Significance
ROS1 fusion kinases are critical oncogenes in several malignancies, suggesting that ROS1 inhibitors are likely to be effective molecularly targeted therapies in these patients. Although phase I/II clinical trials using the ALK/ROS1 inhibitor crizotinib to treat ROS1 fusion-harboring non–small-cell lung cancer patients demonstrate early success, evidence of clinical resistance to crizotinib due to the acquired ROS1G2032R mutation was recently reported. Here, we demonstrate that foretinib is a more potent ROS1 inhibitor than crizotinib in vitro and in vivo and remains effective against crizotinib-resistant ROS1 kinase domain mutations, including ROS1 G2032R. Taken together, our findings establish foretinib as a highly promising therapeutic candidate for treating patients with ROS1-driven malignancies and provide rationale for rapid clinical translation.
Abstract
The rapidly growing recognition of the role of oncogenic ROS1 fusion proteins in the malignant transformation of multiple cancers, including lung adenocarcinoma, cholangiocarcinoma, and glioblastoma, is driving efforts to develop effective ROS1 inhibitors for use as molecularly targeted therapy. Using a multidisciplinary approach involving small molecule screening in combination with in vitro and in vivo tumor models, we show that foretinib (GSK1363089) is a more potent ROS1 inhibitor than crizotinib (PF-02341066), an ALK/ROS inhibitor currently in clinical evaluation for lung cancer patients harboring ROS1 rearrangements. Whereas crizotinib has demonstrated promising early results in patients with ROS1-rearranged non–small-cell lung carcinoma, recently emerging clinical evidence suggests that patients may develop crizotinib resistance due to acquired point mutations in the kinase domain of ROS1, thus necessitating identification of additional potent ROS1 inhibitors for therapeutic intervention. We confirm that the ROS1G2032R mutant, recently reported in clinical resistance to crizotinib, retains foretinib sensitivity at concentrations below safe, clinically achievable levels. Furthermore, we use an accelerated mutagenesis screen to preemptively identify mutations in the ROS1 kinase domain that confer resistance to crizotinib and demonstrate that these mutants also remain foretinib sensitive. Taken together, our data strongly suggest that foretinib is a highly effective ROS1 inhibitor, and further clinical investigation to evaluate its potential therapeutic benefit for patients with ROS1-driven malignancies is warranted.
Receptor tyrosine kinases (RTKs) are critical mediators of extracellular signals that control key cell growth, survival, and motility pathways. Conversely, deregulated and constitutive RTK activation is responsible for the initiation and progression of many cancers. Multiple mechanisms contribute to aberrant RTK activation including chromosomal rearrangements, point mutations, and gene amplification. Oncogenic activation of the orphan RTK c-ros oncogene 1 (ROS1) is observed in a subset of patients with glioblastoma, non–small-cell lung cancer (NSCLC), and cholangiocarcinoma (1–5). In most cases, ROS1 signaling is activated by interchromosomal translocation or intrachromosomal deletion that results in N-terminal ROS1 fusion genes. Several ROS1 kinase fusion proteins have been identified, including the Fused in Glioblastoma–ROS1 (FIG–ROS) that was first discovered in a human glioblastoma cell line (2) and more recently in patients with NSCLC (4), cholangiocarcinoma (3), and serous ovarian carcinoma (6). The SLC34A2–ROS1 (SLC–ROS) fusion is present in a subset of patients with NSCLC (1, 7) and gastric cancer (8). Other ROS1 fusions include CD74–ROS1, EZR–ROS1, LRIG3–ROS1, SDC4–ROS1, and TPM3–ROS1 (5).
Given the recent success of molecularly targeted therapies in treating cancers driven by oncogenic kinases, there is acute clinical momentum to identify inhibitors that selectively target ROS1 fusions. Because the ROS1 and Anaplastic Lymphoma Kinase (ALK) domains are partially homologous, the Food and Drug Administration (FDA)-approved ALK/MET kinase inhibitor crizotinib is being investigated via phase I/II clinical trials for its efficacy in ROS1-driven lung cancer patients (9). Although early results appear promising, consistent with the clinical experience of crizotinib in patients with ALK-positive lung cancer to date, as well as prior experience with kinase inhibitors in many other malignancies (10–13), recent evidence suggests that a subset of patients with crizotinib-treated ROS1 fusion-positive may acquire ROS1 kinase domain mutations that confer drug resistance, thus necessitating alternative therapeutic approaches.
To identify additional and potentially more efficacious ROS1 inhibitors, we used an unbiased, high-throughput kinase inhibitor screening assay and discovered that foretinib (GSK1363089) and Gö6976 are potent inhibitors of ROS1. Foretinib selectively suppresses the growth of the SLC–ROS-driven human NSCLC cell line HCC78 and of FIG–ROS-driven murine cholangiocarcinoma, but not of EGFR-driven NSCLC or phosphatase and tensin homolog (PTEN)-suppressed murine cholangiocarcinoma cells. Further, treatment of tumor-bearing mice with foretinib resulted in specific and dramatic regression of FIG–ROS-driven tumors in contrast to non-FIG–ROS tumors that share similar histopathological features. Importantly, we also use a cell-based in vitro resistance screen to preemptively identify several ROS1 kinase domain point mutations that confer resistance to crizotinib and show that these crizotinib-resistant ROS1 mutants remain sensitive to foretinib. These data suggest that foretinib may provide an alternative front-line treatment for ROS1-positive tumors and an effective second-line approach for patients that develop crizotinib-resistant disease.
Results
High-Throughput Kinase Inhibitor Screen Identifies Foretinib and Gö6976 as Potent ROS1 Inhibitors.
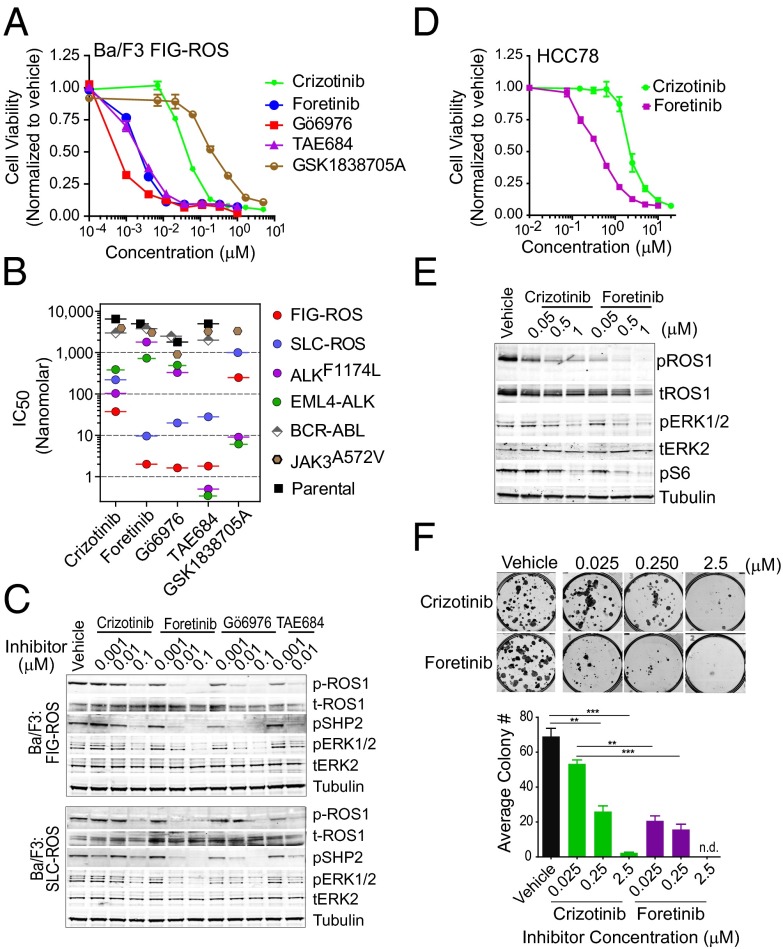
The FIG–ROS fusion protein transforms a murine interleukin-3 dependent pro-B lineage cell line, Ba/F3, and National Institutes of Health (NIH) 3T3 cells and promotes tumor formation in vivo (3, 5). To identify ROS1 inhibitors that selectively block the growth of Ba/F3 FIG–ROS cells, we used a cell-based high-throughput kinase inhibitor screen encompassing graded concentrations of 104 unique inhibitors seeded in 384-well plates (Dataset S1). The following criteria were used to select potential ROS1 inhibitors for further evaluation: selective sensitivity of the Ba/F3 FIG–ROS cells at an inhibitor concentration ≤10 nM and a ≥5-fold lower 50% growth inhibitory constant (IC50) compared with Ba/F3 cells expressing BCR–ABL or Ba/F3 parental cells. Using this assay, we identified foretinib and Gö6976 as two potent inhibitors of Ba/F3 FIG–ROS cell growth with IC50s of 2 nM and 1 nM, respectively (Fig. 1 A and B). Human ROS1 and ALK share 42–48% kinase domain homology, and consequently the ALK/MET inhibitor crizotinib and the ALK inhibitor NVP–TAE684 (TAE684) have recently been reported to show activity against ROS1 fusions in Ba/F3 cells (3, 4) and in the SLC–ROS-expressing human NSCLC cell line HCC78 (1, 7). Consistent with these published studies and providing validation of our screening approach, we identified both crizotinib and TAE684 as inhibitors of Ba/F3 FIG–ROS cell growth, with IC50s of 38 nM and 2 nM, respectively (Fig. 1 A and B).
Fig. 1.
Foretinib inhibits ROS1 fusions in Ba/F3 and HCC78 cells. (A) Growth of Ba/F3 FIG–ROS cells after 72 h exposure with varying concentrations of crizotinib, foretinib, Gö6976, TAE684, and GSK1838705A as normalized to vehicle-treated cells. The values are means ± SEM from three independent experiments performed in triplicate. (B) IC50 values for the Ba/F3 FIG–ROS, SLC–ROS, EML4–ALK, ALKF1174L, BCR–ABL, JAK3A572V, and parental cells treated with the indicated inhibitors as determined from nonlinear regression curve fit analysis of the dose–response curves. (C) Immunoblot analysis of ROS1 fusion proteins and their downstream effectors after inhibitor treatment. (D) Growth of HCC78 cells after 72 h exposure with varying concentrations of crizotinib and foretinib. The normalized values are means ± SEM from three independent experiments. (E) Immunoblot analysis of SLC–ROS, ERK1/2, and protein S6 phosphorylation after inhibitor treatment in HCC78 cells. (F) Representative images and quantification of the number of colonies formed by HCC78 cells plated with or without increasing concentration of crizotinib and foretinib after 10 d. Immunoblots shown in C and E are cropped images representative of three independent experiments. Where indicated, **P < 0.01 and ***P < 0.001 by t test.
Given current efforts to treat ROS1-driven cancers with ALK inhibitors (14), we directly compared the efficacy of foretinib and Gö6976 to the previously known ALK inhibitors, crizotinib, TAE684, and GSK1838705A (15). For this, we used Ba/F3 cells expressing ROS1 fusions, the EML4–ALK fusion, or the activating ALKF1174L point mutant (16). To determine whether the targeting efficiency is comparable for different ROS1 fusions, we used Ba/F3 cells expressing either SLC–ROS or FIG–ROS. Foretinib demonstrated potent inhibition of both the FIG–ROS and SLC–ROS fusions (IC50: 2 nM and 10 nM, respectively), representing ∼20-fold increased inhibitory potency compared with crizotinib (IC50: 38 nM and 220 nM, respectively). Ba/F3 cells expressing either EML4–ALK or ALKF1174L were relatively insensitive to foretinib and Gö6976 but robustly inhibited by the ALK-targeted compounds (Fig. 1B). In agreement with these findings, foretinib and Gö6976 effectively induced apoptosis in Ba/F3 FIG–ROS and SLC–ROS but not in EML4–ALK- or ALKF1174L-expressing cells (Fig. S1 A–C).
Foretinib is a multikinase inhibitor with reported efficacy for MET and VEGFR2 (17). To confirm the selectivity of foretinib for inhibiting the growth of cancer cells expressing ROS1 fusions, we compared the effect of foretinib on Ba/F3 cells harboring either FIG–ROS or SLC–ROS to Ba/F3 cells expressing BCR–ABL (18) or JAK3A572V (19), as well as IL3-dependent Ba/F3 parental cells. Consistent with the results from the unbiased inhibitor screen, Ba/F3 cells expressing ROS1 fusions demonstrated superior sensitivity to foretinib, Gö6976, and TAE684 compared with crizotinib (Fig. 1B). This finding was paralleled by a dose-dependent decrease in phosphorylation of ROS1 and its downstream effectors, SHP2 and ERK1/2 (Fig. 1C). Conversely, the Ba/F3 BCR–ABL and JAK3A572V-expressing cells as well as the parental cells were insensitive to foretinib, Gö6976, and ALK inhibitors, suggesting there is minimal toxicity of these compounds due to off-target effects (Fig. 1B).
Among these inhibitors, foretinib and crizotinib are the only two compounds currently in clinical development. Although we found TAE684 and Gö6976 to also be potent ROS1 inhibitors, these are research-purpose compounds that do not have a clear path for clinical translation. As foretinib has an established safety and pharmacokinetic profile and is currently being evaluated in multiple phase I/II trials (20, 21), we chose to restrict further analysis to the comparison of foretinib and crizotinib.
In NIH 3T3 cells transformed with FIG–ROS or SLC–ROS, foretinib more robustly suppressed ROS1 autophosphorylation as well as anchorage-independent colony formation compared with crizotinib (Fig. S2). To determine whether these results could be recapitulated in human cancer cells, we exposed HCC78 cells, a NSCLC cell line expressing SLC–ROS, to increasing concentrations of foretinib or crizotinib. Consistent with our studies in Ba/F3 cells, foretinib inhibited the proliferation of HCC78 cells and abolished SLC–ROS and ROS1-effector protein phosphorylation at significantly lower doses than crizotinib (Fig. 1 D and E). Foretinib also showed greater efficacy than crizotinib in blocking colony formation (Fig. 1F) and migration of HCC78 cells (Fig. S3A). By contrast, the overall selectivity of foretinib to block the growth of ROS1 fusion-driven NSCLC compared with other NSCLC cells was demonstrated by marked resistance of EGFR-driven NSCLC PC9 and HCC4011 cells to foretinib and crizotinib, but sensitivity to the EGFR inhibitor erlotinib, as expected (Fig. S3B). Additionally, consistent with our findings for Ba/F3 cells expressing activated ALK, human lymphoma cell lines harboring the NPM–ALK fusion (KARPAS-299 and SUDHL1) exhibited the expected response to crizotinib, but were relatively insensitive to foretinib. To further rule out the possibility that the inhibitory effect of crizotinib and foretinib on HCC78 cells might be due to their efficacy as MET inhibitors, we tested additional MET kinase inhibitors (MGCD-265, SGX-523, and JNJ38877605). HCC78 cells were insensitive to these compounds, implying that MET inhibition could not account for the effect of foretinib and crizotinib on these cells (Fig. S3C).
Foretinib Is a Potent Inhibitor of FIG–ROS-Driven Cholangiocarcinoma.
Cholangiocarcinoma is a cancer of the bile duct and the second most common hepatic malignancy with a median survival of less than 2 y from diagnosis. The recent discovery of FIG–ROS kinase fusions in ∼9% of patients with cholangiocarcinoma provides a potential opportunity for the effective implementation of ROS1 inhibitors in this subgroup of patients (3). To test whether crizotinib or foretinib are viable candidates for treating FIG–ROS-driven cholangiocarcinoma, we used a murine cholangiocarcinoma model (22) to generate tumor cell lines that were either driven by expression of the FIG–ROS fusion or by short hairpin RNA-mediated loss of the tumor suppressor Pten (shPten). Whereas the viability of shPten-expressing cell lines was minimally affected by either crizotinib or foretinib, FIG–ROS-expressing tumor cell lines were significantly inhibited in the presence of either drug (Fig. 2A). In line with our data from Ba/F3 and HCC78 cells, foretinib was more potent than crizotinib at inhibiting cell proliferation. Foretinib, and to a lesser extent crizotinib, also significantly decreased the propensity of FIG–ROS but not shPten cholangiocarcinoma tumor cell lines to form anchorage-independent colonies in soft agar (Fig. 2B). Direct assessment of FIG–ROS signaling activity by immunoblot analysis revealed near complete inhibition of FIG–ROS autophosphorylation and SHP2 and ERK1/2 phosphorylation by foretinib at 100 nM, whereas crizotinib had only a moderate effect at the same concentration (Fig. 2C).
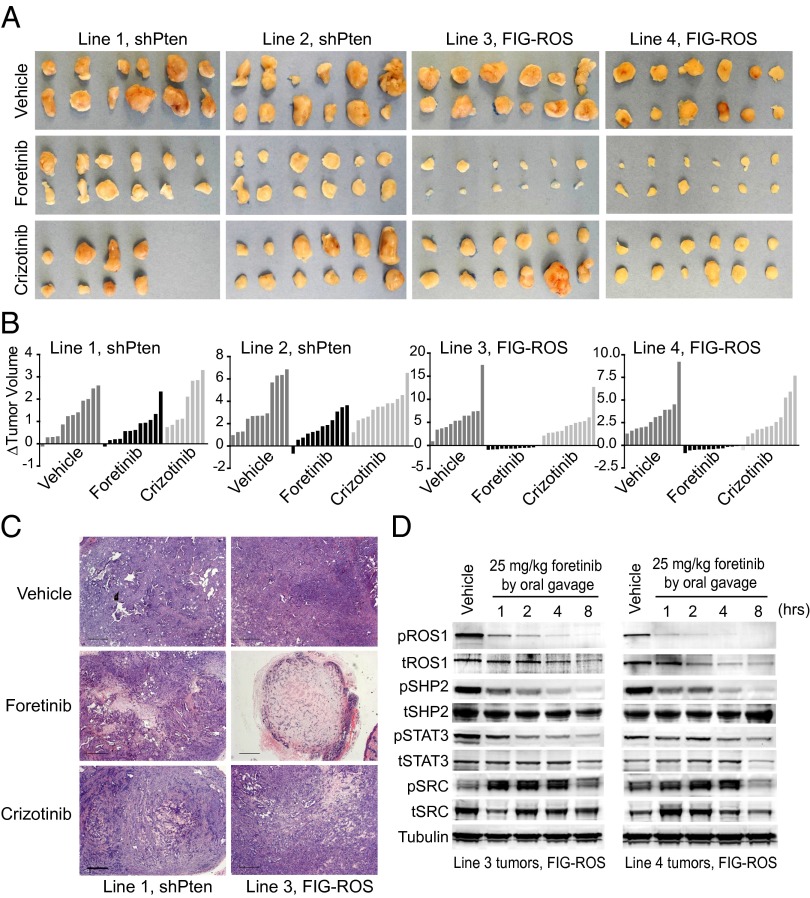
Fig. 2.
Foretinib and crizotinib inhibit FIG–ROS and effector signaling in murine cholangiocarcinoma tumor cell lines. (A) IC50 values for shPten- (lines 1 and 2) and FIG–ROS (lines 3 and 4)-driven murine cholangiocarcinoma cell lines treated with indicated inhibitors as determined from nonlinear regression curve fit analysis of the dose–response curves. (B) Analysis of anchorage-independent colony formation of FIG–ROS (line 4) and shPten (line 2) cholangiocarcinoma cell lines treated with inhibitors at the indicated concentrations. The numbers of colonies formed in the presence of foretinib are shown as a percentage of the number of colonies with vehicle treatment. Colony number is an average obtained using three independent wells normalized to vehicle control. (C) Immunoblot analysis of FIG–ROS fusion protein phosphorylation and downstream effector modulation after varying doses of crizotinib and foretinib. Cropped images representative of three independent experiments are shown.
Based on this robust in vitro activity, we evaluated the efficacy of foretinib on established tumors in vivo. Mice harboring FIG–ROS or shPten-driven s.c. tumors with a diameter of 4–6 mm were treated by oral gavage with vehicle, foretinib (25 mg/kg) or crizotinib (25 mg/kg) once per day for 9 consecutive days. Tumor volume was assessed before and posttreatment. Foretinib significantly reduced FIG–ROS-driven tumor volume compared with shPten tumors (lines 3 and 4: P < 0.0001; lines 1 and 2: P = 0.068 and 0.058, respectively; Fig. 3 A–C and Fig. S4 A and B). Equal dosing with crizotinib had a moderate effect on FIG–ROS tumor volume, but the differences were not statistically significant. To confirm that this oral dose of foretinib sufficiently blocked ROS1 activation in the treated tumors, we harvested FIG–ROS tumors at 1, 2, 4, and 8 h after a single treatment with vehicle or 25 mg/kg foretinib. We observed robust and time-dependent inhibition of FIG–ROS phosphorylation in foretinib-treated tumors compared with vehicle-treated controls (Fig. 3D). Concomitant with diminished FIG–ROS activation, phosphorylation of SHP2 and STAT3, but not SRC, was decreased in the treated tumors, suggesting FIG–ROS actively signals through SHP2 and STAT3 pathways (23). The transiently enhanced phosphorylation of SRC in foretinib-treated tumors further supports its selective activity for FIG–ROS-regulated pathways rather than attenuation of global signaling due to cytotoxic effects. Overall, these data demonstrate that foretinib is significantly more effective than crizotinib in blocking the growth of ROS1 fusion-driven cancers and highlights the potential for foretinib as a promising treatment for tumors carrying ROS1 fusions.
Fig. 3.
Foretinib significantly reduces FIG–ROS-driven tumor burden and inhibits intratumoral FIG–ROS phosphorylation and effector pathway activation. (A) Images of explanted FIG–ROS- and shPten-driven s.c. tumors after 9 d of foretinib (25 mg/kg) or crizotinib (25 mg/kg) treatment by oral gavage. (B) Waterfall plots showing change in FIG–ROS and shPten tumor volume from treatment initiation to 24 h after administration of the last dose. Relative tumor growth was calculated as described in Methods. (C) Representative H&E stained tumor sections from line 1 (shPten) and line 3 (FIG–ROS) after 9 d of inhibitor treatment. (Scale bar, 500 μm.) (D) Immunoblot analysis of intratumoral FIG–ROS phosphorylation and downstream effector modulation in tumors harvested from vehicle- or foretinib (25 mg/kg)-treated mice at 1, 2, 4, and 8 h after oral gavage. Cropped images representative of three independent experiments are shown.
Crizotinib-Resistant ROS1 Mutants Remain Sensitive to Foretinib.
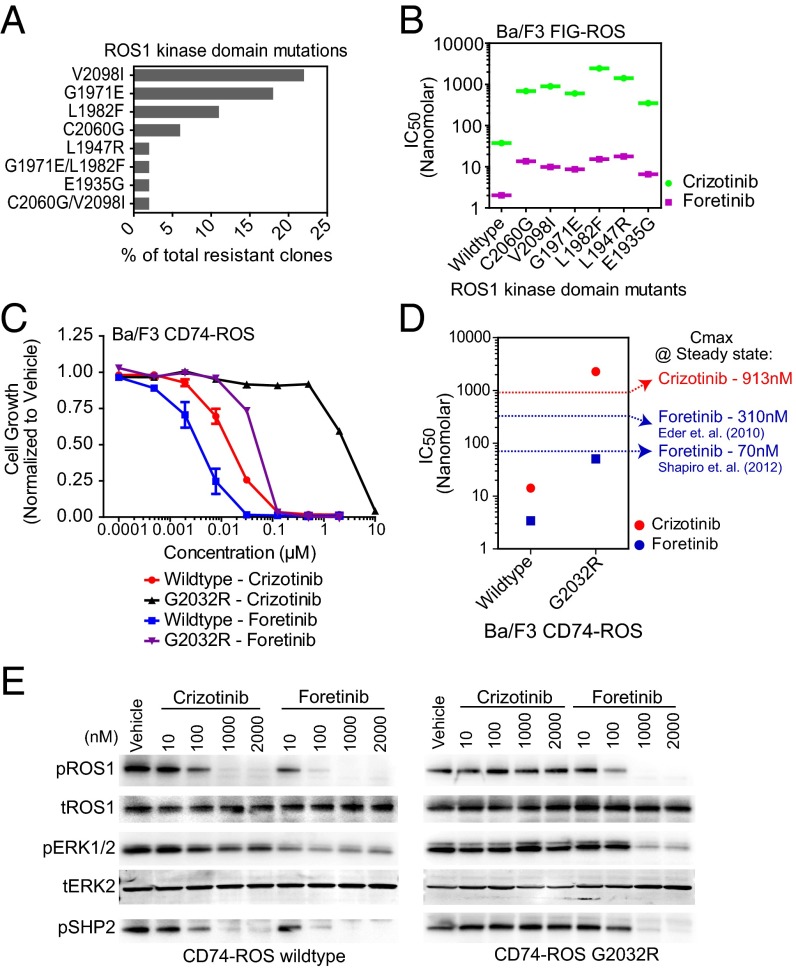
Despite the clinical benefit of molecularly targeted tyrosine kinase inhibitor therapies, development of therapeutic resistance remains a challenge. For example, acquisition of point mutations in the BCR–ABL and EGFR kinase domains that interfere with inhibitor binding frequently result in resistance to imatinib in chronic myeloid leukemia and to erlotinib in NSCLC, respectively (24, 25). Whereas clinical data from the initial phase I trial using crizotinib to treat patients with ROS1-rearranged lung cancer are promising (9), a subset of these patients with crizotinib-treated ROS1 expression may become resistant to therapy, as observed for patients with ALK fusion-bearing lung cancer (26, 27). To preemptively identify ROS1 kinase domain mutations conferring resistance to crizotinib, we performed an N-ethyl-N-nitrosourea (ENU)-assisted accelerated mutagenesis screen (28) using Ba/F3 FIG–ROS cells in the presence of increasing concentrations of crizotinib (500–2,500 nM). We observed a crizotinib concentration-dependent reduction in the percentage of wells with outgrowth, and the recovered clones were submitted for subsequent ROS1 kinase domain sequence analysis (Fig. S5 A and B). Six different ROS1 kinase domain point mutations were identified either alone or in combination, the two most frequent being V2098I and G1971E (Fig. 4A). Notably, all recovered clones remained sensitive to depletion of FIG–ROS by RNA interference, confirming that the ENU-based mutagenesis did not induce additional mutational events that confer FIG–ROS independence (Fig. S5C).
Fig. 4.
Inhibitory efficacy of foretinib for crizotinib-resistant ROS1 kinase domain mutations discovered from accelerated mutagenesis screening and from a crizotinib-resistant patient. (A) Percent frequency of ROS1 kinase domain mutations found in the ENU-treated Ba/F3 FIG–ROS clones isolated after growth and clonal expansion in the presence of 1,000 nM crizotinib. (B) IC50 values for Ba/F3 FIG–ROS wild-type and FIG–ROS kinase domain mutant cells treated with foretinib and crizotinib as determined from nonlinear regression curve fit analysis of the dose–response curves. (C) Growth of Ba/F3 CD74–ROS wild-type and G2032R mutant cells after 72 h exposure with graded concentrations of crizotinib or foretinib. Data are shown as normalized to vehicle-treated cells. The values are means ± SEM from three independent experiments performed in triplicate. (D) IC50 values for Ba/F3 CD74–ROS wild-type and G2032R mutant cells treated with foretinib and crizotinib as determined from nonlinear regression curve fit analysis of the dose–response curves. Dotted lines indicate the previously reported steady-state Cmax for crizotinib (red) and foretinib (blue). (E) Immunoblot analysis of CD74–ROS (wild type versus G2032R) phosphorylation and downstream effector modulation after treatment of cells with varying doses of crizotinib and foretinib as shown. Cropped images representative of three independent experiments are shown.
To understand how these mutations might confer drug resistance, we constructed homology models of the ROS1 kinase domain bound to crizotinib and foretinib based on reported crystallographic data for crizotinib:ALK- and foretinib:MET-bound complexes (Fig. S5D). The L1947R, L1982F, and V2098I mutations map close to structural features that are implicated in inhibitor binding (nucleotide binding loop, helix αC, and activation loop, respectively). Importantly, mutations of L1152 in ALK, a residue homologous to L1982 in ROS1, confer crizotinib resistance (29) and this mutation was recently identified in a patient with crizotinib-treated NSCLC with clinical resistance (26). Also, DFG motif-proximal mutations in ALK exhibit increased resistance to crizotinib (29). The mutations G1971E and C2060G map to locations that are distant to the inhibitor binding site in the N-terminal and C-terminal lobes, respectively.
We generated each of the six most frequently recovered crizotinib-resistant mutants (V2098I, G1971E, L1982F, C2060G, L1947R, and E1935G) in Ba/F3 FIG–ROS cells and confirmed their ability to confer IL-3 independence similar to wild-type FIG–ROS (Fig. S5E). As expected, all tested FIG–ROS mutants exhibited increased resistance to crizotinib (IC50 range: 350–2,450 nM; Fig. 4B and Fig. S6 A and C). IC50 values of the FIG–ROS mutants for crizotinib were close to or exceeded the clinically achievable maximum plasma concentration (411 ng/mL; 913 nM), suggesting that some of these mutants may be inefficiently inhibited at physiologically relevant concentrations. Notably, whereas the FIG–ROS kinase domain mutants exhibited slightly reduced sensitivity to foretinib (IC50 range: 7–18 nM), all were inhibited at concentrations below the recently described safe and tolerated at clinically achievable plasma levels (Fig. 4B and Fig. S6 B and C) (20, 30). The differential sensitivity of these mutants to crizotinib was further supported by immunoblot analysis that showed decreased inhibition of FIG–ROS autophosphorylation in mutant compared with wild-type cells as well as foretinib-treated cells (Fig. S6D).
While our manuscript was under review, therapeutic resistance to crizotinib in a patient with CD74–ROS-rearranged lung adenocarcinoma was reported, resulting from the acquisition of a G2032R kinase domain point mutation which precludes crizotinib binding due to steric hindrance (31). To assess whether the G2032R mutation retained sensitivity to foretinib similarly to the ROS1 mutations recovered in the accelerated mutagenesis screen, we evaluated the efficiency of foretinib to suppress ROS1 phosphorylation and ROS1-dependent cell growth in stably transformed Ba/F3 CD74–ROS wild-type and G2032R mutant cell lines. Ba/F3 CD74–ROSG2032R cells were highly resistant to crizotinib (IC50: ∼2200 nM) compared with wild-type CD74–ROS (IC50: 14 nM) (Fig. 4C). Despite being less sensitive to foretinib than the wild-type CD74–ROS fusion, the CD74–ROSG2032R mutant kinase remained sensitive to foretinib (IC50: 50 nM) at concentrations below plasma levels that can be safely achieved in patients with oral dosing. Thus, foretinib may offer an efficient second-line therapy in patients who are treatment refractory due to the acquisition of ROS1 mutations that confer resistance to crizotinib.
Discussion
The growing recognition of patients harboring ROS1 fusions as a distinct subgroup within multiple cancer entities provides impetus for the development of ROS1-directed therapeutic strategies. ROS1 fusions lead to constitutive kinase activation and can function either as dominant oncogenes or in cooperation with other genetic aberrations. Our overall goal was to take advantage of a high-throughput unbiased screening approach to identify small-molecule kinase inhibitors with potent activity against ROS1. Using this methodology, we found that foretinib is a highly effective inhibitor of ROS1 in human and murine model systems, demonstrating greater potency compared with crizotinib both in vitro and in vivo. To date, putative membrane-associated as well as cytoplasmic ROS1 fusions have been identified (5, 14). Our cellular model systems consistently revealed reduced sensitivity of the putative membrane-associated SLC–ROS compared with cytoplasmic FIG–ROS, suggesting the intriguing possibility that subcellular localization may modulate inhibitor sensitivity for ROS1 fusions, which should be taken into consideration for future efforts directed at the development of novel ROS1 inhibitors.
Despite the reported binding affinity (Kd) of crizotinib for ROS1 being lower than that of foretinib (4.4 nM and 14 nM, respectively; Dataset S2), we found foretinib to be a significantly more potent ROS1 inhibitor in cell-based assays, accentuating that the in vitro binding affinity does not directly translate to inhibitory efficacy in the cellular context. Thus, functional assays are indispensable for preclinical drug discovery of lead compounds. Whereas ALK inhibitors are at the forefront of ROS1-targeted therapy investigation, we note that the ALK inhibitor GSK1838705A does not effectively inhibit ROS1. These data highlight that not all ALK and ROS1 inhibitors exhibit unequivocal inhibitory reciprocity.
Emerging clinical data establish that the multikinase inhibitor crizotinib is an effective therapeutic agent in patients with lung adenocarcinoma harboring oncogenic ROS1 fusions (1, 7, 9, 31). Even with these promising results from clinical trials evaluating crizotinib, previous experience with the clinical use of kinase inhibitors to treat cancer suggests that resistance is a common obstacle, frequently occurring through acquisition of mutations in the targeted kinase that interfere with drug binding and lead to reactivation of oncogenic signaling. Indeed, Awad et al. recently reported on clinical resistance to crizotinib in a patient with CD74–ROS-harboring lung adenocarcinoma that resulted in disease progression and death (31). Sequencing of the pretreated and posttreatment tumor sample from this patient revealed the acquisition of a G2032R mutation in the ROS1 kinase domain that conferred resistance to crizotinib. Here we demonstrate that foretinib effectively inhibits the CD74–ROSG2032R mutant kinase (IC50: 50 nM) at clinically feasible concentrations. In an effort to further anticipate additional ROS1 kinase domain mutations that may confer resistance to crizotinib and test the inhibitory efficacy of foretinib in this context, we used a well-established in vitro cell-based accelerated mutagenesis screen. Crizotinib-resistant clones were recovered at concentrations up to 1,500 nM, which is above clinically achievable levels of this drug in patients, although each of these mutations alone was potently inhibited by foretinib (IC50s ≤ 20 nM) at physiologically relevant concentrations. Specifically, Eder et al. (30) report that the mean steady state Cmax of foretinib in patient plasma after administering the maximum tolerated dose of 3.6 mg/kg (median: 240 mg for 5 consecutive days every 2 wk) is 340 nM. In a second study with modified dose structure (80 mg once daily for 2 wk), the steady-state peak concentration was 46 ng/mL (72 nM) (20). These previous pharmacokinetic studies of foretinib strongly suggest that this inhibitor is poised for further clinical evaluation for the treatment of ROS1-driven malignancies.
In conclusion, our results establish that foretinib is a significantly more potent ROS1 inhibitor compared with crizotinib. Recently, the FDA granted accelerated approval to crizotinib for ALK-positive, locally advanced or metastatic NSCLC. Our preclinical data suggest that foretinib may be similarly promising either as first-line therapy for cancer patients harboring ROS1 rearrangements or as a second-line approach for patients failing crizotinib therapy due to the acquisition of resistant mutations.
Methods
Additional methods are in SI Methods.
Cell Lines/Culture.
Parental Ba/F3 cells (American Type Culture Collection, ATCC) were cultured in RPMI medium 1640 with 10% (vol/vol) FBS, l-glutamine, penicillin/streptomycin, and 15% (vol/vol) conditioned media produced from the WEHI-3 myelomonocytic IL-3 secreting cells. Ba/F3 cell lines were maintained at densities of 0.5 × 106 to 1.5 × 106/mL and infected with retrovirus-encoding human FIG–ROS–S (indicated FIG–ROS) or SLC–ROS–S (indicated SLC–ROS) fusion genes. Stable cell lines were generated after sorting for GFP expression on a FACS Aria flow cytometer. Cells were counted daily using a Guava Personal Cell Analysis flow cytometer. NIH 3T3 cells (obtained from ATCC) were cultured in DMEM-high glucose medium supplemented with 10% FCS, l-glutamine, and penicillin/streptomycin. Early passage NIH 3T3 cells were infected with retrovirus and sorted for GFP expression as described for Ba/F3 cells. HCC78 cells [obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DMSZ) cell bank], were cultured in RPMI medium 1640 with 10% FBS, l-glutamine, and penicillin/streptomycin; this media formulation is referred to as RPMI-10% FBS hereon. The commercially obtained cell lines were passaged no more than six times and used for fewer than 6 mo after receipt.
Cell Viability Assays.
Cell lines were distributed in 96-well plates and incubated with escalating concentrations of inhibitors for 72 h. HCC78 and murine cholangiocarcinoma (mCC) cells were seeded at 12–18 h before adding inhibitors. The following cell densities were used: Ba/F3 cells – 4 × 103 cells per well; HCC78 and mCC cells – 1.5 × 103 cells per well. Cell viability was measured using a methanethiosulfonate (MTS)-based assay (CellTiter96 Aqueous One Solution; Promega) and absorbance (490 nm) was read at 30 min and 1 h after adding reagent using a BioTek Synergy 2 plate reader. To facilitate comparison between experiments, the raw MTS absorbance of vehicle (0.1% DMSO)-treated cells was set to 1 and absorbance from inhibitor-treated wells was normalized to the vehicle-treated value. Each experiment had a minimum of three replicate wells per condition and the average and SEM was plotted for curve fit analysis. Data were normalized using Microsoft Excel and further nonlinear regression curve fit analysis of the normalized data for determination of IC50 values was performed using GraphPad Prism software.
Accelerated Cell-Based Mutagenesis Screen.
Ba/F3 cells expressing FIG–ROS were treated overnight ENU (50 μg/mL), pelleted, resuspended in fresh media, and distributed into 96-well plates at a density of 1 × 105 cells per well in 200 μL complete media supplemented with crizotinib, ranging from 500 nM to 2,500 nM. The wells were observed for cell growth under an inverted microscope and media color change every 2 d for 1 mo. The contents of wells exhibiting cell outgrowth were transferred to a 24-well plate containing 2 mL RPMI-10% FBS media supplemented with crizotinib at the same concentration as in the initial 96-well plate. At confluency, cells in 24-well plates were collected by centrifugation. To focus on identification of mutations conferring high-level resistance to crizotinib, DNA was extracted from the cell pellets of expanded clones from 750 nM to 1,500 nM crizotinib-treated plates using a DNEasy Tissue kit (Qiagen); no outgrowth was observed in the number of wells surveyed at concentrations of 2,000 nM and above. The FIG–ROS kinase domain was amplified using primers FIG–ROS M13-Kin3F (5′ GTA AAA CGA CGG CCA GTG CGA GAC TAG CTG CCA AGT AC) and ROS M13-Kin1 Rev (5′ CAG GAA ACA GCT ATG ACC GCC ATC TTC ACC TTC AAA GC), and the PCR products were bidirectionally sequenced using M13F (5′ GTA AAA CGA CGG CCA GTG) and M13 Rev (5′ CAG GAA ACA GCT ATG ACC) primers. The chromatographs were analyzed for mutations using Mutation Surveyor software (SoftGenetics).
Statistical Analysis.
Where indicated, the Student t test (Microsoft Excel or GraphPad Prism) was used to determine statistical significance and comparisons; P values less than 0.05 were deemed significant. Asterisks in figures and the description of asterisks in figure legends indicate level of statistical significance.
Supplementary Material
Acknowledgments
The authors thank Dr. Rani George (Harvard Medical School) and Dr. Thomas Weisner (Memorial Sloan Kettering Cancer Center) for generously sharing the pMSCV-ALK-F1174L and pMIG-EML4-ALK retroviral constructs, respectively, and Dr. Lukas Dow (Memorial Sloan-Kettering Cancer Center) for critical reading of the manuscript. A.S. is a recipient of German Research Foundation Postdoctoral Fellowship SA2278/1-1. J.E. is a recipient of predoctoral training Grant 12PRE11470005 by the American Heart Association. U.P.S. is a recipient of National Science Foundation career Award MCB0746589.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1319583110/-/DCSupplemental.
References
- 1.Bergethon K, et al. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol. 2012;30(8):863–870. doi: 10.1200/JCO.2011.35.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Charest A, et al. Fusion of FIG to the receptor tyrosine kinase ROS in a glioblastoma with an interstitial del(6)(q21q21) Genes Chromosomes Cancer. 2003;37(1):58–71. doi: 10.1002/gcc.10207. [DOI] [PubMed] [Google Scholar]
- 3.Gu TL, et al. Survey of tyrosine kinase signaling reveals ROS kinase fusions in human cholangiocarcinoma. PLoS ONE. 2011;6(1):e15640. doi: 10.1371/journal.pone.0015640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rimkunas VM, et al. Analysis of receptor tyrosine kinase ROS1-positive tumors in non-small cell lung cancer: Identification of a FIG-ROS1 fusion. Clin Cancer Res. 2012;18(16):4449–4457. doi: 10.1158/1078-0432.CCR-11-3351. [DOI] [PubMed] [Google Scholar]
- 5.Takeuchi K, et al. RET, ROS1 and ALK fusions in lung cancer. Nat Med. 2012;18(3):378–381. doi: 10.1038/nm.2658. [DOI] [PubMed] [Google Scholar]
- 6.Birch AH, et al. Chromosome 3 anomalies investigated by genome wide SNP analysis of benign, low malignant potential and low grade ovarian serous tumours. PLoS ONE. 2011;6(12):e28250. doi: 10.1371/journal.pone.0028250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davies KD, et al. Identifying and targeting ROS1 gene fusions in non-small cell lung cancer. Clin Cancer Res. 2012;18(17):4570–4579. doi: 10.1158/1078-0432.CCR-12-0550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee J, et al. Identification of ROS1 rearrangement in gastric adenocarcinoma. Cancer. 2013;119(9):1627–1635. doi: 10.1002/cncr.27967. [DOI] [PubMed] [Google Scholar]
- 9.Shaw AT, et al. 2012 ASCO Annual Meeting, Chicago. Alexandria, VA: American Society of Clinical Oncology; 2012. Clinical activity of crizotinib in advanced non-small cell lung cancer (NSCLC) harboring ROS1 gene rearrangement. [Google Scholar]
- 10.Garraway LA, Jänne PA. Circumventing cancer drug resistance in the era of personalized medicine. Cancer Discov. 2012;2(3):214–226. doi: 10.1158/2159-8290.CD-12-0012. [DOI] [PubMed] [Google Scholar]
- 11.Lovly CM, Pao W. Escaping ALK inhibition: Mechanisms of and strategies to overcome resistance. Sci Transl Med. 2012;4(120):ps2. doi: 10.1126/scitranslmed.3003728. [DOI] [PubMed] [Google Scholar]
- 12.O’Hare T, et al. AP24534, a pan-BCR-ABL inhibitor for chronic myeloid leukemia, potently inhibits the T315I mutant and overcomes mutation-based resistance. Cancer Cell. 2009;16(5):401–412. doi: 10.1016/j.ccr.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oxnard GR, et al. Acquired resistance to EGFR tyrosine kinase inhibitors in EGFR-mutant lung cancer: Distinct natural history of patients with tumors harboring the T790M mutation. Clin Cancer Res. 2011;17(6):1616–1622. doi: 10.1158/1078-0432.CCR-10-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ou SH, Tan J, Yen Y, Soo RA. ROS1 as a ‘druggable’ receptor tyrosine kinase: Lessons learned from inhibiting the ALK pathway. Expert Rev Anticancer Ther. 2012;12(4):447–456. doi: 10.1586/era.12.17. [DOI] [PubMed] [Google Scholar]
- 15.Sabbatini P, et al. GSK1838705A inhibits the insulin-like growth factor-1 receptor and anaplastic lymphoma kinase and shows antitumor activity in experimental models of human cancers. Mol Cancer Ther. 2009;8(10):2811–2820. doi: 10.1158/1535-7163.MCT-09-0423. [DOI] [PubMed] [Google Scholar]
- 16.Heukamp LC, et al. Targeted expression of mutated ALK induces neuroblastoma in transgenic mice. Sci Transl Med. 2012;4(141):41ra91. doi: 10.1126/scitranslmed.3003967. [DOI] [PubMed] [Google Scholar]
- 17.Qian F, et al. Inhibition of tumor cell growth, invasion, and metastasis by EXEL-2880 (XL880, GSK1363089), a novel inhibitor of HGF and VEGF receptor tyrosine kinases. Cancer Res. 2009;69(20):8009–8016. doi: 10.1158/0008-5472.CAN-08-4889. [DOI] [PubMed] [Google Scholar]
- 18.O’Dwyer ME, Druker BJ. STI571: An inhibitor of the BCR-ABL tyrosine kinase for the treatment of chronic myelogenous leukaemia. Lancet Oncol. 2000;1:207–211. doi: 10.1016/s1470-2045(00)00149-2. [DOI] [PubMed] [Google Scholar]
- 19.Walters DK, et al. Activating alleles of JAK3 in acute megakaryoblastic leukemia. Cancer Cell. 2006;10(1):65–75. doi: 10.1016/j.ccr.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 20.Shapiro GI, et al. A Phase 1 dose-escalation study of the safety and pharmacokinetics of once-daily oral foretinib, a multi-kinase inhibitor, in patients with solid tumors. Invest New Drugs. 2013;31(3):742–750. doi: 10.1007/s10637-012-9881-z. [DOI] [PubMed] [Google Scholar]
- 21.Naing A, et al. A comparison of the pharmacokinetics of the anticancer MET inhibitor foretinib free base tablet formulation to bisphosphate salt capsule formulation in patients with solid tumors. Invest New Drugs. 2012;30(1):327–334. doi: 10.1007/s10637-010-9536-x. [DOI] [PubMed] [Google Scholar]
- 22.Saborowski A, et al. Mouse model of intrahepatic cholangiocarcinoma validates FIG–ROS as a potent fusion oncogene and therapeutic target. Proc Natl Acad Sci USA. 2013;110:19513–19518. doi: 10.1073/pnas.1311707110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Charest A, et al. ROS fusion tyrosine kinase activates a SH2 domain-containing phosphatase-2/phosphatidylinositol 3-kinase/mammalian target of rapamycin signaling axis to form glioblastoma in mice. Cancer Res. 2006;66(15):7473–7481. doi: 10.1158/0008-5472.CAN-06-1193. [DOI] [PubMed] [Google Scholar]
- 24.Pao W, Chmielecki J. Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nat Rev Cancer. 2010;10(11):760–774. doi: 10.1038/nrc2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O’Hare T, Deininger MW, Eide CA, Clackson T, Druker BJ. Targeting the BCR-ABL signaling pathway in therapy-resistant Philadelphia chromosome-positive leukemia. Clin Cancer Res. 2011;17(2):212–221. doi: 10.1158/1078-0432.CCR-09-3314. [DOI] [PubMed] [Google Scholar]
- 26.Katayama R, et al. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung Cancers. Sci Transl Med. 2012;4(120):20ra17. doi: 10.1126/scitranslmed.3003316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doebele RC, et al. Mechanisms of resistance to crizotinib in patients with ALK gene rearranged non-small cell lung cancer. Clin Cancer Res. 2012;18(5):1472–1482. doi: 10.1158/1078-0432.CCR-11-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bradeen HA, et al. Comparison of imatinib mesylate, dasatinib (BMS-354825), and nilotinib (AMN107) in an N-ethyl-N-nitrosourea (ENU)-based mutagenesis screen: high efficacy of drug combinations. Blood. 2006;108(7):2332–2338. doi: 10.1182/blood-2006-02-004580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang S, et al. Crizotinib-resistant mutants of EML4-ALK identified through an accelerated mutagenesis screen. Chem Biol Drug Des. 2011;78(6):999–1005. doi: 10.1111/j.1747-0285.2011.01239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eder JP, et al. A phase I study of foretinib, a multi-targeted inhibitor of c-Met and vascular endothelial growth factor receptor 2. Clin Cancer Res. 2010;16(13):3507–3516. doi: 10.1158/1078-0432.CCR-10-0574. [DOI] [PubMed] [Google Scholar]
- 31.Awad MM, et al. Acquired resistance to crizotinib from a mutation in CD74-ROS1. N Engl J Med. 2013;368(25):2395–2401. doi: 10.1056/NEJMoa1215530. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.