Significance
This paper discusses speciation by allopolyploidy of tetraploid wheat, which in turn is a milestone event in establishing hexaploid bread wheat. The results suggested that karyotype stabilization together with variation in copy number of coding genes and localized changes in genomic repeats may have contributed to the establishment of tetraploid wheat as successful species. These observations are of relevance to the plant-breeding community for developing unique wheat cultivars by wide-hybridization and chromosomal engineering.
Keywords: intergenomic rearrangement, genomic shock, polyploid speciation
Abstract
Polyploidy or whole-genome duplication is recurrent in plant evolution, yet only a small fraction of whole-genome duplications has led to successful speciation. A major challenge in the establishment of nascent polyploids is sustained karyotype instability, which compromises fitness. The three putative diploid progenitors of bread wheat, with AA, SS (S ∼ B), and DD genomes occurred sympatrically, and their cross-fertilization in different combinations may have resulted in fertile allotetraploids with various genomic constitutions. However, only SSAA or closely related genome combinations have led to the speciation of tetraploid wheats like Triticum turgidum and Triticum timopheevii. We analyzed early generations of four newly synthesized allotetraploid wheats with genome compositions SshSshAmAm, SlSlAA, SbSbDD, and AADD by combined fluorescence and genomic in situ hybridization-based karyotyping. Results of karyotype analyses showed that although SshSshAmAm and SlSlAA are characterized by immediate and persistent karyotype stability, massive aneuploidy and extensive chromosome restructuring are associated with SbSbDD and AADD in which parental subgenomes showed markedly different propensities for chromosome gain/loss and rearrangements. Although compensating aneuploidy and reciprocal translocation between homeologs prevailed, reproductive fitness was substantially compromised due to chromosome instability. Strikingly, localized genomic changes in repetitive DNA and copy-number variations in gene homologs occurred in both chromosome stable lines, SshSshAmAm and SlSlAA. Our data demonstrated that immediate and persistent karyotype stability is intrinsic to newly formed allotetraploid wheat with genome combinations analogous to natural tetraploid wheats. This property, coupled with rapid gene copy-number variations, may have laid the foundation of tetraploid wheat establishment.
Polyploidy or whole-genome duplication (WGD) is a driving force in plant and vertebrate evolution (1–5). However, recent molecular phylogenetic studies have argued against a general creative role of WGD in plant evolution (6–8). This incongruence in opinions is long standing; in fact Stebbins in his seminal work in 1970s stated, “polyploidy has contributed little to progressive evolution” (9). Clearly, the two schools of thought regarding the roles of WGD in plant evolution have existed for years, and can converge if we accept the idea that WGDs have occurred frequently in nature but only a small fraction thereof have contributed to successful speciation (7, 10). However, the genetic and genomic factors determining one of the two fundamental fates for a nascent polyploid remained elusive.
The Triticum–Aegilops complex includes both diploid and polyploid species with phylogenetically well-defined organismal relationships (11). This plant group is therefore suitable to address the issue raised above. This is especially so because all polyploid species of the Triticum–Aegilops complex represent examples of evolutionarily recent successful speciation events via allopolyploidization, i.e., hybridization and doubling of genomes from Triticum/Aegilops species (11). Speciation of allohexaploid common wheat, Triticum aestivum L., the founder crop for the initial establishment of Middle-Eastern and European agriculture, is characterized by two sequential polyploidization events, i.e., allotetraploidization and allohexaploidization. The former event involved hybridization of two diploid species, Triticum urartu (genome AA) and a yet-unknown or extinct goat-grass species of the genus Aegilops section Sitopsis (genome SS ∼ BB). This event led to the speciation of allotetraploid emmer wheat, Triticum turgidum ssp. dicoccoides (12). The latter event that involved hybridization of a primitive domesticated form of T. turgidum (genome BBAA) with a goat-grass species Aegilops tauschii (genome DD) led to the speciation of common wheat (11). In parallel, allohexaploidization by hybridization of another allotetraploid wheat, Triticum timopheevii (genome GGAA) (13) and Einkorn wheat, Triticum monococcum (genome AmAm) has resulted in the speciation of Triticum zhukovskyi (genome GGAAAmAm) (14) (Fig. S1A). Thus, the natural hexaploid bread wheat has three diploid progenitors (13) (Fig. S1A) that have diverged from a common ancestor only about 2.5–4.5 Mya (15). Therefore, it is perhaps not surprising that allotetraploidization by hybridization of any two of these three diploid species can still be accomplished artificially to produce fertile tetraploid plants (16, 17) (Fig. S1B). Furthermore, the diploid progenitor species are known to exist sympatrically across several geographical areas in the Eastern Mediterranean region and the Near East (13). These features of the diploid progenitor species of polyploid wheat raise an intriguing question: Why has only the genome combination of SSAA or closely related ones led to successful speciation of the two natural allotetraploid wheat species, T. turgidum and T. timopheevii? This question is germane to the more general issue as to why only a small fraction of WGDs have led to successful speciation in the evolutionary history of angiosperms.
A major challenge for the establishment of newly formed allopolyploids as new species is sustained karyotype instability, in particular, whole-chromosome aneuploidy, which, at the organismal level, is associated with compromised cellular metabolism and reduced fitness (18). In newly formed allohexaploid wheat, extensive whole-chromosome aneuploidy was reported (19, 20). However, the immediate chromosomal consequences associated with formation of allotetraploid wheat have remained uninvestigated.
Here, we conducted in-depth molecular cytogenetic analyses of karyotype stability using fluorescence and genomic in situ hybridization (FISH and GISH) techniques and performed locus-specific molecular genetic analysis of gene copy-number variations (CNVs) in a set of protein-coding genes using newly synthesized allotetraploid wheats. Different genomic combinations of diploid Triticum/Aegilops species that are parsimoniously representing the diploid progenitors of tetraploid and hexaploid wheats were used in this study. We document that dramatic differences in karyotype stability of both chromosome number and structure existed between the SshSshAmAm/SlSlAA versus the SbSbDD/AADD genome combinations. Remarkably, localized loss or gain of DNA repeats and CNVs of gene homologs occurred widely in both chromosome stable lines, SshSshAmAm and SlSlAA, which, in their genomic constitutions, mimic natural tetraploid wheats, T. turgidum and T. timopheevii. We propose that persistent karyotype stability coupled with localized genomic changes and rapid CNVs of gene homologs, have laid the foundation for successful tetraploid wheat establishment and speciation.
Results
Karyotyping by FISH and GISH.
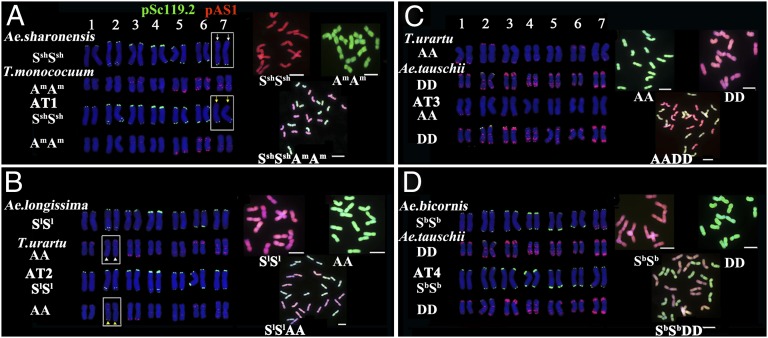
Four synthetic allotetraploid wheats (2n = 4x = 28) namely AT1 (genome SshSshAmAm), AT2 (genome SlSlAA), AT3 (genome AADD), and AT4 (genome SbSbDD) were used for karyotyping (Table S1; SI Materials and Methods). These synthetic wheat genotypes were derived by hybridization of the following diploid wheat progenitors: wheat with A or Am genome, Aegilops species of Sitopsis section with B-related genomes, and A. tauschii with D genome. Thus, these genotypes represent all known or putative progenitors of natural tetraploid and hexaploid wheats (Fig. S1A). For karyotype analyses, we used a two-step procedure based on sequential FISH with two DNA repeats (pSc119.2 and pAS1) followed by GISH. The procedure enabled characterization of parental subgenomes and allowed unequivocal discrimination of all chromosomes in each of the four synthetic wheats (Fig. 1 A–D; Fig. S2). Chromosome identifications were independently validated using three additional FISH probes, namely 5S ribosomal DNA (rDNA), 45S rDNA, and (GAA)n oligonucleotide, where in all cases the results were found confirmatory (Fig. S3). Using this robust molecular cytogenetic procedure, we obtained full karyotypes of a total of 489 individual plants from the four synthetic lines: 92 of AT1, 96 of AT2, 205 of AT3 and 96 of AT4 (Table S1). The plants of each line were sampled from several successive generations (Table S1), and therefore allowed assessment of trangenerational chromosome dynamics, i.e., stability versus instability.
Fig. 1.
Sequential FISH- and GISH-based karyotypes in the four synthetic allotetraploid wheats and their diploid parental species (SI Materials and Methods). The boxed chromosomes are those showing localized, persistent genomic changes in the synthetic tetraploid wheats relative to their diploid parents. A, B, C, and D respectively represent FISH and GISH images of AT1, AT2, AT3, and AT4 along with their corresponding diploid parental species. The probes used in FISH are pSc119.2 (green) and pAS1 (red). (Scale bars, 10 μm.)
Immediate and Transgenerational Karyotype Stability Showed by Synthetic Allotetraploids with SshSshAmAm and SlSlAA Genomes.
Of the 92 karyotyped AT1 plants (SshSshAmAm), only 2 (2.2%) plants were numerical aneuploids showing monosomy for a single chromosome 1Am (2n = 27), whereas the remaining plants were euploids (2n = 28), containing full chromosome complements of both parents (Fig. 2 A and C; Dataset S1). Of the 96 karyotyped AT2 plants (SlSlAA), 5 (5.3%) plants showed aneuploidy and only 1 (∼1%) of the 5 plants was a numerical aneuploid (3A monosomic, 2n = 27). The remaining 4 plants showed “compensated aneuploidy” (21), containing reciprocal loss and gain of one or more of the homeologous chromosome groups, and hence maintaining a balanced chromosome number of 2n = 28 (Fig. 2 B and D; Dataset S1). Of these 4 aneuploid plants, 3 are monosomics/trisomics of the group-2 chromosomes (2Sl/2A), and 1 is double monosomic/trisomic for group-2 and -3 chromosomes (Dataset S1). Thus, both SshSshAmAm and SlSlAA are intrinsically stable as euploids from the onset of allotetraploidization, and the stability is persistent across the four to eight successive generations (Table S1; Dataset S1).
Fig. 2.
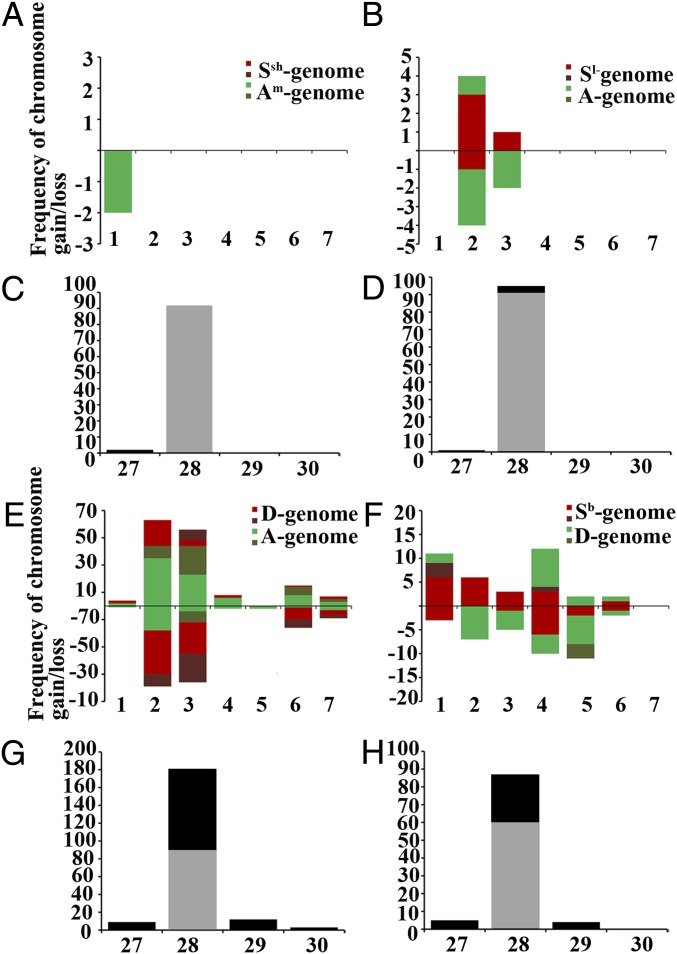
Numerical chromosome changes in a total of 489 individuals in the four synthetic allotetraploid wheats based on sequential FISH/GISH karyotyping. (A, B, E, and F) Propensities for gain/loss of individual chromosomes of each of the seven homeologous groups (bars 1–7 on the x axis) and the numbers of aneuploid chromosomes of each of the seven homeologous groups observed in the four combinations are shown on the y axis. Depiction via stacked bar chart in A, B, E, and F is according to ref. 21, whereby chromosome loss and gain of either one homolog (lighter colors) or both homologs (darker colors) are shown below and above the x axis, respectively. (C, D, G, and H ) The x axis represents the chromosome counts in euploids/compensated aneuploids (2n = 28) and other aneuploids with 2n = 27, 29, and 30, and the y axis represents total number of plants showing a particular ploidy. The black and gray portions of the 2n = 28 bars denote for compensated aneuploidy and euploidy, respectively. A and C, B and D, E and G, and F and H are AT1, AT2, AT3, and AT4, respectively.
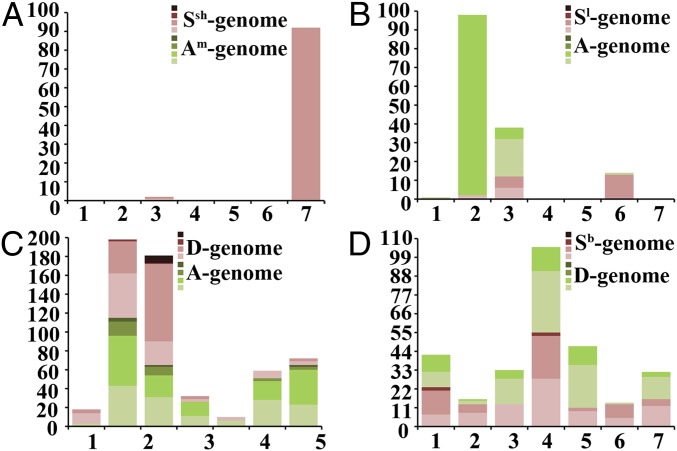
We next examined structural variations in the chromosomes in these two lines. We found that none of the 92 karyotyped AT1 plants showed large intergenomic rearrangements detectable by FISH and GISH analyses. However, in all analyzed plants, a loss of the pSc119.2 signal from the proximal end of the short arm of chromosome 7Ssh was observed (Figs. 1A and 3A; Fig. S2A; Dataset S1). However, the loss was undetectable by GISH analysis (Fig. 1B; Fig. S4A). As this signal was present in both homologs of chromosome 7Ssh of the diploid parental species Aegilops sharonensis (accession TH02) (Fig. 1A), its loss from both homologs of 7Ssh in all AT1 plants (Figs. 1A and 3A) suggested that it is a localized genomic alteration that probably took place instantaneously following allotetraploidization and was thereafter maintained. Similar localized loss of pSc119.2 signals was also detected from the long arm of chromosome 3Ssh (Fig. S4A), but it occurred in only 2 of the 92 plants (∼2.2%) (Fig. 3A; Dataset S1).
Fig. 3.
Structural chromosome changes in a total of 489 individuals in the four synthetic allotetraploid wheats based on sequential FISH/GISH karyotyping. Propensities for restructuring (intergenomic reciprocal rearrangements or unidirectional homeologous transfer) by each of the seven homeologous groups (bars 1–7 on the x axis; numbers of restructured chromosomes appear on the y axis) are shown in the stacked bar chart. The severity of restructuring for a given chromosome is indicated by color intensity at four scales, from lightest to darkest denoting the number of restructured copies for a given chromosome ranging from one copy (a single homolog restructured), two (both homologs restructured), three (all three homologs in trisomy restructured), and four (all four homologs in tetrasomy restructured). A, B, C, and D are AT1, AT2, AT3, and AT4, respectively.
Likewise, a localized genomic change was detected in all 96 analyzed plants of AT2 (Fig. 3B; Fig. S2B; Dataset S1). However, in this case it was a gain of the pSc119.2 signal at the proximal end of the chromosome arm 2AL, which was also detectable by GISH (Fig. 1B; Fig. S4B). This signal was nonexistent in the diploid parental species T. urartu accession TMU38 (Fig. 1B). Therefore, this gain of segment by 2A homologs (Figs. 1B and 3B) in all analyzed plants represented a localized but persistent genomic change. Apart from this, we also detected similar small-scale changes in 46 of the 96 plants (47.9%) (Fig. 3B; Dataset S1), which showed either gain or loss of pSc119.2 FISH signals. Additionally, most of these changes were also detectable by GISH (Figs. S4B and S5A). These localized genomic changes appeared to be either reciprocal exchanges between 2Sl and 2A or 3Sl and 3A, or unidirectional transfer of small chromosome segments from the corresponding homeologs to chromosomes 1A, 3A, 6A, 2Sl, 3Sl, and 6Sl (Figs. S2B, S4B and S5A). The majority (58.7%) of these changes involved only one homolog (Fig. 3B; Dataset S1). Similar to AT1, no large-scale intergenomic rearrangements were detected in AT2 (Fig. S4B).
By taking both numerical and structural chromosomal data together, it is clear that a prominent feature of SshSshAmAm and SlSlAA (which are analogous to the naturally formed tetraploid wheats T. turgidum and T. timopheevii) is that they have intrinsically stable karyotypes; both lines are virtually stabilized euploids harboring limited small-scale intergenomic rearrangements from the beginning of their genesis. Strikingly, both lines contained a unique localized genomic change either involving loss or gain of the pSc119.2 DNA repeat, which occurred in all analyzed plants of both lines.
AADD and SbSbDD Allotetraploid Wheats Are Associated with Massive and Persistent Whole-Chromosome Aneuploidy.
Of the 205 karyotyped AT3 plants with AADD, genome 24 (11.7%) showed numerical aneuploidy with 2n = 27, 29 or 30, while the remaining 181 plants (88.3%) were either euploids or compensated aneuploids (Fig. 2 E and G; Dataset S1). Of the 9 plants with 2n = 27, 3 were monosomics for chromosomes 4A, 7A, or 6D, and the remaining 6 plants showed more complex chromosome compositions which involved aneuploid chromosomes of three different homeologous groups (Fig. S6A; Dataset S1). Fifteen plants with 2n = 29 or 30 chromosomes contained even more complex chromosome compositions, with aneuploid chromosomes ranging from nullisomics to tetrasomics, involving up to three homeologous groups (Fig. 2E; Fig. S6A; Dataset S1). Notably, of the 181 plants with 2n = 28 chromosomes, only 90 (50%) were bona fide euploids, containing fully additive parental chromosome complements, while the remaining 91 were compensated aneuploids. Thus, the proportion of aneuploids in AT3, including the 24 cases with numerical changes, is 57% (115/205) (Fig. 2 E and G; Dataset S1). Of these 91 plants, 3 contained highly unbalanced chromosome compositions with monosomy–trisomy, occurring within one subgenome (Dataset S1). The other 88 plants are fully compensated aneuploids containing reciprocal gain and loss of homeologs involving one to three chromosome groups (Fig. S6A; Dataset S1). Chromosomes of both subgenomes in AT3 showed variable propensities for gain or loss (chi-squared test, i.e., prop. test in R, P < 2.2e−16) (Fig. 2E). Group-2 and -3 chromosomes showed the highest frequencies of aneuploidy, and all were reciprocal gains/losses of homeologs (Fig. 2E). Groups 6 and 7 showed the same kind of reciprocal numerical changes but at lower frequencies (Fig. 2E). The rest of chromosome groups, i.e., 1, 4, and 5, are highly stable (Fig. 2E). In summary, the A subgenome has a tendency to gain rather than lose chromosomes (prop. test, P < 2.2e−16), while the contrary is true for D subgenome (prop. test, P < 2.2e−16) (Fig. 2 E and G).
Of the 96 karyotyped AT4 plants (genome SbSbDD), 9 (9.4%) plants showed numerical aneuploidy with 2n = 27 or 29, while the remaining 87 plants (90.6%) were either euploids or compensated aneuploids (Fig. 2 F and H; Dataset S1). The five plants with 2n = 27 chromosomes were mainly monosomics for chromosomes 1Sb, 3Sb, 4Sb, and 2D, while 1 plant showed trisomy with double monosomy for three different chromosomes (Fig. S6B; Dataset S1). All 4 plants with 2n = 29 were trisomics for chromosome 4D (Fig. S6B; Dataset S1). Of the 87 plants with 2n = 28, 60 plants were bona fide euploids, while the remaining 27 were compensated aneuploids (Fig. 2 F and H; Fig. S6B). Thus, the frequency of aneuploids in AT4, including the nine cases of numerical aneuploidy, is 37.5% (36 of 96 plants). Similar to the situation in AT3, a substantial proportion of plants with compensated aneuploidy showed reciprocal gain or loss of chromosomes from homeologous groups and are hence unbalanced in their gene contents (Fig. S6B; Dataset S1). Both subgenomes in AT4 also showed different propensities for gain or loss of chromosomes (Fig. 2F) (prop. test, P = 0.01368 and 0.0001851, respectively). The chromosomes of groups 1, 2, 4, and 5 showed higher levels of aneuploidy in comparison with the three other groups (3, 6, and 7). In addition, chromosomes 7Sb and 7D showed no aneuploidy. Chromosomes of groups 2 and 4 mainly showed reciprocal gain or loss of homeologs, while those of groups 1 and 5 displayed predominantly nonreciprocal concurrent gain or loss of homeologs (Fig. 2F). In summary, the Sb subgenome is more prone to gain than loss of chromosomes (prop. test, P = 0.0001851), while the contrary is true for the D subgenome (prop. test, P = 0.01368) (Fig. 2F).
AADD and SbSbDD Allotetraploid Wheats Underwent Extensive Chromosome Restructuring.
Of the 205 AT3 plants karyotyped, 195 (95.1%) plants harbored at least one rearrangement (Fig. 3C; Dataset S1). Among the 195 plants with chromosome rearrangements, 54 (27.7%) contained reciprocally translocated segments (Fig. 3C; Figs. S2C and S5B; Dataset S1), which were large and readily detectable by GISH with breakpoints occurring mainly at termini (but also interstitial) of either long or short arms (Fig. S4C). Notably, in general chromosomes showing higher propensities for aneuploidy (Fig. 2E) are more prone to rearrangements, although groups 6 and 7 showed the opposite trend (Figs. 2E and 3C). All seven chromosome groups showed intergenomic rearrangements but at markedly variable frequencies (prop. test, P < 2.2e−16) (Fig. 3C). Chromosome groups 2 and 3 showed the highest frequencies of rearrangements followed by groups 6 and 7, while groups 1 and 5 showed the lowest (Fig. 3C). A common feature for groups 2 and 3 is that most of the rearrangements are reciprocal translocations between homeologs, while those of groups 1, 4, 5, 6 and 7 are mainly unidirectional homeologous segment transfer (Fig. 3C; Figs. S2C, S4C, and S5B).
Of the 96 karyotyped plants of AT4, 94 (97.9%) harbor at least one chromosome rearrangement (Fig. 3D; Dataset S1). Among the 94 plants showing chromosome rearrangements, 37 (38.5%) contained reciprocally translocated segments (Fig. 3D; Figs. S2D and S5C; Dataset S1), with features similar to those of AT3 (Figs. S2D and Fig. S4D). Unexpectedly, two reciprocal rearrangements in AT4 occurred between two unrelated chromosome pairs, i.e., between 1Sb and 5D and between 1D and 5Sb (Figs. S2D and S5C). All seven chromosome groups of AT4 showed intergenomic rearrangements at variable frequencies (prop. test, P < 2.2e−16) (Fig. 3D), including either or both reciprocal and unidirectional rearrangements (Figs. S2D, S4D, and S5C). Also similar to AT3, in general the chromosomes showing higher propensities for aneuploidy (Fig. 2F) were more prone to rearrangements (Fig. 3D). However, group 7 chromosomes were exceptions, which showed no aneuploidy (Fig. 2F) but exhibited high incidence of rearrangements (Fig. 3D). For chromosome groups 1, 3, 4, and 7, the two parental subgenomes showed more or less equal propensities for rearrangements, but chromosomes from the rest three groups were highly biased toward one subgenome, either Sb (groups 2 and 6) or D (group 5) (Fig. 3D).
Small-scale genomic changes of the pSc119.2 repeat were also detected in AT4, involving chromosome groups 3, 4, and 7, and chromosomes 5Sb and 6Sb (Figs. S2D and S4D), but none of these were present in all plants, and therefore were not persistent (Dataset S1). Interestingly, all signal losses occurred to the Sb-subgenome chromosomes, whereas all signal gains occurred in the D-subgenome chromosomes (Figs. S2D and S4D), which was consistent with the localized persistent changes found in AT1 and AT2 (Fig. 1 A and B).
To summarize, chromosomes of both AT3 and AT4 have a strong propensity to rearrange. As a result, >95% of plants from these two lines contain at least one rearranged chromosome. Moreover, multiple restructured chromosomes tend to occur in a single plant, resulting in extensively remodeled karyotypes. The chromosome rearrangements coupled with massive aneuploidy in these plants (Fig. 2; Fig. S5; Dataset S1) may render both of these lines into a state of genomic chaos.
Rapid CNV Occurred to Gene Homologs of the SshSshAmAm and SlSlAA Synthetic Allotetraploid Wheats.
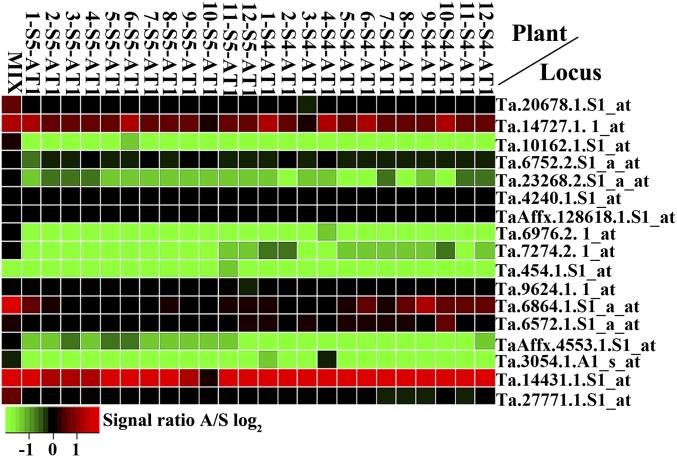
As described above, despite karyotype stability in both chromosome number and structure in AT1 (SshSshAmAm) and AT2 (SlSlAA), a localized genomic change of either loss or gain of the pSc119.2 DNA repeat occurred in both lines (Fig. 1 A and B; Fig. S2 A and B). This striking observation promoted us to test whether genomic changes have also occurred to protein-coding genes in these two lines. For this purpose, we quantified the relative CNV of homologs of each of the 17 and 31 genes, respectively, in AT1 and AT2, for which specific pyrosequencing primers capable of distinguishing parental subgenomes were designed (Dataset S2; SI Materials and Methods). We performed the analysis for 24 individual plants from two generations of AT1 and three generations of AT2, respectively (Table S2). We found that compared with the parental relative homolog ratio (RHR) for each gene in the 1:1 parental DNA mixes, significant alteration in RHR (t test, P < 0.05) due to loss of one or more of homologous copies (SI Materials and Methods) occurred at most of the 408 data points (17 genes × 24 plants) of AT1 and 744 data points (31 genes × 24 plants) of AT2 (Fig. 4; Fig. S7; Dataset S3). In comparison, analysis of fluctuations in RHRs (due to any number of reasons, including imprecise measurements of DNA quantity, pipetting errors, etc.) across a set of 10 independent 1:1 DNA mixes of parental species each of AT1 and AT2 (SI Materials and Methods) never reached a statistically significant level. Notably, in contrast to the complete loss of the pSc119.2 DNA repeat in AT1 (Fig. 1A), no case of total loss of a gene homolog(s) was found in either AT1 or AT2 (Fig. 4; Fig. S7). These results indicated that rapid CNVs in homologs of coding genes have occurred in both synthetic allotetraploid wheats although they are highly stable in chromosome number and structure.
Fig. 4.
CNVs recorded in 24 individual AT1 plants for a set of 17 genes (Dataset S2) using locus-specific pyrosequencing primers. The RHR of the 408 data points (17 genes × 24 plants) was calculated based on three biological replications, and its significant deviation from that of parental 1:1 DNA mixes was computed (for details, see SI Materials and Methods).
Seed Set Is Compromised by Extensive Karyotype Instability.
We determined the number of grains produced by two synthetic allotetraploid wheat lines, AT2 (SlSlAA), which was karyotype stable, and AT3 (AADD), which showed massive aneuploidy and rearrangements. We found that no significant difference (t test, P > 0.05) in seed set existed among selected euploid plants of AT3, aneuploid plants of AT3 (with chromosome numbers of 2n = 29 or 30), or euploid plants of AT2 (Fig. S8). However, both aneuploids with 2n = 27 or compensated aneuploids with 2n = 28 of AT3 showed significant reductions in seed production compared with the other plant groups (Fig. S8). Given the persistent karyotype instability in both chromosome number and structure in AT3 and AT4, reproductive fitness of both lines and derived populations is expected to suffer further deterioration with progression of generations.
Discussion
Polyploidy or WGD is a game-changing event in the life history of an organism, either to fuel evolution or to cause extinction, and hence bearing a profound impact on evolutionary trajectories of organisms involved (3). Enhanced evolvability of polyploids in comparison with their diploid progenitors is regarded as a major advantage of WGD (3, 22). Greater genomic plasticity coupled with the stronger buffering capacity due to WGD render polyploids capable of generating greater and faster heritable genetic variations than their diploid progenitors, and enable polyploids to adapt to novel ecological niches or adjust in situ to the changing environments (3, 4). However, genetic variations need to occur in a timely fashion for evolution to act before nascent polyploids are outcompeted by their diploid progenitors or relatives (3). Numerous recent studies in diverse newly formed plant allopolyploids have indeed documented that rapid and extensive genomic changes and gene-expression repatterning are hallmarks of nascent WGD (1, 2, 4, 11, 23).
Karyotype instability in chromosome number and/or structure is frequently associated with nascent WGD, and these changes are often interlaced with molecular level changes as demonstrated in yeast (24), Brassica (25, 26), and Tragopogon (21, 27). Nevertheless, as pointed out by Otto (3), nascent WGD-induced genetic variability can fuel evolution only if individuals can survive the onslaught of genomic changes. Given the large and generally negative effects on organismal fitness by aneuploidy and large-scale rearrangements, it is conceivable that if these events are too traumatic, extinction will be the outcome. Therefore, it is likely that the survived and eventually established nascent polyploids are those that are able to fine-tune the balance of mutability and karyotype stability.
We recently reported that whole-chromosome aneuploidy generally occurs in newly synthesized allohexaploid wheat but that intergenomic rearrangements are rare (20). An interesting finding is that reproductive fitness is not significantly affected by most types of aneuploidy, probably due to a strong compensating capacity of being hexaploid (20). Especially in cases where extensive and persistent whole-chromosome aneuploidy occurred, they often involve only one homolog (i.e., heterozygous condition), and hence are potentially revertible to euploidy. These features led to the proposition that newly formed allohexaploid wheat may be able to survive over a protracted period for a karyotype stabilization mechanism to evolve, leading to its establishment (20). Nevertheless, information regarding the immediate chromosomal consequences of nascent allotetraploidization in wheat has been absent.
Here, we have conducted in-depth molecular cytogenetic analyses of four synthetic allotetraploid wheat lines. Two lines (AT1 and AT2) with genomes SshSshAmAm and SlSlAA are analogous to natural tetraploid wheat (genome BBAA), while the other two lines (AT3 and AT4) with genomes AADD and SbSbDD have no extant (natural) counterparts. However, the diploid parental species involved in these combinations are known to distribute sympatrically (13). Therefore, we have addressed the question why only the genome combination of SSAA or closely related ones has led to successful speciation of the two natural allotetraploid wheat species, T. turgidum and T. timopheevii. Our results found stunning differences in karyotype stability of both chromosome number and structure between SshSshAmAm/SlSlAA and AADD/SbSbDD. Whereas the former is characterized by initiation and transgenerational karyotype stability, massive aneuploidy and extensive chromosome restructuring are persistently associated with the latter. Remarkably, both SshSshAmAm and SlSlAA showed more localized rapid genomic changes involving loss or gain of the pSc119.2 repeat. The fact that this type of genomic change occurred to both homologs of the relevant chromosome of all karyotyped plants originating from several independent S2 individuals in each line points to an earlier occurrence of the events and their trangenerational perseverance. It is tempting to speculate that such localized systemic genomic changes, being likely under selection, may have played a role in karyotype stabilization, which merits further investigation.
As proposed, rapid genomic changes are probably essential for nascent polyploids to survive and establish (3, 11). The observations that homolog(s) of coding genes showed rapid CNVs in both SshSshAmAm and SlSlAA has provided strong evidence to support this hypothesis and demonstrate that the nascent allopolyploids possess a strong capacity to generate immediate molecular genetic variations. Given the vast number and kind of genes sensitive to dosage stoichiometry in plants (28), it can be imagined that at least some of the CNVs may cause substantial plasticity in gene expression and their products, and by extension, phenotypic novelties for Darwinian selection. The rapid occurrence of CNVs is consistent with earlier findings of rapid gene loss and elimination of noncoding DNA in wheat (17, 29) and Tragopogon (27).
The genetic basis underlying the dramatic differences in karyotype stability between the synthetic allotetraploid lines is not clear. The fact that all aneuploid individuals are organismal rather than cellular (no mosaic somatic aneuploidy identified) indicates failures of proper chromosome segregation in meiosis as a major cause for the genesis of aneuploidy in AADD and SbSbDD. Meiosis is fine-tuned cellular machinery (30), which in newly formed allotetraploids is inherited from divergent diploid ancestors where it is optimized over evolutionary time. Conceivably, incompatibility occurring to any critical component of meiosis may cause chromosome missegregation (31). Moreover, WGD alone could result in asynchronization of meiosis (32), resulting in structural chromosomal changes due to homeologous chromosome pairing and illegitimate (or nonhomologous) recombination. In this respect, the findings of Kumar et al. (33) are illuminating. The authors showed that certain accessions of diploid Aegilops with the S genome carry suppressors of homeologous pairing with immediate effects on the chromosome pairing in the newly formed allopolyploids (33). In light of these findings, it is perhaps not surprising to see extremely low incidence of chromosome rearrangements in genotypes with SshSshAmAm and SlSlAA genomes if the parental accessions of A. sharonensis and Aegilops longissima carry suppressors of homeologous chromosome pairing. In contrast, the rampant rearrangements detected in AADD and SbSbDD could be explained if the parental accessions either lacked these suppressors or carried enhancers of homeologous chromosome pairing. Regardless, the severity of the rapid karyotype repatterning in genotypes with AADD and SbSbDD genomes is striking, with >95% of plants carrying multiple chromosome rearrangements. A similar phenomenon, but to a lesser extent, of chromosome restructuring was observed in newly formed allotetraploids of Brassica (26) and Tragopogon (21). Conceivably, the massive aneuploidy and extensive restructuring may constitute “snowballing” effects leading to a vicious cycle of chromosomal instability culminating in extinction (24). As a result, negative effect(s) on fitness can be immediate (as witnessed by reduced seed set) and/or rapidly increase with each generation. Indeed, a synthetic allotetraploid wheat (genome AADD) produced earlier by Sears was found to have reasonably high fertility (76%) in earlier generations (16) but reduced fertility, only 1–3%, in later generations (34).
The most unexpected finding of this investigation is the high degree of karyotype stability in both chromosome number and structure in the two synthetic lines with genomes SshSshAmAm and SlSlAA, which in their genome constitutions mimic that of natural tetraploid wheats, T. turgidum and T. timopheevii. This finding may have implications for the evolution of meiotic chromosome stability in polyploid wheat. For example, it is generally believed that in both natural tetra- and hexaploid wheats, the Pairing homeologous 1 (Ph1) gene (35) is essential for maintaining meiotic chromosome stability by controlling exclusive homologous pairing and bivalent formation. Our results suggest that immediate Ph1 functionality is likely dispensable for this purpose in the newly formed allotetraploid wheats with genomes SshSshAmAm or SlSlAA; instead the scenario of alternative homeologous pairing suppressors that are introduced from diploid parental species and exert immediate effects on chromosome pairing in the resultant allopolyploids (33) seems more plausible. Irrespective of mechanisms, we propose that the intrinsic karyotype stability together with rapid CNVs in homologs of coding genes may lay the foundation of natural tetraploid wheat establishment and speciation.
Materials and Methods
Plant Materials.
Four sets of synthetic allotetraploid wheat lines produced in Moshe Feldman's laboratory (Weizmann Institute of Science, Rehovot, Israel) were used in this study (SI Materials and Methods).
Karyotyping by Sequential GISH and FISH.
The protocols were followed essentially as described in ref. 36, with minor modifications (20). For further details and probe information, see SI Materials and Methods.
Pyrosequencing.
The protocol was essentially as reported in ref. 20. For further detailed, see SI Materials and Methods.
Seed Setting.
Greenhouse-grown plants of two allotetraploid wheat lines, AT2 (SbSbAA) and AT3 (AADD), were used to score seed setting (for details, see SI Materials and Methods).
Statistical Analyses.
Statistical analyses were performed using the data analysis function of Microsoft Excel and R (version 2.15.0). For details of data collection and statistical analyses, see SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Moshe Feldman for providing the initial seeds of the synthesized allotetraploids and the parental genotypes. This work was supported by grants from National Natural Science Foundation of China (30120243), 863 (2010AA1000686001), the Program for Introducing Talents to Universities (B07017), and the Joint Center for Plant Genetic Research between Northeast Normal University (China) and Washington State University.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1319598110/-/DCSupplemental.
References
- 1.Adams KL, Wendel JF. Polyploidy and genome evolution in plants. Curr Opin Plant Biol. 2005;8(2):135–141. doi: 10.1016/j.pbi.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 2.Chen ZJ. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu Rev Plant Biol. 2007;58:377–406. doi: 10.1146/annurev.arplant.58.032806.103835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Otto SP. The evolutionary consequences of polyploidy. Cell. 2007;131(3):452–462. doi: 10.1016/j.cell.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 4.Soltis PS, Soltis DE. The role of hybridization in plant speciation. Annu Rev Plant Biol. 2009;60:561–588. doi: 10.1146/annurev.arplant.043008.092039. [DOI] [PubMed] [Google Scholar]
- 5.Jiao Y, et al. Ancestral polyploidy in seed plants and angiosperms. Nature. 2011;473(7345):97–100. doi: 10.1038/nature09916. [DOI] [PubMed] [Google Scholar]
- 6.Wood TE, et al. The frequency of polyploid speciation in vascular plants. Proc Natl Acad Sci USA. 2009;106(33):13875–13879. doi: 10.1073/pnas.0811575106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mayrose I, et al. Recently formed polyploid plants diversify at lower rates. Science. 2011;333(6047):1257. doi: 10.1126/science.1207205. [DOI] [PubMed] [Google Scholar]
- 8.Arrigo N, Barker MS. Rarely successful polyploids and their legacy in plant genomes. Curr Opin Plant Biol. 2012;15(2):140–146. doi: 10.1016/j.pbi.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 9.Stebbins GL. Chromosomal Evolution in Higher Plants. London: Edward Arnold; 1971. [Google Scholar]
- 10.Madlung A. Polyploidy and its effect on evolutionary success: Old questions revisited with new tools. Heredity (Edinb) 2013;110(2):99–104. doi: 10.1038/hdy.2012.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feldman M, Levy AA. Genome evolution due to allopolyploidization in wheat. Genetics. 2012;192(3):763–774. doi: 10.1534/genetics.112.146316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dvorak J. The relationship between the genome of Triticum urartu and the A and B genomes of Triticum aestivum. Can J Genet Cytol. 1976;18(2):371–377. [Google Scholar]
- 13.Kilian B, et al. Independent wheat B and G genome origins in outcrossing Aegilops progenitor haplotypes. Mol Biol Evol. 2007;24(1):217–227. doi: 10.1093/molbev/msl151. [DOI] [PubMed] [Google Scholar]
- 14.Dvorák J, Terlizzi P, Zhang HB, Resta P. The evolution of polyploid wheats: Identification of the A genome donor species. Genome. 1993;36(1):21–31. doi: 10.1139/g93-004. [DOI] [PubMed] [Google Scholar]
- 15.Huang S, et al. Genes encoding plastid acetyl-CoA carboxylase and 3-phosphoglycerate kinase of the Triticum/Aegilops complex and the evolutionary history of polyploid wheat. Proc Natl Acad Sci USA. 2002;99(12):8133–8138. doi: 10.1073/pnas.072223799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sears ER. Chromosome pairing and fertility in hybrids and amphidiploids in the Triticinae. Res Bull Missouri Ag Exp Sta. 1941;337:1–20. [Google Scholar]
- 17.Ozkan H, Levy AA, Feldman M. Allopolyploidy-induced rapid genome evolution in the wheat (Aegilops-Triticum) group. Plant Cell. 2001;13(8):1735–1747. doi: 10.1105/TPC.010082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Birchler JA. Insights from paleogenomic and population studies into the consequences of dosage sensitive gene expression in plants. Curr Opin Plant Biol. 2012;15(5):544–548. doi: 10.1016/j.pbi.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 19.Mestiri I, et al. Newly synthesized wheat allohexaploids display progenitor-dependent meiotic stability and aneuploidy but structural genomic additivity. New Phytol. 2010;186(1):86–101. doi: 10.1111/j.1469-8137.2010.03186.x. [DOI] [PubMed] [Google Scholar]
- 20.Zhang H, et al. Persistent whole-chromosome aneuploidy is generally associated with nascent allohexaploid wheat. Proc Natl Acad Sci USA. 2013;110(9):3447–3452. doi: 10.1073/pnas.1300153110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chester M, et al. Extensive chromosomal variation in a recently formed natural allopolyploid species, Tragopogon miscellus (Asteraceae) Proc Natl Acad Sci USA. 2012;109(4):1176–1181. doi: 10.1073/pnas.1112041109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dubcovsky J, Dvorak J. Genome plasticity a key factor in the success of polyploid wheat under domestication. Science. 2007;316(5833):1862–1866. doi: 10.1126/science.1143986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doyle JJ, et al. Evolutionary genetics of genome merger and doubling in plants. Annu Rev Genet. 2008;42:443–461. doi: 10.1146/annurev.genet.42.110807.091524. [DOI] [PubMed] [Google Scholar]
- 24.Sheltzer JM, et al. Aneuploidy drives genomic instability in yeast. Science. 2011;333(6045):1026–1030. doi: 10.1126/science.1206412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gaeta RT, Pires JC, Iniguez-Luy F, Leon E, Osborn TC. Genomic changes in resynthesized Brassica napus and their effect on gene expression and phenotype. Plant Cell. 2007;19(11):3403–3417. doi: 10.1105/tpc.107.054346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiong Z, Gaeta RT, Pires JC. Homoeologous shuffling and chromosome compensation maintain genome balance in resynthesized allopolyploid Brassica napus. Proc Natl Acad Sci USA. 2011;108(19):7908–7913. doi: 10.1073/pnas.1014138108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buggs RJA, et al. Rapid, repeated, and clustered loss of duplicate genes in allopolyploid plant populations of independent origin. Curr Biol. 2012;22(3):248–252. doi: 10.1016/j.cub.2011.12.027. [DOI] [PubMed] [Google Scholar]
- 28.Birchler JA, Veitia RA. Gene balance hypothesis: Connecting issues of dosage sensitivity across biological disciplines. Proc Natl Acad Sci USA. 2012;109(37):14746–14753. doi: 10.1073/pnas.1207726109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kashkush K, Feldman M, Levy AA. Gene loss, silencing and activation in a newly synthesized wheat allotetraploid. Genetics. 2002;160(4):1651–1659. doi: 10.1093/genetics/160.4.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.von Wettstein D, Rasmussen SW, Holm PB. The synaptonemal complex in genetic segregation. Annu Rev Genet. 1984;18:331–413. doi: 10.1146/annurev.ge.18.120184.001555. [DOI] [PubMed] [Google Scholar]
- 31.Cifuentes M, Grandont L, Moore G, Chèvre AM, Jenczewski E. Genetic regulation of meiosis in polyploid species: New insights into an old question. New Phytol. 2010;186(1):29–36. doi: 10.1111/j.1469-8137.2009.03084.x. [DOI] [PubMed] [Google Scholar]
- 32.Storchová Z, et al. Genome-wide genetic analysis of polyploidy in yeast. Nature. 2006;443(7111):541–547. doi: 10.1038/nature05178. [DOI] [PubMed] [Google Scholar]
- 33.Kumar S, Friebe B, Gill BS. Fate of Aegilops speltoides-derived, repetitive DNA sequences in diploid Aegilops species, wheat-Aegilops amphiploids and derived chromosome addition lines. Cytogenet Genome Res. 2010;129(1-3):47–54. doi: 10.1159/000314552. [DOI] [PubMed] [Google Scholar]
- 34.Siddiqui KA. Extraction of AADD component of Triticum aestivum (AABBDD) Hereditas. 1971;68(1):151–158. [Google Scholar]
- 35.Sears ER. Genetic control of chromosome pairing in wheat. Annu Rev Genet. 1976;10:31–51. doi: 10.1146/annurev.ge.10.120176.000335. [DOI] [PubMed] [Google Scholar]
- 36.Kato A, Lamb JC, Birchler JA. Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc Natl Acad Sci USA. 2004;101(37):13554–13559. doi: 10.1073/pnas.0403659101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.