Significance
An endoplasmic reticulum stress- and osmotic stress-induced cell death pathway has emerged as a relevant adaptive response of plant cells to multiple environmental stimuli. We identified a unique component of this integrated circuit of stress-induced cell death, the GmNAC30 transcriptional factor, which binds to GmNAC81 in the nucleus of plant cells to coordinately regulate the expression of the vacuolar processing enzyme, a plant-specific executioner of cell death. In addition to describing a plant-specific endoplasmic reticulum stress cell death response that communicates with other environmental stimuli, the study deciphered the regulation of vacuolar processing enzyme expression that has been shown to be involved in several events of cell death in plants.
Keywords: NAC transcriptional factors, cell death signaling, abiotic stresses, ER stress
Abstract
Prolonged endoplasmic reticulum and osmotic stress synergistically activate the stress-induced N-rich protein-mediated signaling that transduces a cell death signal by inducing GmNAC81 (GmNAC6) in soybean. To identify novel regulators of the stress-induced programmed cell death (PCD) response, we screened a two-hybrid library for partners of GmNAC81. We discovered another member of the NAC (NAM-ATAF1,2-CUC2) family, GmNAC30, which binds to GmNAC81 in the nucleus of plant cells to coordinately regulate common target promoters that harbor the core cis-regulatory element TGTG[TGC]. We found that GmNAC81 and GmNAC30 can function either as transcriptional repressors or activators and cooperate to enhance the transcriptional regulation of common target promoters, suggesting that heterodimerization may be required for the full regulation of gene expression. Accordingly, GmNAC81 and GmNAC30 display overlapping expression profiles in response to multiple environmental and developmental stimuli. Consistent with a role in PCD, GmNAC81 and GmNAC30 bind in vivo to and transactivate hydrolytic enzyme promoters in soybean protoplasts. A GmNAC81/GmNAC30 binding site is located in the promoter of the caspase-1–like vacuolar processing enzyme (VPE) gene, which is involved in PCD in plants. We demonstrated that the expression of GmNAC81 and GmNAC30 fully transactivates the VPE gene in soybean protoplasts and that this transactivation was associated with an increase in caspase-1–like activity. Collectively, our results indicate that the stress-induced GmNAC30 cooperates with GmNAC81 to activate PCD through the induction of the cell death executioner VPE.
The NAC (NAM-ATAF1,2-CUC2) domain-containing proteins constitute a large family of plant-specific transcription factors (TF) that is represented by at least 151 members in Oryza sativa, 126 members in Arabidopsis thaliana, and 153 members in Glycine max (1, 2). The NAC proteins are highly conserved in their N termini, which bind specifically to target DNA through a unique type of TF fold consisting of a twisted six-stranded β-sheet that is surrounded by a few helical elements (3). Their C-terminal regions are divergent in sequence and length and function as activators or repressors of transcription (4, 5). Biological functions involved in development and in the stress response have been assigned to the members of this family (6, 7). A growing body of evidence has demonstrated a pivotal role for the NAC genes in the regulation of developmental programmed leaf senescence and programmed cell death. Several NAC genes from different species have been shown to be highly expressed during leaf senescence and to play a pivotal role in leaf senescence (8). In soybean, at least three NAC genes are associated with senescence (9). A subset of these senescence-induced GmNAC has emerged as regulators of stress-induced senescence and cell death (9, 10). Among these genes, the soybean GmNAC6 [Glyma12g02540.1, recently designated GmNAC081 (1)] potentially integrates multiple stress signaling pathways into a programmed cell death (PCD) response (10).
GmNAC6, hereafter designated GmNAC81 for standardization with the literature (1), has been identified as a component of the endoplasmic reticulum (ER) stress- and osmotic stress-induced cell death response, which is mediated by the N-rich proteins (NRP) in soybean, which harbors a development and cell death domain (DCD) at the C terminus (10, 11). The NRP/DCD-mediated signaling pathway integrates a cell death signal generated from prolonged ER and osmotic stress into a synergistic and convergent response (11, 12). The current model for this integrative pathway states that, under stress conditions, GmERD15, an ER stress- and osmotic stress-induced transcriptional activator, up-regulates NRP/DCD expression, which in turn induces GmNAC81 to promote a cell death response resembling a PCD event (10, 11, 13). Accordingly, the overexpression of either NRP/DCD or GmNAC81 in soybean protoplasts induces caspase-3–like activity and promotes extensive DNA fragmentation. Furthermore, the transient expression of NRP/DCD or GmNAC81 (GmNAC6) in planta causes leaf yellowing, chlorophyll loss, malondialdehyde production, and the induction of senescence marker genes, which are hallmarks of leaf senescence.
As a branch of the ER stress response that connects with other environmentally induced responses, the stress-induced NRP/DCD-mediated cell death signaling pathway may allow for the versatile adaptation of cells to different stresses (14). Accordingly, the modulation of this pathway by the constitutive expression of the ER molecular chaperone binding protein (BiP) has been shown to promote a better adaptation of transgenic lines to drought (15, 16). Despite its conceptual relevance in plant adaptation to adverse conditions, several key players of this stress-induced signaling pathway are unknown, and the molecular events downstream of GmNAC81 remain to be determined. Here, we isolated an NAC domain-containing protein from soybean, GmNAC30, by its capacity to bind to GmNAC81 in yeast. We showed that GmNAC81 and GmNAC30 interact with each other in vitro and in vivo, bind to common cis-regulatory sequences in target promoters, and synergistically regulate these target genes. One such target gene, the vacuolar processing enzyme (VPE), may be responsible for the execution of the cell death program that is induced by ER and osmotic stress.
Results
Identification of GmNAC30, a Unique Soybean NAC Domain-Containing TF, as a GmNAC81-Specific Interactor.
To identify the potential targets of GmNAC81 in the N-rich–mediated cell death response, we screened a soybean cDNA library (13) with the full-length GmNAC81 ORF as bait. From ∼5 × 105 clones that were screened, three positive clones harbored cDNA fragments from the soybean gene GmNAC30 (previously designated GmNAC32 by ref. 17). We confirmed that GmNAC81 was able to interact with the full-length GmNAC30 protein in yeast (Fig. S1).
There are six predicted homologs of GmNAC30 in the soybean genome (76–63% sequence identity) that cluster together in a phylogenetic tree of soybean NAC domain-containing proteins (Fig. S1D) and are most closely related to the ATAF group of the NAC family from Arabidopsis (Fig. S1E). The deduced protein sequence of GmNAC30 displays a highly conserved NAC domain in the N terminus, which is divided into the five NAC subdomains (A–E) of conserved blocks (Fig. S1F) (18). Consistent with the presence of a predicted nuclear localization signal within the GmNAC30 D subdomain (Fig. S1F, in red), the fluorescence of a GmNAC30–GFP fusion protein was concentrated in the nucleus of electroporated soybean protoplasts that were also stained with the nuclear marker, DAPI (Fig. S2A). GmNAC81 has also been localized to the nucleus (9).
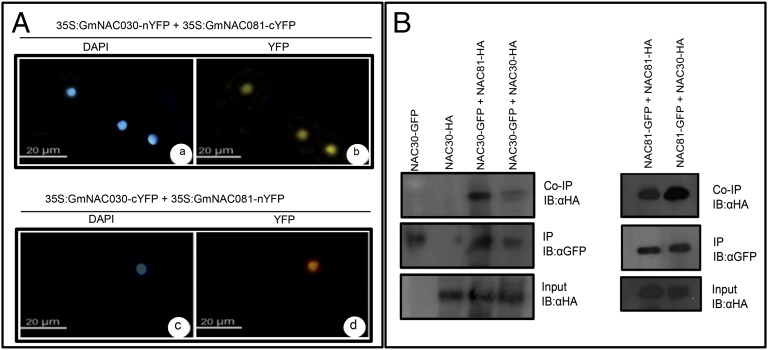
We next used the bimolecular fluorescence complementation (BiFC) assay to determine whether GmNAC81 and GmNAC30 would interact in the nucleus of plant cells. The complementation of the fluorescent YFP fragments that were mediated by the interaction between GmNAC30 and GmNAC81 was detected in the nuclei of transfected leaf protoplasts coincident with the DAPI signal (Fig. 1A, a and b). Additionally, the formation of a GmNAC81–GmNAC30 complex occurred in vivo independent of the orientation of the GmNAC81 or GmNAC30 fusions (N terminus or C terminus of YFP) (Fig. 1A, c and d), and the reconstituted fluorescent signal was much higher than the background levels (control panels with combination of the protein fusions with empty vectors) (Fig. S2D). The accumulation of the GmNAC81 and GmNAC30 transcripts in the protoplasts that were electroporated with different combinations of the constructs was confirmed by quantitative RT-PCR (qRT-PCR) (Fig. S2 E and F). Collectively, these results indicate that GmNAC81 interacts with GmNAC30 in the nucleus of plant cells.
Fig. 1.
GmNAC30 binds to GmNAC81 in the nucleus of plant cells. (A) The in vivo interaction between GmNAC81 and GmNAC30 by BiFC analysis. The fluorescence (YFP) images were taken from the soybean leaf protoplasts that coexpressed 35S:GmNAC30-nYFP + 35S:GmNAC81-cYFP and 35S:GmNAC30-cYFP + 35S:GmNAC81-nYFP fusion proteins 36 h after the electroporation of the protoplasts with the indicated DNA constructs. (Scale bars, 20 µm.) (B) Homo- and heterodimerization between GmNAC30 and GmNAC81. The immunoprecipitation of transiently expressed proteins was performed using anti-GFP matrix, and coimmunoprecipitated proteins were detected using anti-HA antibody. (Top) Immunoblots of coimmunoprecipitated GmNAC30-HA and GmNAC81-HA (Co-IP); (Middle) immunoprecipitated GmNAC30-GFP and GmNAC81-GFP (IP); (Bottom) total protein extracts (input HA) from transiently expressed proteins in N. benthamiana leaves. The expression levels of the input HA-fused proteins were assessed by anti-HA of crude extracts. These results were replicated twice from independent experiments.
To investigate whether GmNAC30 harbors a functional transactivator domain, the full-length GmNAC30 coding region was fused to the binding domain of GAL4 (BD-GAL4) and expressed as a fusion protein in a yeast strain harboring the reporter genes HIS3 and lacZ (β-galactosidase) under the control of the GAL1 promoter. We also included in the assay GmNAC35 (GmNAC2 in ref. 9) and NIG [an unrelated Arabidopsis protein that exhibits transactivation activity in yeast (19)] as positive controls and the full-length GmNAC81 as a negative control (9). The expression of BD-GAL4 fused to GmNAC30 promoted the growth of yeast in the absence of histidine and in the presence of 3-aminotriazole (Fig. S2B) and induced lacZ expression as measured by β-galactosidase activity (Fig. S2C). These results demonstrate that GmNAC30 exhibits transactivation activity in yeast and are consistent with the presence of the v-conserved transactivation motif EVQSDEPKW in its transcriptional activation region (TAR) (Fig. S1F). As previously demonstrated, the full-length GmNAC81 fused to BD-GAL4 was not able to activate histidine/adenine auxotrophy (Fig. S2B) or induce high levels of β-galactosidase expression (Fig. S2C); however, the expression of its carboxyl domain promoted the transactivation of the reporter genes in yeast (Fig. S2B). These results also confirmed previous observations that the N-terminal region of some NAC domain-containing proteins interferes negatively with the transactivation activity in their C terminus, because the deletion of their N terminus renders the BD-fused truncated protein a potent transactivator in yeast (20, 21). With such a potential conformational hindrance, GmNAC81 may depend on specific interactions with other transfactors to activate their target genes as heterodimers, which favors the argument that the interaction of GmNAC81 and GmNAC30 may be biologically relevant.
As a GmNAC81-Specific Partner, GmNAC30 May Act as a Downstream Target of the Osmotic Stress- and ER Stress-Induced N-Rich–Mediated Cell Death Signaling Pathway.
The NRP-mediated signaling pathway has been shown to integrate and transduce a cell death signal that was derived from prolonged ER and osmotic stress (14). Upon activation, NRPs trigger a signaling cascade that culminates with the enhanced expression of GmNAC81 to promote a programmed cell death response (10). As a putative downstream component of the ER stress- and osmotic stress-integrating signaling pathway, we compared the expression of GmNAC30 and GmNAC81 under conditions that activate this specific cell death program (Fig. 2). We also monitored the expression of NRP-B, another member of the ER stress- and osmotic stress-induced cell death signaling pathway, as additional evidence for the activation of the cell death program. Controls for the effectiveness of the osmotic stress treatment, such as the seed maturation protein (SMP), and the ER stress treatment, such as calnexin and BiP, were also included in the assay. As with the transcripts of GmNAC81 and NRP-B, the treatment of soybean seedlings with the osmotic stress-inducer polyethylene glycol (PEG) and the ER stress-inducer tunicamycin (which blocks protein glycosylation in the organelle) induced the accumulation of the GmNAC30 transcript (Fig. 2 A and B). Similarly, the treatment of soybean cells with cycloheximide, a potent inducer of cell death in soybean protoplasts, strongly up-regulated GmNAC30 gene expression, indicating that GmNAC30 may be involved in the cell death events (Fig. 2C) (10, 11). This interpretation was further confirmed by applying the TUNEL assay for the in situ detection of DNA fragmentation in the GmNAC30-expressing protoplasts (Fig. 2E). The extensive cleavage of nuclear DNA into oligonucleosome-sized fragments is one feature of active cell death (PCD). The nuclei of the control cells that were transformed with the empty vector fluoresced intensely with DAPI and exhibited only TUNEL-negative nuclei. In contrast, the GmNAC30-expressing samples had TUNEL-positive nuclei that showed the same degree of staining as the DNase-treated positive controls and the GmNAC81- or NRP-B–expressing protoplasts (Fig. S3). These results suggest that GmNAC30 promotes cell death when it is transiently expressed in protoplasts and displays a pattern of expression similar to that of GmNAC81 and NRPs, which are components of the osmotic stress- and ER stress-induced cell death signaling pathway. We have recently demonstrated that the transient expression of NRP-A or NRP-B in leaf protoplasts activates the GmNAC81 promoter and induces the accumulation of GmNAC81 transcripts (10). Here we showed that the overexpression of GmNRP-A or GmNRP-B in leaf protoplasts also caused a significant increase in GmNAC30 transcript accumulation (Fig. 2D). Collectively, these results favor the argument that GmNAC30, as a GmNAC81 partner, acts downstream of the NRPs in the stress-induced cell death signaling pathway.
Fig. 2.
GmNAC30 may be a downstream component of the ER stress- and osmotic stress-induced NRP-mediated cell death signaling pathway. (A–C) GmNAC30 is induced by osmotic stress, ER stress, and cell death inducers. The soybean seedlings were treated with the osmotic stress-inducer PEG for 10 h (A), the ER stress-inducer tunicamycin for 16 h (B), or the cell death-inducer cycloheximide (C) for 6 h and 12 h, and the expression of the indicated genes was monitored by qRT-PCR. The gene expression was calculated using the 2−ΔCT method and the RNA helicase as an endogenous control. GmSMP is a seed-maturation protein that was used as a control for the PEG treatment. GmCNX (calnexin) and GmBiPD (binding protein) are ER markers. GmNAC81 and GmNRP-B are components of the ER stress- and osmotic stress-integrating cell death pathway. The values are given as the mean ± confidence interval of three independent experiments. The asterisks indicate significant differences from the controls by the t test at P ≤ 0.05. (D) GmNRP-A and GmNRP-B induce the expression of the GmNAC30 gene. The plasmids containing the GmNRP-A and GmNRP-B expression cassettes or the empty vector were electroporated into the soybean protoplasts, and the gene expression of the GmNAC genes was monitored by qRT-PCR as in A–C. (E) DNA fragmentation was promoted by GmNAC30 expression. The cells were sampled 36 h postelectroporation of the soybean protoplasts with the empty vector and GmNAC30 expression cassettes, and submitted to TUNEL labeling. As a positive control, the untransfected cells were also treated with DNase. The nuclei were stained with DAPI. The magnification was 1.3-fold higher than in Fig. 1A.
GmNAC81 and GmNAC30 Bind in Vivo to Common Target Promoters and Regulate the Expression of the Target Genes.
To identify the potential target promoters of GmNAC30 and GmNAC81, we used a small-scale ChIP sequencing assay in which the precipitated DNA from individual GmNAC81- or GmNAC30-expressing tissues were cloned and sequenced. From 25 sequences targeted by each TF, we found 17 common sequences that mapped to the 5′ transcriptional regulatory regions of soybean genes (Table S1). The expression of these genes was monitored by qRT-PCR in soybean protoplasts that were electroporated with 35S:GmNAC30 or 35S:GmNAC81. Both GmNAC30 and GmNAC81 were efficiently expressed in the transfected soybean protoplasts (Fig. S4 A and B). The transient expression of either GmNAC81 or GmNAC30 promoted a significant induction of genes encoding a dual specificity protein phosphatase, a predicted endo-1,3-β-glucanase, a d-alanyl-d-alanine carboxypeptidase, and a Cdc2-related protein kinase, in addition to inducing Gm20g22140 expression (Fig. S4 C–G), but did not alter the expression of the negative control gene AT5G05800 (Fig. S4H). The expression of GmNAC81 and GmNAC30 down-regulated the expression of another subset of the selected genes (Fig. S5), indicating that GmNAC81 and GmNAC30 may coordinately function as both activators and repressors of gene expression in soybean.
GmNAC81 and GmNAC30 Are Required for the Full Transactivation or Repression of Target Promoters and Bind to the Common Consensus Sequence Element TGTGT[T/C/G].
To provide further evidence for the regulation of these target genes by GmNAC81 or GmNAC30, we performed a GUS (glucuronidase) transactivation assay in soybean protoplasts using the 1- to 2-kb 5′ flanking sequences of an up-regulated (d-alanyl-d-alanine carboxypeptidase) gene and a down-regulated [-aminocyclopropane-1-carboxylase synthase (ACC) synthase] gene fused to the GUS reporter (Fig. 3). The accumulation of the GmNAC81 and GmNAC30 transcripts in the electroporated protoplasts was confirmed by qRT-PCR (Fig. S4I). The results of transactivation or repression are shown by β-galactosidase activity (Fig. 3 A and B), as well as by the quantitation of the reporter GUS transcript accumulation (Fig. 3C). Consistent with the gene-expression profile, GmNAC81 and GmNAC30 specifically activated and repressed the ACPase (carboxypeptidase) promoter and ACC promoter, respectively. The expression of an unrelated TF from the MyB family, At5G05800, did not target either NAC-regulated promoter. The cotransfection of the soybean protoplasts with both GmNAC81 and GmNAC30 promoted an enhanced activation of the ACPase promoter (Fig. 3 A and C), as well as an increased repression of the ACC promoter (Fig. 3 B and C) compared with the regulation of the gene reporters by the individual expression of the transfactors. Collectively, these results indicate that GmNAC81 and GmNAC30 function as transcriptional activators or repressors of common target genes that require both transfactors for full regulation. Therefore, GmNAC81 and GmNAC30 may form a heterodimer to coordinately regulate common target promoters.
Fig. 3.
GmNAC81 and GmNAC30 determine the full activation or repression of target promoters and bind specifically to the core sequence TGTGTT in vitro. (A) GmNAC30 and GmNAC81 repress an ACC synthase promoter. The soybean protoplasts were coelectroporated with plasmids carrying the 1000pACCpro:β-GUS gene and either the 35S:GmNAC81 or 35S:GmNAC30 DNA constructs or a combination of both DNA constructs. After 48 h, the β-GUS activity (nmol⋅min⋅mg protein) was measured from the total protein extracts of the transfected soybean cells. An unrelated MyB transfactor, 35S:At5g05800, was used as a negative control. The error bars represent the confidence interval (α = 0.05) of three biological replicates. (B) The full activation of the carboxypeptidase (ACPase) promoter requires both GmNAC81 and GmNAC30. The soybean protoplasts were coelectroporated with plasmids carrying the 2000pACPasepro:β-GUS gene and the same combinations of DNA constructs, as described in A, and the samples were processed for β-GUS activity, as in A. (C) Quantitation of the GUS reporter gene expression by qRT-PCR. The soybean protoplasts were electroporated with the same DNA constructs as described in A and B, and the GUS transcript levels were monitored by qRT-PCR. The relative quantitation as demonstrated by a log2 scale of gene expression was calculated using the 2−ΔΔCt method and helicase as an endogenous control. The values are relative to the control treatment (empty vector), and the error bars represent the confidence interval (α = 0.05) of three biological replicates. An unrelated MyB transfactor, 35S::At5g05800, was used as a negative control. (D) The specific binding of GmNAC81 and GmNAC30 to their core DNA binding sites. An 18-bp biotin-labeled fragment harboring the directly repeated core sequence TGTGTT was incubated with an E. coli-produced and purified GST, GST-tagged GmNAC30 (N30), GST-GmNAC81 (N81), and both NAC proteins (N30+N81) for 20 min at room temperature in the absence and presence of a 100-fold molar excess of unlabeled probe, as indicated in the figure. The products were separated by electrophoresis in a 4% (wt/vol) polyacrylamide gel in TB buffer. The arrow indicates the DNA:protein complexes.
Our data suggest that GmNAC81 and GmNAC30 may form homo- or heterodimers to regulate target promoters in planta. Nevertheless, in yeast the transactivation activity of full-length GmNAC81 was impaired, suggesting that GmNAC81 activity may require heterodimerization with GmNAC30. In fact, expression of GmNAC81 or GmNAC30 separately in soybean protoplasts led to the induction of the other TF partner providing the means for heterodimerization (Fig. S4J). To examine this hypothesis further, we transiently expressed GmNAC30 and GmNAC81 in Nicotiana benthamiana leaves and checked all possible combinations by coimmunoprecipitation assays (Fig. 1B). These TFs could form homo- and heterodimers in vivo, but nevertheless the GmNAC30/GmNAC81 complex signal was stronger, suggesting a higher affinity for the heterodimer interactions. These results complemented our promoter transactivation assay data, which revealed a stronger NAC-mediated target promoter regulation when both TFs were provided in trans (Fig. 3).
The sequences that were coimmunoprecipitated by the ChIP assay share the consensus sequence TGTGTT, which was found as a derivative form (TGTGT[T/G/C]) in GmNAC81- and GmNAC30-regulated promoters. Except for the serine/threonine protein kinase and nonspecific serine/threonine protein kinase promoters, which do not harbor a derivative consensus sequence, the core consensus sequence was found in all other common target genes within 1- to 2-kb 5′ flanking sequences (Table S1). The relative proximity of the putative cis-element to the promoter core of the NAC-regulated genes suggests that the consensus sequence functions as a specific binding site for GmNAC81 and GmNAC30. To confirm this hypothesis, we examined whether Escherichia coli-produced and -purified GmNAC81 and GmNAC30 were able to bind to the consensus sequence in vitro (Fig. 3D). Both purified GST-GmNAC81 and GST-GmNAC30 bound to the labeled probe harboring directly repeated TGTGTT core sequences, but not to a mutated sequence in which both Gs were replaced by Cs. A GST-purified protein did not bind to the core consensus sequence, confirming that the in vitro DNA:protein complex formation was caused by interactions between the NAC TFs and the core consensus sequence. These interactions were specific because a 100-fold molar excess of unlabeled probe efficiently competed for the binding. Collectively, these results demonstrate that both GmNAC81 and GmNAC30 bind specifically to the core consensus sequence TGTGTT.
GmNAC81 and GmNAC30 Transactivate the Expression of the VPE.
The repertoire of genes that are up-regulated by GmNAC81 and GmNAC30 and were detected by the ChIP assay includes predominantly hydrolytic enzymes (carboxypeptidase, phosphatase, and glucanase) that may be involved in plant PCD. A plant-specific strategy for PCD consists of the up-regulation of a variety of vacuolar hydrolytic enzymes that may be released into the cytosol to hydrolyze organelles and nuclear DNA, leading to cell death (22). The VPE has been shown to trigger vacuolar collapse-mediated PCD in pathogenesis and development (23, 24). The VPE family in soybean is represented by five homologs. Except for the Glyma17g34900 promoter, which does not harbor a GmNAC81/GmNAC30 binding site, the remaining VPE promoters contain one to three copies of the cis-regulatory element. These observations prompted us to examine whether GmNAC81 and GmNAC30 could control VPE expression in soybean protoplasts. We first examined whether GmNAC81 and GmNAC30 bind to a VPE promoter (Glyma14g10620) in vivo. We performed ChIP experiments in which a TGTGTT-containing fragment of the VPE promoter was amplified from the precipitated DNA of individual GmNAC81- or GmNAC30-expressing tissues, but not from the precipitated DNA of GFP-expressing tissues (Fig. 4A). Then, we used a luciferase transactivation assay in agroinfiltrated tobacco leaves and we showed that GmNAC81 and GmNAC30 transactivated the VPE promoter fused to the reporter luciferase (Fig. 4B). Furthermore, the ectopic expression of either GmNAC81 or GmNAC30 strongly induced the expression of the VPE gene Glyma14g10620 (Fig. 4C) and the up-regulation of the soybean VPE homolog was associated with a 2.5- to 3.0-fold higher expression of caspase-1 activity (Fig. 4D). The inclusion of a caspase-1–specific inhibitor reduced the enzyme activity to basal levels. We also demonstrated that GmNAC81 overexpression in three independently transformed soybean lines (Fig. S6A) led to induction of GmNAC30 (Fig. S6B) and was associated with an induction of VPE expression (Fig. S6C) and an increase in caspase-1 activity (Fig. S6D). Given that VPE is a key executioner of cell death in plant cells, the presence of a GmNAC81/GmNAC30 binding site in the VPE promoter, along with the capacity of the TFs to directly transactivate its expression, suggest that GmNCA81/GmNAC30 and VPE constitute a regulatory cascade for stress-induced cell death in soybean.
Fig. 4.
GmNAC81 and GmNAC30 bind to VPE promoter in vivo, transactivate VPE expression and induce caspase-1–like activity. (A) GmanC81 and GmNAC30 bind to the VPE promoter in vivo. ChIP assay was performed with GmNAC30-GFP–, GmNAC81-GFP–, or GFP-expressing leaves using anti-GFP antibodies. The 103-bp fragment of VPE 5′ flanking region was detected by PCR amplification when DNA was immunoprecipitated from GmNAC30-GFP– or GmNAC81-GFP–expressing leaves but not from GFP-expressing leaves. “C” represents amplification of control plasmid. (Right) Amplifications from input total DNA. (B) GmNAC30 and GmNAC81 transactivate the VPE promoter. Tobacco leaves were agroinfiltrated with a construct harboring the 2000pVPE::Luciferase gene alone or in combination with 35S::GmNAC30 or 35S::GmNAC81. Luciferase activity was determined 36 h postagroinoculation. (C) GmNAC81 and GmNAC30 induce VPE expression in the soybean protoplasts. The transcript levels of VPE were quantified by qRT-PCR 36 h postelectroporation of the soybean protoplasts with GmNAC81, GmNAC30, or the empty vector. The gene expression was calculated using the 2−ΔCT method and RNA helicase as an endogenous control. The error bars represent the confidence interval (α = 0.05) of three biological replicates. (D) GmNAC30 and GmNAC81 induce caspase-1–like activity in the soybean protoplasts. Caspase-1–like activity was determined from GmNAC81- and GmNAC30-expressing protoplasts in the absence and presence of a specific competitor. The asterisks indicate significant differences by the t test at P ≤ 0.05 (n = 3).
Discussion
GmNAC30 as a Downstream Component of the NRP-Mediated Cell Death Signaling Pathway.
In this study, we characterized another member of the NAC domain-containing proteins of plant-specific transfactors, GmNAC30, as a possible component of the ER stress- and osmotic stress-induced NRP-mediated cell death signaling pathway. Our hypothesis that GmNAC30 functions in this stress-induced cell death-integrated circuit is supported by the following observations. First, GmNAC30 was identified by its capacity to bind to GmNAC81, a downstream component of the signaling pathway, in the nucleus of plant cells. Second, like GmNAC81, GmNAC30 is induced by ER and osmotic stress with similar kinetics, and is also induced by the ectopic expression of NRP-A and NRP-B. Third, like GmNAC81, GmNAC30 is up-regulated by the cell death-inducer cycloheximide and promotes cell death when it is transiently expressed in soybean protoplasts. Fourth, as evidenced by the ChIP assay, GmNAC30 and GmNAC81 were found to bind in vivo to common target promoters and regulate their expression in a coordinated manner. The target promoters were regulated by the individual expression of each TF, but they required both TFs for their full induction or repression. Finally, GmNAC81 and GmNAC30 are coordinately regulated by multiple environmental and developmental stimuli, as would be expected for TFs that act in concert as heterodimers to regulate gene expression.
GmNAC30 and GMNAC81 Bind in Vivo to Common Target Promoters and Function as Repressors or Activators of Gene Expression.
In general, NAC domain-containing proteins have been described as repressors or activators of gene expression (4, 5). For example, Arabidopsis ATAF1 and ATAF2 act as transcriptional repressors of genes involved in pathogenesis, whereas OsNAC6, a rice ortholog of ATAF1, is a positive contributor to disease resistance and salinity tolerance (25–27). Contrasting results have been reported for ATAF1 in drought tolerance, as it may function as a positive or negative regulator (27, 28). Our data revealed that GmNAC81 and GmNAC30 are capable of suppressing or activating transcription through the coordinated action on common target promoters. GmNAC81 and GmNAC30 specifically bind in vitro to the core DNA binding element TGTG[T/G/C] and in vivo to TGTG[T/G/C]-containing promoters. Furthermore, the GmNAC81- or GmNAC30-mediated transactivation or repression of the target promoters is enhanced when both TFs are provided in trans. These results indicate that these TFs may bind to their target promoters as heterodimers. Consistent with this interpretation, we showed that, although GmNAC81 and GmNAC30 could form homo- and heterodimers in vivo by coimmunoprecipitation, the immunosignals of the GmNAC30/GmNAC81 complex were stronger than the homodimer signals, suggesting more affinity and stability of heterodimers.
Further supporting this hypothesis, we demonstrated that GmNAC81 interacts with GmNAC30 in the nucleus of transfected cells. The heterodimerization of these TFs may explain their ability to repress or activate gene expression through the same core DNA binding sequence. It has been shown in a yeast one-hybrid assay that the conformational interactions between the N-terminal NAC domain harboring a NAC repression domain (NARD) and the C-terminal TAR of NAC proteins are crucial for their activating or repressing activities (5) (see also commentary to Fig. S2). The current model for the transcriptional repressing or activating function of NAC proteins holds that the strength of a NARD function over a TAR function, or vice versa, renders the NAC protein as a repressor or activator. According to this model, the capacity of GmNAC81 and GmNAC30 to function as transcriptional repressors or activators would depend on the conformational assembly of these TFs at their binding site. Different assemblies could be driven by the homo- or heterodimerization of GmNAC30 and GmNAC81, as well as by the promoter context that would provide different stabilization contacts for the DNA:protein interaction.
GmNAC30 Cooperates with GmNAC81 to Induce PCD via the Activation of VPE.
Since the discovery of the stress-induced NRP-mediated cell death signaling pathway, which integrates a PCD signal derived from prolonged ER and osmotic stress, the progress toward the elucidation of the components and activation mechanisms of the pathway has been limited (14). The stress-induced NRP-mediated cell death signaling pathway has been shown to induce a cell death response, with the hallmarks of leaf senescence and PCD (10, 11). Apart from the knowledge that NRPs induce the expression of GmNAC81, a regulator of the cell death response, the events downstream of GmNAC81 that could account for the execution of the cell death program are totally unknown. In the present investigation, in addition to describing GmNAC30 as a molecular partner of GmNAC81, we identified the downstream targets of this interaction. Significantly, we showed that GmNAC81 and GmNAC30 induce the expression of caspase-1–like VPE, underlying a mechanism for the execution of the ER stress- and osmotic-stress induced cell death program (Fig. S6E). In this model, prolonged ER and osmotic stress induce the expression of the transcriptional activator GmERD15 to target the NRP promoter. The up-regulation of NRPs leads to the induction of GmNAC81 and GmNAC30, which cooperate with each other to activate the VPE promoter and expression. VPE was originally identified as a vacuolar enzyme responsible for the maturation of seed-storage proteins and various vacuolar proteins (29). VPE was then associated with the Tobacco mosaic virus-induced hypersensitive cell death and, more recently, with developmental PCD (22, 23). VPE is a cysteine protease that exhibits caspase-1–like activity and cleaves a peptide bond at the C-terminal side of asparagine and aspartic acid (22). Because VPE acts as a processing enzyme to activate various vacuolar proteins, it has been proposed that VPE might also convert inactive hydrolytic enzymes to their active forms, which are involved in the disintegration of vacuoles, initiating the proteolytic cascade in plant PCD. As vacuole-triggered PCD is unique to plants, it is not surprising that a stress-induced plant-specific signal transduction pathway serves as a regulatory circuit leading to the activation of VPE.
Materials and Methods
A detailed description of materials and methods is provided in SI Materials and Methods.
Plasmid Construction, Yeast Two-Hybrid Screening, and Transactivation Assays in Yeast.
The recombinant plasmids were obtained through the GATEWAY system (Invitrogen Life Technologies). The two-hybrid screening and transactivation assays in yeast cells we performed as described previously (13).
Transient Expression in Protoplasts and qRT-PCR.
The protoplasts were prepared directly from soybean leaves and the transient expression assays were performed by the electroporation (250 V, 250 μF) as previously described (11). The subcellular localization of GFP fusions and BiFC assays were examined by confocal microscopy. See Table S2 for primers used.
ChIP Assay.
The ChIP assay was performed using a chromatin immunoprecipitation kit (Imprint ChIP kit; Sigma), according to the manufacturer’s instructions.
EMSA.
The EMSAs were performed as previously described (13).
Supplementary Material
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 19189.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1311729110/-/DCSupplemental.
References
- 1.Le DT, et al. Genome-wide survey and expression analysis of the plant-specific NAC transcription factor family in soybean during development and dehydration stress. DNA Res. 2011;18(4):263–276. doi: 10.1093/dnares/dsr015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nuruzzaman M, et al. Genome-wide analysis of NAC transcription factor family in rice. Gene. 2010;465(1–2):30–44. doi: 10.1016/j.gene.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 3.Chen Q, Wang Q, Xiong L, Lou Z. A structural view of the conserved domain of rice stress-responsive NAC1. Protein Cell. 2011;2(1):55–63. doi: 10.1007/s13238-011-1010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fujita M, et al. A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 2004;39(6):863–876. doi: 10.1111/j.1365-313X.2004.02171.x. [DOI] [PubMed] [Google Scholar]
- 5.Tran LS, et al. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell. 2004;16(9):2481–2498. doi: 10.1105/tpc.104.022699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakashima K, Takasaki H, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. NAC transcription factors in plant abiotic stress responses. Biochim Biophys Acta. 2012;1819(2):97–103. doi: 10.1016/j.bbagrm.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 7.Olsen AN, Ernst HA, Leggio LL, Skriver K. NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 2005;10(2):79–87. doi: 10.1016/j.tplants.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 8.Ricachenevsky FK, Menguer PK, Sperotto RA. kNACking on heaven’s door: How important are NAC transcription factors for leaf senescence and Fe/Zn remobilization to seeds? Front Plant Sci. 2013;4:226. doi: 10.3389/fpls.2013.00226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pinheiro GL, et al. Complete inventory of soybean NAC transcription factors: Sequence conservation and expression analysis uncover their distinct roles in stress response. Gene. 2009;444(1-2):10–23. doi: 10.1016/j.gene.2009.05.012. [DOI] [PubMed] [Google Scholar]
- 10.Faria JA, et al. The NAC domain-containing protein, GmNAC6, is a downstream component of the ER stress- and osmotic stress-induced NRP-mediated cell-death signaling pathway. BMC Plant Biol. 2011;11:129. doi: 10.1186/1471-2229-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costa MDL, et al. A new branch of endoplasmic reticulum stress signaling and the osmotic signal converge on plant-specific asparagine-rich proteins to promote cell death. J Biol Chem. 2008;283(29):20209–20219. doi: 10.1074/jbc.M802654200. [DOI] [PubMed] [Google Scholar]
- 12.Irsigler AS, et al. Expression profiling on soybean leaves reveals integration of ER- and osmotic-stress pathways. BMC Genomics. 2007;8:431. doi: 10.1186/1471-2164-8-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alves MS, et al. A novel transcription factor, ERD15 (Early Responsive to Dehydration 15), connects endoplasmic reticulum stress with an osmotic stress-induced cell death signal. J Biol Chem. 2011;286(22):20020–20030. doi: 10.1074/jbc.M111.233494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reis PAB, Fontes EPB. N-rich protein (NRP)-mediated cell death signaling: A new branch of the ER stress response with implications for plant biotechnology. Plant Signal Behav. 2012;7(6):628–632. doi: 10.4161/psb.20111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reis PAB, et al. The binding protein BiP attenuates stress-induced cell death in soybean via modulation of the N-rich protein-mediated signaling pathway. Plant Physiol. 2011;157(4):1853–1865. doi: 10.1104/pp.111.179697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valente MA, et al. The ER luminal binding protein (BiP) mediates an increase in drought tolerance in soybean and delays drought-induced leaf senescence in soybean and tobacco. J Exp Bot. 2009;60(2):533–546. doi: 10.1093/jxb/ern296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hao YJ, et al. Plant NAC-type transcription factor proteins contain a NARD domain for repression of transcriptional activation. Planta. 2010;232(5):1033–1043. doi: 10.1007/s00425-010-1238-2. [DOI] [PubMed] [Google Scholar]
- 18.Ooka H, et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003;10(6):239–247. doi: 10.1093/dnares/10.6.239. [DOI] [PubMed] [Google Scholar]
- 19.Carvalho CM, et al. A novel nucleocytoplasmic traffic GTPase identified as a functional target of the bipartite geminivirus nuclear shuttle protein. Plant J. 2008;55(5):869–880. doi: 10.1111/j.1365-313X.2008.03556.x. [DOI] [PubMed] [Google Scholar]
- 20.Tran LS, et al. Molecular characterization of stress-inducible GmNAC genes in soybean. Mol Genet Genomics. 2009;281(6):647–664. doi: 10.1007/s00438-009-0436-8. [DOI] [PubMed] [Google Scholar]
- 21.Xie Q, Frugis G, Colgan D, Chua NH. Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 2000;14(23):3024–3036. doi: 10.1101/gad.852200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hara-Nishimura I, Hatsugai N, Nakaune S, Kuroyanagi M, Nishimura M. Vacuolar processing enzyme: An executor of plant cell death. Curr Opin Plant Biol. 2005;8(4):404–408. doi: 10.1016/j.pbi.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 23.Hatsugai N, et al. A plant vacuolar protease, VPE, mediates virus-induced hypersensitive cell death. Science. 2004;305(5685):855–858. doi: 10.1126/science.1099859. [DOI] [PubMed] [Google Scholar]
- 24.Zhang H, et al. The role of vacuolar processing enzyme (VPE) from Nicotiana benthamiana in the elicitor-triggered hypersensitive response and stomatal closure. J Exp Bot. 2010;61(13):3799–3812. doi: 10.1093/jxb/erq189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delessert C, et al. The transcription factor ATAF2 represses the expression of pathogenesis-related genes in Arabidopsis. Plant J. 2005;43(5):745–757. doi: 10.1111/j.1365-313X.2005.02488.x. [DOI] [PubMed] [Google Scholar]
- 26.Nakashima K, et al. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 2007;51(4):617–630. doi: 10.1111/j.1365-313X.2007.03168.x. [DOI] [PubMed] [Google Scholar]
- 27.Wu Y, et al. Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Res. 2009;19(11):1279–1290. doi: 10.1038/cr.2009.108. [DOI] [PubMed] [Google Scholar]
- 28.Lu PL, et al. A novel drought-inducible gene, ATAF1, encodes a NAC family protein that negatively regulates the expression of stress-responsive genes in Arabidopsis. Plant Mol Biol. 2007;63(2):289–305. doi: 10.1007/s11103-006-9089-8. [DOI] [PubMed] [Google Scholar]
- 29.Hara-Nishimura I, Inoue K, Nishimura M. A unique vacuolar processing enzyme responsible for conversion of several proprotein precursors into the mature forms. FEBS Lett. 1991;294(1-2):89–93. doi: 10.1016/0014-5793(91)81349-d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.