Significance
Members of the AT-HOOK MOTIF CONTAINING NUCLEAR LOCALIZED (AHL) family are involved in various plant biological processes. Our findings reveal a molecular model whereby the AHLs interact with each other via the plant and prokaryote conserved (PPC)/domain of unknown function #296 (DUF296) domain to form homo-/hetero-complexes, possibly trimers. The AHL complex also interacts with other nuclear proteins to form a macromolecular complex that modulates plant growth and development. The coordinated action of AHLs requires an AT-hook motif capable of binding AT-rich DNA, as well as a PPC/DUF296 domain containing a conserved six-amino-acid region. Our proposed model provides a better understanding of the roles of AHL genes in regulating plant growth and development, which may in turn lead to better seedling establishment and increased yield.
Keywords: enhanceosome, seedling establishment
Abstract
The Arabidopsis thaliana genome encodes 29 AT-HOOK MOTIF CONTAINING NUCLEAR LOCALIZED (AHL) genes, which evolved into two phylogenic clades. The AHL proteins contain one or two AT-hook motif(s) and one plant and prokaryote conserved (PPC)/domain of unknown function #296 (DUF296) domain. Seedlings lacking both SOB3/AHL29 and ESC/AHL27 confer a subtle long-hypocotyl phenotype compared with the WT or either single-null mutant. In contrast, the missense allele sob3-6 confers a dramatic long-hypocotyl phenotype in the light. In this study, we examined the dominant-negative feature of sob3-6 and found that it encodes a protein with a disrupted AT-hook motif that abolishes binding to AT-rich DNA. A loss-of-function approach demonstrated different, yet redundant, contributions of additional AHL genes in suppressing hypocotyl elongation in the light. We showed that AHL proteins interact with each other and themselves via the PPC/DUF296 domain. AHLs also share interactions with other nuclear proteins, such as transcription factors, suggesting that these interactions also contribute to the functional redundancy within this gene family. The coordinated action of AHLs requires an AT-hook motif capable of binding AT-rich DNA, as well as a PPC/DUF296 domain containing a conserved Gly-Arg-Phe-Glu-Ile-Leu region. Alteration of this region abolished SOB3/AHL29’s physical interaction with transcription factors and resulted in a dominant-negative allele in planta that was phenotypically similar to sob3-6. We propose a molecular model where AHLs interact with each other and themselves, as well as other nuclear proteins, to form complexes which modulate plant growth and development.
Since the colonization of land plants, plant genomes have rapidly expanded and diversified from their common ancestor through the process of evolution. During the long period of natural selection that shaped the morphology and adaptivity of plants to their environment, some genes that regulate important growth and developmental processes of ancient plant species have been amplified and gradually expanded into multimember gene families (1–4). One such family, the AT-HOOK MOTIF CONTAINING NUCLEAR LOCALIZED (AHL) genes, exists in all plant species that have been sequenced thus far, from the moss Physcomitrella patens (5) to both modern dicot and monocot plants, such as Arabidopsis thaliana (6), Oryza sativa (7, 8), Sorghum bicolor (9), and Populus trichocarpa (10). This high conservation through evolution suggests that the AHL gene family is important for plant growth and development.
The AHL proteins contain two conserved structural units, the AT-hook motif and the plant and prokaryote conserved (PPC) domain, the latter being also annotated as the domain of unknown function #296 (DUF296). The AT-hook motif enables binding to AT-rich DNA and has been identified in various gene families both in prokaryotes and eukaryotes including the high mobility group A (HMGA) proteins in mammals (11). The AT-hook motif contains a conserved palindromic core sequence, Arg-Gly-Arg, which binds the minor groove of AT-rich B-form DNA (12). On binding with DNA, the Arg-Gly-Arg core sequence adopts a concave conformation with close proximity to the backbone of the DNA, and the side chains of both arginine residues firmly insert into the minor groove (12).
The PPC/DUF296 domain is ∼120 amino acids in length and exists as a single protein in Bacteria and Archaea (13). Crystal structures of several bacterial and archaeal PPC/DUF296 proteins have been determined (14, 15). All of the above prokaryotic PPC/DUF296 proteins share the same tertiary structure with five β-strands forming an antiparallel β-sheet, which partially surrounds a single α-helix. The solved crystal structures suggest that the prokaryotic PPC/DUF296 proteins form a trimer (14). In land plants, the PPC/DUF296 domain is found in AHL proteins, where it is located at the carboxyl end relative to the AT-hook motif(s) (13). In Arabidopsis AHL1, a hydrophobic region at the C-terminal end of its PPC/DUF296 domain is essential for its nuclear localization (13). However, there are currently no known biological functions for this domain in regulating plant growth and development.
Based on gene overexpression and biochemical studies, members of the AHL family have been proposed to regulate diverse aspects of growth and development in plants. Overexpression of either of two Arabidopsis AHL genes, SUPPRESSOR OF PHYTOCHROME B-4 #3 (SOB3/AHL29) or ESCAROLA (ESC/AHL27) represses hypocotyl elongation for seedlings grown in the light, but not in darkness (16). As adults, these overexpression plants develop larger organs, including expanded leaves and enlarged flowers and fruits, together with delayed flowering and senescence (16). The AHL genes modulate several aspects of plant growth and development, including hypocotyl elongation in the light (AHL22, ESC/AHL27, and SOB3/AHL29) (16, 17), flower development (16–20), and root growth (21). Overexpression of other AHL gene members, such as HERCULES (HRC/AHL25), also enhances adult leaf and stem growth (22). Several AHL genes are involved in the homeostasis of phytohormones or the mediation of their responses. For example, overexpression of either AT-HOOK PROTEIN OF GA FEEDBACK REGULATION 1 (AGF1/AHL25) or AGF2/AHL15 suggests that these genes play a role in maintaining the negative feedback of GA 3-oxidase in gibberellin signaling (23). Based on yeast-one-hybrid and transactivation analysis, Catharanthus roseus AHLs have been shown to regulate the jasmonic acid response of AP2 transcription factors (24). Transcript accumulation of AHL21 was dramatically suppressed in cytokinin-treated Arabidopsis roots (25). Overexpression studies also implicate AHL genes (ESC/AHL27 and AHL20) in the regulation of plant innate immune responses (19, 26). Gene overexpression and related biochemical analyses, however, can be misleading due to neomorphic or hypermorphic phenotypes; in other words, those reactions caused by gene misexpression or biologically irrelevant protein levels. Therefore, it is important to complement gene overexpression and biochemical studies with loss-of-function genetic analysis to unveil the biological functions of the AHL genes in plants.
Our previous loss-of-function study showed that two Arabidopsis AHLs, SOB3/AHL29 and ESC/AHL27, repress hypocotyl growth redundantly in light-grown seedlings (16). Single loss-of-function mutants for either SOB3/AHL29 (sob3-4) or ESC/AHL27 (esc-8) have WT phenotypes. In contrast, the sob3-4 esc-8 double mutant exhibits slightly increased hypocotyl growth under continuous white, red, far-red, and blue light, providing the first AHL loss-of-function analysis and demonstrating that at least SOB3/AHL29 and ESC/AHL27 act as negative regulators of hypocotyl elongation. In support of these observations, an AHL18-AHL22-ESC/AHL27-SOB3/AHL29 RNAi line also exhibits a long-hypocotyl phenotype in white, blue, red, and far-red light (17). These studies suggest functional redundancy among multiple AHL genes in Arabidopsis. In contrast, genetic analyses of single AHL loss-of-function mutants in monocots demonstrated that they regulate inflorescence and floral organ growth, as well as palea formation (27, 28). Although we are beginning to understand the roles of AHL genes in plant growth and development, especially with regard to hypocotyl elongation, the molecular mechanism underlying their functional redundancy in Arabidopsis and other plants is not known.
In contrast to the subtle phenotypes shown by sob3-4 esc-8 (16) and the quadruple RNAi line (17), the missense activation-tagged allele sob3-6, which was identified in our previous study, confers a much more severe long-hypocotyl phenotype in the light (16). Analysis of this dominant-negative sob3-6 allele, which is caused by a single amino acid change in the conserved core of the AT-hook, has led to the hypothesis that this DNA-binding domain, as well as protein-protein interactions, is necessary for the biological function of this gene family.
In this study, we show that the sob3-6 allele encodes a full-length protein that no longer binds AT-rich DNA. Loss-of-function analysis with triple- and quadruple-null mutants demonstrates that multiple AHL genes, but not all, suppress hypocotyl growth in the light. We also show that multiple WT AHLs, as well as the SOB3-6 mutant protein, can associate with each other in the nucleus and that the PPC/DUF296 domain mediates this interaction. Overexpression of this domain also leads to a dominant-negative long hypocotyl phenotype, which is similar to that of sob3-6. In addition, a yeast two-hybrid (Y2H) library screen identified non-AHL, nuclear-localized, DNA-binding proteins that physically interact with AHLs. We demonstrate that a conserved six-amino-acid region in the PPC/DUF296 domain of SOB3/AHL29 is necessary for transcriptional activation in yeast and interaction with at least one transcription factor. Removal of this region also confers a dominant-negative long hypocotyl phenotype when expressed in plants. Based on these results, we propose a molecular model whereby AHLs interact with themselves and each other, as well as other nuclear proteins, to form complexes similar to the human enhanceosome, which modulate plant growth and development.
Results
AHL Gene Family in A. thaliana.
The A. thaliana genome encodes 29 AHL genes (Fig. 1A). A Bayesian phylogenetic analysis showed that these 29 Arabidopsis AHL genes evolved into two phylogenetic clades (clades A and B; Fig. 1B). Clade A AHLs are intronless with only one AT-hook motif and a single PPC/DUF296 domain. Clade B consists of intron-containing AHLs with either one or two AT-hook motif(s) and a single PPC/DUF296 domain (Fig. 1B). The type 1 and 2 AT-hook motifs are distinguished by the sequences that flank the conserved Arg-Gly-Arg core, especially those at the carboxyl end. The consensus sequence at the carboxyl end of the conserved core in type 1 AT-hook motifs in AHL proteins is Gly-Ser-Lys-Asn-Lys, whereas Arg-Lys-Tyr-X is found at the carboxyl end of the core in type 2 AT-hook motifs (Fig. 1 C and D). Within each clade, the Arabidopsis AHLs tend to coexpress with other members (Fig. S1) (29, 30).
Fig. 1.
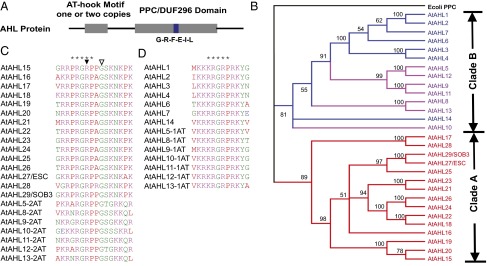
The AHL gene family in A. thaliana. (A) Topologies of the AHL proteins. A conserved six-amino-acid region in the PPC/DUF296 domain is highlighted by the blue box. (B) Phylogeny of the AHL gene family using Bayesian analysis. Numbers near branches indicate Bayesian posterior probabilities for given clades. AHL genes containing only one type 1 AT-hook motif are shown in red. AHL genes containing one type 2 AT-hook motif are shown in blue. AHL genes containing two AT-hook motifs (one each of type 1 and 2) are shown in purple. Two forms of the AT-hook motif, types 1 (C) and 2 (D), were identified in the Arabidopsis AHL proteins. The stars indicate the sequences of the AT-hook motifs. The arrowhead and open arrowhead represent the sob3-6 mutation (R-to-H) and sob3-5 mutation (G-to-Q) in SOB3/AHL29, respectively.
The PPC/DUF296 domain of SOB3/AHL29 is predicted to share the same order of secondary structural elements, β1-α-β2-β3-β4-β5-β6-β7-β8-β9, as observed in their prokaryotic counterparts (Fig. S2). We predicted the tertiary structure of the PPC/DUF296 domain from SOB3/AHL29 via homology modeling (Fig. S3). It is also predicted to adopt a similar tertiary structure as observed in the crystal structures of prokaryotic PPC/DUF296 proteins. The PPC/DUF296 proteins form a trimer, where the β-sheets mediate interactions among the three PPC monomers (14). This structure suggests that the PPC/DUF296 domains in Arabidopsis AHL proteins could associate with each other as their counterparts do in prokaryotes.
sob3-6 Is a Dominant-Negative Allele Disrupting AHL Protein Interactions with DNA.
To obtain loss-of-function alleles, we treated activation-tagged SOB3-D seeds with ethyl methanesulfonate (EMS) and screened for intragenic suppressors of the short-hypocotyl phenotype conferred by overexpression of SOB3/AHL29 (16). In addition to the sob3-4 null allele, we identified two additional missense mutant alleles, sob3-5 and sob3-6, which produced longer hypocotyls in white light than the WT (Fig. 2A) (16). The sob3-5 allele has a Gly80 to Gln mutation, which is adjacent to the carboxyl end of the core sequence of the type 1 AT-hook motif (Fig. 1C). The sob3-6 mutation is at the second arginine (Arg77 to His) in the conserved AT-hook core motif (Fig. 1C). Mutations in this Arg-Gly-Arg core motif have been shown to abolish DNA binding by the AT-hook motif in non-AHL proteins (31, 32).
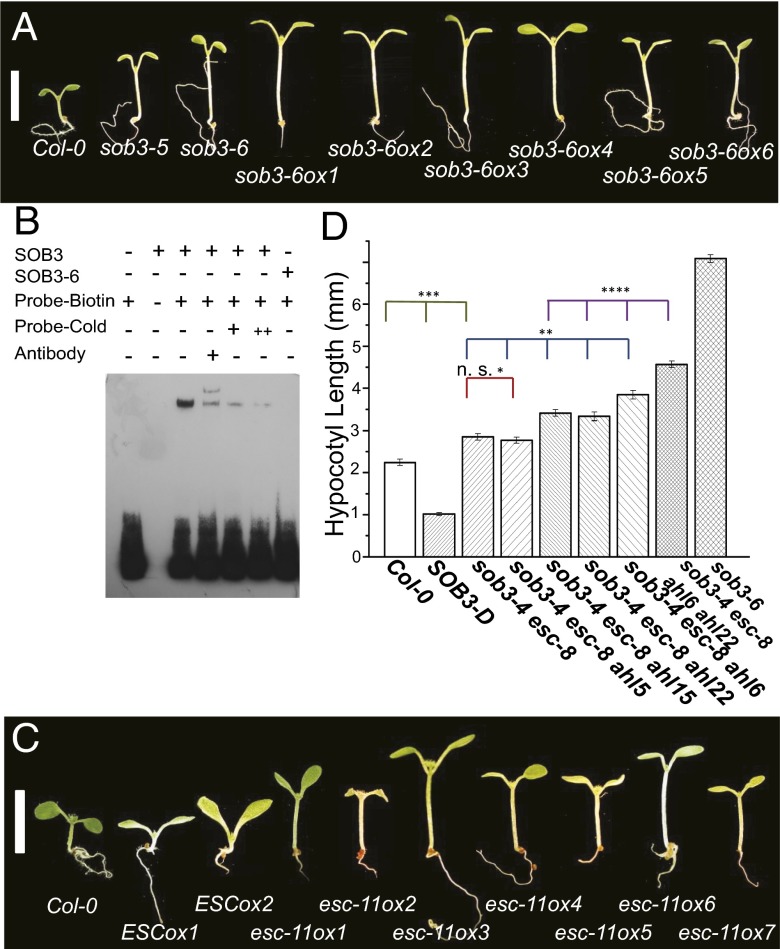
Fig. 2.
sob3-6 is a dominant-negative allele involved in regulating hypocotyl elongation in light growth. (A) Hypocotyl growth of WT Col-0, sob3-5, sob3-6, and multiple independent transgenic lines overexpressing the sob3-6 allele, growing in 30 μmol⋅s−1⋅m−2 white light. (B) SOB3 bound to the biotin-labeled pea PRA2 probe in EMSA. A super shift was detected with the addition of the anti-SOB3 antibody. With increasing concentrations of cold probe, the binding of PRA2 probe by SOB3 could be competed away. However, SOB3-6 did not bind to the PRA2 probe in EMSA. (C) Hypocotyl growth of Col-0, ESC/AHL27 overexpression lines (ESCox1 and ESCox2) and multiple esc-11 overexpression lines, growing in 30 μmol⋅s−1⋅m−2 white light. (Scale bar, 5 mm.) (D) Six-day-old Arabidopsis seedlings were grown in 25 μmol⋅m−2⋅s−1 white light at 25 °C. The error bar denotes the SEM. In a Student t test (unpaired two-tailed t test with unequal variance). *not significant with P = 0.39. **P < 4.5E-4. ***P < 7.5E-7. ****P < 3.2E-7. Col-0, n = 24; SOB3-D, n = 29; sob3-4 esc-8, n = 34; sob3-4 esc-8 ahl5, n = 50; sob3-4 esc-8 ahl15, n = 47; sob3-4 esc-8 ahl22, n = 32; sob3-4 esc-8 ahl6, n = 31; quadruple null, n = 38; sob3-6, n = 32.
Due to the nature of the mutation, the SOB3-6 protein is likely to have minimal or no AT-rich DNA binding capability. To test this hypothesis, we performed an EMSA with SOB3 and SOB3-6 proteins using a probe sequence taken from the AT-rich PRA2 promoter from pea (Pisum sativum) (17, 19). SOB3 readily binds with the biotin-labeled oligonucleotide PRA2 probe (Fig. 2B). Incubation with an anti-SOB3 antibody resulted in a supershift of the PRA2 probe, indicating the presence of SOB3 in the protein-DNA complex. In addition, binding of the PRA2 probe by SOB3 could be competed away by adding a cold (unlabeled) competitive probe. In contrast, no DNA-binding could be detected when using SOB3-6 protein instead of SOB3 (Fig. 2B and Fig. S4A).
sob3-6 was identified as an intragenic suppressor of the short-hypocotyl phenotype conferred by the gain-of-function SOB3-D mutant, leading to a light-grown hypocotyl that is even longer than the WT (16). The observation that the sob3-6 allele confers the opposite phenotype of SOB3-D suggests that the Arg77 to His missense mutation disrupts AT-hook motif function, rendering sob3-6 a negative allele. The sob3-6 negative allele was initially identified as exhibiting a long-hypocotyl phenotype in white light–grown heterozygous sob3-6/SOB3-D plants (16). Because one copy of sob3-6 was sufficient to suppress the SOB3-D gain-of-function phenotype, we hypothesized that this missense mutation functions as a dominant-negative allele. We tested this hypothesis by overexpressing the sob3-6 cDNA under the constitutive Cauliflower Mosaic Virus (CaMV) 35S promoter in WT Arabidopsis. Multiple independent T1 (hemizygous) and T2 transgenic lines recapitulated the long-hypocotyl phenotype of the original sob3-6 allele when grown in white light, with some lines conferring an even longer hypocotyl phenotype than the original sob3-6 mutant line (Fig. 2A).
We also overexpressed another AHL, ESC/AHL27, driven by the CaMV 35S promoter in Arabidopsis. The ESC/AHL27 overexpression lines exhibited similar suppressed hypocotyl growth as seen in SOB3-D seedlings (16) (Fig. 2C). We further created the same sob3-6-like mutation (Arg91 to His) in the AT-hook motif of the ESC/AHL27 gene (designated as esc-11). Multiple independent T1 and T2 esc-11 overexpression lines (also driven by the CaMV35S promoter) exhibited long hypocotyls in white light, recapitulating the sob3-6 phenotype (Fig. 2C) and demonstrating that multiple AHLs with a sob3-6–like mutation function as dominant-negative alleles.
AHL Genes Contribute Differently to the Suppression of Hypocotyl Elongation in White Light.
The sob3-6 and esc-11 overexpression lines both exhibited longer hypocotyls than the sob3-4 esc-8 double-null mutant. This observation suggests that other AHLs with similar functions are also affected by these dominant-negative alleles. To test this hypothesis and further examine the function of additional AHLs with regard to suppression of hypocotyl elongation in the light, we generated four AHL triple-null mutant combinations (Fig. 2D and Fig. S4 B–E). Three triple-null mutants (ahl6 sob3-4 esc-8, ahl15 sob3-4 esc-8, and ahl22 sob3-4 esc-8) exhibited longer hypocotyls than the sob3-4 esc-8 double-null, although they varied with regard to the severity of the phenotype (P < 4.5E-4). A fourth triple-null mutant, sob3-4 esc-8 ahl5, had a similar light-grown hypocotyl length as the sob3-4 esc-8 double-null (Fig. 2D). We also made the sob3-4 esc-8 ahl6 ahl22 quadruple-null mutant. The sob3-4 esc-8 ahl6 ahl22 quadruple null conferred an even longer hypocotyl phenotype than both triple-null lines when grown in white light, although this phenotype was still shorter than that of sob3-6 (P < 3.2E-7; Fig. 2D). Together these data demonstrate that these AHL genes contribute differently to white light–mediated suppression of hypocotyl elongation.
AHL Proteins Form Homo- and Hetero-Complexes.
The dominant-negative long-hypocotyl phenotype of sob3-6 and esc-11 overexpressing seedlings suggests a model whereby these mutant proteins physically interact with other AHL family members and/or other shared interacting partners, rendering the complex functionally inactive due to abolished DNA binding by the mutated AT-hook motif. To test this hypothesis, we first performed a targeted GAL4-based Y2H assay (Fig. 3A). Using the lowest concentration of 3-amino-1, 2, 4-triazol (3-AT) that prevented transcriptional autoactivation by the AHL baits, we demonstrated that SOB3/AHL29 and ESC/AHL27 interact with themselves as well as with each other (Fig. 3A and Fig. S5A).
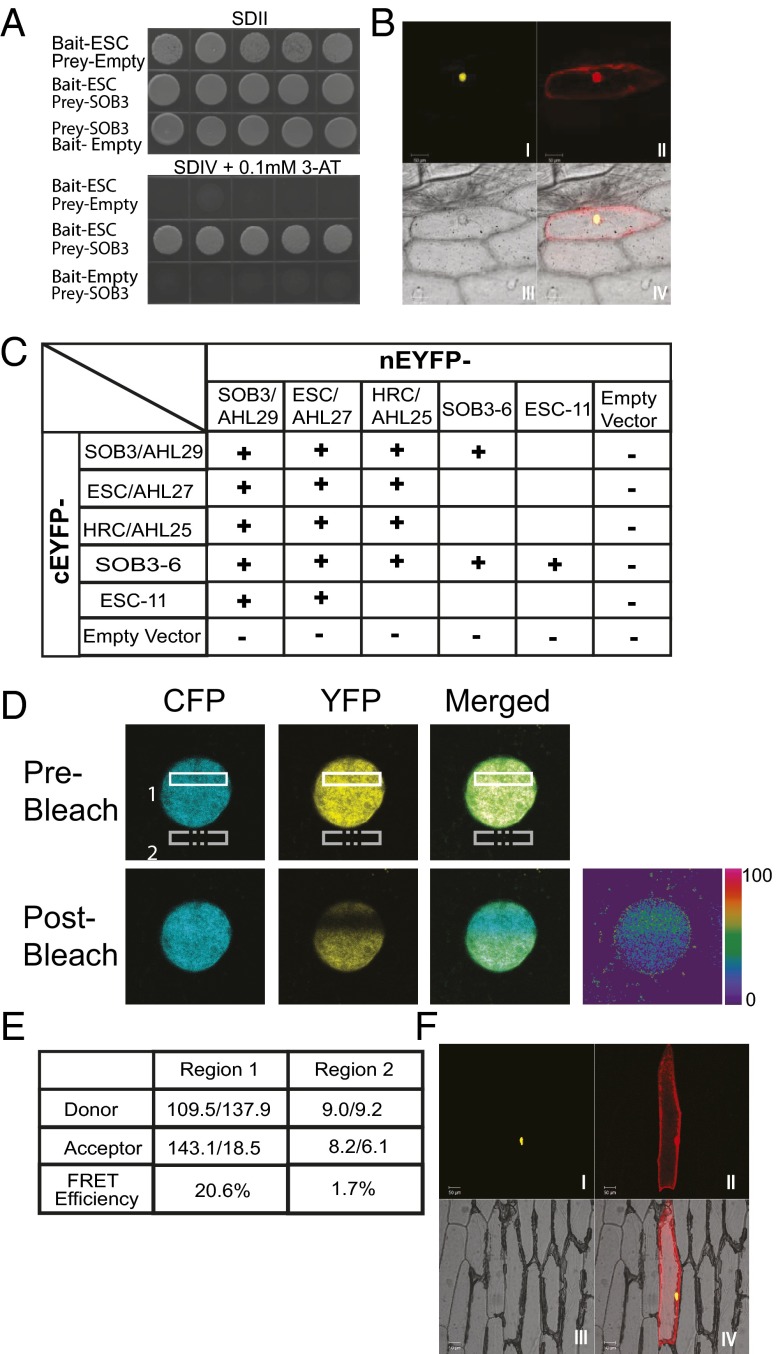
Fig. 3.
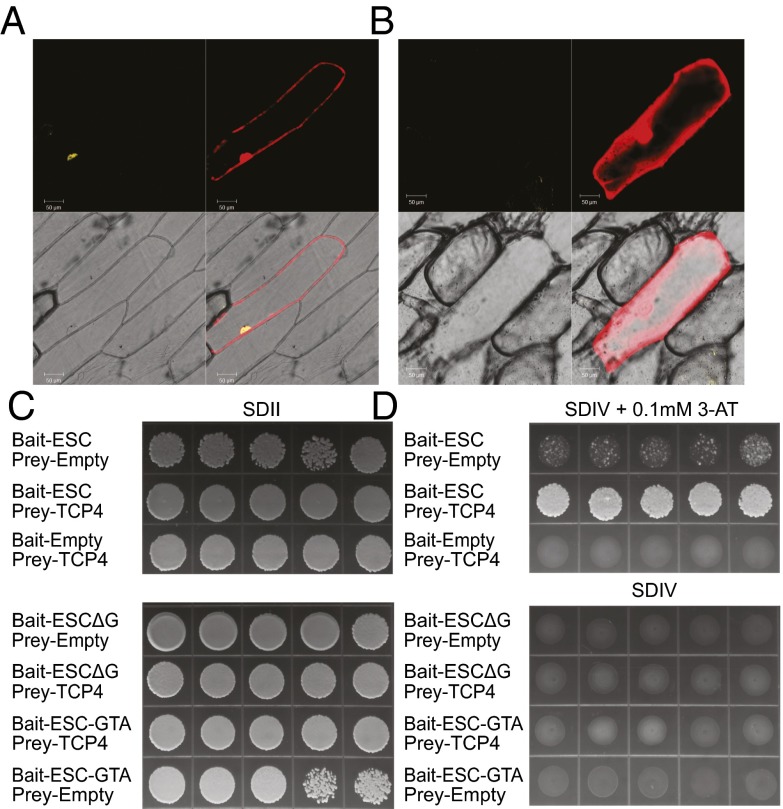
The AHL proteins interact with each other and themselves. SOB3/AHL29 interacts with ESC/AHL27 in Y2H (A) and BiFC (B) assays. Yeast transformed with the indicated prey and bait plasmids were plated on SDII, as well as on SDIV media (supplemented with 3-AT of indicated concentration), and checked for growth after 5 d in 30 °C. Five independent biological replicates are shown. Onion epidermal cells, transformed with plasmids expressing nEYFP-SOB3 and cEYFP-ESC, were checked for reconstructed yellow fluorescence signal (I) and red fluorescence signal (II), observed in bright field (III) and merged (IV). The protein-protein interactions tested by BiFC and/or Y2H were summarized as in C. Interaction among three copies of SOB3/AHL29 proteins was examined by BiFC-FRET assay (D). Onion epidermal cells were transformed with plasmids expressing nEYFP-SOB3, cEYFP-SOB3, and CFP-SOB3. (E) Fluorescent intensities (before bleach/after bleach) were examined individually in region 1 and control region 2 with laser bleaching at the acceptor’s (YFP) excitation wavelength. (F) SOB3-6 and ESC-11 interact with each other in the BiFC assay.
We further examined the AHL-AHL interactions in planta using a bimolecular fluorescence complementation (BiFC) assay via transient expression in onion epidermal cells (33, 34). In this experiment, two halves of the enhanced YFP (EYFP) (nEYFP, N terminus half of EYFP; cEYFP, C terminus half of EYFP) were translationally fused to the 5′ end of SOB3/AHL29 and ESC/AHL27 (Fig. 3B and Fig. S5 B–F). A separate plasmid expressing a monomeric red fluorescent protein (mRFP) was also cotransformed with the nEYFP and cEYFP fusions as an indicator of successful transformation. The mRFP protein was expressed in the cytoplasm and in the nucleus. The reconstructed yellow fluorescence signal could be detected in the transformed onion epidermal cells and colocalized with the red fluorescent signal only in the nucleus (Fig. 3B). No yellow fluorescence was observed with the negative controls using empty BiFC vectors (Fig. S5 G–L).
We also used the BiFC assay to demonstrate interactions among SOB3/AHL29, ESC/AHL27, and another clade A AHL, HRC/AHL25 (Fig. 3C). In addition, we used the Y2H and BiFC assays to test and demonstrate that two clade B AHLs, AHL5 and AHL12, interact with SOB3/AHL29 and ESC/AHL27, as well as with themselves (Fig. S5 M–P). AHL5 and AHL12 each contain two AT-hook motifs (one each of type 1 and 2) and one PPC/DUF296 domain. Thus, the tested AHL proteins in both clades can associate with each other and with themselves, regardless of the number and type of the AT-hook motif(s).
To further characterize the AHL protein complex in planta, we performed a BiFC-FRET assay in onion epidermal cells (Fig. 3D). Three copies of SOB3/AHL29 were fused separately to the two halves of the EYFP, as well as to intact cyan fluorescent protein (CFP). The resulting three plasmids were simultaneously transformed into onion epidermal cells (Fig. 3D). Yellow fluorescence could be observed in the nucleus due to reconstitution of EYFP by the physical interaction between the two SOB3/AHL29 proteins that were each fused either to nEYFP or cEYFP. Cyan fluorescent signal could also be detected in the nucleus due to the nuclear localization of SOB3/AHL29. A laser, set at the acceptor’s excitation wavelength, was then used to bleach a selected region in the nucleus. Following this treatment, the intensity of yellow fluorescence diminished, whereas the cyan fluorescence increased, suggesting that, before bleaching, FRET was occurring between the donor (CFP) and the acceptor (reconstructed EYFP) (Fig. 3E). The high efficiency of FRET indicated that the three SOB3/AHL29 proteins form at least a trimer complex in planta.
sob3-6 and esc-11 Mutations Do Not Abolish AHL Physical Interactions or Nuclear Localization.
We further used the Y2H and BiFC assays to test whether SOB3-6 and ESC-11 proteins interact with their WT forms in yeast and in planta. In both assays, the mutant proteins interacted with WT SOB3 and ESC proteins and with themselves (Fig. 3F and Fig. S5Q). In addition, these interactions still occurred in the nucleus (Fig. 3F), demonstrating that the dominant-negative effect of these alleles is not acting through the disruption of nuclear localization and further that disruption of the AT-hook motif did not abolish the ability of AHLs to form homo- or hetero-complexes.
AHLs Interact via the PPC/DUF296 Domain.
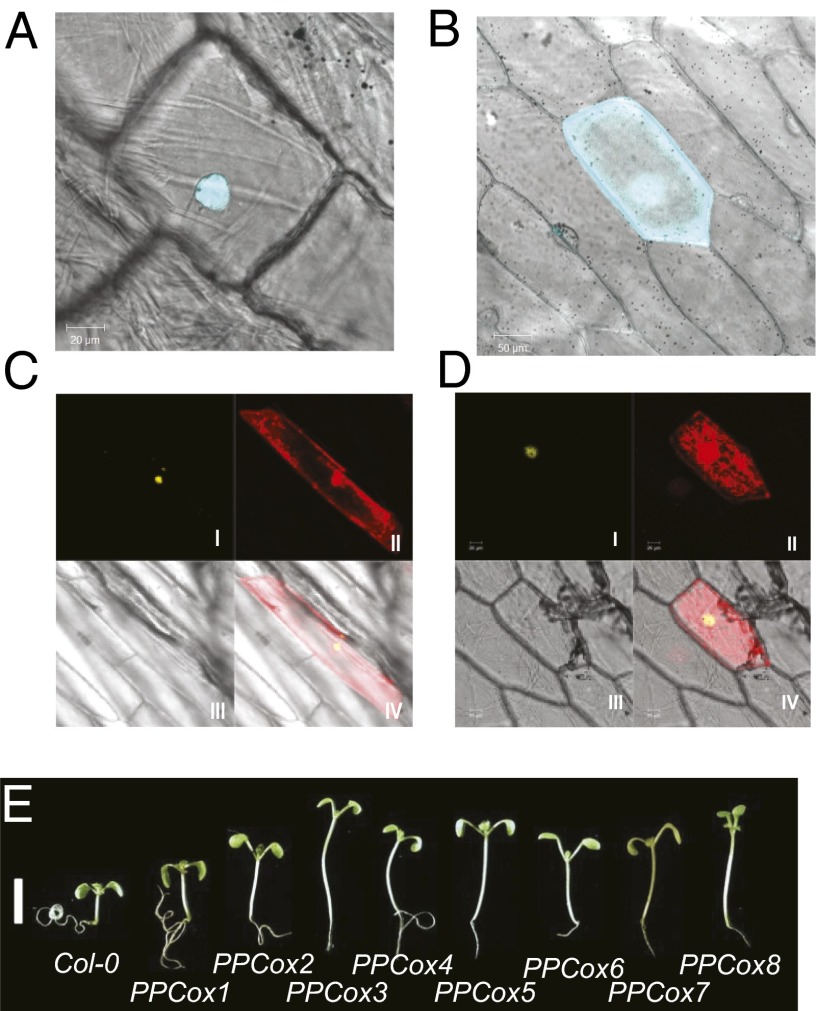
We fused the PPC/DUF296 domain from SOB3/AHL29 to the carboxyl end of CFP to examine its subcellular localization via transient expression in onion epidermal cells. The PPC/DUF296 domain of SOB3/AHL29 localized to the nucleus of onion epidermal cells (Fig. 4 A and B). This finding is consistent with the report that the C-terminal hydrophobic region of the PPC/DUF296 domain is essential for the nuclear localization of AHL1 (13). The SOB3/AHL29 PPC/DUF296 domain also interacted with full-length SOB3/AHL29 (Fig. S5 R and S) and ESC/AHL27 (Fig. S5T) proteins in Y2H assays. A role for the PPC/DUF296 domain in mediating protein-protein interactions in planta was further supported by BiFC analysis (Fig. 4C). The SOB3/AHL29 PPC/DUF296 domain also interacted with SOB3-6 and ESC-11 mutant proteins in both assays (Fig. 4D and Fig. S5 U and V).
Fig. 4.
The PPC/DUF296 domain mediates interactions between AHL proteins. The SOB3 PPC/DUF296 domain tagged with CFP localizes to the nucleus in transformed onion epidermal cells (A) in contrast to free CFP (B). The SOB3 PPC/DUF296 domain interacts with SOB3 in the BiFC assay (C). The PPC/DUF296 domain of SOB3/AHL29 also interacts with SOB3-6, which contains a disrupted AT-hook motif, in the BiFC assay (D). (E) Compared with WT Col-0 (Left), overexpression of the PPC/DUF296 domain in SOB3 leads to longer hypocotyl growth in the light in multiple independent transformation events.
To test the hypothesis that the dominant-negative nature of sob3-6 is caused by the association of its PPC/DUF296 domain with other AHL proteins, we overexpressed this domain under the control of the constitutive CaMV 35S promoter in WT Arabidopsis and examined the hypocotyl phenotype in white light (Fig. 4E). Multiple independent T1 seedlings exhibited a long-hypocotyl phenotype in white light. Many lines recapitulated the phenotypes of sob3-6/esc-11 overexpression, as well as the original sob3-6 line (Fig. 2 A and C), demonstrating that overexpression of the SOB3 PPC/DUF296 domain alone functions in a dominant-negative manner (Fig. 4E). Together these results support the hypothesis that the AHLs suppress hypocotyl elongation in Arabidopsis seedlings by associating with each other via the PPC/DUF296 domain and that this suppression requires functional AT-hook motifs that recognize AT-rich chromosomal regions.
AHLs Interact with Other Nuclear-Localized Non-AHL Proteins.
To further test the hypothesis that AHL proteins interact with each other and possibly have shared interacting partners, we performed a blind Y2H library screen using multiple AHL proteins as the bait. We identified multiple AHLs as interacting partners (Table 1 and Table S1) supporting the AHL-AHL physical interactions identified in our study. Interestingly, using just the SOB3/AHL29 PPC/DUF296 domain alone as bait in the library screen, we identified SOB3/AHL29, as well as AHL3, as interaction partners. In addition, we also identified several transcription factors that interact with the AHLs (Table 1, Table S1, and Fig. S6 A–H), suggesting that the redundancy among AHLs is not only attributable to their physical interactions with each other but also due to other common interacting partners. We also found that core histones (histones H2B, H3, and H4) in nucleosomes interact with multiple AHLs in the Y2H analysis (Fig. S6 I–K and Table S1).
Table 1.
Combined interactors identified in Y2H library screens and/or targeted Y2H assays
| Interaction | AGI number |
| AHLs interacted with AHLs | |
| AHL1 (as bait)—AHL8 | |
| AHL5 (as bait)—AHL3 | |
| AHL27 (as bait)—AHL23 | |
| AHL27 (as bait)—AHL20 | |
| PPC/DUF296 of SOB3 (as bait)—AHL29/SOB3 | |
| PPC/DUF296 of SOB3 (as bait)—AHL3 | |
| Transcription factors interacted with AHLs | |
| AP2/EREBP | At5G61890 |
| NAC transcription factor (ATAF2) | At5G08790 |
| Zinc-finger protein | At1G09520, At2G36930 |
| TCP transcription factor 4 | At3G15030 |
| TCP transcription factor 13 | At3G02150 |
| TCP transcription factor 14 | At3G47620 |
| Transcriptional coactivator (KIWI) | At5G09250 |
| Methyltransferase interacted with AHLs | |
| Methyltransferase | AT5G10830 |
| Histones interacted with AHLs | |
| Histone H2B | At1G07790 |
| Histone H3.3 | At5G10980, At4G40040 |
| Histone H4 | At3G53730 |
Conserved Region in the PPC/DUF296 Domain of SOB3/AHL29 Is Essential for Its Activation of Transcription and Physical Interaction with Non-AHL Interactors.
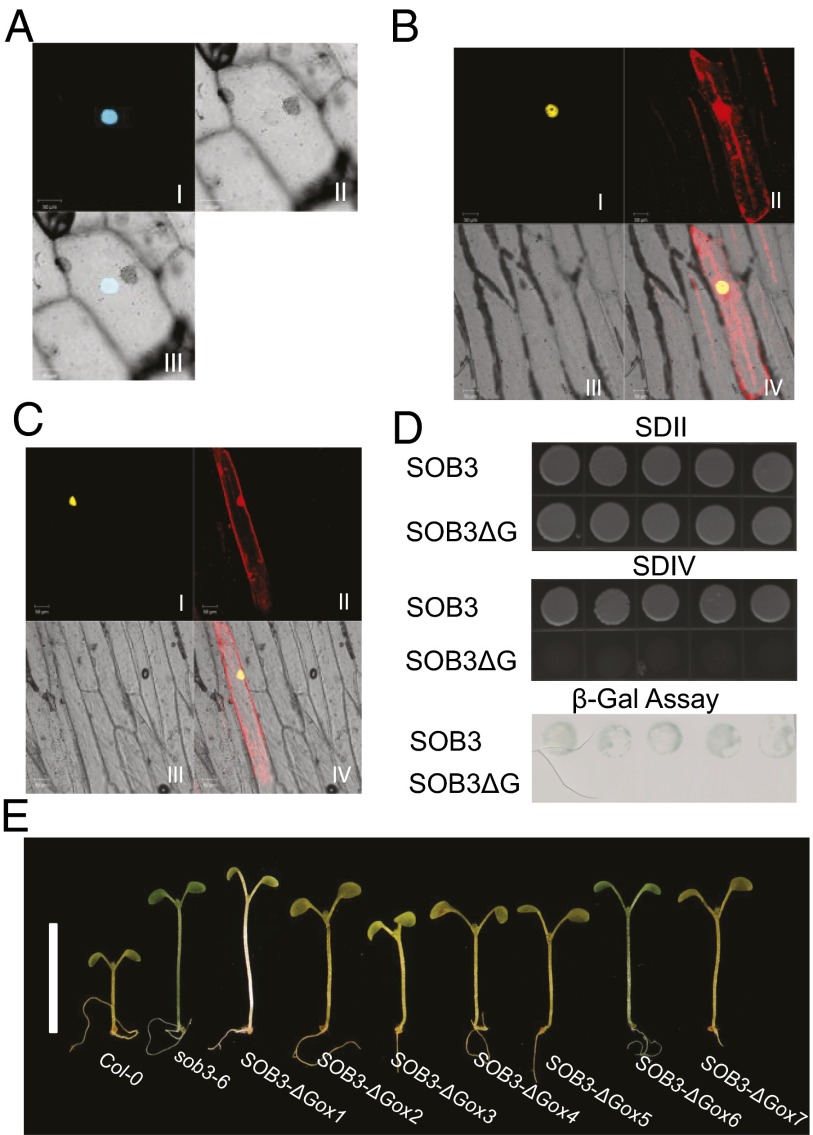
Alignment of amino acid sequences of the PPC/DUF296 domain from the AHLs of several sequenced plant genomes reveals a conserved six-amino-acid region, Gly-Arg-Phe-Glu-Ile-Leu, which is predicted to exist as one β-strand in its secondary structure (Fig. S2). We deleted this six-amino-acid region from the coding sequence of SOB3/AHL29 and designated it as sob3-ΔG. The deletion did not affect the nuclear localization of the mutant SOB3-ΔG protein (Fig. 5A). In addition, it did not disrupt interactions with WT SOB3/AHL29 or ESC/AHL27 (Fig. 5B and Fig. S6 L and M). The PPC/DUF296 domain of SOB3/AHL29 also interacted with SOB3-ΔG in both assays (Fig. 5C and Fig. S6 N and O). These results, together with the observed interactions of SOB3-ΔG with both SOB3-6 and the ESC protein with the same deletion (ESC-ΔG) (Fig. S6 P–R), suggest that protein stability, nuclear localization, and association among AHL proteins do not depend on this region.
Fig. 5.
Transcription activation by SOB3/AHL29 depends on a conserved region in the PPC/DUF296 domain. (A) Subcellular localization of SOB3-ΔG protein in onion epidermal cells. Protein-protein interaction analysis of SOB3-ΔG with full-length SOB3/AHL29 in BiFC (B). Protein-protein interaction analysis of SOB3-ΔG with the SOB3/AHL29 PPC/DUF296 domain in BiFC (C). (D) Comparison of transcription activation by SOB3/AHL29- and SOB3-ΔG-containing yeast growing on SDIV plates and in a β-gal assay. Five independent biological replicates are shown. (E) Multiple T2 generation, 6-d-old seedlings overexpressing SOB3-ΔG under the CaMV 35S promoter exhibited a longer hypocotyl phenotype in 23 μmol⋅s−1⋅m−2 white light. (Scale bar, 1 cm.)
AHL25 from Arabidopsis has been shown to activate transcription in a yeast one-hybrid assay (23). We tested SOB3/AHL29 for transcriptional activation in yeast (Fig. 5D and Fig. S7A). With no 3-AT supplemented in synthetic dropout IV (SDIV) minimal media, yeast expressing SOB3/AHL29 as a Y2H bait exhibited strong autoactivation activity (Fig. 5D). Interestingly, the SOB3/AHL29 PPC/DUF296 domain alone tagged with the DNA-binding domain used in the Y2H assay also conferred autoactivation (Fig. S5S). This observation suggests that the PPC/DUF296 domain is responsible for transcriptional activation by the AHL proteins in yeast.
In contrast, SOB3-ΔG demonstrated a complete loss of transcriptional activation compared with the WT SOB3 (Fig. 5D and Fig. S7A). Yeast transformed with WT SOB3/AHL29 or SOB3-ΔG survived on synthetic dropout II (SDII) media. When transferred to SDIV media without 3-AT, only the yeast transformed with WT SOB3/AHL29 survived, due to activation of transcription by the AHL protein. Neither growth on SDIV media nor β-gal activity was observed in the five biological yeast replicates that were transformed with SOB3-ΔG (Fig. 5D). We also created another mutant allele (designated as SOB3-GTA) in which the six amino acids were substituted with alanine residues. SOB3-GTA also lost transcriptional activity in yeast (Fig. S7A). Similar alleles created for ESC/AHL27 (ESC-ΔG and ESC-GTA) also demonstrated loss of transcriptional activity in yeast in contrast with the WT (Fig. S7B). Therefore, this six-amino-acid region in the PPC/DUF296 domain is essential for the activation of transcription by AHL proteins.
If transcriptional activation is necessary for biological function, then removal of this six-amino-acid region would alleviate, or even abolish, SOB3/AHL29 suppression of hypocotyl elongation in the light. To test this hypothesis, we overexpressed SOB3-ΔG driven by the CaMV 35S promoter in WT Arabidopsis. Surprisingly, we observed a long-hypocotyl phenotype in multiple independent hemizygous lines (Fig. 5E), which is similar to seedlings overexpressing sob3-6, esc-11, or the SOB3 PPC/DUF296 domain alone (Figs. 2 A and C and 4E). Alleles of sob3-6 and SOB3-ΔG, as well as the SOB3 PPC/DUF296 domain alone, are likely to have similar efficacy with regard to promoting hypocotyl growth in the light (Fig. S7C). Overexpression of SOB3-GTA by the CaMV 35S promoter also recapitulated a similar long-hypocotyl phenotype (Fig. S7D). Thus, SOB3-ΔG and SOB3-GTA function as dominant-negative alleles, even though the mutant proteins still bear functional AT-hook motifs and could bind with the AT-rich DNA PRA2 probe (Fig. S7E). This finding suggests that interactions with transcription factors mediated by this Gly-Arg-Phe-Glu-Ile-Leu region in the PPC/DUF296 domain are necessary for the biological function of AHL proteins.
The dependence on this six-amino-acid region in the PPC/DUF296 domain of SOB3/AHL29 for transcriptional activation in yeast, together with the observed interactions with transcription factors identified in this study, implies that this region may play a key role in mediating the physical interaction between SOB3/AHL29 and transcription factor(s) in planta. To test this hypothesis, we first used Y2H and BiFC assays to examine the physical interaction with the transcription factor TCP4. TCP4 interacted with WT SOB3/AHL29 in both the BiFC (Fig. 6A) and Y2H assays (Fig. S6C). However, TCP4 no longer interacted with SOB3-ΔG (Fig. 6B). TCP4 also interacted with WT ESC/AHL27 but not with ESC-ΔG or ESC-GTA (Fig. 6 C and D and Fig. S6D), even though all three forms of ESC were expressed at similar levels (Fig. S8A). Similarly, we also tested interactions with another transcription factor, TCP13 (previously also known as TCP10). TCP13 has been shown to localize in the nucleus (35), as well as in the chloroplast (36). From the results of our BiFC assay, as well as targeted Y2H analysis, TCP13 interacted with the WT SOB3/AHL29 protein and ESC/AHL27 (Figs. S6 A and B and S8 B and C). However, TCP13 no longer interacted with SOB3-ΔG (Fig. S8 D and E).
Fig. 6.
Interactions of AHLs with transcription factors depends on a six-amino-acid conserved region in the PPC/DUF296 domain. (A) SOB3/AHL29 interacted with the transcription factor TCP4, and (B) this interaction was abolished when the conserved six amino acids in the PPC/DUF296 domain was removed in the BiFC assay. (C and D) ESC/AHL27 interacted with TCP4, and the interaction was abolished when the same six-amino-acid region was removed or substituted with alanine residues in a Y2H assay.
Non-AHL Interactors Modulate Hypocotyl Growth in the Light.
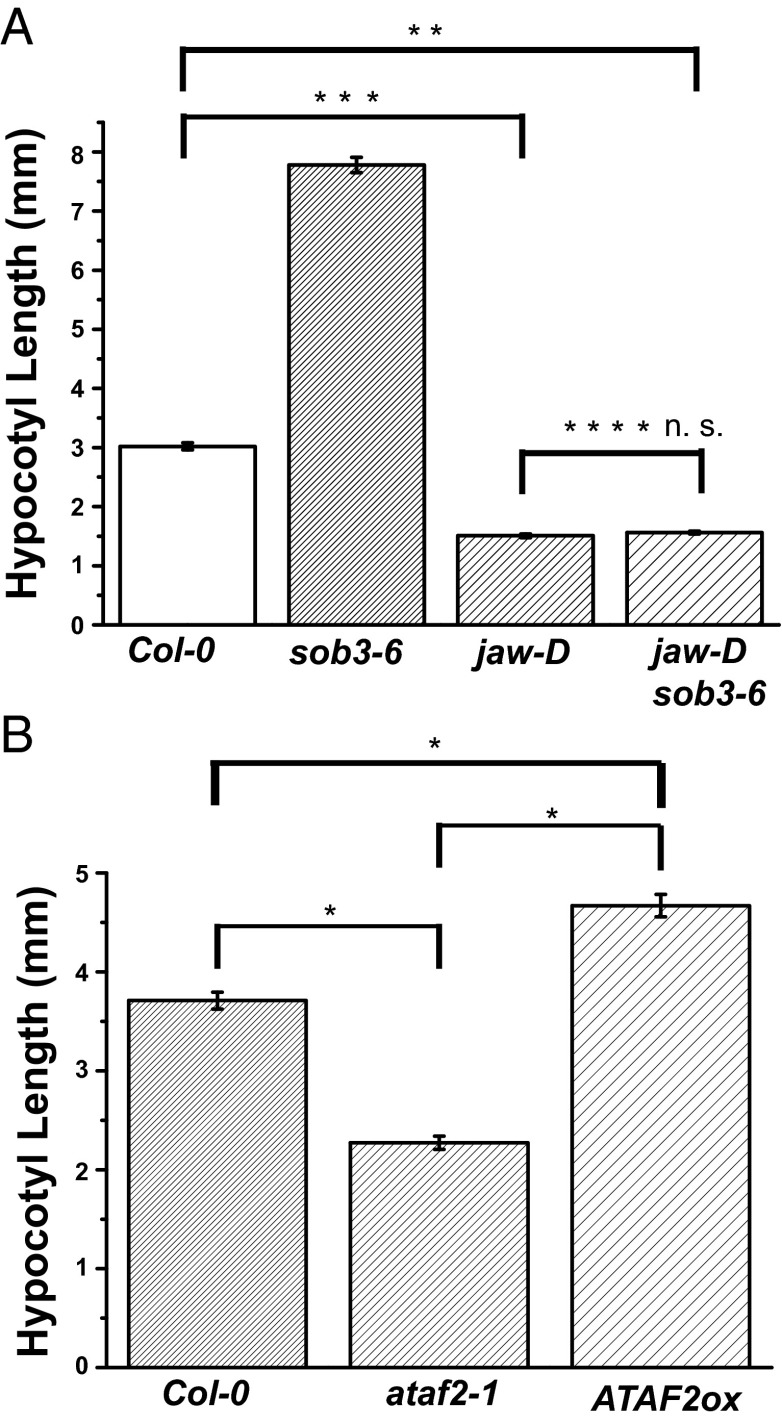
To understand the roles of the AHL-interacting transcription factors in hypocotyl elongation in the light, we first examined members of the TCP gene family. Jaw-D, in which several TCP genes were specifically knocked down (especially TCP4) (37), exhibited a shorter hypocotyl phenotype (Fig. 7A), which is consistent with a previous report (38), suggesting TCPs play a role in regulating hypocotyl elongation. We also examined roles of other AHL interactors in hypocotyl elongation in the light. We overexpressed the transcription factor ATAF2, one member of the plant-specific NAC gene family, driven by the CaMV 35S promoter in WT Arabidopsis. The homozygous single-locus insertion overexpression line (ATAF2ox) confers a longer hypocotyl than the WT in the light, whereas the loss-of-function line, ataf2-1, showed suppressed hypocotyl growth in the light (Fig. 7B). Another plant-specific AHL interacting partner that was identified in this study, Gibberellic Acid-Stimulated Arabidopsis 4 (GASA4), also has been implicated in regulating hypocotyl growth in the light (39).
Fig. 7.
Hypocotyl analysis of seedlings grown in continuous white light. (A) Six-day-old seeedlings were grown in 20 μmol⋅m−2⋅s−1 of white light at 25 °C. Col-0, n = 147. sob3-6, n = 63. jaw-D, n = 70. jaw-D sob3-6, n = 232. (B) Seedlings (3.5 d old) were grown in 10 μmol⋅m−2⋅s−1 of white light at 25 °C. Col-0, n = 48. ataf2-1, n = 48. ATAF2ox, n = 48. The error bar denotes SEM. In a Student t test (unpaired two-tailed t test with unequal variance): *P < 1.6E-9. **P < 1.7E-54. ***P < 1.3E-55. ****Not significant, P = 0.13.
To further understand the biological meaning of the physical interactions between the TCPs and AHLs, we crossed jaw-D with the activation-tagged sob3-6 allele. The jaw-D sob3-6 double mutant line exhibited a similar hypocotyl length as jaw-D (P = 0.13; Fig. 7A). This observation suggests that the long-hypocotyl phenotype conferred by the activation-tagged sob3-6 allele in the light requires the presence of functional TCP proteins. We examined transcript accumulation of SOB3 (or sob3-6) in these lines and observed similar levels of gene expression in each case. These data indicate that the observed genetic epistasis is not due to TCPs affecting transcription accumulation of these alleles of sob3 (Fig. S9 A and B). Collectively, these observations demonstrate a direct role for the participation of physical interacting partners in AHL-mediated hypocotyl elongation in the light.
Molecular Model for AHL Proteins.
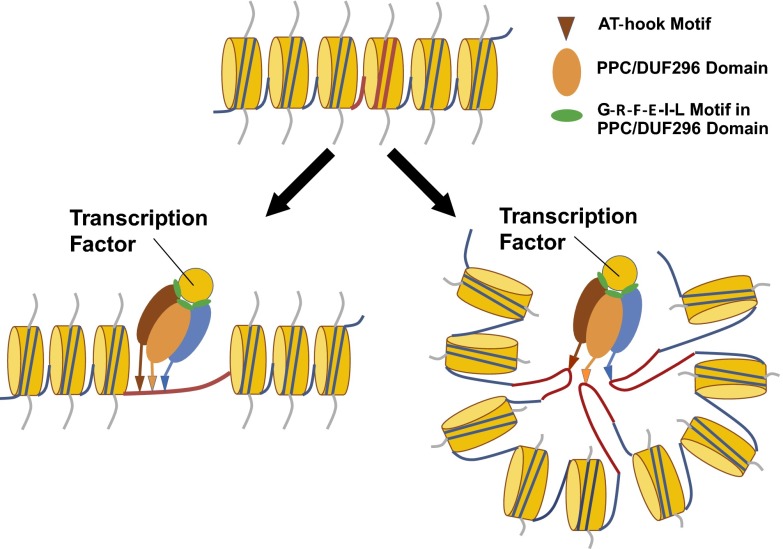
Based on our results, as well as those in the literature, we propose a working model that describes the physical interactions among AHLs, other nuclear proteins, and AT-rich chromosomal DNA (Fig. 8). The AHL proteins form a homo-/hetero-complex via the PPC/DUF296 domain, possibly resulting in a trimer (Fig. 8). The multiple AT-hook motifs of the AHL homo-/hetero-complex then allow it to bind AT-rich DNA regions of the same or different chromosomes. Finally, the AT-rich DNA-anchored AHL complex, aided by the Gly-Arg-Phe-Glu-Ile-Leu region, recruits other nuclear non-AHL proteins, such as transcription factors, to form a DNA-AHL-transcription factor (DNA-AHL-TF) complex, which modulates hypocotyl growth in the light.
Fig. 8.
The AHL gene family regulates plant growth and development by forming an AHL complex. The AHL proteins associate with themselves and each other to form a homo-/hetero-trimeric complex via the PPC/DUF296 domains. The complex uses its AT-hook motifs to anchor itself to AT-rich DNA regions (red-color region) and recruit either transcription factors or other non-AHL proteins in order to regulate plant growth and development. In the proposed molecular model, the hypocotyl-growth-promoting functions of associated transcription factors are rendered nonfunctional by their physical interaction with the DNA-AHL-TF complex.
Discussion
A unique missense allele, sob3-6, caused by an amino acid change in the core of the AT-hook motif, confers a dramatic long-hypocotyl phenotype in the light (16). In this study, we determined that sob3-6 functions as a dominant-negative allele due to abolished AT-rich DNA-binding ability (Fig. 2). Overexpression of the SOB3/AHL29 PPC/DUF296 domain also led to a similar long-hypocotyl phenotype in light-grown seedlings through a dominant-negative process (Fig. 4). These dominant-negative alleles demonstrated the importance of the AT-hook motif(s) in AHL proteins and implied a biological function for the PPC/DUF296 domain. The severity of the phenotype conferred by these dominant-negative mutations also suggests that other AHL genes are involved in regulating hypocotyl growth in Arabidopsis seedlings.
Our previous study showed that SOB3/AHL29 and ESC/AHL27 redundantly suppress hypocotyl elongation in light-grown Arabidopsis seedlings (16). As a part of this study, we characterized hypocotyl growth in four triple-null lines, each in which a different AHL gene (AHL5, AHL6, AHL15, or AHL22) was removed from the sob3-4 esc-8 double mutant background (Fig. 2D). We determined that the removal of AHL5 (unlike the other three AHL genes) did not have an impact on hypocotyl elongation. Of the other three AHLs, removal of AHL6 had the greatest impact, demonstrating that AHL genes have different quantitative contributions to seedling elongation in the light. The sob3-4 esc-8 ahl6 ahl22 quadruple-null line exhibited an even longer hypocotyl than the relevant triple-null controls, although it was still shorter than the dominant-negative sob3-6 mutant (Fig. 2D). This difference suggests that there are additional proteins, possibly AHLs, involved in this developmental process.
Given that not all Arabidopsis AHLs contribute to light-grown hypocotyl elongation (e.g., AHL5), and the probability that some may act redundantly, a dominant-negative approach may be useful for revealing the biological role of other members in this gene family. It is important to note, however, that it would be best if the dominant-negative proteins were expressed according to their endogenous pattern, using the native promoter (possibly with enhancer elements), or via TILLING (40), TALEN (41), or CRISPR/Cas (42) approaches. Such genetic approaches could be very powerful in the case of genomes with even larger AHL gene families, such as Glycine max (43) and Brassica rapa (44), which each have >50 AHLs.
In our proposed molecular model, we suggest that the genetic redundancy among AHL proteins is not only due to interactions among themselves, mediated by the PPC/DUF296 domain, but also due to shared, nuclear-localized, non-AHL interactors (Fig. 8). The predicted tertiary structure of the PPC/DUF296 domain of SOB3/AHL29 suggests trimeric oligomerization, which is similar to that observed in seven determined crystal structures of this domain from Bacteria and Archaea [Protein Data Bank (PDB) ID: 2H6L, 2DT4, 2HX0, 2NMU, 2P6Y, 3HTN, and 3HWU] (14, 15). Trimer formation in a proposed AHL complex is also supported by the physical interactions observed among AHLs in this study (Figs. 3 and 4 and Fig. S5). The 29 AHL genes encoded by the A. thaliana genome allow flexibility in complex formation and subsequent regulation of the related biological processes in plant growth and development.
According to our proposed model, a trimeric AHL complex would have three or more (up to six) AT-hook motifs, depending on the composition of its subunits. The number of AT-hook motifs, as well as their types (type 1 or 2), would determine the AHL complex’s affinity for AT-rich DNA. It is important to note that the AT-hook motifs in AHL proteins are identical to the ones in human HMGA proteins. The human genome encodes three HMGA proteins, each of which contains three AT-hook motifs (45). Among the three AT-hook motifs in an HMGA protein, the middle one is identical to the AT-hook motif in SOB3/AHL29 (Fig. S10). These AT-hook motifs also confer the highest affinity to AT-rich DNA (12, 46). The flanking AT-hook motifs of HMGAs are similar to the type 2 AT-hook motif of AHLs. These flanking HMGA AT-hook motifs both have decreased DNA-binding affinity compared with type 1 (12). With these different affinities in mind, trimeric AHL complexes could possess a variety of combinations of AT-hook motifs that in turn could impart to the complex a wide range of binding affinities for AT-rich chromosomal regions. Such AHL complexes could regulate a variety of biological processes.
The human HMGA proteins have been proposed to promote the assembly of an enhanceosome by binding to the AT-rich promoter region of genes and further recruiting other nuclear proteins, such as chromatin remodeling machinery and transcription factors, to positively or negatively regulate gene expression (47–49). In the case of virus-induced transcription of the human IFN-β gene, HMGA has been proposed to first bind AT-rich DNA in a 1:1 (protein:DNA) ratio and subsequently alter chromatin status, as well as serve as a platform for the recruitment of eight transcription factors. This process leads to the assembly of the nuclear macromolecular complex (50, 51). The 1:1 HMGA:DNA binding ratio indicates that three AT-hook motifs of one HMGA are necessary to target HMGA to the corresponding chromosome region and induce assembly of the enhanceosome.
Similar promoter binding capabilities have been reported for several AHL proteins. AHL15 and AHL25 bind to the promoter of GA 3-oxidase 1 in Arabidopsis (23). AHL21 binds to the promoter of the ETTIN/auxin response factor 3 gene (20). AHL22 binds to the promoter of the Flowering Locus T gene (18). In Catharanthus roseus, five AHLs bind to the promoter of the APETALA2 (AP2) transcription factor ORCA3 gene (24). These observations suggest that trimeric AHL complexes could also be targeted to promoter regions through their AT-hook motifs.
The three HMGA AT-hook motifs necessary for AT-rich DNA binding correspond to the three motifs (at minimum) in a proposed trimeric AHL complex. This molecular model explains the dominant-negative long-hypocotyl phenotypes conferred by the sob3-6 allele (which does not bind AT-rich DNA) or expression of the PPC domain alone (which lacks an AT-hook motif). Both dominant-negative alleles encode proteins containing an intact PPC/DUF296 domain, which could still physically interact with WT AHL proteins and form an AHL complex with attenuated DNA-binding activity (Figs. 2 and 4). A similar dominant-negative phenomenon has also been observed with a human HMGA protein bearing a nonfunctional AT-hook motif (32), which resembles the sob3-6 allele in this study.
We demonstrated that the conserved Gly-Arg-Phe-Glu-Ile-Leu region of the PPC/DUF296 domain is necessary for physical interaction with the transcription factors TCP4 and TCP13 (Fig. 6 and Figs. S6 A–D and S8). We also showed that expression of SOB3 alleles that are mutated in this region confers a dominant-negative long-hypocotyl phenotype (Fig. 5E and Fig. S7D). Homology modeling of the SOB3/AHL29 PPC/DUF296 domain suggests that this conserved region is located at the exterior of each monomer and could combine to form an exposed quaternary domain after trimerization (Fig. S3D). This prediction suggests that the quaternary domain composed of these three six-amino-acid regions could serve as the site of physical interaction between the AHL complex and transcription factors (Fig. 8). However, the exact biological importance of this domain with regard to physical interactions with transcription factors is still unknown.
Some members of the AHL gene family are negative regulators of hypocotyl elongation in the light (16, 17) (Fig. 2D). In contrast, we identified AHL-interacting transcription factors (ATAF2 and TCPs) that are positive regulators of this event (37, 38) (Fig. 7). In the proposed molecular model, the hypocotyl growth-promoting function of associated transcription factors is rendered nonfunctional by their physical interaction in the DNA-AHL-TF complex. For example, the overexpression of WT AHLs (e.g., SOB3-D, SOB3ox, and ESCox) leads to an increased accumulation of DNA-AHL-TF complexes resulting in a short hypocotyl phenotype (16) (Fig. 2). When the relevant AHL genes are knocked out in higher-order combinations (e.g., sob3-4 esc-8 ahl6 ahl22), or lose their ability to bind DNA (e.g., sob3-6 and esc-11), this leads to a decreased accumulation of DNA-AHL-TF complexes resulting in a long hypocotyl phenotype (16) (Fig. 2 A and C), possibly by releasing the transcription factors from the complex and allowing them to promote hypocotyl elongation.
Similarly, mutating the conserved Gly-Arg-Phe-Glu-Ile-Leu region of the PPC/DUF296 domain (e.g., SOB3-ΔG and SOB3-GTA) disrupts interactions with certain transcription factors and also leads to a decreased accumulation of DNA-AHL-TF complexes resulting in a long hypocotyl phenotype via growth promotion from the free transcription factors (Fig. 5E and Fig. S7D). The jaw-D sob3-6 double mutant demonstrates the importance of some TCP transcription factors in contributing to the long-hypocotyl phenotype conferred by the sob3-6 disruption of the DNA-AHL-TF complexes (Fig. 7A), presumably due to the lack of free TCPs for promoting hypocotyl elongation. Hence, the sob3-6 phenotype is gone and the double mutant exhibits a similar hypocotyl length as observed in the jaw-D single mutant.
How exactly the physical interaction with the DNA-AHL complex impairs the functions of their associated transcription factors remains to be explored. It is possible that the association with DNA-AHL complex interferes with the normal binding of transcription factors to the promoter regions of the genes that they regulate, subsequently impairing their biological functions. It is also possible that the association with the DNA-AHL complex alters posttranslational modifications associated with the transcription factors, in turn rendering the associated transcription factors nonfunctional. On the other hand, in the sob3-6 and esc-11 dominant-negative lines, the abolishment of DNA-binding ability for the AHL complex possibly destabilizes the association between AHLs and transcription factors; therefore, transcription factors that would normally be part of the DNA-AHL-TF complexes are released and function to promote hypocotyl growth in the light.
Through a combination of overexpression and loss-of-function analysis, we determined that multiple members of the AHL gene family suppress hypocotyl elongation redundantly in light-grown seedlings. The physical interactions among the AHL proteins via the PPC/DUF296 domain, as well as their interactions with other common nuclear proteins, such as transcription factors, may be important for their biological functions. However, biological functions of each AHL gene in the Arabidopsis genome still need to be determined. Multiple lines of evidence suggest that functional similarities exist between the HMGA and AHL proteins in that they both form nuclear macromolecular complexes on AT-rich chromatin regions. Further experiments should be conducted to reveal binding sites of AHL proteins in the Arabidopsis genome. The biological importance of the protein-protein interactions between the AHL complex and other nuclear proteins also needs to be further determined.
Materials and Methods
Detailed descriptions of plant materials, seedling growth conditions and hypocotyl measurement, plasmid construction, phylogenetic analysis, protein expression and purification, EMSA, bimolecular fluorescent complementation assay, and yeast two-hybrid assays are provided in SI Materials and Methods. Briefly, all Arabidopsis lines are in Columbia background. Sequences of the AHL genes and the transfer DNA insertion lines were obtained from the Arabidopsis Biological Research Center. Surface-sterilized Arabidopsis seeds were sown on media containing 1.0% (wt/vol) phytagel, 1.5% (wt/vol) sucrose, and 0.5× Linsmaier and Skoog–modified basal medium and incubated for 5 d at 25 °C in continuous white light under the indicated intensity. EMSA was performed using the LightShift Chemiluminescent EMSA Kit (Thermo Scientific).
Supplementary Material
Acknowledgments
We thank Dr. Raymond Reeves (Washington State University) and the members in the M.M.N. laboratory for comments on the manuscript. We thank the Franceschi Microscopy and Imaging Center at Washington State University for technical support, Drs. Hanjo Hellmann and Sutton Mooney (Washington State University) for the Y2H plasmids and Y2H library, Dr. Ian Street (Dartmouth College) for developing the SOB3/AHL29 antibody used in this study, and Dr. Amit Dhingra (Washington State University) for using the PDS-1000/He system (Bio-Rad). This work was supported by Chemical Sciences, Geosciences and Biosciences Division, Office of Basic Energy Sciences, Office of Sciences, US Department of Energy Grant DE-PS02-09ER09-02 (to M.M.N.). Analysis of ATAF2 was supported by the United States National Science Foundation, Division of Integrative Organismal Systems Grant 0758411 (to M.M.N.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1219277110/-/DCSupplemental.
References
- 1.Lynch M, Conery JS. The origins of genome complexity. Science. 2003;302(5649):1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- 2.Bowman JL, Floyd SK. The ancestral developmental tool kit of land plants. Int J Plant Sci. 2007;168(1):1–35. [Google Scholar]
- 3.Bowman JL, Floyd SK, Sakakibara K. Green genes-comparative genomics of the green branch of life. Cell. 2007;129(2):229–234. doi: 10.1016/j.cell.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 4. Arabidopsis Interactome Mapping C; Arabidopsis Interactome Mapping Consortium (2011) Evidence for network evolution in an Arabidopsis interactome map. Science 333(6042):601–607. [DOI] [PMC free article] [PubMed]
- 5.Rensing SA, et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science. 2008;319(5859):64–69. doi: 10.1126/science.1150646. [DOI] [PubMed] [Google Scholar]
- 6.Arabidopsis Genome Initiative Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408(6814):796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 7.Yu J, et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296(5565):79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- 8.Goff SA, et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica) Science. 2002;296(5565):92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- 9.Paterson AH, et al. The Sorghum bicolor genome and the diversification of grasses. Nature. 2009;457(7229):551–556. doi: 10.1038/nature07723. [DOI] [PubMed] [Google Scholar]
- 10.Tuskan GA, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313(5793):1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- 11.Aravind L, Landsman D. AT-hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids Res. 1998;26(19):4413–4421. doi: 10.1093/nar/26.19.4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huth JR, et al. The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif. Nat Struct Biol. 1997;4(8):657–665. doi: 10.1038/nsb0897-657. [DOI] [PubMed] [Google Scholar]
- 13.Fujimoto S, et al. Identification of a novel plant MAR DNA binding protein localized on chromosomal surfaces. Plant Mol Biol. 2004;56(2):225–239. doi: 10.1007/s11103-004-3249-5. [DOI] [PubMed] [Google Scholar]
- 14.Lin LY, et al. Crystal structure of Pyrococcus horikoshii PPC protein at 1.60 A resolution. Proteins. 2007;67(2):505–507. doi: 10.1002/prot.21270. [DOI] [PubMed] [Google Scholar]
- 15.Lin LY, et al. Crystallization and preliminary X-ray crystallographic analysis of a conserved domain in plants and prokaryotes from Pyrococcus horikoshii OT3. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2005;61(Pt 4):414–416. doi: 10.1107/S1744309105007815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Street IH, Shah PK, Smith AM, Avery N, Neff MM. The AT-hook-containing proteins SOB3/AHL29 and ESC/AHL27 are negative modulators of hypocotyl growth in Arabidopsis. Plant J. 2008;54(1):1–14. doi: 10.1111/j.1365-313X.2007.03393.x. [DOI] [PubMed] [Google Scholar]
- 17.Xiao C, Chen F, Yu X, Lin C, Fu YF. Over-expression of an AT-hook gene, AHL22, delays flowering and inhibits the elongation of the hypocotyl in Arabidopsis thaliana. Plant Mol Biol. 2009;71(1-2):39–50. doi: 10.1007/s11103-009-9507-9. [DOI] [PubMed] [Google Scholar]
- 18.Yun J, Kim YS, Jung JH, Seo PJ, Park CM. The AT-hook motif-containing protein AHL22 regulates flowering initiation by modifying FLOWERING LOCUS T chromatin in Arabidopsis. J Biol Chem. 2012;287(19):15307–15316. doi: 10.1074/jbc.M111.318477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lim PO, et al. Overexpression of a chromatin architecture-controlling AT-hook protein extends leaf longevity and increases the post-harvest storage life of plants. Plant J. 2007;52(6):1140–1153. doi: 10.1111/j.1365-313X.2007.03317.x. [DOI] [PubMed] [Google Scholar]
- 20.Ng KH, Yu H, Ito T. AGAMOUS controls GIANT KILLER, a multifunctional chromatin modifier in reproductive organ patterning and differentiation. PLoS Biol. 2009;7(11):e1000251. doi: 10.1371/journal.pbio.1000251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou J, Wang X, Lee JY, Lee JY. Cell-to-cell movement of two interacting AT-hook factors in Arabidopsis root vascular tissue patterning. Plant Cell. 2013;25(1):187–201. doi: 10.1105/tpc.112.102210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jiang C (2004) US Patent 6,717,034 B2.
- 23.Matsushita A, Furumoto T, Ishida S, Takahashi Y. AGF1, an AT-hook protein, is necessary for the negative feedback of AtGA3ox1 encoding GA 3-oxidase. Plant Physiol. 2007;143(3):1152–1162. doi: 10.1104/pp.106.093542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vom Endt D, Soares e Silva M, Kijne JW, Pasquali G, Memelink J. Identification of a bipartite jasmonate-responsive promoter element in the Catharanthus roseus ORCA3 transcription factor gene that interacts specifically with AT-Hook DNA-binding proteins. Plant Physiol. 2007;144(3):1680–1689. doi: 10.1104/pp.107.096115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rashotte AM, Carson SD, To JP, Kieber JJ. Expression profiling of cytokinin action in Arabidopsis. Plant Physiol. 2003;132(4):1998–2011. doi: 10.1104/pp.103.021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu H, Zou Y, Feng N. Overexpression of AHL20 negatively regulates defenses in Arabidopsis. J Integr Plant Biol. 2010;52(9):801–808. doi: 10.1111/j.1744-7909.2010.00969.x. [DOI] [PubMed] [Google Scholar]
- 27.Gallavotti A, et al. BARREN STALK FASTIGIATE1 is an AT-hook protein required for the formation of maize ears. Plant Cell. 2011;23(5):1756–1771. doi: 10.1105/tpc.111.084590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin Y, et al. An AT-hook gene is required for palea formation and floral organ number control in rice. Dev Biol. 2011;359(2):277–288. doi: 10.1016/j.ydbio.2011.08.023. [DOI] [PubMed] [Google Scholar]
- 29.Obayashi T, Hayashi S, Saeki M, Ohta H, Kinoshita K. ATTED-II provides coexpressed gene networks for Arabidopsis. Nucleic Acids Res. 2009;37(Database issue):D987–D991. doi: 10.1093/nar/gkn807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Obayashi T, Nishida K, Kasahara K, Kinoshita K. ATTED-II updates: Condition-specific gene coexpression to extend coexpression analyses and applications to a broad range of flowering plants. Plant Cell Physiol. 2011;52(2):213–219. doi: 10.1093/pcp/pcq203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gordon BR, et al. Structural basis for recognition of AT-rich DNA by unrelated xenogeneic silencing proteins. Proc Natl Acad Sci USA. 2011;108(26):10690–10695. doi: 10.1073/pnas.1102544108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Himes SR, et al. The role of high-mobility group I(Y) proteins in expression of IL-2 and T cell proliferation. J Immunol. 2000;164(6):3157–3168. doi: 10.4049/jimmunol.164.6.3157. [DOI] [PubMed] [Google Scholar]
- 33.Citovsky V, Gafni Y, Tzfira T. Localizing protein-protein interactions by bimolecular fluorescence complementation in planta. Methods. 2008;45(3):196–206. doi: 10.1016/j.ymeth.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 34.Citovsky V, et al. Subcellular localization of interacting proteins by bimolecular fluorescence complementation in planta. J Mol Biol. 2006;362(5):1120–1131. doi: 10.1016/j.jmb.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 35.Suzuki T, Sakurai K, Ueguchi C, Mizuno T. Two types of putative nuclear factors that physically interact with histidine-containing phosphotransfer (Hpt) domains, signaling mediators in His-to-Asp phosphorelay, in Arabidopsis thaliana. Plant Cell Physiol. 2001;42(1):37–45. doi: 10.1093/pcp/pce011. [DOI] [PubMed] [Google Scholar]
- 36.Baba K, Nakano T, Yamagishi K, Yoshida S. Involvement of a nuclear-encoded basic helix-loop-helix protein in transcription of the light-responsive promoter of psbD. Plant Physiol. 2001;125(2):595–603. doi: 10.1104/pp.125.2.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palatnik JF, et al. Control of leaf morphogenesis by microRNAs. Nature. 2003;425(6955):257–263. doi: 10.1038/nature01958. [DOI] [PubMed] [Google Scholar]
- 38.Schommer C, et al. Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 2008;6(9):e230. doi: 10.1371/journal.pbio.0060230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen IC, Lee SC, Pan SM, Hsieh HL. GASA4, a GA-stimulated gene, participates in light signaling in Arabidopsis. Plant Sci. 2007;172(6):1062–1071. [Google Scholar]
- 40.Till BJ, Zerr T, Comai L, Henikoff S. A protocol for TILLING and Ecotilling in plants and animals. Nat Protoc. 2006;1(5):2465–2477. doi: 10.1038/nprot.2006.329. [DOI] [PubMed] [Google Scholar]
- 41.Christian M, et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186(2):757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feng Z, et al. Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 2013;23(10):1229–1232. doi: 10.1038/cr.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmutz J, et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463(7278):178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- 44.Wang X, et al. Brassica rapa Genome Sequencing Project Consortium The genome of the mesopolyploid crop species Brassica rapa. Nat Genet. 2011;43(10):1035–1039. doi: 10.1038/ng.919. [DOI] [PubMed] [Google Scholar]
- 45.Reeves R. HMGA proteins: Isolation, biochemical modifications, and nucleosome interactions. Methods Enzymol. 2004;375:297–322. doi: 10.1016/s0076-6879(03)75020-4. [DOI] [PubMed] [Google Scholar]
- 46.Dragan AI, Liggins JR, Crane-Robinson C, Privalov PL. The energetics of specific binding of AT-hooks from HMGA1 to target DNA. J Mol Biol. 2003;327(2):393–411. doi: 10.1016/s0022-2836(03)00050-0. [DOI] [PubMed] [Google Scholar]
- 47.Panne D, Maniatis T, Harrison SC. An atomic model of the interferon-beta enhanceosome. Cell. 2007;129(6):1111–1123. doi: 10.1016/j.cell.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lomvardas S, Thanos D. Modifying gene expression programs by altering core promoter chromatin architecture. Cell. 2002;110(2):261–271. doi: 10.1016/s0092-8674(02)00822-x. [DOI] [PubMed] [Google Scholar]
- 49.Martinez Hoyos J, et al. Identification of the genes up- and down-regulated by the high mobility group A1 (HMGA1) proteins: Tissue specificity of the HMGA1-dependent gene regulation. Cancer Res. 2004;64(16):5728–5735. doi: 10.1158/0008-5472.CAN-04-1410. [DOI] [PubMed] [Google Scholar]
- 50.Dragan AI, Carrillo R, Gerasimova TI, Privalov PL. Assembling the human IFN-beta enhanceosome in solution. J Mol Biol. 2008;384(2):335–348. doi: 10.1016/j.jmb.2008.09.015. [DOI] [PubMed] [Google Scholar]
- 51.Fonfría-Subirós E, et al. Crystal structure of a complex of DNA with one AT-hook of HMGA1. PLoS ONE. 2012;7(5):e37120. doi: 10.1371/journal.pone.0037120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.