:8785–8795. doi: 10.4049/jimmunol.177.12.8785. [DOI] [PubMed] [Google Scholar]
- 89.Lupp C, et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2:204. doi: 10.1016/j.chom.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 90.Stecher B, et al. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 2007;5:2177–2189. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Farage M, Maibach H. Lifetime changes in the vulva and vagina. Arch Gynecol Obstet. 2006;273:195–202. doi: 10.1007/s00404-005-0079-x. [DOI] [PubMed] [Google Scholar]
- 92.Hickey RJ, Zhou X, Pierson JD, Ravel J, Forney LJ. Understanding vaginal microbiome complexity from an ecological perspective. Transl Res. 2012;160:267–282. doi: 10.1016/j.trsl.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Antonio MA, Hawes SE, Hillier SL. The identification of vaginal Lactobacillus species and the demographic and microbiologic characteristics of women colonized by these species. J Infect Dis. 1999;180:1950–1956. doi: 10.1086/315109. [DOI] [PubMed] [Google Scholar]
- 94.Spurbeck RR, Arvidson CG. Lactobacilli at the front line of defense against vaginally acquired infections. Future Microbiol. 2011;6:567–582. doi: 10.2217/fmb.11.36. [DOI] [PubMed] [Google Scholar]
- 95.Rose WA, II, et al. Commensal bacteria modulate innate immune responses of vaginal epithelial cell multilayer cultures. PLoS ONE. 2012;7:e32728. doi: 10.1371/journal.pone.0032728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhou X, et al. Characterization of vaginal microbial communities in adult healthy women using cultivation-independent methods. Microbiology. 2004;150:2565–2573. doi: 10.1099/mic.0.26905-0. [DOI] [PubMed] [Google Scholar]
- 97.Witkin SS, Alvi S, Bongiovanni AM, Linhares IM, Ledger WJ. Lactic acid stimulates interleukin-23 production by peripheral blood mononuclear cells exposed to bacterial lipopolysaccharide. FEMS Immunol Med Microbiol. 2011;61:153–158. doi: 10.1111/j.1574-695X.2010.00757.x. [DOI] [PubMed] [Google Scholar]
- 98.Genc MR, et al. Polymorphism in intron 2 of the interleukin-1 receptor antagonist gene, local midtrimester cytokine response to vaginal flora, and subsequent preterm birth. Am J Obstet Gynecol. 2004;191:1324–1330. doi: 10.1016/j.ajog.2004.05.074. [DOI] [PubMed] [Google Scholar]
- 99.Gabryszewski SJ, et al. Lactobacillus-mediated priming of the respiratory mucosa protects against lethal pneumovirus infection. J Immunol. 2011;186:1151–1161. doi: 10.4049/jimmunol.1001751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Garcia-Crespo KE, et al. Lactobacillus priming of the respiratory tract: Heterologous immunity and protection against lethal pneumovirus infection. Antiviral Res. 2013;97:270–279. doi: 10.1016/j.antiviral.2012.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Herbst T, et al. Dysregulation of allergic airway inflammation in the absence of microbial colonization. Am J Respir Crit Care Med. 2011;184:198–205. doi: 10.1164/rccm.201010-1574OC. [DOI] [PubMed] [Google Scholar]
- 102.Hand TW, et al. Acute gastrointestinal infection induces long-lived microbiota-specific T cell responses. Science. 2012;337:1553–1556. doi: 10.1126/science.1220961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Haas A, et al. Systemic antibody responses to gut commensal bacteria during chronic HIV-1 infection. Gut. 2011;60:1506–1519. doi: 10.1136/gut.2010.224774. [DOI] [PubMed] [Google Scholar]


