Significance
HIV-1 is a lentivirus and replicates through a replication cycle involving several DNA forms including ssDNA. Here we report that synthetic DNA oligos corresponding to DNA forms of the lentivirus replication cycle as well as viral DNA are detected by the immunological DNA sensor IFN-inducible protein 16 (IFI16) and stimulate innate immune responses through a pathway dependent on stimulator of IFN genes (STING). Moreover, we show that replication of HIV-1 is elevated in cells with decreased expression of IFI16 or STING. We suggest IFI16 is a sensor for lentivirus DNA in macrophages stimulating innate immune responses, which contribute to early control of the virus.
Keywords: innate immunity, antiviral defense, DNA sensing
Abstract
Replication of lentiviruses generates different DNA forms, including RNA:DNA hybrids, ssDNA, and dsDNA. Nucleic acids stimulate innate immune responses, and pattern recognition receptors detecting dsDNA have been identified. However, sensors for ssDNA have not been reported, and the ability of RNA:DNA hybrids to stimulate innate immune responses is controversial. Using ssDNAs derived from HIV-1 proviral DNA, we report that this DNA form potently induces the expression of IFNs in primary human macrophages. This response was stimulated by stem regions in the DNA structure and was dependent on IFN-inducible protein 16 (IFI16), which bound immunostimulatory DNA directly and activated the stimulator of IFN genes –TANK-binding kinase 1 - IFN regulatory factors 3/7 (STING–TBK1–IRF3/7) pathway. Importantly, IFI16 colocalized and associated with lentiviral DNA in the cytoplasm in macrophages, and IFI16 knockdown in this cell type augmented lentiviral transduction and also HIV-1 replication. Thus, IFI16 is a sensor for DNA forms produced during the lentiviral replication cycle and regulates HIV-1 replication in macrophages.
The innate immune system senses virus infection largely via recognition of viral nucleic acid structures, leading to the induction of interferons (IFNs) and IFN-stimulated genes (ISGs) (1–5). IFNs constitute an essential part of early defense against viral infections and also are involved in the subsequent activation of adaptive immune responses (6, 7). HIV-1 and other lentiviruses replicate through a life cycle involving steps in which the genomic information is carried in the form of single-stranded (ss) RNAs, an RNA:DNA hybrid, ssDNA, and dsDNA (8). Nucleic acids are potent stimulators of innate immune responses, and pattern-recognition receptors (PRRs) sensing HIV-1 RNA have been reported in cell types not normally permissive to HIV-1 infection (5, 9, 10). Recently, a DNA sensor for HIV DNA also has been reported in a monocytic cell line and in macrophages rendered hypersensitive to retroviral DNA through Vpx-mediated degradation of the deoxynucleoside triphosphate triphosphohydrolase sterile alpha motif (SAM) domain and the histidine-aspartic (HD) domain-containing protein 1 (SAMHD1) (11). However, there is limited knowledge about how the innate immune system senses HIV infection in permissive cell types, such as macrophages, and how this sensing affects control of infection. Moreover, whether HIV nucleic acids are protected by the capsid complex in the cytoplasm is a matter of discussion, and it has been proposed that uncompleted reverse transcription (RT) may lead to cytosolic accumulation of viral DNA fragments available for innate immune recognition (12).
Most known nucleic acid-recognizing PRRs induce type I (α/β) and III (λ) IFNs, at least partly explaining the importance of these PRRs in antiviral defense. In addition, PRRs can induce ISGs directly, thereby exerting antiviral activity independent of IFN production (13, 14). Toll-like receptors (TLRs) detect viral nucleic acids in endosomes, with TLR3, -7/8, and -9 detecting dsRNA, ssRNA, and unmethylated DNA, respectively (5, 15, 16). In the cytosol, dsRNA is recognized by retinoic acid-inducible gene I (RIG-I), melanoma differentiation-associated gene 5, and DEAD box helicase 1 (DDX1)/DDX21/DHX36, and the response is amplified by additional features such as 5′-triphosphorylation and higher-order RNA structures (17–20). Finally, dsDNA also is a potent stimulator of innate immunity, and about 10 DNA sensors now have been proposed (3, 4, 21–26). dsDNA-induced IFN responses have been reported to proceed through a pathway involving stimulator of IFN genes (STING), TANK-binding kinase (TBK1), and IFN regulatory factor (IRF) 3 and 7 (4, 24, 27, 28). To date there is no knowledge about intracellular sensors for ssDNA, although accumulating data suggest that such molecules have key roles in host defense and autoimmune diseases (29–31). Moreover, the ability of RNA:DNA hybrids to stimulate innate immune responses is unresolved (30, 32). In addition to the proposed role for ssDNA in evoking innate immune responses during HIV-1 infection (30), it has been reported that an AT-rich stem-loop DNA motif, which is abundantly present in the Plasmodium falciparum genome, is responsible for stimulating IFN responses during infection with the malaria parasite in mice (31). Beyond infections, evidence suggests that lack of 3′ repair nuclease 1 (Trex1) in mice and humans leads to the accumulation of DNA species, including endogenous retroelements, in the cytoplasm and to the development of IFN-driven autoimmune diseases, including Aicardi–Goutières syndrome and chilblain lupus (29, 33).
In this work, we report that DNA-containing nucleic acid forms produced during lentiviral RT induced IFN expression to varying degrees. The immunostimulatory motif in the ssDNAs derived from the HIV cDNA was harbored in the stem regions generated by internal folding of the DNA strands, which induced type I and III IFN responses through a STING-dependent pathway in macrophages. The DNA sensor IFN-inducible protein 16 (IFI16) interacted directly with stem-rich DNA and was responsible for the IFN response to this pathogen-associated molecular pattern (PAMP). Importantly, IFI16 was recruited to cytosolic regions of lentiviral DNA accumulation, and IFI16 associated with lentiviral DNA, and knockdown of IFI16 in macrophages reduced HIV-induced expression of ISGs and promoted viral replication. Therefore we conclude that IFI16 is a sensor for lentiviral RT products and restricts HIV-1 replication in human macrophages.
Results
Stem Structures of ssDNA Stimulate Type I and III IFN Responses in Human Macrophages.
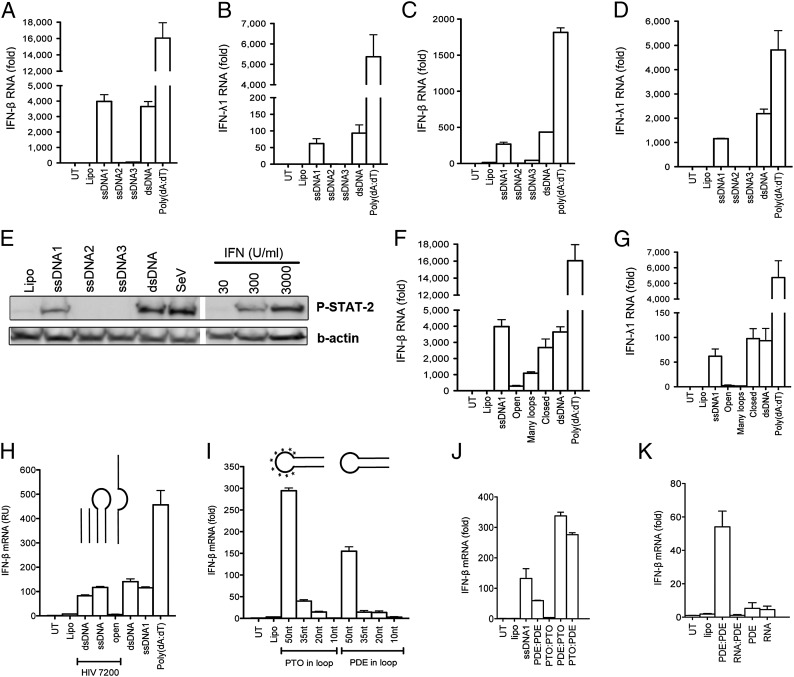
The immunostimulatory potential of dsDNA is well described, but knowledge about immune activation by ssDNA and DNA:RNA hybrids remains limited. To investigate the innate immunological response to ssDNA, we generated long synthetic ssDNA fragments (100–125 nt) derived from three different areas of the 5′ UTR region [+1 to the AUG start codon of group-specific antigen (Gag) ORF] of the HIV-1 Bal genome (Table S1) and compared the ability of these oligos (ssDNA1–3) to stimulate IFN responses in primary human monocyte-derived macrophages (hMDMs) and in a human macrophage-like cell line [phorbol12-myristate13-acetate (PMA)-differentiated THP1 cells]. Cells were transfected with ssDNA1–3, dsDNA, or poly(deoxyadenylic-thymidylic) acid [poly(dA:dT)], and the induction of IFN-β or IFN-λ1 gene expression was evaluated after 6 h. In hMDMs from four different donors, dsDNA, poly(dA:dT), and ssDNA1 robustly induced synthesis of both IFN-β and IFN-λ1 mRNA (Fig. 1 A and B). ssDNA2 and -3 stimulated very limited induction of either response. Similar observations also were made in PMA-differentiated macrophage-like THP1 cells (Fig. 1 C and D). To exclude any delayed response in cells stimulated with ssDNA2 and ssDNA3, we conducted kinetic experiments evaluating the IFN-β induction over 24 h (Fig. S1A). Again, only ssDNA1 and dsDNA induced strong IFN-β expression, which peaked at about 12 h poststimulation. To exclude the possibility that the limited IFN induction by ssDNA2 and ssDNA3 was caused by poor transfection efficacy, all three ssDNAs were fluorescently labeled, and transfection was verified by confocal microscopy. We observed no differences between the ssDNA oligos with respect to percentage of DNA-positive cells, number of DNA foci per cell, and size of the DNA foci. Finally, the differential induction of IFNs by the ssDNA oligos also was reflected in the ability of supernatants from THP1 cells to induce IFN bioactivity as measured by STAT2 phosphorylation and induction of ISG expression (Fig. 1E and Fig. S1B). These results suggest the existence of a cellular sensor for ssDNA in human macrophages and also that different ssDNAs differentially stimulate this sensor.
Fig. 1.
ssDNA induces type I and III IFN responses in human macrophages. (A–D) hMDMs (A and B; n = 4) and THP1 cells (C and D; n = 5) were transfected with three ssDNA oligo nucleotides (ssDNA1–3) derived from HIV-1, dsDNA, or poly(dA:dT), respectively. RNA was collected after 6 h of incubation and analyzed for levels of IFN-β (A and C) and IFN-λ1 (B and D) mRNA by RT-qPCR. (E) Supernatants from cells treated as in C and D for 20 h were used to stimulate parental THP1 cells for 50 min. Cell lysates were analyzed for STAT2 phosphorylation WB. Recombinant IFN-α was used as standard, and Sendia virus (SeV) was used as control. (F and G) Variants of ssDNA1 (open, many loops, and closed) were transfected into hMDMs, and RNA was isolated and analyzed for IFN-β (F) and IFN-λ1 (G) mRNA expression (n = 4). (H–K) THP1 cells were transfected with the indicated DNA oligos. RNA was collected after 6 h of incubation and was analyzed for IFN-β mRNA expression by RT-qPCR (n = 2–4). Asterisks in I indicate stem-loop DNA with PTO in the loop region. All data are presented as a representative dataset from one of multiple independent experiments performed in duplicate (mean ± SEM). Lipo, lipofectamine; UT, untreated.
To investigate the ssDNA-induced innate response further, we measured levels of IL-6, TNF-α, and IL-1β in the supernatants from PMA-differentiated THP1 cells transfected with ssDNA1–3, dsDNA, and poly(dA:dT) for 16 h. ssDNA1 induced modest production of TNF-α and IL-6 in the macrophages and also stimulated IL-1β secretion in cells pretreated with LPS, although not to the extent seen with dsDNA or poly(dA:dT) stimulation (Fig. S1 C–E). ssDNA2 and ssDNA3 stimulated little or no production of the cytokines measured. A characterization of the ability of increasing doses of ssDNA1 and dsDNA to induce IFN-β mRNA expression revealed that the two immunostimulatory DNA forms induced IFN-β expression with very similar dependence on dose (Fig. S1F). Collectively, these data demonstrate that immunostimulatory ssDNA1 induces a cytokine profile similar to that induced by dsDNA in macrophages.
The observed differential ability of the ssDNAs to induce IFN responses could result from either sequence-specific recognition of the DNA or the secondary structures in the DNAs. To assess this difference, we used the mFOLD algorithm to predict the secondary structures of ssDNA1–3. SsDNA1 was predicted to contain long stem structures with terminal loop regions; ssDNA2 and -3 were predicted to be more open in structure than ssDNA1, and the stem-loop of ssDNA3 was predicted to be relatively stable based on the Gibbs energy level (Fig. S2A). Thus, the predicted secondary structure of the ssDNAs suggests a correlation between the extent of stem structures in the ssDNA and activation of IFN responses. Finally, we investigated an ssDNA 70mer previously reported to be a poor inducer of IFN responses (4) and found that it was predicted not to contain a significant secondary structure, supporting our observations from ssDNA1–3 (Fig. S2B). To test further the idea that the secondary structure determined the immunostimulatory activity of sDNA, we mutated different regions within ssDNA1 to change the secondary structure (Fig. S2C). Interestingly, induction of IFN gene expression was strongly reduced if the stem regions were replaced with less structured or more loop-rich regions, although we did note that the ssDNA1 mutant with many loops still induced some degree of IFN-β expression (Fig. 1 F and G).
To exclude the possibility that the superior induction of IFNs by ssDNA1 was caused by a sequence-specific motif, we used a randomly picked 50-nucleotide sequence from within the HIV-1 genome to design an ssDNA with a 50mer stem and a 10-nucleotide loop that was either closed or completely open in its structure and a version in which the loop was removed. Importantly, like ssDNA1, HIV7200_dsDNA and HIV7200_ssDNA induced IFN-β mRNA, whereas HIV7200_open did not induce any IFN response (Fig. 1H). To evaluate further the role of the stem in the induction of IFN-β expression by stem-loop DNA folded from ssDNA strands, we next designed a series of variants of the HIV7200 oligo. We designed two sets of oligos: one with a natural phosphodiester (PDE) backbone in the loop, and one with a DNase-insensitive phosphothioester (PTO) backbone in the loop. Furthermore, we decreased the stem region of these oligos from 50 to 35, 20, or 10 bp. When transfected into macrophages, we found that the ability of the oligos to stimulate IFN responses was highly dependent on the length of the stem; the response was reduced strongly when the stem was reduced from 50 to 35 nucleotides (Fig. 1I). The oligos with the PTO backbone in the loop gave rise to stronger IFN-β expression but also exhibited lower response when the stem was reduced. In the next series of experiments we wanted to determine which part of the DNA duplex triggers the IFN response. For this purpose, we used two ssDNA oligos with either PDE or PTO backbones and hybridized them to generate four combinations of dsDNAs (PDE:PDE, PTO:PTO, PDE:PTO, and PTO:PDE). We observed that dsDNA-induced IFN-β induction was abrogated when the natural PDE backbone was replaced with PTO on both DNA strands (Fig. 1J). However, dsDNAs with one PDE and one PTO strand induced significantly more IFN-β expression than the natural PDE duplex.
Finally we examined whether the RNA:DNA hybrids induced IFN-β expression in macrophages after transfection. As shown in Fig. 1K, this form of nucleic acid, which could mimic the RNA:DNA duplex of the lentiviral replication cycle, did not elevate the levels of IFN-β mRNA significantly. The short AT-rich ssDNA oligodeoxyribonucleotide previously reported to induce IFN responses in murine macrophages (31) did also not induce expression of IFN-β in THP1 cells (Fig. S1G). Taken together, these data suggest that, in addition to dsDNA, ssDNA is detected by intracellular sensors that recognize the PDE DNA backbone in stable stem regions in the DNA secondary structure and trigger IFN responses. DNA:RNA duplexes or small ssDNA fragments do not induce IFN responses in human macrophages.
Stem-Rich DNA Induces IFN-β via IFI16 Through a STING–TBK1–IRF3/7 Pathway.
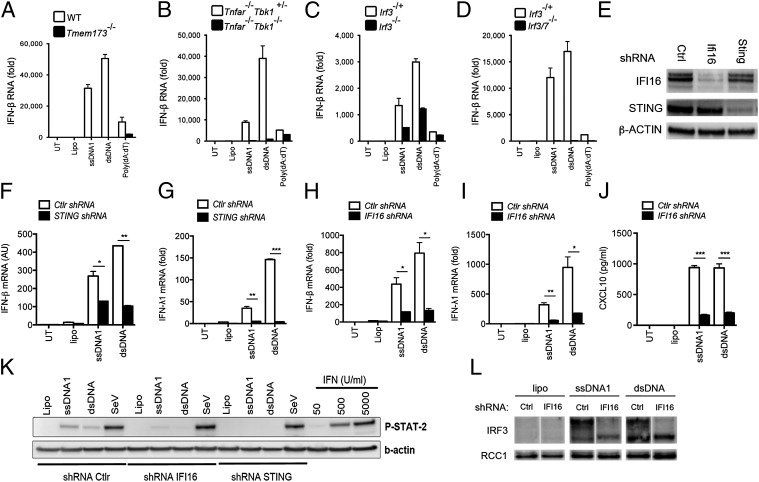
To investigate the signaling pathway for IFN induction by stem-rich ssDNA, we transfected different wild-type and knockout murine bone marrow–derived macrophages (BMDMs) with ssDNA1 or dsDNA. The response to ssDNA1 and dsDNA was abrogated in transmembrane protein 173 double-knockout (Tmem173−/−) BMDMs (deficient in STING) compared with wild-type cells (Fig. 2A). The TLR7 agonist ssRNA40 induced unaltered TNF-α responses in Tmem173−/− macrophages (Fig. S3 A–D). Similar to the response in Tmem173−/− BMDMs, macrophages lacking TBK1 were unable to induce the synthesis of IFN-β in response to ssDNA1 and dsDNA (Fig. 2B). Next we tested the role of IRF3 and observed partial reduction in the IFN-β response to ssDNA and dsDNA in cells lacking IRF3 (Fig. 2C). To investigate a potential role for IRF7 in the IRF3-independent IFN response, we examined IRF3/7 double-deficient BMDMs and observed that the IFN response was totally abrogated in the absence of both IRF3 and 7 (Fig. 2D). Of note, we observed that ssDNA2 and ssDNA3, which did not potently stimulate IFN expression in human macrophages, did evoke strong responses in murine macrophages, and this response was induced through a STING–TBK1–IRF3/7-dependent pathway (Fig. S3 E–L). Thus, stem-rich DNA induces innate responses in murine macrophages through a pathway dependent on STING, TBK1, and IRF3/7, whereas ssDNAs with less secondary structure induce responses only in murine macrophages.
Fig. 2.
HIV-derived ssDNA induces IFN responses through a pathway dependent on IFI16, STING, TBK1, and IRF3/7. (A–D) BMDMs from mice with homozygous deletions of the genes encoding (A) Tmem173 (i.e., STING) (n = 3), (B) TBK1 (n = 2), (C) IRF3 (n = 2), or (D) IRF3/IRF7 (n = 2) and their respective controls were transfected with 2 µg/mL of ssDNA1 or dsDNA and were incubated for 6 h. RNA was isolated and analyzed for induction of IFN-β by RT-qPCR. (E) Western blot of IFI16, STING, and β-actin on extracts from THP1-derived cells stably transduced with lentiviral shRNAs [control (Ctrl), IFI16, and STING]. (F–I) RNA was isolated and analyzed for induction of IFN-β (F and H) and IFN-λ1 (G and I) by RT-qPCR. ssDNA1 or dsDNA (2 µg/mL) was transfected into THP1 control and STING shRNA-knockdown cells (n = 3) (F and G) or into THP1 control and IFI16 shRNA-knockdown cells (H and I) followed by 6 h of incubation (n = 4). (J) Culture supernatants from THP1 control and IFI16 shRNA-knockdown cells treated for 16 h were analyzed for CXCL10 secretion by ELISA (n = 3). (K) Culture supernatants from THP1 control, IFI16, and STING shRNA-knockdown cells were used to stimulate parental THP1 cells for 50 min. Cell lysates were subjected to Western blotting and probed with anti–P-STAT2 antibody. (L) Total IRF3 in nuclear extracts from THP1 control and IFI16 shRNA-knockdown cells treated for 2 h with 2 µg/mL of ssDNA1 or dsDNA. Levels of IRF3 in the nucleus were detected by Western blotting. Regulator of chromosome condensation 1 (RCC1) was used as loading control. A–D and F–K are representative datasets from one of multiple independent experiments performed in duplicate (mean ± SEM). *P < 0.05; **P < 0.01; ***P < 0.001; Student’s t test.
To investigate whether human macrophages also responded to ssDNA1 through a STING-dependent pathway, we generated THP1-derived cell lines with stable knockdown of STING by multiple shRNA lentiviral transductions. We observed strong reduction in the levels of STING in the transduced cell lines (Fig. 2E), which retained the ability to respond to Sendai viral RNA (Fig. S4 A, C, and D). In the cells with reduced levels of STING, the induction of IFN-β and IFN-λ1 mRNA expression and of IFN bioactivity by ssDNA1 and dsDNA was reduced significantly (Fig. 2 F, G, and K and Fig. S4 C and D). Thus, ssDNA induces IFN responses in both human and murine cells in a STING-dependent manner.
Several cytosolic DNA sensors, including IFI16, DDX41, and cyclic GMP-AMP synthase (cGAS), induce type I IFN through a STING-dependent pathway (4, 26, 34). We investigated the possible role for these proteins in sensing of stem-rich DNA. Undifferentiated THP1 cells, recently used to demonstrate a role for cGAS in HIV-induced IFN-β expression (11), have been reported to express only low constitutive levels of IFI16, and PMA-differentiated THP1 cells have been reported to express high levels of IFI16 (4, 35, 36). We confirmed these previous findings in our cells (Fig. S4E). Next, using the lentiviral shRNA transduction system, we generated a THP1-derived IFI16 shRNA cell line and observed potent knockdown of IFI16 protein but unaltered response to Sendai virus infection in the PMA-differentiated cells (Fig. 2E and Fig. S4B). Importantly, the induction of IFN-β and IFN-λ1 mRNA expression by ssDNA1 and dsDNA, as well as the DNA-driven induction of IFN bioactivity and ISG expression, was reduced markedly in IFI16-knockdown cells (Fig. 2 H–K), and this reduction was also observed for dsDNAs composed of hybrids of PDE and PTO strands (Fig. S4F). Of note, we found that the pronounced impairment of IFN-β induction after transfection with ssDNA or dsDNA in IFI16-knockdown cells also was observed in cells with knockdown of DDX41 or cGAS (Fig. S4 G–I), possibly indicating an as yet unidentified mechanistic interaction among these three proposed DNA sensors. Finally, using Western blotting on nuclear extracts, we investigated the nuclear translocation of IRF3 in control and IFI16-knockdown THP1 cells treated with Lipofectamine alone or stimulated with ssDNA1 or dsDNA for 2 h. Treatment with either ssDNA1 or dsDNA induced high levels of IRF3 in the nucleus, which were decreased in IFI16-knockdown cells (Fig. 2L). The multiple bands observed in the Western blot after probing with anti–IRF-3 likely reflected different phosphorylated forms of IRF-3 (36).
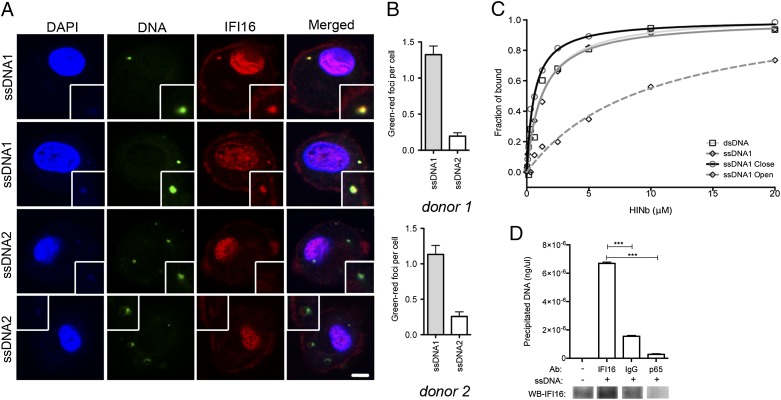
Immunofluorescence imaging of hMDMs with FITC-labeled ssDNAs further demonstrated that ssDNA1 localized together with cytoplasmic endogenous IFI16 in distinct foci, whereas the nonstimulatory ssDNA2 barely colocalized with IFI16 (Fig. 3 A and B). We also observed that ssDNA1 colocalized with STING in distinct foci in the cytoplasm (Fig. S5), similar to the staining pattern for ssDNA1 and IFI16 (Fig. 3A). To determine whether IFI16 is able to bind the ssDNA directly, we conducted an in vitro binding assay using a purified IFI16 HINb (hematopoietic interferon-inducible nuclear b) domain and FITC-labeled DNAs. We observed that ssDNA1 and dsDNA bound IFI16 with comparable strength (Fig. 3C), whereas ssDNA1-closed (Fig. 1 F and G and Fig. S2C) displayed a slightly increased binding capacity. Importantly, ssDNA1-open, which was a poor inducer of IFN expression and was predicted not to contain significant stem regions (Fig. S2C), was bound by IFI16 HINb domain with a markedly reduced affinity. Finally, we immunoprecipitated IFI16 from cells transfected with ssDNA1 and quantified the amount of ssDNA1 coprecipitated with IFI16 by quantitative PCR (qPCR). As shown in Fig. 3D, the immunostimulatory HIV-derived DNA indeed was precipitated together with IFI16. Taken together, these results suggest that IFI16 directly binds to DNA structural foldings and works as a sensor for dsDNA and stem-rich ssDNA, stimulating IFN expression through a STING-dependent pathway.
Fig. 3.
Immunostimulatory ssDNA colocalizes with IFI16 and interacts with the IFI16 HINb domain. (A) Confocal microscopy images of hMDMs transfected for 1 h with FITC-labeled ssDNA1 and ssDNA2 after protein staining for IFI16. Insets illustrate closeup of foci formations. (Scale bar, 10 um.) (B) Quantification of DNA-IFI16 colocalization in hMDMs from two different donors transfected for 1 h with ssDNA1 or ssDNA2. (C) Fluorescence polarization assays for FAM-labeled ssDNA1 upon binding to the IFI16 HINb domain. The binding isotherms between the IFI16 HINb domain and the FAM-labeled DNAs are graphed as shown. The apparent Kd values derived from the fluorescence polarization assays are dsDNA, 1.299 ± 0.404 μM; ssDNA1, 1.198 ± 0.122 µM; ssDNA1 closed, 0.605 ± 0.071 µM; and ssDNA1 open, 9.878 ± 2.225 μM. (D) THP1 cells were transfected with 2 µg/mL of ssDNA1 for 2 h. Cytoplasmic lysates were subjected to immunoprecipitation using beads coupled to the indicated antibodies. (Upper) Coprecipitated ssDNA1 was amplified by qPCR and quantified by comparison with a standard curve using ssDNA1. (Lower) Western blotting was used to quantify the amount of endogenous IFI16 bound to the beads. ***P < 0.001; Student's t test.
Trex1 Counteracts IFI16-Dependent Induction of IFN Expression by Stem-Rich DNA.
The exonuclease Trex1 has been reported to digest endogenous DNA in the cytosol, thereby preventing the development of autoimmune diseases (29). More recently, Yan et al. (30) have demonstrated that HIV-1 may escape innate immune recognition of viral DNA in a manner dependent on Trex1. Therefore, we wanted to investigate if IFI16-dependent sensing of DNA was more apparent in settings where DNA accumulated in the cytosol because of the absence of Trex1.
We first explored the DNA-induced IFN responses in Trex1-deficient murine macrophages and found that type I IFN induction by HIV-1–derived ssDNA1 was significantly elevated in Trex1-deficient murine macrophages compared with wild-type cells at both the RNA and protein levels (Fig. S6 A and B). In contrast, the response to dsDNA was not or was only marginally affected by Trex1 deficiency. Finally, Trex1 deficiency did not affect the induction of TNF-α by ssDNA1 or dsDNA (Fig. S6C).
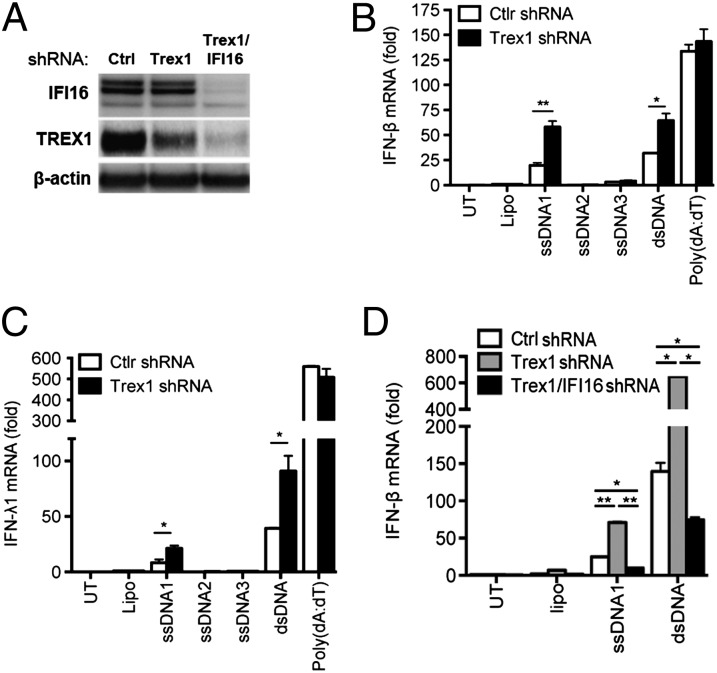
To investigate the role of Trex1 in human cells, we used THP1 cells, which were transduced with lentiviral vectors encoding Trex1 shRNA. With this method we were able to achieve knockdown of Trex1 (Fig. 4A). Transfection with ssDNA1 resulted in the induction of IFN-β and IFN-λ1 expression, which was elevated further in the Trex1-knockdown cells (Fig. 4 B and C). In contrast ssDNA2 and -3 remained unable to induce IFN-β expression in cells with reduced Trex1 expression (Fig. 4 B and C), indicating that the differential stimulatory potential of the ssDNAs tested was not caused by a Trex1-mediated degradation of the DNAs with less secondary structure. We also observed that induction of IFN-β and IFN-λ1 by poly(dA:dT) was not affected by Trex1 knockdown, thus demonstrating that the IFN response was not globally elevated and also that not all DNA species exhibited the same sensitivity to Trex1-mediated degradation. Next, we generated a Trex1-IFI16 double-knockdown cell line to evaluate the effect of IFI16 on a background of Trex1 depletion. Interestingly, we reproducibly observed significantly more efficient knockdown of Trex1 in cells in which IFI16 was already knocked down (Fig. 4A). More importantly, we found that ssDNA-induced expression of IFN-β, which was elevated in Trex1-knockdown cells, was largely abrogated when IFI16 expression was reduced (Fig. 4D). Thus, Trex1 counteracts the IFI16-dependent induction of IFN expression by stem-rich DNA.
Fig. 4.
Trex1 counteracts IFI16-dependent induction of IFN expression by stem-rich DNA. (A) Western blot for IFI16, Trex1, and β-actin in THP1 control, Trex1, and Trex1/IFI16 shRNA-knockdown cells. (B and C) THP1 Trex1-knockdown cells were transfected with ssDNA1, ssDNA2, ssDNA3, dsDNA, or Poly(dA:dT) (2 µg/mL). RNA was isolated 6 h after treatment and analyzed for IFN-β (B) and IFN-λ (C) by RT-qPCR (n = 2). (D) Control shRNA, Trex1 shRNA, and Trex1/IFI16 shRNA knockdown THP1 cells were stimulated for 6 h with 2 µg/mL of ssDNA1 or dsDNA. RNA was isolated and analyzed for IFN-β by RT-qPCR (n = 3). Data are shown as one dataset representative of multiple independent experiments performed in duplicate (mean ± SEM). *P < 0.05; **P < 0.01; Student’s t test.
Replication of HIV and Lentiviral Transduction Is Elevated in the Absence of IFI16.
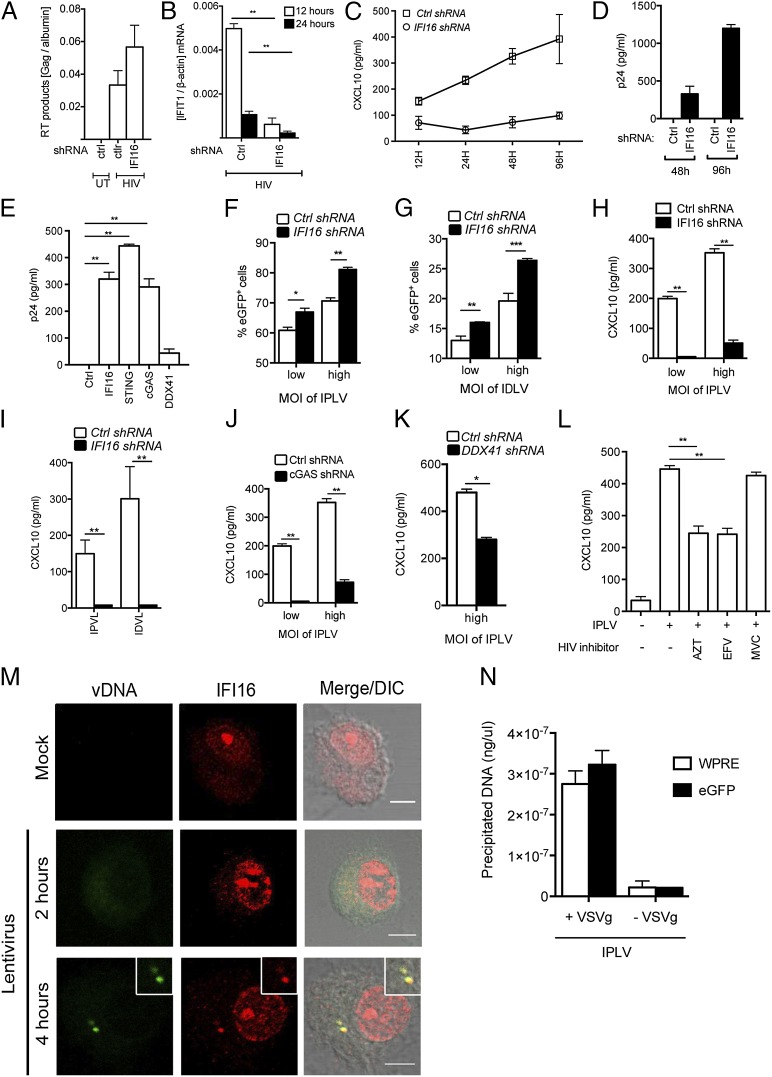
IFNs and numerous ISGs are known to restrict viral replication, and they can block both early and late stages of HIV infection (37–40). Therefore we wanted to explore whether IFI16 may have a role in restricting HIV replication. To test this idea, we challenged THP1 control and IFI16 shRNA-knockdown cells with an HIV-1 Bal replication-competent virus in a 5-d infection protocol. We first examined the accumulation of RT products in the infected cells as measured by the accumulation of HIV Gag DNA 12 h after infection (Fig. 5A). We observed that HIV-1 RT products did accumulate to a comparable degree in control and IFI16 shRNA THP1-derived cells, with a trend toward more viral DNA in IFI16-knockdown cells. Importantly, expression of the highly inducible ISG, interferon-induced protein with tetratricopeptide repeats 1 (IFIT1), 12 h after HIV-1 infection was reduced significantly in cells with IFI16 knockdown (Fig. 5B). Production of other ISGs also was observed to be dependent on IFI16, because reduced expression of this DNA sensor led to impaired accumulation of C-X-C motif chemokine 10 (CXCL10) in supernatants from cells infected with HIV-1 (Fig. 5C). Finally, we examined HIV-1 replication in THP1 cells as measured by the accumulation of p24 in the culture supernatant. In the THP1 control shRNA cell line, the p24 levels did not increase above background levels (Fig. 5D), consistent with previous reports (41). Importantly, in cells with IFI16 knockdown, the virus gained the capacity to replicate (Fig. 5D). Similar findings were made in cells with stable knockdown of cGAS or STING but not in cells with knockdown of DDX41 (Fig. 5E), thus indicating that IFI16 exerts its anti-HIV effect in a manner dependent on the cGAS–STING pathway.
Fig. 5.
IFI16 has an important role in controlling HIV-1 and lentiviral vectors. (A) THP1 control and IFI16-knockdown cells were infected with HIV-1 Bal. HIV-1 RT DNA products were measured by PCR on HIV-1 Gag DNA 12 h postinfection. (B) Cellular RNA was isolated 12 or 24 h after infection and IFIT1 levels were analyzed by RT-qPCR. (C and D) Cells were infected with HIV-1 Bal and culture supernatants were harvested at the indicated time points and analyzed for levels of CXCL10 (C) and HIV-1 p24 (D). (E) THP1 control, IFI16, STING, cGAS, and DDX41 knockdown cells were infected with HIV-1 Bal for 96 h, and p24 levels in the supernatant were measured by ELISA. (F–K) THP1 control, IFI16, DDX41, and cGAS knockdown cells were infected with VSV-G pseudotype eGFP lentiviruses expressing either proficient or deficient viral integrase. (F and G) eGPF-positive cells were evaluated by flow cytometry 96 h after infection. (H–K) CXCL10 levels in the supernatant were measured on samples harvested 24 h after infection. (L) THP1 cells were treated with AZT (5 μM), EFV (5 μM), or MVC (5 μM) and infected with IPLVs for 24 h. Levels of CXCL10 in the supernatants were measured by ELISA. Results in A–L are shown as one representative dataset from two to three independent experiments performed in triplicate (mean ± SEM). (M) hMDMs were infected with lentivirus pseudoparticles for 2 or 4 h, and viral DNA was visualized by specific FISH probes [viral DNA (vDNA); green] and was costained with anti-IFI16–specific antibodies (IFI16; red). (Scale bar, 5 μm.) The mock sample was fixed after 4 h of incubation. Data from two other donors are shown in Fig. S7. DIC, differential interference contrast. (N) THP1 cells were treated with entry-proficient (+VSV-G) or entry-deficient (−VSV-G) IPLVs for 4 h. Cytoplasmic lysates were subjected to immunoprecipitation using anti-IFI16–coupled beads. Coprecipitated viral DNA was amplified by PCRs targeting WPRE and cGFP in the IPLV genome and was quantified by comparison with a standard curve. *P < 0.05; **P < 0.01; Student’s t test.
We speculated that IFI16 potentially could affect viral replication at a preintegration step and addressed this possibility using a single-round infection model of lentivirus pseudoparticles carrying an eGFP-vector signal. THP1 cells infected with eGPF viruses were evaluated for the expression of eGFP 96 h after infection. DNase treatment of the virus preparations did not affect eGFP expression and marginally reduced the CXCL10 response induced by the lentiviruses (Fig. S8). In an integrase-proficient lentivirus system (IPLV), IFI16 knockdown cells showed a significant increase in the percentage of cells positive for eGFP as compared with control cells, and this finding was consistent over different viral titers (Fig. 5F). Interestingly, this increase was also found when we used an integrase-deficient lentivirus (IDLV), suggesting that IFI16 is involved in the activation of antiretroviral mechanisms acting before the integration of viral DNA into the host cell genome (Fig. 5G). Both the integrase-proficient and -deficient viruses induced CXCL10 expression in a manner strongly dependent on IFI16 (Fig. 5 H and I), and the IPLV-induced CXCL10 expression was reduced partially in cells with DDX41 knockdown and strongly in cells with cGAS knockdown (Fig. 5 J and K), thus demonstrating an inverse correlation between lentiviral transduction and the induction of ISG expression. The IPLV-induced CXCL10 response was significantly inhibited by the RT inhibitor AZT and the nonnucleotide RT inhibitor efavirenz (EFV) but not by the C-C motif chemokine receptor 5 (CCR5) antagonist maraviroc (MCV) (Fig. 5L). Treatment of cells with the integrase inhibitor raltegravir alone stimulated CXCL10 expression, so we were not able to analyze the effect this inhibitor may have on lentivirus-driven ISG expression. However, together with the finding that lentiviruses devoid of integrase activity induced CXCL10 expression (Fig. 5I), these data suggest that the lentiviral RT products are the PAMPs stimulating the innate immune responses in an IFI16-dependent manner.
To explore further the role of IFI16 in recognizing retroviral RT products, we performed DNA FISH with lentivirus-specific probes targeting the U5 region of HIV on hMDMs infected with the IPLV described above. At 2 h after infection, accumulation of viral DNA in the cytoplasm was not found, but after 4 h of infection, we observed clear FISH-positive staining in the infected but not in the uninfected cells (Fig. 5M), indicating that lentiviral RT products do accumulate in the cytosol of infected hMDMs. The staining exhibited a clear punctuate nature, resembling what is seen after DNA transfection (Fig. 3A) and with the accumulation of herpesvirus DNA (42). Evaluated over three donors, FISH-positive staining was seen in 10–20% of the cells after 4 h of infection (see Fig. S7 for data from two other donors). Importantly, the great majority of FISH-positive spots also stained positive for IFI16, with clear colocalization. Finally, we treated macrophages with entry-proficient [vesicular stomatitis virus envelope G glycoprotein-positive (VSV-G+)] or entry-deficient (VSV-G−) lentiviruses for 4 h and generated cytoplasmic lysates from which IFI16 was immunoprecipitated. Importantly, we found that viral DNA coprecipitated with IFI16 from cells treated with entry-proficient virus but not from cells treated with entry-deficient virus (Fig. 5N).
Collectively, these data suggest that IFI16 senses lentiviral RT products in the cytoplasm in macrophages and that this event is required to induce antiviral functions with the ability to control the virus before integration.
IFI16 Is Essential for Control of HIV Replication in Primary Human Macrophages.
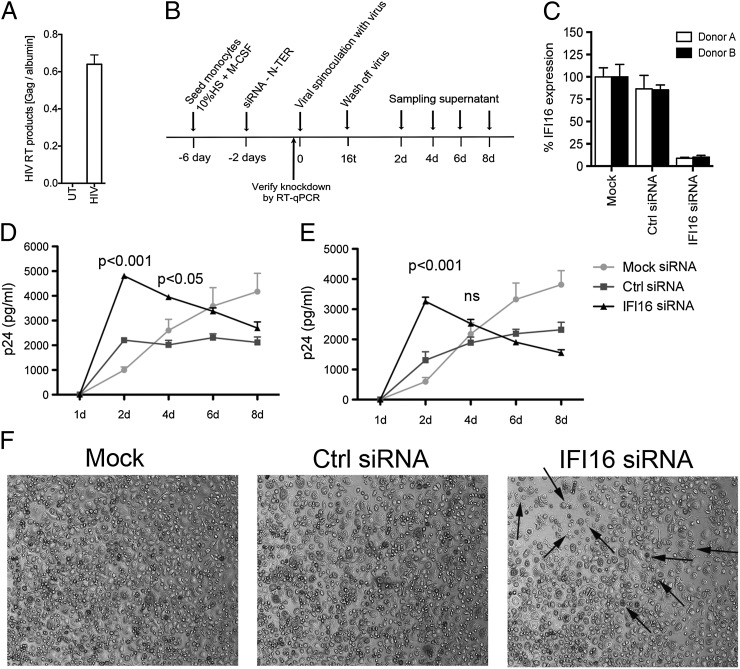
The viral experiments presented above were conducted in macrophage cell lines or with the use of VSV-G pseudotype lentiviruses. In a final series of experiments, we wanted to address the role of IFI16 in hMDMs, infected with replication-competent macrophage-tropic HIV strains. Challenge with HIV-Bal resulted in high levels of RT products in the cells at 12 h after infection (Fig. 6A), reflecting the calculated multiplicity of infection (MOI) of 1.5 added to the cells. Next, we treated hMDMs with scramble siRNA or specific siRNA against IFI16 and infected the treated cells with HIV-1 using the scheme shown in Fig. 6B. Knockdown was verified on the day of infection with HIV-Bal, and we consistently observed more than 90% knockdown of IFI16 (Fig. 6C). The siRNA treatment did not lead to the induction of IFN/ISGs as measured by expression of CXCL10 and IFN-β mRNA (Fig. S9). Importantly, we measured significantly higher levels of HIV-1 p24 in the supernatants from two different donors with IFI16 knockdown than in the cells treated with scramble siRNA (Fig. 6 D and E). In both donors, the elevated accumulation of p24 was observed at the early time points after infection. In IFI16 siRNA-treated cells the early elevated p24 production was replaced by decreased p24 levels at day 4 and thereafter. Interestingly, HIV-1 caused significantly more cytopathic effects in the IFI16-depleted hMDMs (arrows in Fig. 6F), and the degree of cell damage tended to correlate with lower p24 production in the IFI16-knockdown cells at later time points. We observed no cell death in hMDMs treated with IFI16 siRNA without HIV infection. In summary, these data support a role for IFI16 in the innate sensing of HIV-1 RT products and in the regulation of viral replication in macrophages. Lack of IFI16 leads to uncontrolled replication, which in turn leads either to exhaustion of the cell or cell death.
Fig. 6.
IFI16 is essential for control of HIV-1 infection in primary human macrophages. (A) Levels of HIV-1 Gag DNA in hMDMs infected with HIV-1 Bal for 12 h were measured by qPCR (n = 2). (B) Summary of experimental setup in hMDMs. Primary monocytes were differentiated into macrophages and treated with siRNA (165 nM) before HIV-1 Bal infection (MOI of 1 calculated on Tzmbl titration and X-Gal straining). (C) Levels of IFI16 knockdown by a scramble and specific siRNA were measured by RT-qPCR on RNA harvested on the day of viral infection. Results represent four independent siRNA transfections made in cells from both donors (mean ± SEM). (D and E) hMDMs from donors A (D) and B (E) treated with control or IFI16 siRNA were infected with HIV-1 Bal (MOI of 1.0). Cultures were harvested at the indicated time points, and p24 levels were measured by ELISA (n = 4; two-way ANOVA with Bonferroni multiple comparison). (F) Representative images of hMDMs from donor A at day 4 after viral infection. Arrows indicate cells exhibiting apoptotic morphology.
Discussion
In recent years it has been recognized that that the innate immune system plays an important role in controlling HIV infection and in the pathogenesis of AIDS (43). Nucleic acids are potent stimulators of innate immune responses (44), and many nucleic acid PAMPs have been described, including dsRNA, 5′ triphosphate (5′-PPP) RNA, and dsDNA (45–49). Recently, it also has been reported that ssDNA stimulates the innate immune system (29–31), but PRRs sensing ssDNA remain to be described. HIV-1 and other lentiviruses replicate through a life cycle that involves an RT step converting the ssRNA genome into a cDNA strand, which first is annealed with the RNA template strand, afterwards exhibiting some degree of single-strandedness (notably the 5′ end of the cDNA strand) (8). This cDNA template then is used to generate the second DNA strand, generating a proviral genome consisting of dsDNA that can be integrated into the genome of permissive host cells. However, although these events represent the productive replication cycle, RT is a very error-prone process, and most RT products do not give rise to a proviral DNA that can be integrated in the nucleus (50). Thus, cytosolic accumulation of viral DNA fragments from abortive viral replication can be sensed by PRRs to trigger innate immune responses (12).
Using ssDNA derived from the 5′ UTR region of the HIV viral cDNA, we here report that, like dsDNA, ssDNA is a potent PAMP in human macrophages and that the immunostimulatory motif is the stem regions in the secondary structure arising from hybridization of complementary regions within the DNA strand. However, RNA:DNA duplexes do not stimulate innate immune responses in human macrophages. IFI16 recognizes stem-rich DNA and induces IFN responses through a STING–TBK1–IRF3/7 pathway, and lack of the 5′–3′ exonuclease Trex1 augments this IFI16-dependent response. In the macrophage-like THP1cell line, which is believed to be nonpermissive for HIV-1 replication, knockdown of IFI16, cGAS, or STING led to the expression of p24 by replication-competent HIV-1. Knockdown of IFI16 also led to elevated expression of GFP from an LVDP, thus suggesting that IFI16 inhibits lentivirus replication before integration. Finally, primary human macrophages with IFI16 knockdown exhibited augmented early replication and accelerated virus-mediated cytopathic effects after HIV-1 infection. Thus, we propose that IFI16 is a sensor for ssDNA and dsDNA in the lentiviral replication cycle and exerts early control of HIV infection in macrophages, preventing virus-induced cytopathic effects.
A central issue in the field of nucleic acid recognition is identifying the structures being sensed by PRRs. This issue is particularly relevant for the more flexible nucleic acid structures, such as ssRNA and ssDNA, because they may fold in different conformations, depending on the presence of complementary strands, nucleic acid-interacting proteins, and other factors. For RNA it has been reported that RIG-I recognizes 5′-PPP RNA presented in a panhandle structure (51). In our work we found that the ability of ssDNA to induce IFN responses in human macrophages and to interact with the IFI16 HINb domain correlated with the proportion of stem regions in the DNA and the length of the stem regions. We further suggest that the immunostimulatory potential for stem-rich DNA likewise depends on the length and stability of the stem. The observed differential ability of ssDNA1 versus ssDNA2/3 to stimulate IFN expression could not be explained by differential sensitivity toward Trex1-mediated degradation, because ssDNA2/3 remained unable to induce innate responses in cells in which Trex1 expression was reduced. In mice, the situation may be more complex. An AT-rich-loop DNA with a 5-nt stem region has been reported previously to induce IFN responses through a STING–TBK1–IRF3 pathway in mice (31). However, this DNA oligo did not stimulate IFN responses in the THP1 cells and hMDMs used in the present study. Moreover, in sharp contrast to our observations in human macrophages, the HIV-derived ssDNA2 and -3 did, in fact, induce strong IFN responses in murine macrophages. Therefore, like human macrophages, murine cells sense stem-rich DNA but may also be equipped with potent systems to sense linear or looped ssDNA. This issue needs further clarification but potentially raises some concerns regarding the use of murine cells and pseudotyped viruses to study HIV and innate immunology.
A key finding of the present work is that IFI16 is a sensor of immunostimulatory lentiviral DNA. We found that IFI16 colocalized with the immunostimulatory HIV-derived ssDNA1 in macrophages but not with the nonstimulating ssDNA2. Second, we found that the HINb domain of IFI16 interacted directly with ssDNA1 and did so with higher affinity than a less stimulatory, modified form of ssDNA1. Third, we demonstrated coprecipitation of endogenous IFI16 with synthetic ssDNA1 or lentiviral DNA in cytoplasmic lysates from macrophages either transfected with DNA or infected with lentiviruses. Fourth, in hMDMs we observed recruitment of IFI16 to areas accumulating HIV RT products. Finally, we found that knockdown of IFI16 reduced the induction of IFN responses by synthetic and lentiviral DNA. It has been reported previously that IFI16 binds ssDNA and also cruciform DNA in vitro (52); in this report we show that ssDNA and lentiviral DNA in the cytosol is bound to and sensed by IFI16 and stimulates innate immune responses. The HINb domain of IFI16 is the main DNA-binding domain of IFI16 (4) and has been reported to interact with the DNA backbone upon binding (53). The present finding that PDE:PTO duplexes induce more IFN-β than PDE:PDE duplexes suggests that PDE:PTO duplexes still bind IFI16, because only one of the two nonbridging phosphate oxygen atoms is replaced by sulfur in PTO, and the PTO sulfur can form hydrogen bonds similar to the diester oxygen isotope. The thioester backbone may also confer less susceptibility to nuclease degradation. In addition to IFI16, we also tested the role of two other proposed DNA sensors, namely DDX41 and cGAS, in the innate response to ssDNA and lentivirus infections. Interestingly, we found that IFN/ISG expression induced by ssDNA1, dsDNA, or lentivirus is dependent on IFI16 and cGAS and, to a lesser extent dependent on DDX41. These data suggest crosstalk among IFI16, DDX41, and cGAS in the DNA-activated STING-dependent pathway. Whether these proteins act in the same or parallel pathways remains unresolved at present. Previous work by Chen and colleagues (26, 54) on cGAS, which produces cyclic GMP-AMP (cGAMP) mediating STING activation, has been performed mainly in undifferentiated THP1 cells, which, unlike PMA-differentiated THP1 cells and monocyte-derived macrophages, do not express IFI16. The same group recently reported cGAS to be a sensor of HIV-1 DNA in undifferentiated THP1 cells, stimulating cGAMP production and the accumulation of IFN-β mRNA (11). In that study, no data were provided on the role for cGAS in controlling HIV-1 replication. When we examined the role of the DNA sensors in controlling HIV-1 replication, we found that reduced expression of IFI16 and cGAS, but not of DDX41, led to significantly elevated HIV-1 replication. Such data are compatible with at least two models. In one model the different DNA sensors work in separate pathways, merging along the signal transduction pathway. In the other model the DNA-sensing machinery acts through one pathway in a cassette-like fashion involving several proteins, all of which are required for maximal immune activation but not for suboptimal responses. The latter model is supported further by data demonstrating the reconstitution of a DNA-activated pathway in HEK293T cells by ectopic expression of cGAS and STING (55) and by the reconstitution of a DNA-activated pathway in HEK293 cells, which harbor endogenous STING expression, by ectopic expression of IFI16 (35). Knowledge regarding the interplay between the proposed DNA sensors and STING during HIV infection, and in infections in general, is important for understanding how the host mounts protective immune responses.
HIV-1 infection induces only weak innate immune responses in cell culture (56), probably because of several factors, including a relatively weak immunostimulatory capacity of the viral genomic RNA (9, 10), the Trex1-dependent elimination of excess cDNA from infected cells (30), and direct inhibition of molecules involved in recognition and signaling (9, 57–59). In this work we found that HIV-1 RT products accumulated in infected macrophages and stimulated IFI16-dependent innate immune activation. Using an IDLV vector system, we observed elevated GFP expression in IFI16 shRNA cells as compared with control cells. This finding indicates that IFI16 stimulates antiviral functions acting before integration but does not rule out the possibility that other mechanisms acting at other steps of the replication cycle may also be involved. Accumulating data on ISGs acting as restricting factors [most recently, Schlafen family member 11 (60)] demonstrate that ISGs can target HIV at multiple stages of the replication cycle (43) These ISGs can be induced directly via IRFs, independently of IFN (13). Our finding that lentiviral shRNA knockdown of Trex1 was more efficient on a background of reduced IFI16 levels (shRNA) further suggests that IFI16 has a role in restricting lentiviruses. Although this issue needs further study, one potential implication could be that down-modulation of IFI16 expression may allow more efficient lentiviral gene transfer in human cells.
Two questions raised by the present study are (i) whether HIV-1 DNA is sensed during the productive replication cycle rather than during abortive infection, as indicated in previous studies of T cells (12), and (ii) specifically where in the cell the viral DNA is sensed. IFI16 is expressed in both the cytoplasm and the nucleus and has been proposed to act as a PRR in both subcellular locations (4, 61). HIV DNA is produced in the cytoplasm but also does accumulate in the nucleus either as integrated DNA or as 1- or 2-LTR circles, and in macrophages unintegrated DNA can lead to persistent viral gene transcription (62). Using a FISH system, we observed colocalization between viral DNA and IFI16 in the cytoplasm, and we were able to coprecipitate lentiviral DNA with endogenous IFI16 from cytoplasmic extracts. These data indicate that IFI16 senses the viral DNA in the cytoplasm in macrophages. Further knowledge about where host cells sense lentivirus infections will improve our understanding of the interplay between HIV and the innate immune system and particularly of the potentially different immunomodulatory effects of antiretroviral drugs depending on the step targeted in the viral replication cycle.
In this work we have identified stem-rich secondary structures in ssDNA strands as the immunostimulatory motif of this PAMP and have identified IFI16 as the sensor triggering the innate antiviral response. Our findings, taken together with data on IFN induction by dsDNA but not RNA:DNA duplexes, demonstrate that several DNA forms produced during the replication cycle of lentiviruses can activate the innate immune system via IFI16. Finally, this work demonstrates that IFI16 is actively recruited to sites of accumulation of lentiviral DNA in the cytoplasm in macrophages and activates defense mechanisms against lentiviral vector systems and HIV infection. Because macrophages are believed to be the first cell type infected during HIV-1 infection, these data suggest that IFI16 may be the first PRR sensing the virus after its entry into the cytosol of a permissive cell type.
Materials and Methods
Detailed descriptions of methods are given in SI Materials and Methods.
DNA Stimulation of Cells.
Stimulation of hMDMs, PMA-differentiated THP1, and BMDMs with DNA was conducted as described earlier (42). For details of the ssDNA oligos used, see Table S1; for dsDNA, see ref. 4. For the RNA:DNA hybrid oligo, the RNA strand was synthesized based on the HIV1_7200 sequence and was annealed to the antisense DNA strand in annealing buffer including RNase inhibitors. Poly(dA:dT) was purchased from Sigma (catalog no. P0883-25UN). All DNA oligos were used at a final concentration of 2 μg per stimulation unless specified otherwise. ssRNA40/LyoVec (InvivoGen) was used at 5 μg/mL.
RNA Analysis.
The analyses were performed using pre-made Taqman assays and the RNA-to-Ct one-step kit (Applied Biosystems). qPCR was performed on an MX3005 system (Agilent Technologies, Stratagene). RNA expression was normalized to β-actin and to the relevant untreated control. Data are stated as the mean ± SEM from biological replicates.
Confocal Microscopy.
For visualization of IFI16 after transfection with DNA at the indicated times, cells were fixed and permeabilized with methanol at −20 °C for 5 min and were labeled with antibodies against IFI16 (63) or STING (IMG-6485A; Imgenex) or with DAPI. Images were acquired on a Zeiss LSM 710 confocal microscope using 63× 1.4 oil lenses and were processed using Zen 2010 (Zeiss) and ImageJ (National Institutes of Health).
FISH.
Three different donors were used to generate hMDMs differentiated on coverslips. Cells were treated as indicated for 2–4 h and then fixed in ethanol and acidic acid at −20 °C for 5 min. Thirteen Stellaris FISH probes (Biosearch Technologies) were designed to cover the R′ region of the 5′ LTR and 3′ LTR of HIV Bal. Coverslips were prehybridized for 30 min at 37 °C. Probes were added, and coverslips were held at 37 °C overnight and then were incubated at 95 °C for 2 min. Coverslips were washed in 2× SSC at 20 °C for 5 min and at 60 °C for 5 min. Cells were blocked with 1% (wt/vol) BSA in PBS after labeling with an antibody against IFI16 (63).
Transduction of THP1 Cells with Lentiviral Vectors Encoding eGFP.
For integrase-deficient virus (IDLV) we used the third-generation packaging plasmid pMDlg/p-RRE with mutant D64V. A third-generation self-inactivating lentiviral transfer plasmid encoding eGFP, pCCL-PGK-eGFP, was packed in all viruses. THP1 cells were seeded 20 h before infection with either integrase-proficient viruses (IPLVs) or IDLVs encoding eGFP. Twenty-four hours after transduction supernatant was collected for CXCL10 ELISA, and the cells were supplemented with fresh medium. Ninety-six hours after transduction, cells were analyzed for eGFP fluorescence on a BD FACSCalibur (Becton, Dickinson).
Coprecipitation of IFI16 and DNA.
THP1 cells were transfected with HIV-derived DNA o were infected with IPLV with or without VSV-G pseudotyping and were harvested after 2 or 4 h, respectively. Cells were cross-linked and lysed and then were incubated with Dynabeads protein G (Invitrogen) preconjugated with IFI16, rabbit IgG, or NF-κB p65 rabbit anti-human antibodies. Beads were treated with proteinase K after qPCR for DNA quantification.
HIV Infection.
Full-length HIV-1 provirus consisted of the backbone of the HXB-3 strain of HIV-1 IIIB containing a 2,687-bp SalI-to-BamHI fragment from strain Bal, including Bal Tat, Rev, Vpu, and gp120 sequences plus part of Vpr and gp41 (National Institutes of Health AIDS Research And Reference Reagent Program catalog no. 11414) (64). Virus titer was determined on low-passage-number Tzmbl cells by X-Gal staining as readout. THP1 cells (100,000) were infected with HIV-1 Bal (MOI 1.5) by 1-h spinoculation followed by 6-h infection at 37 °C. Then nonfused virus particles were removed by multiple washes, and cells were used for experiments. Virus production was evaluated by p24 ELISA on cell supernatant. Absolute quantification of HIV DNA was performed by qPCR using primers and probes specific for HIV Gag DNA. Primers and probes specific for albumin were used as an endogenous control to normalize for the amount of input DNA.
Statistical Analysis.
Statistical analyses were performed using the Student’s t test.
Supplementary Material
Acknowledgments
We thank Kirsten S. Petersen for technical support. This work was supported by Danish Medical Research Council Research Grants 09-072636 (to S.R.P.), 11-114457 (to M.R.J.), and 271-08-0864 (to T.H.M.); by Sapere aude Grant 10-081986 (to M.R.J.); by Grants R83-A7598 and R34-3855 from the Lundbeck Foundation; and by the Kathrine og Vigo Skovgaards Fond, the Novo Nordisk Foundation, the Aase og Ejnar Danielsens Fond, and the Aarhus University Research Foundation. R.K.B., R.O.B., and S.B.J. are recipients of PhD fellowships from the Faculty of Health Sciences, Aarhus University. T.S.X. is supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 19183.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1311669110/-/DCSupplemental.
References
- 1.Lund J, Sato A, Akira S, Medzhitov R, Iwasaki A. Toll-like receptor 9-mediated recognition of Herpes simplex virus-2 by plasmacytoid dendritic cells. J Exp Med. 2003;198(3):513–520. doi: 10.1084/jem.20030162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441(7089):101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 3.Takaoka A, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448(7152):501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 4.Unterholzner L, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11(11):997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heil F, et al. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303(5663):1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 6.Paludan SR, Bowie AG, Horan KA, Fitzgerald KA. Recognition of herpesviruses by the innate immune system. Nat Rev Immunol. 2011;11(2):143–154. doi: 10.1038/nri2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.González-Navajas JM, Lee J, David M, Raz E. Immunomodulatory functions of type I interferons. Nat Rev Immunol. 2012;12(2):125–135. doi: 10.1038/nri3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abbink TE, Berkhout B. HIV-1 reverse transcription initiation: A potential target for novel antivirals? Virus Res. 2008;134(1-2):4–18. doi: 10.1016/j.virusres.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 9.Solis M, et al. RIG-I-mediated antiviral signaling is inhibited in HIV-1 infection by a protease-mediated sequestration of RIG-I. J Virol. 2011;85(3):1224–1236. doi: 10.1128/JVI.01635-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berg RK, et al. Genomic HIV RNA induces innate immune responses through RIG-I-dependent sensing of secondary-structured RNA. PLoS ONE. 2012;7(1):e29291. doi: 10.1371/journal.pone.0029291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao D, et al. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science. 2013;341(6148):903–906. doi: 10.1126/science.1240933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doitsh G, et al. Abortive HIV infection mediates CD4 T cell depletion and inflammation in human lymphoid tissue. Cell. 2010;143(5):789–801. doi: 10.1016/j.cell.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nasr N, et al. HIV-1 infection of human macrophages directly induces viperin which inhibits viral production. Blood. 2012;120(4):778–788. doi: 10.1182/blood-2012-01-407395. [DOI] [PubMed] [Google Scholar]
- 14.Hasan M, et al. Trex1 regulates lysosomal biogenesis and interferon-independent activation of antiviral genes. Nat Immunol. 2013;14(1):61–71. doi: 10.1038/ni.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413(6857):732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 16.Hemmi H, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408(6813):740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 17.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5(7):730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 18.Kato H, et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205(7):1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pichlmair A, et al. Activation of MDA5 requires higher-order RNA structures generated during virus infection. J Virol. 2009;83(20):10761–10769. doi: 10.1128/JVI.00770-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Z, et al. DDX1, DDX21, and DHX36 helicases form a complex with the adaptor molecule TRIF to sense dsRNA in dendritic cells. Immunity. 2011;34(6):866–878. doi: 10.1016/j.immuni.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138(3):576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ablasser A, et al. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10(10):1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim T, et al. Aspartate-glutamate-alanine-histidine box motif (DEAH)/RNA helicase A helicases sense microbial DNA in human plasmacytoid dendritic cells. Proc Natl Acad Sci USA. 2010;107(34):15181–15186. doi: 10.1073/pnas.1006539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X, et al. Cutting edge: Ku70 is a novel cytosolic DNA sensor that induces type III rather than type I IFN. J Immunol. 2011;186(8):4541–4545. doi: 10.4049/jimmunol.1003389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hornung V, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458(7237):514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339(6121):786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461(7265):788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paludan SR, Bowie AG. Immune sensing of DNA. Immunity. 2013;38(5):870–880. doi: 10.1016/j.immuni.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134(4):587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan N, Regalado-Magdos AD, Stiggelbout B, Lee-Kirsch MA, Lieberman J. The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nat Immunol. 2010;11(11):1005–1013. doi: 10.1038/ni.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharma S, et al. Innate immune recognition of an AT-rich stem-loop DNA motif in the Plasmodium falciparum genome. Immunity. 2011;35(2):194–207. doi: 10.1016/j.immuni.2011.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takahasi K, et al. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol Cell. 2008;29(4):428–440. doi: 10.1016/j.molcel.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 33.Crow YJ, et al. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 cause Aicardi-Goutières syndrome at the AGS1 locus. Nat Genet. 2006;38(8):917–920. doi: 10.1038/ng1845. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Z, et al. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol. 2011;12(10):959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li T, Diner BA, Chen J, Cristea IM. Acetylation modulates cellular distribution and DNA sensing ability of interferon-inducible protein IFI16. Proc Natl Acad Sci USA. 2012;109(26):10558–10563. doi: 10.1073/pnas.1203447109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith EJ, Marié I, Prakash A, García-Sastre A, Levy DE. IRF3 and IRF7 phosphorylation in virus-infected cells does not require double-stranded RNA-dependent protein kinase R or Ikappa B kinase but is blocked by Vaccinia virus E3L protein. J Biol Chem. 2001;276(12):8951–8957. doi: 10.1074/jbc.M008717200. [DOI] [PubMed] [Google Scholar]
- 37.Gessani S, et al. Induction of beta interferon by human immunodeficiency virus type 1 and its gp120 protein in human monocytes-macrophages: Role of beta interferon in restriction of virus replication. J Virol. 1994;68(3):1983–1986. doi: 10.1128/jvi.68.3.1983-1986.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gessani S, et al. Role of endogenous interferon-beta in the restriction of HIV replication in human monocyte/macrophages. J Leukoc Biol. 1994;56(3):358–361. doi: 10.1002/jlb.56.3.358. [DOI] [PubMed] [Google Scholar]
- 39.Hughes R, Towers G, Noursadeghi M. Innate immune interferon responses to human immunodeficiency virus-1 infection. Rev Med Virol. 2012;22(4):257–266. doi: 10.1002/rmv.1708. [DOI] [PubMed] [Google Scholar]
- 40.Kornbluth RS, Oh PS, Munis JR, Cleveland PH, Richman DD. Interferons and bacterial lipopolysaccharide protect macrophages from productive infection by human immunodeficiency virus in vitro. J Exp Med. 1989;169(3):1137–1151. doi: 10.1084/jem.169.3.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goujon C, Malim MH. Characterization of the alpha interferon-induced postentry block to HIV-1 infection in primary human macrophages and T cells. J Virol. 2010;84(18):9254–9266. doi: 10.1128/JVI.00854-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Horan KA, et al. Proteasomal degradation of herpes simplex virus capsids in macrophages releases DNA to the cytosol for recognition by DNA sensors. J Immunol. 2013;190(5):2311–2319. doi: 10.4049/jimmunol.1202749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iwasaki A. Innate immune recognition of HIV-1. Immunity. 2012;37(3):389–398. doi: 10.1016/j.immuni.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kawasaki T, Kawai T, Akira S. Recognition of nucleic acids by pattern-recognition receptors and its relevance in autoimmunity. Immunol Rev. 2011;243(1):61–73. doi: 10.1111/j.1600-065X.2011.01048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tytell AA, Lampson GP, Field AK, Hilleman MR. Inducers of interferon and host resistance. 3. Double-stranded RNA from reovirus type 3 virions (reo 3-RNA) Proc Natl Acad Sci USA. 1967;58(4):1719–1722. doi: 10.1073/pnas.58.4.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pichlmair A, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314(5801):997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 47.Hornung V, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314(5801):994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 48.Ishii KJ, et al. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat Immunol. 2006;7(1):40–48. doi: 10.1038/ni1282. [DOI] [PubMed] [Google Scholar]
- 49.Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24(1):93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 50.Sloan RD, Wainberg MA. The role of unintegrated DNA in HIV infection. Retrovirology. 2011;8:52. doi: 10.1186/1742-4690-8-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schlee M, et al. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31(1):25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brázda V, Coufal J, Liao JC, Arrowsmith CH. Preferential binding of IFI16 protein to cruciform structure and superhelical DNA. Biochem Biophys Res Commun. 2012;422(4):716–720. doi: 10.1016/j.bbrc.2012.05.065. [DOI] [PubMed] [Google Scholar]
- 53.Jin T, et al. Structures of the HIN domain:DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity. 2012;36(4):561–571. doi: 10.1016/j.immuni.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu J, et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339(6121):826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ablasser A, et al. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498(7454):380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mogensen TH, Melchjorsen J, Larsen CS, Paludan SR. Innate immune recognition and activation during HIV infection. Retrovirology. 2010;7:54. doi: 10.1186/1742-4690-7-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martinelli E, et al. HIV-1 gp120 inhibits TLR9-mediated activation and IFN-alpha secretion in plasmacytoid dendritic cells. Proc Natl Acad Sci USA. 2007;104(9):3396–3401. doi: 10.1073/pnas.0611353104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Doehle BP, Hladik F, McNevin JP, McElrath MJ, Gale M., Jr Human immunodeficiency virus type 1 mediates global disruption of innate antiviral signaling and immune defenses within infected cells. J Virol. 2009;83(20):10395–10405. doi: 10.1128/JVI.00849-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Harman AN, et al. HIV infection of dendritic cells subverts the IFN induction pathway via IRF-1 and inhibits type 1 IFN production. Blood. 2011;118(2):298–308. doi: 10.1182/blood-2010-07-297721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li M, et al. 2012. Codon-usage-based inhibition of HIV protein synthesis by human schlafen 11. Nature 491(7422):125–128.
- 61.Kerur N, et al. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi Sarcoma-associated herpesvirus infection. Cell Host Microbe. 2011;9(5):363–375. doi: 10.1016/j.chom.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kelly J, et al. Human macrophages support persistent transcription from unintegrated HIV-1 DNA. Virology. 2008;372(2):300–312. doi: 10.1016/j.virol.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Costa S, et al. Redistribution of the nuclear protein IFI16 into the cytoplasm of ultraviolet B-exposed keratinocytes as a mechanism of autoantigen processing. Br J Dermatol. 2011;164(2):282–290. doi: 10.1111/j.1365-2133.2010.10097.x. [DOI] [PubMed] [Google Scholar]
- 64.Hwang SS, Boyle TJ, Lyerly HK, Cullen BR. Identification of the envelope V3 loop as the primary determinant of cell tropism in HIV-1. Science. 1991;253(5015):71–74. doi: 10.1126/science.1905842. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.