Viral genomes comprise either RNA or DNA, and sometimes both, as can happen with retroviruses at different stages of their life cycle. Viral nucleic acids are pathogen-associated molecular patterns (PAMPs) that are detected by a variety of pathogen-recognition receptors (PPRs), key components of the innate immune system. A diverse set of PRRs has evolved to sense the genetic material of viruses. Toll-like receptors (TLRs) are archetypical PRRs, and several (TLR3, TLR7/8, and TLR9) recognize various RNA species and DNA CpG motifs in the endosomes. In addition, many cells also have RIG-I–like receptor (RLR) homologs that act as cytosolic sensors of viral RNA species (ssRNA or dsRNA with a 5′ppp group). The ligand specificities of these RLRs and the signaling cascades they induce are relatively well characterized (1). More recently, the discovery that intracellular DNA can induce an IFN response in a TLR-independent manner (2), and the identification of stimulator of IFN genes (STING) as a critical adaptor in the IFN-stimulatory DNA (ISD) pathway (3, 4), have stimulated the discovery of many cytosolic DNA sensors (5). However, our understanding of cytosolic DNA sensors in innate immune sensing is far from clear. Although numerous receptors have been proposed for the STING-dependent ISD pathway, a consistent definition of what constitutes a DNA-based STING-activating ligand is lacking. In PNAS, Jakobsen et al. provide evidence that the candidate DNA sensor IFI16 also serves a sensor of DNA forms generated during the lentiviral replication cycle and may serve to control or restrict HIV replication in myeloid human cell types (6).
Many Sensors for Cytosolic DNA Have Been Proposed
A variety of synthetic DNA ligands and the use of live DNA virus infection have been used to implicate about 10 receptors in the ISD pathway (reviewed in refs. 5 and 7). DNA-dependent activator of IRFs (DAIs) was the first candidate cytosolic DNA sensor to be identified and, as the name implies, interacts with TBK1 and activates IRF3 in a DNA-dependent manner. Nonetheless, DAI-deficient mice did not exhibit the expected deficit phenotype in response to DNA vaccination or DNA virus infection, and the human homolog was dispensable for the ISD pathway in many human cells. DDX41 was identified by an RNAi screen of all extant members of the DExD/H helicase family for defects in the IFN response to transfected poly(dA:dT). Even though the involvement of DDX41 in the STING-dependent ISD pathway was later confirmed with DNA virus infection, the IFN response to poly(dA:dT) can also be induced via the RIG-1 dsRNA-recognition pathway. The rationale for this quirk exemplifies the daunting complexities involved in identifying a bona fide DNA sensor: RNA Pol III can recruit and transcribe AT-rich DNA into dsRNA with 5′-triphosphates, which is then recognized by the RIG-I pathway. Other proposed DNA sensors include DNA damage factors Mre11 and the DNA-PK/Ku70/Ku80 complex and more promiscuous nucleic acid sensors that bind both RNA and DNA ligands, such as high-mobility group box proteins (HMGBs), LSm14A, and LRRFIP1.
IFI16 and cGAS
The remaining two receptors, IFN-γ inducible protein (IFI)16 and cGMP-AMP synthase (cGAS), have a strong body of evidence supporting their role as DNA sensors and are relevant to the present study by Jakobsen et al (6). A slew of high-profile studies this year establish an elegant mechanistic theory for how cGAS might function as a bona fide cytosolic DNA sensor. Z. J. Chen and coworkers discovered that mammalian cytosolic extracts produce cGMP-AMP (cGAMP) from ATP and GTP but only in the presence of DNA (8). cGAMP, a cyclic dinucleotide, serves as a second messenger that is produced by cGAS upon cGAS binding to cytosolic DNA (9). cGAMP then binds to the STING dimeric interface and induces a conformational change that is critical for downstream activation of TBK1 and IRF3 phosphorylation. Three independent groups then confirmed cGAMP as the cognate product of cGAS-DNA interactions and further characterized cGAMP as having an unusual mixed phosphodiester bond [G(2′,5′)pA(3′,5′)p] that results in its high-affinity binding to STING (10–12). Recently, Chen and coworkers reported that cGAS is an innate immune sensor of HIV and other retroviruses, a claim that is made contemporaneously for IFI16 by Jakobsen et al. in the current PNAS study.
IFI16 is a member of the PYHIN family of proteins and has an N-terminal PYrin domain that mediates protein–protein interactions and two C-terminal DNA-binding HIN domains. IFI16 belongs to the AIM2-like receptor (ALR) gene family, and members of the ALR family contribute differentially to activation of the STING-dependent ISD pathway or ASC-dependent inflammasome formation upon detection of intracellular DNA. IFI16 is the only DNA sensor that shuttles between the nucleus and cytoplasm and is predominantly nuclear at steady state. Indeed, many ALRs exhibit steady-state nuclear localization, and it has been suggested that these receptors are positioned there to respond to specific replication intermediates of DNA viruses and retroelements (13). IFI16 has been shown to be required for the STING-dependent ISD pathway in response to transfected DNA or intracellular DNA exposed during HSV-1 virus infection. IFI16 physically binds to dsDNA in a sequence-independent fashion, consistent with structural evidence showing that the HIN domains of IFI16 make contacts with only the sugar-phosphate backbone of dsDNA (14). All of these properties of IFI16 are consistent with the behavior of a bona fide innate immune sensor for intracellular DNA.
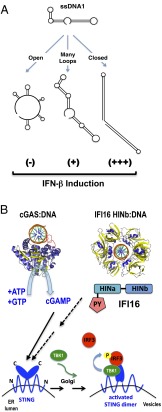
Jakobsen et al. (6) extend the universe of DNA ligands that IFI16 recognizes to include DNA forms that occur during HIV-1 replication. As a retrovirus, the single-stranded, positive-sense RNA genome of HIV-1 must first complete an obligate integration step into the host genome before new viral genomic transcripts can be made to form new viruses. This necessitates multiple stages where viral genomic RNA is reversed-transcribed to form double-stranded cDNA complexes that are competent for nuclear import and chromosomal integration. Along the way, multiple forms of viral-derived nucleic acids (ssRNA, ssDNA, RNA:DNA hybrids, dsDNA) are present in the cytosol and are potentially exposed to multiple PRRs that recognize various forms of viral nucleic acids. Thus, it is not surprising that cytosolic (or even nuclear) DNA sensors can detect at least some forms of HIV-derived nucleic acids. The more significant question is whether sensing HIV-derived DNA forms by IFI16 (or cGAS for that matter) activates the STING-dependent ISD pathway or other inflammatory pathways that impact on HIV replication in a biologically relevant setting. In PNAS, Jakobsen et al. found one ssDNA sequence (ssDNA1) derived from the HIV long-terminal repeat (LTR) region that induces a reproducible IFN-β response in primary human macrophage-derived macrophages (hMDMs) as well as in the PMA- differentiated THP-1 human monocytic cell line. Using different forms of ssDNA1 mutants (length, ∼100 nt) engineered to have divergent secondary structures such as closed, open, or many loops (Fig. 1A), the authors discovered that it was the fortuitous stem–loop structure in ssDNA1 that accounted for its ability to trigger type I and III IFN responses. Further studies confirmed the sequence independence of this DNA-ligand triggered IFN response and revealed that the stability and length (at least 50 nt) of the dsDNA stem was the critical determinant of the IFN responses. In a series of loss-of-function studies, the authors show that “HIV-derived ssDNA1” induces IFN responses via canonical members of the STING-dependent ISD pathway, which include STING, TBK1, IRF-3/IRF-7, and, now, IFI16 itself. These data reinforce the known properties of IFI16 as a sensor of dsDNA (5). Additionally, the controls are revealing because the “dsDNA” used, a 70-mer duplex, is always a more potent activator of the IFI16-mediated STING-dependent IFN responses than the HIV-derived ssDNA1 (a 50-mer stem loop).
Fig. 1.
Innate immune sensing of DNA substrates. (A) HIV-derived ssDNA1 and mutant sequences that are poor (-), moderate (+) or good (+++) inducers of IFN-beta. (B) Structures of cGAS and IFI16 (HINb domain) bound to dsDNA (orange circle with spokes) indicate sequence-independent binding (14, 15). cGAMP, generated by cGAS upon DNA binding, activates STING directly (solid arrow). It is less clear how IFI16 (PY, Pyrin domain; HINa,b, DNA-binding domains) activates the STING-dependent ISD pathway (dashed arrow).
More significantly, shRNA-mediated knockdown of IFI16 in THP-1 cells and hMDMs resulted in a significant increase in HIV replication, and IFI16 colocalizes with the cognate HIV-derived DNA forms. However, knockdown of STING and cGAS, but not DDX41, also resulted in a similar increase in HIV replication. The impact of the cGAS knockdown on HIV-1 replication is at least consistent with the results of Chen and coworkers indicating that cGAS is an innate immune sensor for HIV (8). Furthermore, as for IFI16, there is similar structural evidence showing that cGAS binds to DNA in a sequence-independent fashion (15) (Fig. 1B). However, whereas Chen and coworkers clearly showed that cGAS is critical for activating STING-dependent type I IFN responses in the context of HIV infection, they did not report the impact of cGAS-mediated sensing on HIV replication, whereas Jakobsen et al. did for IFI16 (6). At minimum, the results of Jakobsen et al. suggest that IFI16 and cGAS are likely to serve nonredundant functions in activating the STING-dependent ISD pathway.
IFI16 and cGAS Clearly Play Nontrivial Roles in the STING-Dependent ISD Pathway
Nonetheless, it is unlikely we will truly discern the specific roles of IFI16 and cGAS in innate immune sensing of HIV-1 until we understand the full palette of functions that these proteins perform and integrate this knowledge with the corpus of data on the biology of HIV. For example, IFI16 was recently reported to be a restriction factor for hCMV: it restricts hCMV replication by displacing Sp1-like transcription factors from the promoter of the hCMV DNA polymerase (16). The HIV core promoter in the LTR has three tandem Sp1-binding sites that are critical for its transcriptional activity. The inhibitory activity of IFI16 on HIV replication, relieved by IFI16 knockdown, may thus also be related to its affect on LTR transcription. The studies reported by Jacobsen et al. reveal the complexity of interpreting the integrative signaling cascades involved in innate immune sensing of HIV but also provide roadmaps for generating testable hypotheses to further our understanding of such fundamental host–pathogen interactions.
Footnotes
The author declares no conflict of interest.
See companion article on page E4571.
References
- 1.Dixit E, Kagan JC. Intracellular pathogen detection by RIG-I-like receptors. Adv Immunol. 2013;117:99–125. doi: 10.1016/B978-0-12-410524-9.00004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24(1):93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 3.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455(7213):674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461(7265):788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Unterholzner L. The interferon response to intracellular DNA: Why so many receptors? Immunobiology. 2013;218(11):1312–1321. doi: 10.1016/j.imbio.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 6.Jakobsen MR, et al. IFI16 senses DNA forms of the lentiviral replication cycle and controls HIV-1 replication. Proc Natl Acad Sci USA. 2013;110:E4571–E4580. doi: 10.1073/pnas.1311669110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cavlar T, Ablasser A, Hornung V. Induction of type I IFNs by intracellular DNA-sensing pathways. Immunol Cell Biol. 2012;90(5):474–482. doi: 10.1038/icb.2012.11. [DOI] [PubMed] [Google Scholar]
- 8.Wu J, et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339(6121):826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339(6121):786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ablasser A, et al. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498(7454):380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao P, et al. Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell. 2013;153(5):1094–1107. doi: 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang X, et al. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol Cell. 2013;51(2):226–235. doi: 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brunette RL, et al. Extensive evolutionary and functional diversity among mammalian AIM2-like receptors. J Exp Med. 2012;209(11):1969–1983. doi: 10.1084/jem.20121960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin T, et al. Structures of the HIN domain:DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity. 2012;36(4):561–571. doi: 10.1016/j.immuni.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Civril F, et al. Structural mechanism of cytosolic DNA sensing by cGAS. Nature. 2013;498(7454):332–337. doi: 10.1038/nature12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gariano GR, et al. The intracellular DNA sensor IFI16 gene acts as restriction factor for human cytomegalovirus replication. PLoS Pathog. 2012;8(1):e1002498. doi: 10.1371/journal.ppat.1002498. [DOI] [PMC free article] [PubMed] [Google Scholar]