Significance
The fate of eukaryotic mRNAs is intimately linked to the complement of proteins that associate with them to form mRNA—protein complexes, the so-called messenger ribonucleoprotein particles (mRNPs). Transitions in the architecture of an mRNP lead to specific functional consequences. DEAD-box proteins are key players in orchestrating these structural rearrangements: They associate with RNA in response to ATP binding and dissociate from it upon ATP hydrolysis. In this paper, we have elucidated the molecular mechanisms by which a DEAD-box protein, which in human cells marks spliced mRNPs for a specialized surveillance pathway, is recognized by the MIF4G domain of a splicing factor. This structure shows how a MIF4G domain can act as a negative regulator of DEAD-box ATPase activity.
Keywords: helicase, mRNP, NMD
Abstract
DEAD-box proteins are involved in all aspects of RNA processing. They bind RNA in an ATP-dependent manner and couple ATP hydrolysis to structural and compositional rearrangements of ribonucleoprotein particles. Conformational control is a major point of regulation for DEAD-box proteins to act on appropriate substrates and in a timely manner in vivo. Binding partners containing a middle domain of translation initiation factor 4G (MIF4G) are emerging as important regulators. Well-known examples are eIF4G and Gle1, which bind and activate the DEAD-box proteins eIF4A and Dbp5. Here, we report the mechanism of an inhibiting MIF4G domain. We determined the 2.0-Å resolution structure of the complex of human eIF4AIII and the MIF4G domain of the splicing factor Complexed With Cef1 (CWC22), an essential prerequisite for exon junction complex assembly by the splicing machinery. The CWC22 MIF4G domain binds both RecA domains of eIF4AIII. The mode of RecA2 recognition is similar to that observed in the activating complexes, yet is specific for eIF4AIII. The way the CWC22 MIF4G domain latches on the eIF4AIII RecA1 domain is markedly different from activating complexes. In the CWC22–eIF4AIII complex, the RNA-binding and ATP-binding motifs of the two RecA domains do not face each other, as would be required in the active state, but are in diametrically opposite positions. The binding mode of CWC22 to eIF4AIII reveals a facet of how MIF4G domains use their versatile structural frameworks to activate or inhibit DEAD-box proteins.
DEAD-box proteins are a large family of RNA-dependent ATPases involved in many aspects of RNA metabolism, including processing, transport, translation, and decay (reviewed in refs. 1 and 2). These proteins generally function to remodel ribonucleoprotein particles (RNPs), by locally unwinding the nucleic acid or by displacing and/or recruiting other factors to the nucleic acid they bind to (3–8). Although DEAD-box proteins recognize single-stranded RNAs in a sequence-independent manner in vitro, they act with exquisite specificity in vivo. As an example, the translation initiation factor 4AI (eIF4AI), unwinds RNA secondary structure at the 5′ untranslated region (UTR) of mRNAs, and promotes the recruitment of the small ribosomal subunit (9–11). In contrast, the closely related paralogue eIF4AIII binds tightly on spliced mRNAs as part of the exon junction complex (EJC) (8). The EJC promotes nonsense-mediated mRNA decay (NMD) in human cells (12–14) and the localization of oskar mRNA in the Drosophila embryo (14, 15).
DEAD-box proteins have a common architecture based on two RecA domains connected by a flexible linker (reviewed in ref. 3). A hallmark of these proteins is the conformational plasticity with which they cycle between the active and inactive states of the ATPase reaction (reviewed in ref. 4). In the active state, the two RecA domains adopt a characteristic closed conformation that positions the residues responsible for ATP hydrolysis in the appropriate geometry for catalysis (16–21). In the inactive state, the two domains are in more open configurations (17, 18). Conformational regulation is often used to modulate the ATPase activity of DEAD-box proteins, for example in the activation of the translation initiation factor 4AI (eIF4AI) by the MIF4G domain of eIF4G (22) and in the inhibition of eIF4AI by the MA3 domain of PDCD4 (23, 24). The conformational changes of DEAD-box proteins are important not only for catalysis, but also for protein–protein interactions. In the case of the EJC, for example, the closed RNA-bound conformation of eIF4AIII is required to form a core complex with its binding partners MAGO, Y14, and Barentsz (also known as Metastatic Lymph Node 51; MLN51) (8, 17, 18).
The EJC core is assembled in an ATP-dependent manner on spliced mRNAs typically, but not exclusively, 20–24 nt upstream of exon-exon junctions (25–27). EJC assembly is a complex, stepwise process that is tightly coupled to the splicing reaction (28–30). Recent studies in human cells have shown that EJC assembly requires the binding of eIF4AIII to the splicing factor Complexed With Cef1 (CWC22) (31–33). The interaction of CWC22 with eIF4AIII is incompatible with the interaction with MAGO and Y14 and is necessary for eIF4AIII recruitment to the splicing machinery (31, 32). Consistent with its role in EJC deposition, CWC22 participates to the degradation of an endogenous NMD target in vivo (33). In vitro, CWC22 reduces the ATPase activity of eIF4AIII, suggesting that CWC22 keeps the DEAD-box protein in an inactive state before the EJC assembles at exon ligation (31). CWC22 contains two conserved regions predicted to fold into MIF4G and MA3 domains. Counterintuitively with respect to the known activating properties of MIF4G domains and inhibiting properties of MA3 domains on other DEAD-box proteins (21–24), CWC22 uses its MIF4G domain to bind eIF4AIII (32). In this work, we have elucidated the mechanism with which the MIF4G domain of CWC22 specifically recognizes and negatively regulates eIF4AIII.
Results and Discussion
Domain Requirements for eIF4AIII–CWC22 Complex Formation.
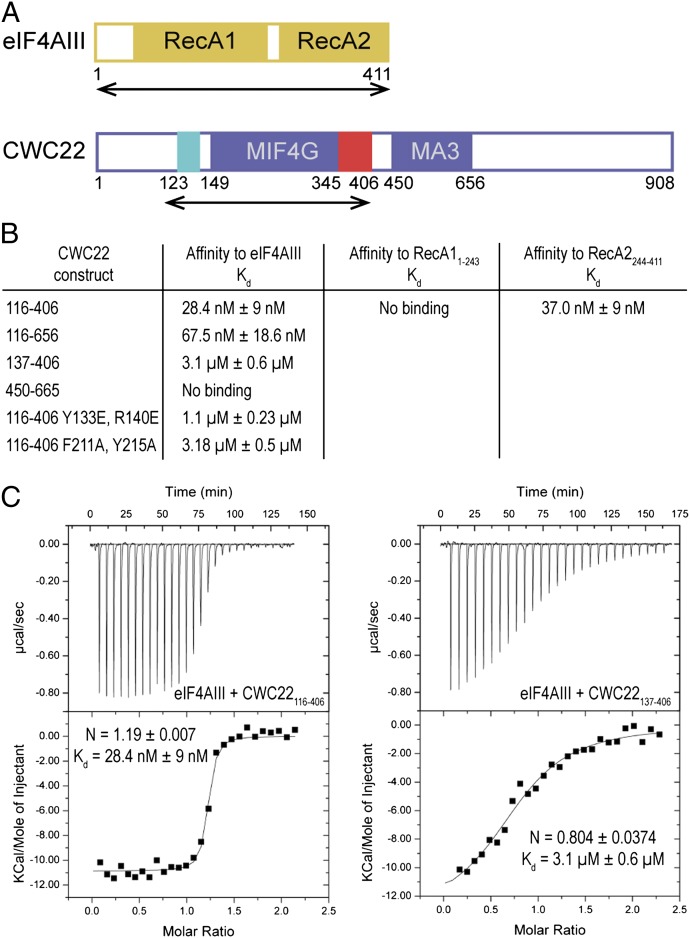
Human eIF4AIII (residues 1–411) contains an N-terminal RecA1 domain and a C-terminal RecA2 domain (17, 18) (Fig. 1A). CWC22 (residues 1–656) contains MIF4G and MA3 domains predicted approximately between residues 145–350 and 450–656, respectively (using the Phyre (34) and HHpred prediction servers; ref. 35) (Fig. 1A). Previous coimmunoprecipitation and pull-down experiments have mapped the interaction between the two proteins to the RecA2 domain of eIF4AIII and to a region of CWC22 containing the MIF4G domain (residues 110–409) (31, 32). We purified different portions of the molecules and assessed their relative binding affinities quantitatively by using isothermal titration calorimetry (ITC).
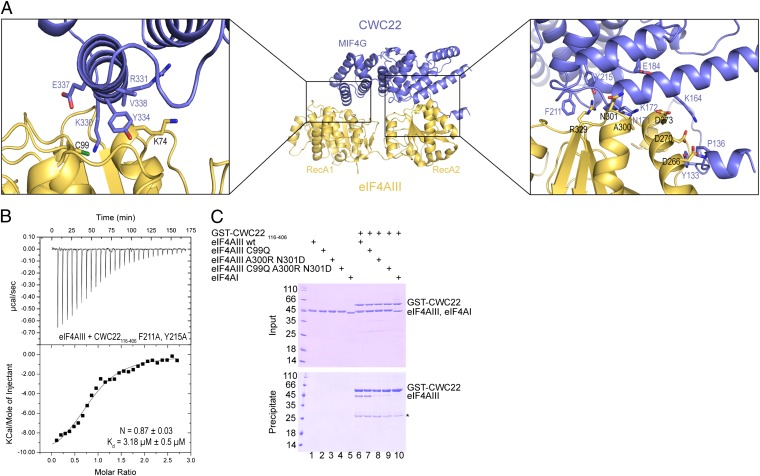
Fig. 1.
Domain requirements for eIF4AIII–CWC22 recognition. (A) Schematic domain organization of human eIF4AIII and CWC22. The RecA domains of eIF4AIII are in yellow. In the case of CWC22, the MIF4G fold is in blue and the N- and C-terminal extension are in cyan and red, respectively. Arrows highlight the regions of the molecules used for crystallization of the complex. The residue numbers of the domain boundaries are indicated (obtained either from the structural analysis in this manuscript or predicted in the case of the MA3 domain). (B) Table summarizing the results of the isothermal calorimetry experiments shown in the manuscript and in Fig. S1. The dissociation constants (Kd) with SDs were calculated with the program Origin. (C) Two representative ITC experiments, showing binding of eIF4AIII full length (f.l.) with different regions of CWC22. The MicroCal cell was filled with eIF4AIII at 25 μM and CWC22 was injected at 250 μM concentration consecutively in 10-μL volumes. Shown in each inset are the number of calculated binding sites (N), and the dissociation constant (Kd) with SDs, as calculated with the program Origin.
An evolutionary conserved region of CWC22 spanning the MIF4G and MA3 domains (residues 116–656) bound full-length (f.l.) eIF4AIII with a Kd of 67.5 nM (Fig. 1B and Fig. S1). The MA3 domain (residues 450–665) showed no detectable binding to eIF4AIII (Fig. 1B and Fig. S1). The presence of the MA3 domain appeared to even have a small negative contribution to eIF4AIII binding: A segment lacking the entire MA3 domain (residues 116–406) resulted in a Kd of 28.4 nM (Fig. 1 B and C). Remarkably, a further 20-residue truncation to the stable structural core of the MIF4G domain identified by limited proteolysis (residues 137–406) decreased binding to eIF4AIII 100-fold (Kd 3.1 μM; Fig. 1 B and C). We concluded that elements both within and outside the MIF4G domain of CWC22 participate in eIF4AIII binding. Upon dissecting eIF4AIII, RecA1 did not show appreciable binding to CWC22 in ITC measurements (Fig. 1B and Fig. S1). RecA2 bound CWC22 with a Kd of 37 nM, slightly higher than that measured in the case of full-length eIF4AIII (Fig. 1B and Fig. S1). Thus, the RecA2 domain of eIF4AIII contains most, albeit not all, of the determinants for CWC22 recognition. A similar relative contribution of the RecA1 and RecA2 domains was reported in the case of the yeast eIF4A–eIF4G complex (22, 36).
Structure of a Human eIF4AIII–CWC22 Complex.
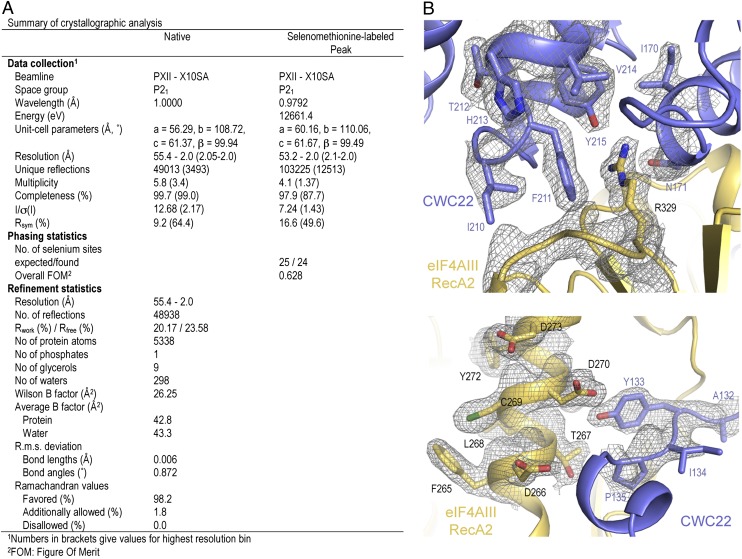
We crystallized the complex of human eIF4AIII (f.l.) and CWC22 (residues 116–406). The crystal structure was solved by a combination of single-wavelength anomalous dispersion (SAD) (using selenomethionine as anomalous scatterer) and molecular replacement (using the structure of human eIF4AIII, PDB ID code 2HXY, as a search model). The structure is refined at 2.0 Å resolution, with a free R factor of 23.58%, working R factor of 20.17% and good stereochemistry (Fig. 2). The final model encompasses the two RecA domains of eIF4AIII (residues 21–411) and residues 123–406 of CWC22. No ordered electron density was present for the N-terminal region of eIF4AIII (residues 1–20) and for the N-terminal residues and a disordered loop of CWC22 (residues 116–122 and 142–148).
Fig. 2.
Crystal structure analysis. (A) Table showing data collection and refinement statistics. (B) Electron density snapshots showing two sections of the complex interface. The 2Fo-Fc map is contoured at 1 σ and shown with the final model superposed.
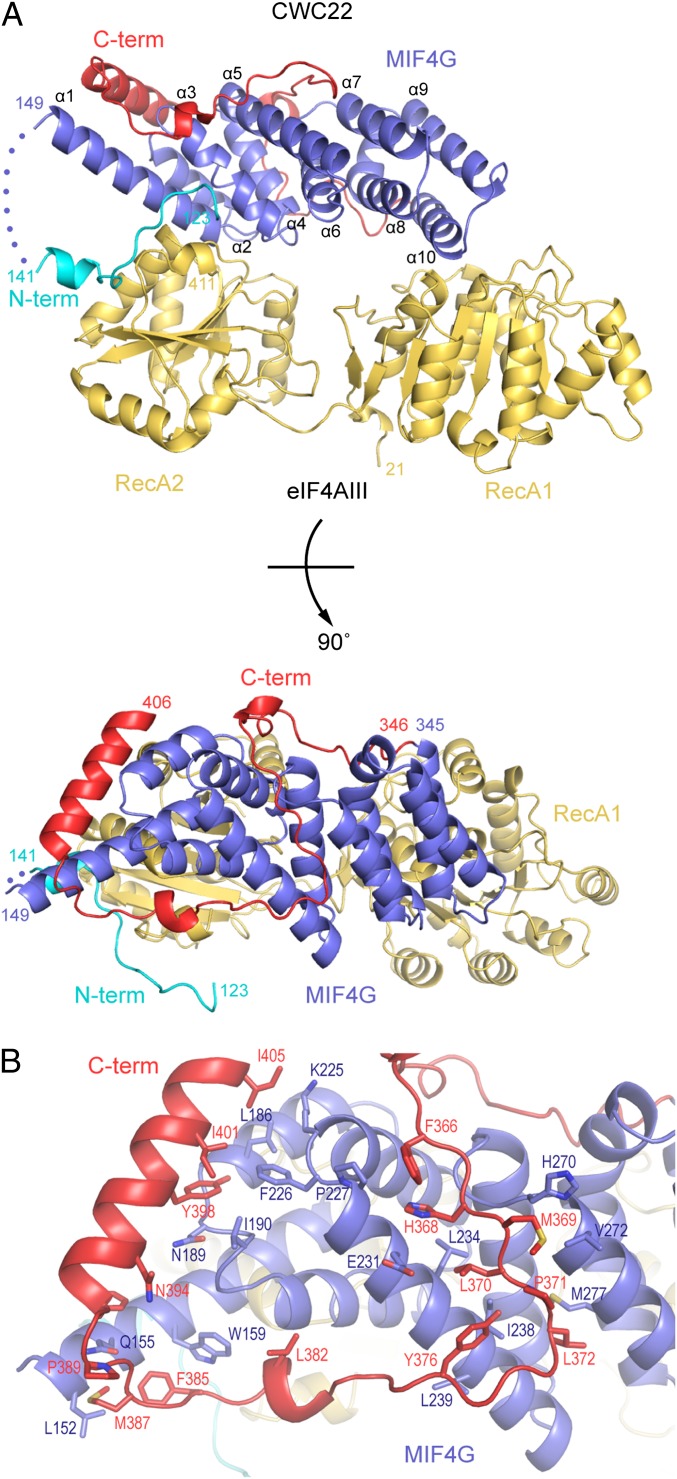
The RecA1 and RecA2 domains of eIF4AIII are each formed by the typical parallel β-sheet surrounded by α-helices (Fig. 3A, Upper). The conformation of the two RecA domains observed in the CWC22-bound structure is rather open (Fig. 3A, Upper), although it is different from the open conformation observed in the apo structure (18) (Fig. S2). We did not detect crystal contacts that would likely impact on this conformation. In addition to CWC22 binding, the N-terminal region of eIF4AIII preceding RecA1 might stabilize the relative conformation of the two RecA domains. In contrast to previous structures (17, 18), the N terminus of eIF4AIII forms an additional β-strand (at residues 28–31), extending the RecA1 β-sheet (Fig. S2). This β-strand interacts with RecA2 (at Arg370, a residue in motif VI of the DEAD-box protein) and approaches the linker that connects RecA1 and RecA2.
Fig. 3.
Structure of the human eIF4AIII–CWC22 complex. (A) The eIF4AIII–CWC22 complex is viewed in two orientations, related by a 90° clockwise rotation around a horizontal axis. The molecules are colored as in the schematics in Fig. 1A. The helices of the five HEAT repeats (blue) are indicated α1–α10. (B) The MIF4G helices (blue) and the C-terminal extension (red) of CWC22 are engaged in a large number of hydrophobic interactions, effectively forming a single structural unit.
CWC22 Has an Atypical MIF4G Domain.
CWC22 comprises a MIF4G fold with N-terminal and C-terminal extensions (Fig. 3A). The MIF4G fold of CWC22 (residues 149–345) consists of five HEAT repeats (Fig. 3A, in blue). Each HEAT motif is formed by two antiparallel α-helices (termed A and B helices). Consecutive repeats pack against each other in a roughly parallel fashion, with 15° rotations between successive repeats (37). This topology results in a crescent-shaped molecule with a convex surface formed by the A helices (α1, α3, α5, α7, α9) and a concave surface formed by the B helices (α2, α4, α6, α8, α10). The MIF4G fold of CWC22 is similar to that of yeast eIF4G (22) and Gle1 (21).
After helix α10, a C-terminal extension of approximately 60 residues stretches over the MIF4G fold (Fig. 3A, in red). Residues 346–390 zig-zag on the edge of the concave surface and across the convex surface toward helix α1, covering a distance of approximately 100 Å with extensive hydrophobic interactions (Fig. 3B). The C-terminal extension ends with an α-helix (α11) that binds the HEAT 1 helices with van der Waals interactions. Given the apolar nature of the intramolecular contacts, the C-terminal extension appears to be an integral part of the CWC22 MIF4G domain. This domain also features an N-terminal extension (residues 123–141; Fig. 3A, in cyan) that is connected with a disordered and presumably flexible linker to helix α1. The N-terminal extension does not engage in intramolecular interactions and is therefore not a structural part of the MIF4G domain, but binds intermolecularly to eIF4AIII.
The N-Terminal Extension of the CWC22 MIF4G Binds at a Hydrophobic Hotspot on RecA2.
The N-terminal extension of the CWC22 MIF4G domain binds eIF4AIII in a surface pocket formed in the RecA2 domain between the N-terminal α-helix and the N-terminal β-strand (Fig. 4). In particular, CWC22 residues 129–140 engage RecA2 with both polar and hydrophobic interactions (Fig. 2B, Lower and 4A), burying more than 1,100 Å2 of surface area. CWC22 Tyr133 and Arg140 contact eIF4AIII Asp270 and Asp266. CWC22 Ala137 and Pro136 pack against eIF4AIII Trp263. We have shown that this binding pocket of eIF4AIII is used for binding UPF3b (38). Remarkably, Tyr133 and Pro135 of CWC22 are at the same structural position as Tyr429 and Pro431 of human UPF3b (Fig. 4 A and C and Fig. S3). This pocket thus appears to be a hotspot used by different binding partners of eIF4AIII.
Fig. 4.
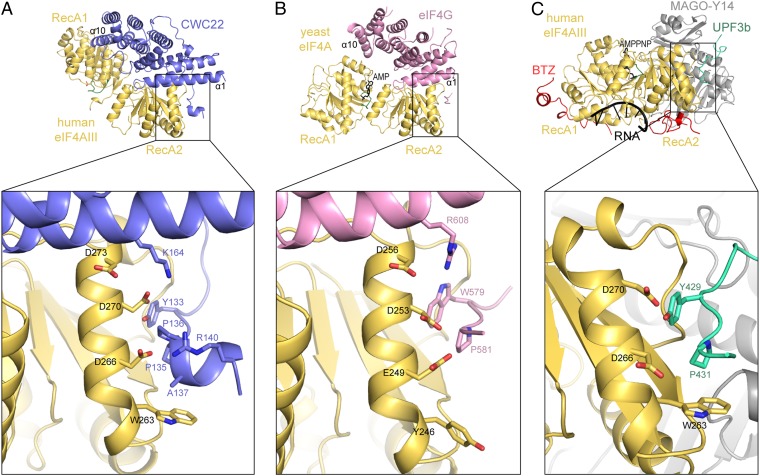
Recognition of a short, flexible motif by the RecA2 domain of eIF4A-like proteins. The structures of eIF4A-like proteins are shown in the same orientation after optimal superposition of their RecA2 domains. The lower images show a zoom-in of the same hydrophobic pocket on RecA2. (A) Structure of the human eIF4AIII–CWC22 complex, shown with eIF4AIII in yellow and CWC22 entirely in blue. (B) Structure of the yeast eIF4A–eIF4G complex (22), shown with eIF4A in yellow and the eIF4G in pink. (C) Structure of human eIF4AIII (yellow) in the EJC complex and bound to UPF3b (in green) (38). BTZ is in red, MAGO and Y14 in gray, and RNA and AMPPNP in black.
A corresponding binding pocket also exists in yeast eIF4A and is used to recognize a flexible N-terminal extension of the eIF4G MIF4G domain (22) (Fig. 4B). Trp579 and Pro581 of yeast eIF4G are at the equivalent position of Tyr133 and Pro135 of human CWC22. In the case of yeast eIF4G, mutation of Trp579 has been shown to significantly decrease the binding affinity for yeast eIF4A and to lead to a temperature-sensitive phenotype (22). We evaluated the contribution of this binding site in the case of the human eIF4AIII–CWC22 complex. A Tyr133Glu and Arg140Glu mutant of CWC22 interacted with eIF4AIII with a Kd of 1.1 μM, 100-fold lower than the wild type (Fig. 1B and Fig. S1). We concluded that both human CWC22 and yeast eIF4G feature a flexible element that is connected to the N terminus of their MIF4G domains and that docks onto a prominent pocket present in the RecA2 domains of their respective binding partners. Binding to this pocket in RecA2 provides an important contribution to complex formation in both the eIF4A–eIF4G (22) and eIF4AIII–CWC22 complexes.
The MIF4G Domain of CWC22 Interacts with the Two RecA Domains of eIF4AIII.
The MIF4G domain of CWC22 binds eIF4AIII at two sites. At one end of the MIF4G crescent, HEAT 1 and 2 contact RecA2 (Fig. 3A). Here, CWC22 Phe211, Tyr215, and Asn171 (on helices α1 and α4) surround eIF4AIII Arg329, whereas CWC22 Lys164 and Lys172 form hydrogen bonds with eIF4AIII Asp270, Asp273, and Ala300 main-chain oxygen (Fig. 5A, Right and Fig. S4A). Consistently, it has been shown that mutation of Asn171 and Lys172 in CWC22 impairs the coprecipitation of endogenous eIF4AIII from cell lysates and that mutation of Asp270 and Asp273 in eIF4AIII impairs the coprecipitation of endogenous CWC22 (32). A Phe211Ala, Tyr215Ala mutation in CWC22, engineered to selectively weaken this interaction site, decreased the binding affinity for eIF4AIII of a hundredfold, to a Kd of 3.18 μM (Figs. 1B and 5B). At the other end of the MIF4G crescent, helix α10 in HEAT 5 contacts RecA1 (Fig. 3A). CWC22 uses Lys330, Tyr334, and Val338 to interact with Cys99 and to approach the top of the N-terminal helices of eIF4AIII (Fig. 5A, Left and Fig. S4B). The interaction of the MIF4G domain involves 3 salt bridges and 26 hydrogen bonds, and buries a surface area of more than 1,200 Å2 of RecA2 and 700 Å2 of RecA1. The extent of the interactions of the MIF4G domain and of the N-terminal extension identified from the structural analysis is in line with the larger energetic contribution of the RecA2 domain observed in ITC measurements (Fig. 1B).
Fig. 5.
Interactions of the CWC22 MIF4G domain with eIF4AIII RecA1 and RecA2. (A) Two zoom-in views showing the interactions of the CWC22 MIF4G domain at RecA1 (Left) and RecA2 (Right). Included are conserved residues involved in the interaction as well as residues that differ in eIF4AI (Ala300, Asn301 in RecA2 and Cys99 in RecA1). (B) ITC experiment measuring the affinity between human eIF4AIII and a mutant of CWC22 with Phe211Ala and Tyr215Ala substitutions. Perturbing the hotspot of interactions between RecA2 and the MIF4G domain of CWC22 results in a significant decrease in affinity. (C) Protein coprecipitations by GST pull-down assays. GST-tagged CWC22 (residues 116–406) was incubated with eIF4AIII wild type and mutants in a buffer containing 200 mM NaCl before coprecipitation with glutathione-Sepharose beads, as indicated. One-fifth of the input and the entire glutathione eluates were analyzed on Coomassie-stained 13.5% SDS/PAGE. The mutants were designed to mimic residues in eIF4AI. Precipitation with eIF4AI was added as a control. The asterisk indicates an impurity (GST).
The eIF4AIII-interacting residues of CWC22 are conserved in metazoans and are also in part conserved in NOM1, an eIF4AIII-interacting protein involved in prerRNA processing (39) (Fig. S5). The CWC22-interacting residues of eIF4AIII are also conserved to a large extent in the paralogue eIF4AI (Fig. S6). However, CWC22 does not bind eIF4AI (31). Mutants with several amino acid substitutions on RecA2 have been shown to weaken the interaction with CWC22 (32). Inspection of the structure reveals that only a few of the CWC22-interacting residues differ between eIF4AIII and eIF4AI: Cys99 at RecA1 (Gln in eIF4AI), and Ala300, Asn301 at RecA2 (Arg and Asp in eIF4AI) (Fig. 5A and sequence alignment in Fig. S6). Based on the structural analysis, we made selective substitutions of eIF4AIII and tested the ability of the recombinant mutant proteins to interact with CWC22 in GST–pull-down assays. As controls, GST-CWC22 was able to precipitate wild-type eIF4AIII but not eIF4AI (Fig. 5C, lanes 6 and 10). Mutation of Cys99Gln (on RecA1) did not appreciably affect the interaction with CWC22 (Fig. 5C, lane 7). In contrast, mutation of Ala300Arg and Asn301Asp (on RecA2) weakened the interaction with CWC22 (Fig. 5C, lane 8). These two substitutions are predicted to make unfavorable electrostatic interactions with an incoming CWC22 (at Lys172 and Glu184) (Fig. 5A, Right). The results are in line with the notion that the RecA2 domain provides the major contribution to CWC22 binding and indicate that small differences on the surface of DEAD-box proteins can be sufficient to impart specificity.
CWC22 Holds eIF4AIII in an Inactive Conformation.
The bidentate mode of MIF4G interaction in the eIF4AIII–CWC22 complex is reminiscent of that observed in the eIF4A–eIF4G complex (22, 36). At the N-terminal end of the MIF4G domain, the interactions of helices α1 and α4 of CWC22 with the RecA2 domain of eIF4AIII are strikingly similar to the interactions observed in the eIF4A–eIF4G complex (Fig. 6 A and B and Fig. S7A). At the C-terminal end of the MIF4G domain, both CWC22 and eIF4G use helix α10 to contact RecA1, but the interaction site on RecA1 differs between the two complexes (Fig. 6 A and B and Fig. S7). Whereas eIF4G contacts the side of the N-terminal helices of eIF4A, CWC22 contacts the top of the corresponding helices of eIF4AIII. Superposition of the structures of yeast eIF4A–eIF4G and human eIF4AIII–CWC22 at their RecA2-MIF4G units shows a large difference in the relative orientations of their RecA1 domains, which are rotated approximately 90° with respect to each other (Fig. S7A).
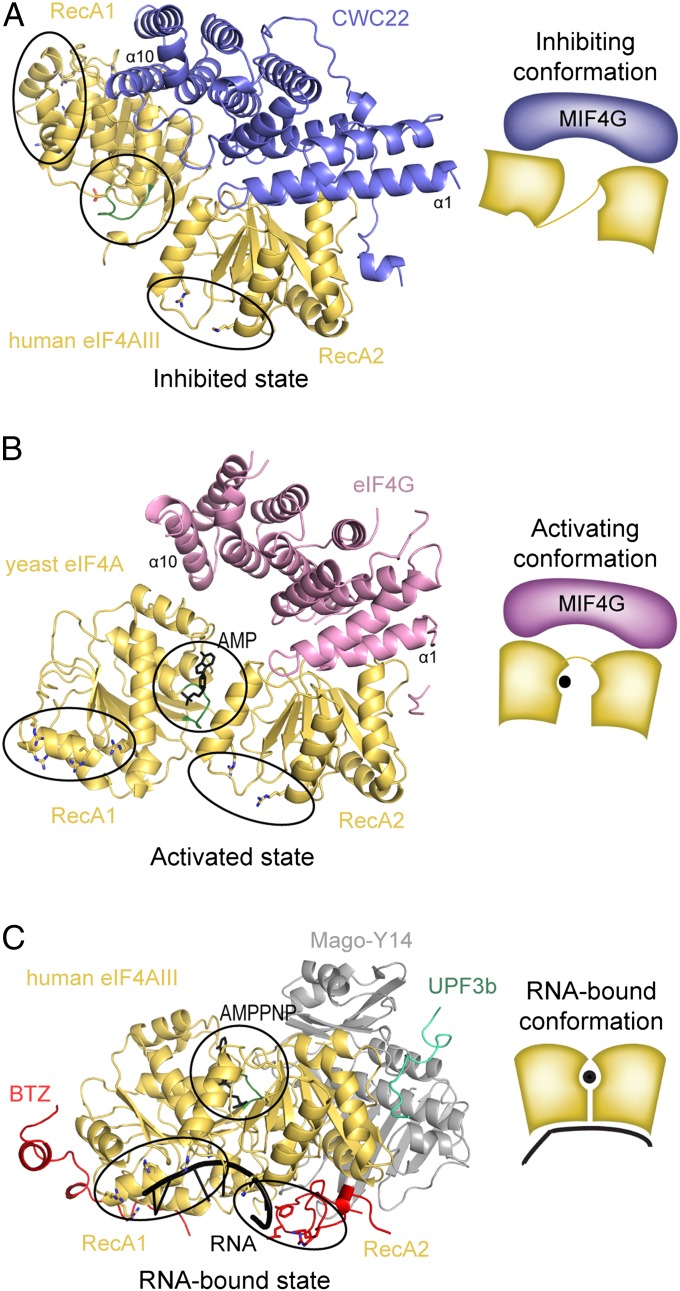
Fig. 6.
Conformational regulation of eIF4A-like proteins. The left images show the structures of the human eIF4AIII–CWC22 complex (A), yeast eIF4A–eIF4G complex (22) (B), and human eIF4AIII in the EJC complex and bound to UPF3b (38) (C) in the same orientation and colors as in Fig. 4. RNA-binding residues and ATP-binding residues of eIF4A-like proteins are boxed with an ellipse and a circle, respectively. The ATP-binding P loop in RecA1 is shown in green. In A, a phosphate ion from the crystallization buffer is shown bound to the P loop. The right images show schematic representations of the structures in the left images, highlighting the differences in the relative conformation of the two RecA domains in the inhibited (A), activated (B), and RNA-ATP-bound (C) states.
The different conformation of RecA1 with respect to RecA2 has significant consequences. In the case of the yeast eIF4A–eIF4G complex, the DEAD-box motifs for RNA and ATP binding present in the RecA1 and RecA2 domains face each other, approaching the conformation required for the active state of the DEAD-box protein (Fig. 6 B and C, compare the positions of ellipses and circles). A similar conformation has been reported for another helicase–MIF4G complex, that of Dbp5 and Gle1 (21). In both cases, binding of the MIF4G domain is known to boost the ATPase activity of the DEAD-box protein (36, 40). Mechanistically, the MIF4G domains of eIF4G and Gle1 are thought to restrict the conformational space of the RecA domains they bind to, thereby increasing the on and off rates of the reaction (36, 40). In the case of the eIF4AIII–CWC22 complex, the RNA-binding residues of RecA1 are far from the RNA-binding site on RecA2 (Fig. 6 A and C, compare position of the ellipses). The ATP-binding loop (P-loop, in green in Fig. 6A) does not point toward RecA2 as it does in the active conformation, but is in a diametrically opposite position (Fig. 6, compare position of the circles). Thus, CWC22 holds eIF4AIII in an inactive conformation.
We note that having a strong anchoring interaction on the RecA2 domain and a weak latching interaction on the RecA1 domain would allow the MIF4G domain to let go of RecA1 without significantly affecting the overall binding affinity. From a structural standpoint, it would therefore in principle be possible for eIF4AIII to change the conformation of RecA1 and achieve a closed RNA–ATP-bound conformation even when in complex with CWC22. However, eIF4AIII has a weak RNA-dependent ATPase activity in the absence of Barentsz (8). We speculate that the weak RNA-binding properties of eIF4AIII in the absence of Barentsz or other proteins is not sufficient to drive the RecA1 domain away from the interactions of the inhibiting conformation. In this situation, CWC22 would be able to prevent eIF4AIII from binding RNA before the EJC is assembled onto spliced mRNAs.
Conclusions
MIF4G domains (such as in CWC22, eIF4G, and Gle1) are emerging as widespread regulators of RNA-dependent ATPases. At the structural level, MIF4G domains look overall rather similar, as do the RecA domains of the DEAD-box proteins they bind to (e.g., eIF4AIII, eIF4A, and Dbp5). At the molecular level, the eIF4AIII–CWC22, eIF4A–eIF4G, and Dbp5–Gle1 complexes all use the N-terminal helices of the MIF4G domain to anchor the RecA2 domain of the ATPase and the C-terminal helices of the MIF4G domain to latch on the RecA1 domain. Although the anchoring mechanism is remarkably similar in the three complexes, the latching interactions differ. As a result, these MIF4G proteins stabilize the RecA domains of their respective DEAD-box protein either in an activated or in an inhibited conformation. The opposite functional readout is imparted by subtle changes in the intermolecular contacts, which, even with hindsight, would be difficult to predict. In the case of other MIF4G domains, ATPase rates will have to be experimentally measured to assess whether they up-regulate, down-regulate, or have no effect on the activity of the DEAD-box protein they bind to. Subtle changes also allow MIF4G domains to discriminate between a binding partner and very similar proteins in the cell. The current structural information suggests that MIF4G domains can fine tune the specificity toward paralogous DEAD-box proteins by a discrimination mechanism based on a few substitutions causing electrostatic or steric clashes.
Experimental Procedures
Protein Expression and Purification.
Human full-length eIF4AIII (411 residues), human CWC22 (MIF4G domain 116–406), and constructs thereof were expressed as recombinant GST-tagged proteins by using Escherichia coli BL21-Gold (DE3) pLysS cells (Stratagene), Terrific Broth (TB) medium, and overnight induction at 16 °C. Cells were lysed in 40 mM Tris at pH 7.5, 500 mM NaCl, 10% glycerol, 2 mM DTT supplemented with protease inhibitors (Roche), and the GST-tagged proteins were purified by affinity chromatography on glutathione resin (Clontech). After cleaving the GST tag with PreScission protease, proteins were further purified by ion exchange chromatography (MonoQ) and size-exclusion chromatography (Superdex; GE Healthcare) using 40 mM Tris at pH 7.5, 200 mM NaCl, 10% glycerol, and 2 mM DTT as gel-filtration buffer. The eIF4AIII–CWC22 complex was formed by incubating the two purified proteins in an estimated 1:1 ratio for 30 min at room temperature. The complex was purified by size exclusion chromatography (Superdex S200; GE Healthcare). Mutants of eIF4AIII and CWC22 were generated by using the QuikChange Site-Directed Mutagenesis kit (Stratagene), and expressed and purified as described for the wild-type proteins.
Crystallization and Structure Determination.
Crystals were grown at 18 °C by vapor diffusion from sitting nanodrops formed by equal volumes of protein (3 mg/mL) and crystallization buffer [12% (wt/vol) polyethylene glycol (PEG) 20000, 0.1 M Na-K phosphate at pH 6.5). Crystals (40 × 30 × 10 µm3) appeared after 10 d. Selenomethionine-substituted crystals were obtained in similar conditions. Crystals were transferred to a stabilization solution [14% (wt/vol) PEG 20000 and 0.1 M Na-K phosphate at pH 6.5] containing 25% (vol/vol) glycerol as a cryoprotectant. All data were collected at Swiss Light Source beamline PXII. Data were processed and scaled with the XDS package (41) (Fig. 2A). The crystals belong to the monoclinic space group P21 and contain one eIF4AIII–CWC22 complex in the asymmetric unit with a solvent content of 46%.
The structure was determined by SAD (from data collected at selenium peak wavelength) combined with molecular replacement in PHASER (42) using the apo eIF4AIII structure (Protein Data Bank ID code 2HXY) as a search model. The initial electron density was of good quality and allowed most of the model to be automatically built with AutoSol, as implemented in PHENIX (43), and to be manually completed by using the program COOT (44). Refinement was carried out with PHENIX (43). The Ramachandran plot statistics for the final model in Fig. 2A were calculated with the program Molprobity (45).
ITC.
ITC was carried out by using a VP-ITC Microcal calorimeter (Microcal; GE Healthcare). All samples were dialyzed in a buffer containing 40 mM Hepes at pH 7.5, 200 mM NaCl, 1.5 mM Tris(2-chlorethyl)phosphate. The eIF4AIII samples (full-length and fragments) were concentrated to 25 µM and CWC22 samples (fragments and mutants) were concentrated to 250 µM. Titrations were carried out at 23 °C with 25–30 injections of 10 µL of the CWC22 solution into 1.4 mL of eIF4AIII proteins. As control for all ITC measurements, the injectant was titrated into buffer. All data were processed and curves fitted using the Origin software (Microcal).
Pull-Down Assay.
GST-CWC22 (10 µg) together with either human eIF4AIII (wild-type or mutants) or eIF4AI (15 µg) were incubated at room temperature for 30 min. Complexes were applied to glutathione Sepharose beads equilibrated with binding buffer (40 mM Tris at pH 7.5, 200 mM NaCl, 5% glycerol, and 2 mM DTT). Beads were washed three times with binding buffer and eluted with binding buffer containing 30 mM glutathione. Eluted proteins were analyzed by SDS/PAGE.
Supplementary Material
Acknowledgments
We thank the Max Planck Institute (MPI) Crystallization Facility for screenings and optimization; the MPI Core Facility for mass spectrometry analysis; the beamline scientists at the Swiss Light Source for excellent assistance with data collection; Isabelle Barbosa for purified eIF4AI; Debora Makino for help on the structure determination; and members of our laboratories for discussion and critical reading of the manuscript. This work was supported by the Max Planck Gesellschaft; the European Research Council Advanced Investigator Grant 294371, Marie Curie Initial Training Network RNPnet 289007, and Deutsche Forschungsgemeinschaft Grants SFB646, SFB1035, GRK1721, FOR1680, and the Center for Integrative Science Munich (to E.C.); and by Centre National de la Recherche Scientifique and the Agence Nationale de la Recherche Grant 2011-BLAN-01801 (to H.L.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4C9B).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1314684110/-/DCSupplemental.
References
- 1.Linder P, Fuller-Pace FV. Looking back on the birth of DEAD-box RNA helicases. Biochim Biophys Acta. 2013;1829(8):750–755. doi: 10.1016/j.bbagrm.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 2.Jankowsky E, Fairman ME. RNA helicases—one fold for many functions. Curr Opin Struct Biol. 2007;17(3):316–324. doi: 10.1016/j.sbi.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 3.Pyle AM. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu Rev Biophys. 2008;37:317–336. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- 4.Henn A, Bradley MJ, De La Cruz EM. ATP utilization and RNA conformational rearrangement by DEAD-box proteins. Annu Rev Biophys. 2012;41:247–267. doi: 10.1146/annurev-biophys-050511-102243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fairman ME, et al. Protein displacement by DExH/D “RNA helicases” without duplex unwinding. Science. 2004;304(5671):730–734. doi: 10.1126/science.1095596. [DOI] [PubMed] [Google Scholar]
- 6.Jankowsky E, Gross CH, Shuman S, Pyle AM. Active disruption of an RNA-protein interaction by a DExH/D RNA helicase. Science. 2001;291(5501):121–125. doi: 10.1126/science.291.5501.121. [DOI] [PubMed] [Google Scholar]
- 7.Lund MK, Guthrie C. The DEAD-box protein Dbp5p is required to dissociate Mex67p from exported mRNPs at the nuclear rim. Mol Cell. 2005;20(4):645–651. doi: 10.1016/j.molcel.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 8.Ballut L, et al. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat Struct Mol Biol. 2005;12(10):861–869. doi: 10.1038/nsmb990. [DOI] [PubMed] [Google Scholar]
- 9.Ray BK, et al. ATP-dependent unwinding of messenger RNA structure by eukaryotic initiation factors. J Biol Chem. 1985;260(12):7651–7658. [PubMed] [Google Scholar]
- 10.Pause A, Méthot N, Svitkin Y, Merrick WC, Sonenberg N. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J. 1994;13(5):1205–1215. doi: 10.1002/j.1460-2075.1994.tb06370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pestova TV, Shatsky IN, Hellen CU. Functional dissection of eukaryotic initiation factor 4F: The 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol Cell Biol. 1996;16(12):6870–6878. doi: 10.1128/mcb.16.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shibuya T, Tange TØ, Sonenberg N, Moore MJ. eIF4AIII binds spliced mRNA in the exon junction complex and is essential for nonsense-mediated decay. Nat Struct Mol Biol. 2004;11(4):346–351. doi: 10.1038/nsmb750. [DOI] [PubMed] [Google Scholar]
- 13.Lykke-Andersen J, Shu MD, Steitz JA. Communication of the position of exon-exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science. 2001;293(5536):1836–1839. doi: 10.1126/science.1062786. [DOI] [PubMed] [Google Scholar]
- 14.Palacios IM, Gatfield D, St Johnston D, Izaurralde E. An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature. 2004;427(6976):753–757. doi: 10.1038/nature02351. [DOI] [PubMed] [Google Scholar]
- 15.Hachet O, Ephrussi A. Drosophila Y14 shuttles to the posterior of the oocyte and is required for oskar mRNA transport. Curr Biol. 2001;11(21):1666–1674. doi: 10.1016/s0960-9822(01)00508-5. [DOI] [PubMed] [Google Scholar]
- 16.Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125(2):287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 17.Bono F, Ebert J, Lorentzen E, Conti E. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell. 2006;126(4):713–725. doi: 10.1016/j.cell.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 18.Andersen CBF, et al. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 2006;313(5795):1968–1972. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- 19.von Moeller H, Basquin C, Conti E. The mRNA export protein DBP5 binds RNA and the cytoplasmic nucleoporin NUP214 in a mutually exclusive manner. Nat Struct Mol Biol. 2009;16(3):247–254. doi: 10.1038/nsmb.1561. [DOI] [PubMed] [Google Scholar]
- 20.Napetschnig J, et al. Structural and functional analysis of the interaction between the nucleoporin Nup214 and the DEAD-box helicase Ddx19. Proc Natl Acad Sci USA. 2009;106(9):3089–3094. doi: 10.1073/pnas.0813267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Montpetit B, et al. A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export. Nature. 2011;472(7342):238–242. doi: 10.1038/nature09862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schütz P, et al. Crystal structure of the yeast eIF4A-eIF4G complex: An RNA-helicase controlled by protein-protein interactions. Proc Natl Acad Sci USA. 2008;105(28):9564–9569. doi: 10.1073/pnas.0800418105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang JH, et al. Crystal structure of the eIF4A-PDCD4 complex. Proc Natl Acad Sci USA. 2009;106(9):3148–3153. doi: 10.1073/pnas.0808275106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loh PG, et al. Structural basis for translational inhibition by the tumour suppressor Pdcd4. EMBO J. 2009;28(3):274–285. doi: 10.1038/emboj.2008.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le Hir H, Izaurralde E, Maquat LE, Moore MJ. The spliceosome deposits multiple proteins 20-24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 2000;19(24):6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saulière J, et al. CLIP-seq of eIF4AIII reveals transcriptome-wide mapping of the human exon junction complex. Nat Struct Mol Biol. 2012;19(11):1124–1131. doi: 10.1038/nsmb.2420. [DOI] [PubMed] [Google Scholar]
- 27.Singh G, et al. The cellular EJC interactome reveals higher-order mRNP structure and an EJC-SR protein nexus. Cell. 2012;151(4):750–764. doi: 10.1016/j.cell.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reichert VL, Le Hir H, Jurica MS, Moore MJ. 5′ exon interactions within the human spliceosome establish a framework for exon junction complex structure and assembly. Genes Dev. 2002;16(21):2778–2791. doi: 10.1101/gad.1030602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Z, Krainer AR. Splicing remodels messenger ribonucleoprotein architecture via eIF4A3-dependent and -independent recruitment of exon junction complex components. Proc Natl Acad Sci USA. 2007;104(28):11574–11579. doi: 10.1073/pnas.0704946104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gehring NH, Lamprinaki S, Hentze MW, Kulozik AE. The hierarchy of exon-junction complex assembly by the spliceosome explains key features of mammalian nonsense-mediated mRNA decay. PLoS Biol. 2009;7(5):e1000120. doi: 10.1371/journal.pbio.1000120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barbosa I, et al. Human CWC22 escorts the helicase eIF4AIII to spliceosomes and promotes exon junction complex assembly. Nat Struct Mol Biol. 2012;19(10):983–990. doi: 10.1038/nsmb.2380. [DOI] [PubMed] [Google Scholar]
- 32.Steckelberg A-L, Boehm V, Gromadzka AM, Gehring NH. CWC22 connects pre-mRNA splicing and exon junction complex assembly. Cell Rep. 2012;2(3):454–461. doi: 10.1016/j.celrep.2012.08.017. [DOI] [PubMed] [Google Scholar]
- 33.Alexandrov A, Colognori D, Shu M-D, Steitz JA. Human spliceosomal protein CWC22 plays a role in coupling splicing to exon junction complex deposition and nonsense-mediated decay. Proc Natl Acad Sci USA. 2012;109(52):21313–21318. doi: 10.1073/pnas.1219725110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kelley LA, Sternberg MJE. Protein structure prediction on the Web: A case study using the Phyre server. Nat Protoc. 2009;4(3):363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 35.Söding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33(Web Server issue):W244-8. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hilbert M, Kebbel F, Gubaev A, Klostermeier D. eIF4G stimulates the activity of the DEAD box protein eIF4A by a conformational guidance mechanism. Nucleic Acids Res. 2011;39(6):2260–2270. doi: 10.1093/nar/gkq1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andrade MA, Perez-Iratxeta C, Ponting CP. Protein repeats: Structures, functions, and evolution. J Struct Biol. 2001;134(2-3):117–131. doi: 10.1006/jsbi.2001.4392. [DOI] [PubMed] [Google Scholar]
- 38.Buchwald G, et al. Insights into the recruitment of the NMD machinery from the crystal structure of a core EJC-UPF3b complex. Proc Natl Acad Sci USA. 2010;107(22):10050–10055. doi: 10.1073/pnas.1000993107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alexandrov A, Colognori D, Steitz JA. Human eIF4AIII interacts with an eIF4G-like partner, NOM1, revealing an evolutionarily conserved function outside the exon junction complex. Genes Dev. 2011;25(10):1078–1090. doi: 10.1101/gad.2045411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weirich CS, et al. Activation of the DExD/H-box protein Dbp5 by the nuclear-pore protein Gle1 and its coactivator InsP6 is required for mRNA export. Nat Cell Biol. 2006;8(7):668–676. doi: 10.1038/ncb1424. [DOI] [PubMed] [Google Scholar]
- 41.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr. 2007;63(Pt 1):32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 45.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.