Significance
We have identified a phage-encoded inhibitor of the major cytoskeletal protein in bacterial division. The inhibition is shown to confer T7 bacteriophage with a significant growth advantage in dividing hosts. Our studies thus indicate a strategy in bacteriophages to maximize their progeny number by inhibiting escape of one of the daughter cells of an infected bacterium.
Keywords: host takeover, bacterial division, bacteriophage biology
Abstract
Bacteriophages take over host resources primarily via the activity of proteins expressed early in infection. One of these proteins, produced by the Escherichia coli phage T7, is gene product (Gp) 0.4. Here, we show that Gp0.4 is a direct inhibitor of the E. coli filamenting temperature-sensitive mutant Z division protein. A chemically synthesized Gp0.4 binds to purified filamenting temperature-sensitive mutant Z protein and directly inhibits its assembly in vitro. Consequently, expression of Gp0.4 in vivo is lethal to E. coli and results in bacteria that are morphologically elongated. We further show that this inhibition of cell division by Gp0.4 enhances the bacteriophage’s competitive ability. This division inhibition is thus a fascinating example of a strategy in bacteriophages to maximize utilization of their hosts’ cell resources.
The abundance of bacteriophages and their importance to microbial evolution, and consequently to major ecological issues, provide an incentive to study their biology. Bacteriophage T7 and its host, Escherichia coli, provide a model for systematically studying host–virus interactions. The genetics of both the phage and its host have been studied extensively, and the putative functions or tentative physiological roles of over half of the 56 genes of phage T7 and the 4,453 genes of E. coli have been identified (1–3). T7 is a virulent phage which, upon infection of its host E. coli, produces over 100 progeny phage per host in less than 25 min. It is an obligatory lytic phage, meaning that a successful phage growth cycle always results in lysis of the host. The genome of T7 is a 39,937-bp double-stranded DNA molecule (4). Despite extensive knowledge of phage T7, the mechanism by which it takes over the host molecular machinery remains obscure. Specific functions have been attributed to over half of its gene products; all of the phage’s structural gene products have been characterized, as well as those that play roles in phage DNA replication. However, many of the gene products that create favorable conditions for phage growth in the host have not yet been assigned specific functions. One such gene product, which has neither a known function nor a reported phenotype during phage T7 growth, is gene product (Gp) 0.4. Gene 0.4 is a nonessential, 156-bp long gene, producing a peptide which is 51 aa in length. It is transcribed with the early genes ∼2 min after infection (1). A tagged Gp0.4 cloned into a genetically engineered T7 phage was detected in this study during infection by both mass spectrometry and Western blotting. The polycistronic RNA encoding genes 0.3–1.3 is specifically cleaved by RNaseIII upstream of gene 0.3 and downstream of gene 0.4 to yield an mRNA fragment encoding only genes 0.3 and 0.4 (4). In analyses of protein–protein interactions, no interaction of Gp0.4 with other phage proteins was detected (5).
In this study, we identified inhibitory interactions between Gp0.4 and filamenting temperature-sensitive mutant Z (FtsZ), a host protein that is essential for cell division. FtsZ, a homolog of eukaryotic tubulin, is conserved in almost all known bacterial and many archaeal species (6), with some exceptions (7). It assembles the Z ring at the site of cytokinesis, which recruits downstream proteins that remodel the cell wall and form the septum between the daughter cells. Inhibition of FtsZ function thus abolishes the bacterial cell’s ability to divide.
Several endogenous regulators and phage-derived inhibitors of FtsZ have been identified in E. coli. MinC, encoded by E. coli, is an endogenous inhibitor of FtsZ that is responsible, along with MinDE, for the specific localization of active FtsZ in the cell’s center and not in the poles (8). MinC can be activated by DicB, a protein encoded by the E. coli cryptic prophage, Kim, which targets MinC to the Z ring and causes the latter’s disassembly (9, 10). The E. coli-encoded SulA is also an endogenous inhibitor of FtsZ that is induced during DNA damage, thus linking DNA damage to the arrest of cell division (11). The kil gene of the E. coli cryptic prophage, Rac, is another reported FtsZ phage inhibitor (12). Kil toxicity has been shown to be relieved by expression of extra FtsZ; however, no direct interactions of this protein with FtsZ have been shown (12). The kil gene of E. coli λ phage was also implicated in inhibition of cell division, but no direct inhibition of FtsZ was shown (13). Interestingly, very recently, a Bacillus subtilis phage protein was shown to directly interact with FtsZ without inhibiting it. Inducing expression of this protein resulted in cells elongated by ∼50%. It was suggested that this interaction promotes replication of the phage’s DNA (14). Nevertheless, quite remarkably, no physiological role has yet been shown for any of the phage-encoded inhibitors of FtsZ.
Here, we show direct inhibitory interactions between Gp0.4 of the obligatory lytic phage, T7, and FtsZ. These interactions inhibit FtsZ assembly both in vivo and in vitro. We further show that Gp0.4 activity leads to increased competitive ability of the phage by blocking cell division. Our study thus provides a unique example of the physiological role played by a phage FtsZ inhibitor to increase the phage’s propagation capability.
Results
Bacteriophage T7 Gp0.4 Interacts Specifically with FtsZ in Vivo.
To identify the interactions of Gp0.4 with host proteins, we used a tandem-affinity purification (TAP) assay (15). A DNA fragment encoding an IgG-binding protein followed by a calmodulin-binding peptide (CBP) was cloned downstream of gene 0.4 in the T7 genome. The cloning procedures are described in SI Materials and Methods. E. coli hosts were infected with the genetically engineered phage and the infected bacteria, expressing the tagged Gp0.4, were collected. The bacteria were then concentrated and ruptured. The soluble content was affinity-purified on IgG beads and then on calmodulin beads. This procedure yielded the purified Gp0.4 with its interacting proteins along with some nonspecific contaminants. These proteins were identified by mass spectrometry as: FtsZ, the essential division protein described above; PrsA, an essential enzyme that synthesizes the essential cofactor phosphoribosylpyrophosphate; GlpD, a nonessential enzyme catalyzing the synthesis of dihydroxyacetone phosphate, and AceE, a nonessential subunit of the pyruvate dehydrogenase enzyme that catalyzes the synthesis of acetyl-CoA (16).
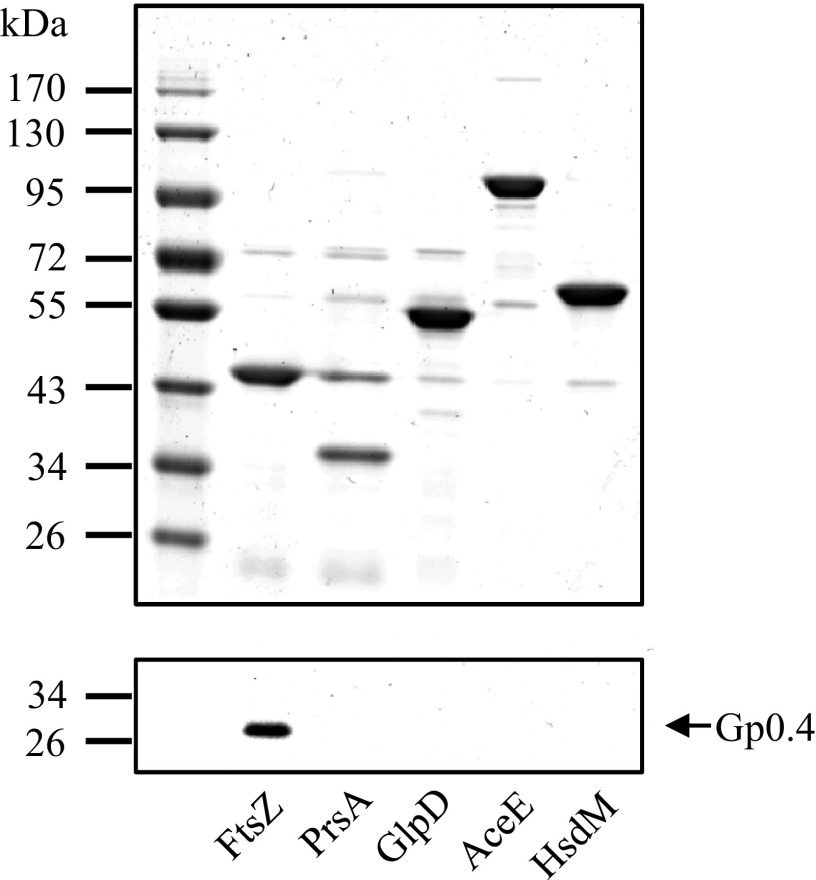
Because TAP occasionally generates false positives, we reciprocally validated the binding of Gp0.4 to each of the four pulled-down proteins and to a negative control, HsdM. We infected bacteria with an engineered T7 phage encoding the tagged Gp0.4, but this time, rather than pulling down Gp0.4, we pulled down each of the identified proteins through its N-terminal His6 tag using Ni-coated beads. The beads were washed and the tagged proteins eluted using imidazole. The total pulled-down proteins were adjusted to similar levels (Fig. 1), and a Western blot was performed to detect Gp0.4 using an antibody against its CBP tag. The only protein found to pull down Gp0.4 was FtsZ (Fig. 1). None of the other proteins, including the negative control, pulled down Gp0.4 at detectable levels. These results indicated that Gp0.4 binds to FtsZ either directly or indirectly. We therefore focused on characterizing the FtsZ–Gp0.4 interaction and did not further examine whether the other identified proteins, PrsA, GlpD, and AceE, indeed bind to Gp0.4 or are false positives from the TAP assay.
Fig. 1.
FtsZ specifically pulls down T7 Gp0.4. Four proteins shown to be pulled down by Gp0.4 [FtsZ (40.3 kDa), PrsA (34.2 kDa), GlpD (56.8 kDa), and AceE (99.7 kDa)] and a negative control [HsdM (59.3 kDa)] were affinity-purified using their His6 tag as described in SI Materials and Methods. The image shows Coomassie-stained 12% polyacrylamide gel of the eluted proteins (Upper). A Western blot to detect pulled-down Gp0.4 was performed using an antibody against the calmodulin binding peptide tag attached to Gp0.4 (Lower). Representative images are shown from one out of three experiments with similar results.
Toxicity of Gp0.4 Is Overcome by Host Mutations in ftsZ.
FtsZ is an essential gene product, and we therefore hypothesized that if Gp0.4 inhibits FtsZ, then its expression will result in bacterial death. Indeed, following expression induction of Gp0.4, using 0.2% l-arabinose, we found a six orders of magnitude drop in bacterial viability (Fig. 2A), indicating that Gp0.4 probably inhibits FtsZ. Notably, similar toxicity was observed in isogenic strains lacking minC or sulA (Fig. S1), the two known endogenous inhibitors of FtsZ, indicating that Gp0.4 inhibits FtsZ, but not through enhancement of these inhibitors’ activity, as exemplified by DicB (10).
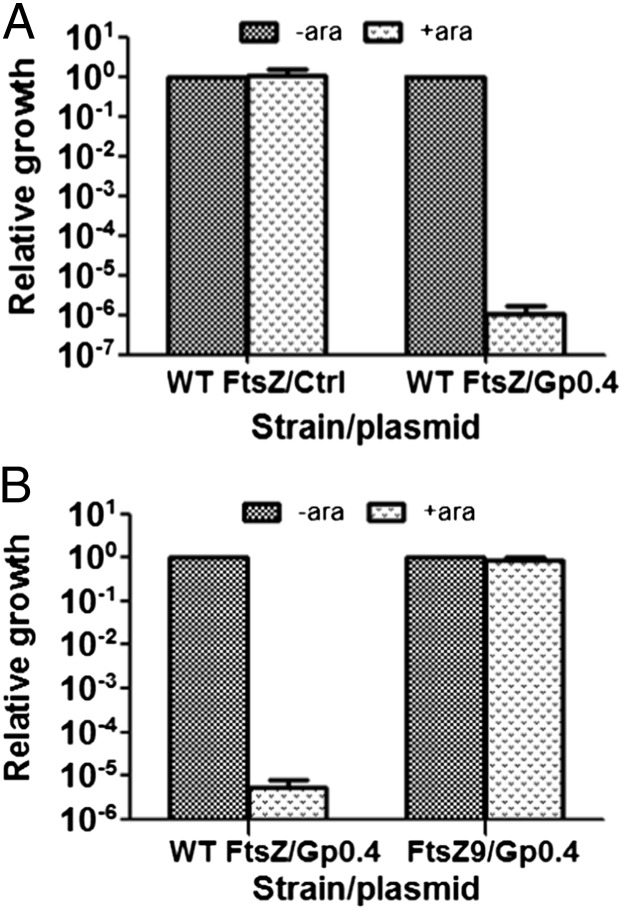
Fig. 2.
In vivo inhibition of bacterial growth by Gp0.4. (A) E. coli bacteria encoding WT FtsZ were transformed with plasmids encoding Gp0.4 (WT FtsZ/Gp0.4) or an empty vector (WT FtsZ/ctrl). Relative growth efficiency was calculated by dividing the cfu obtained in the presence of l-arabinose induction (+ara) by the number of cfu obtained in the absence of induction (-ara). (B) E. coli bacteria encoding the WT FtsZ or the resistant FtsZ9 were both transformed with plasmids encoding Gp0.4. Relative growth efficiency was calculated by dividing the cfu obtained in the presence of arabinose induction (+ara) by the number of cfu obtained in the absence of induction (-ara). Bars represent averages ± SD of three independent experiments.
If toxicity is indeed related to inhibition of FtsZ, then mutants resistant to Gp0.4 expression should arise with mutations in ftsZ. To test this, we isolated nine independent mutants surviving expression of Gp0.4 and sequenced their entire ftsZ gene. In three of these mutants, we identified a 6-nt insertion (TCGGCG) adjacent to a segment that encodes a duplication of this same insertion (TCGGCG|TCGGCG), thus resulting in a triplicate sequence of the 6 nt (TCGGCG|TCGGCG|TCGGCG). It seems likely that the replicating strand briefly denatures from the template and slips backward to anneal to the upstream 6 nt, thus resulting in another copy of these 6 nt. The resulting FtsZ contains a duplication of glycine–valine at position 18. This same insertion mutant, named FtsZ9, was identified previously and shown to overcome inhibition by the FtsZ inhibitors SulA and MinC (17). The ftsZ genes of the other six resistant mutants were also completely sequenced, but no mutations were identified. These mutants may be resistant to Gp0.4 by other genetic changes such as loss of gene 0.4 from the plasmid, loss of sensitivity to arabinose, the expression-inducer reagent (e.g., arabinose transporter mutation), or because of other suppressor mutations. This indicates that the insertion mutation is a dominant mutation, under the tested conditions, that renders the bacteria resistance to Gp0.4 expression. To demonstrate that the mutation in ftsZ renders the bacteria resistant to inhibition by Gp0.4, we used P1 phage to transduce the mutation back into the parental E. coli bacteria that were sensitive to Gp0.4 toxicity. These bacteria were then transformed with a plasmid encoding Gp0.4 and were tested for their susceptibility to Gp0.4 expression. All bacteria that were transduced with the altered ftsZ9 became resistant to Gp0.4 expression, whereas control bacteria that were transduced with the wild-type (WT) ftsZ remained sensitive to Gp0.4 expression (Fig. 2B). These results indicated that Gp0.4 specifically inhibits FtsZ and that this inhibition can be overcome by alteration of FtsZ at a specific site.
Purified Gp0.4 Inhibits Purified FtsZ in Vitro.
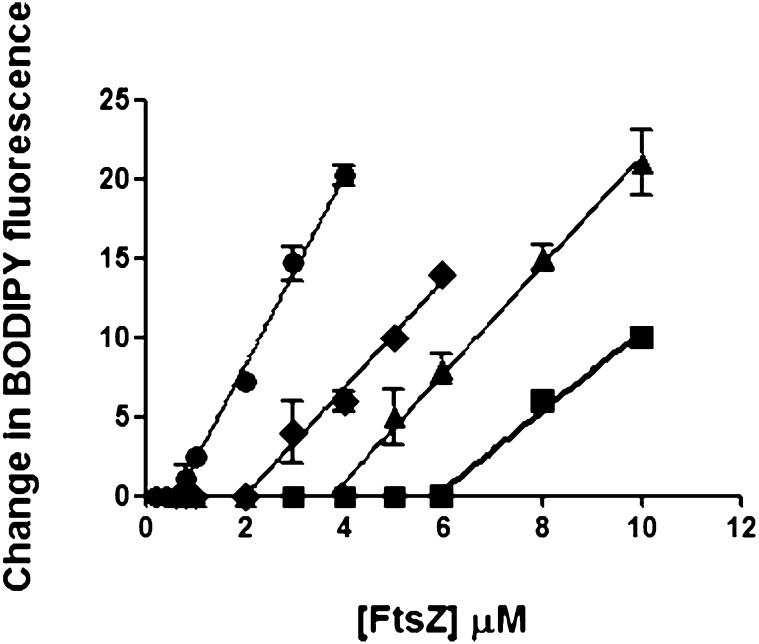
To rule out indirect interactions of Gp0.4 with FtsZ, we purified FtsZ to homogeneity and purchased a chemically synthesized Gp0.4 peptide. Inhibition of FtsZ assembly by Gp0.4 was examined by fluorescence-based assay. This assay monitors filament assembly of an altered FtsZ (S151C/Y222W), via changes in the quenching of the fluorescent dye boron-dipyrromethene (BODIPY) coupled to the cysteine at position 151, as described previously (18). In this assay, there is no assembly until the concentration of FtsZ reaches a critical concentration (Cc), above which there is a linear increase in fluorescence upon assembly. Addition of increasing concentrations of Gp0.4 inhibited assembly of the altered FtsZ filaments, as demonstrated by an upward shift in the apparent Cc (Fig. 3). We also purified to homogeneity the mutant protein FtsZ9. However, its assembly activity, as well as its GTPase activity, could not be detected, consistent with previous reports on the activity of FtsZ9 protein (17). We therefore could not assess its sensitivity to Gp0.4. Nevertheless, the results indicate that Gp0.4 directly blocks the assembly of WT FtsZ in vitro.
Fig. 3.
Gp0.4 effect on FtsZ protofilament assembly. Chemically synthesized Gp0.4 at 0 µM (circle), 1 µM (diamond), 3 µM (triangle), or 5 µM (square) was added to increasing concentrations of purified S151C/Y222W-FtsZ protein. The assembly was measured via a change in the fluorescence monitored by a Shimadzu RF-5301 PC spectrofluorometer. Each sample was monitored in duplicate. Error bars represent the SD of each point. The experiment was repeated twice with similar results.
Gp0.4 Expression Results in Elongation of Bacteria Encoding FtsZ.
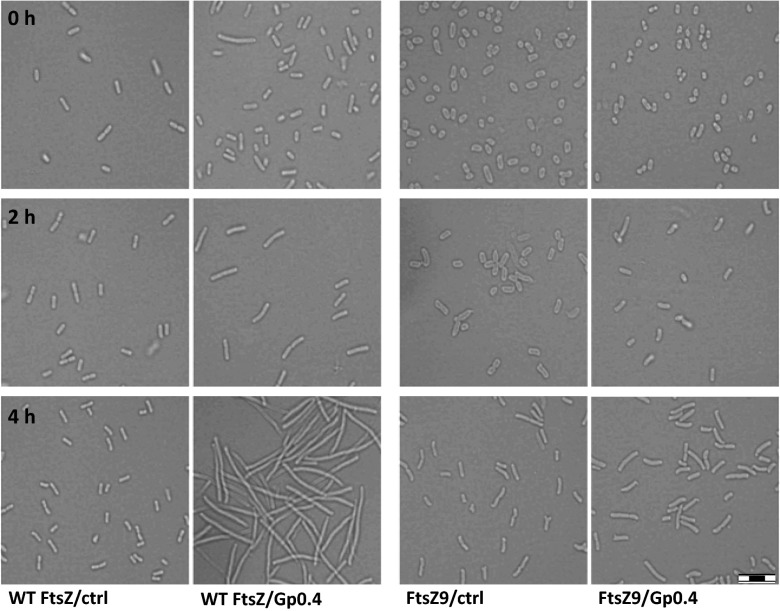
If Gp0.4 indeed directly inhibits FtsZ in vivo, then its expression should result in elongation of the cells. In contrast, bacteria having the Gp0.4-resistant variant FtsZ9 should show a normal morphology upon Gp0.4 expression. To test this, we used light microscopy to monitor bacterial morphology of WT E. coli and an E. coli encoding the ftsZ9 mutant, both induced to express Gp0.4 at low levels (0.001% l-arabinose). Indeed, E. coli bacteria harboring the plasmid encoding Gp0.4, but not those harboring a control empty vector, demonstrated cumulative elongation during the experiment (Fig. 4). E. coli encoding ftsZ9 were significantly less affected by Gp0.4 expression at the expression level used; however, at higher levels of induction (0.2% l-arabinose), these bacteria showed slight filamentation compared to the control bacteria. These results demonstrated that Gp0.4 inhibits division of cells encoding WT FtsZ.
Fig. 4.
Change in morphology of E. coli expressing Gp0.4. E. coli encoding the WT FtsZ or the Gp0.4-resistant FtsZ9 harboring pBAD-0.4 (Gp0.4) or a control pBAD18 (ctrl) plasmid were induced with 0.001% l-arabinose to drive expression from the plasmid promoter, as described in SI Materials and Methods. Images were taken 0, 2, and 4 h after induction. (Scale bar: 8 μm.) Images represent one out of two experiments showing similar results.
Gp0.4 Confers a Competitive Advantage to Bacteriophage T7 by Inhibiting Cell Division.
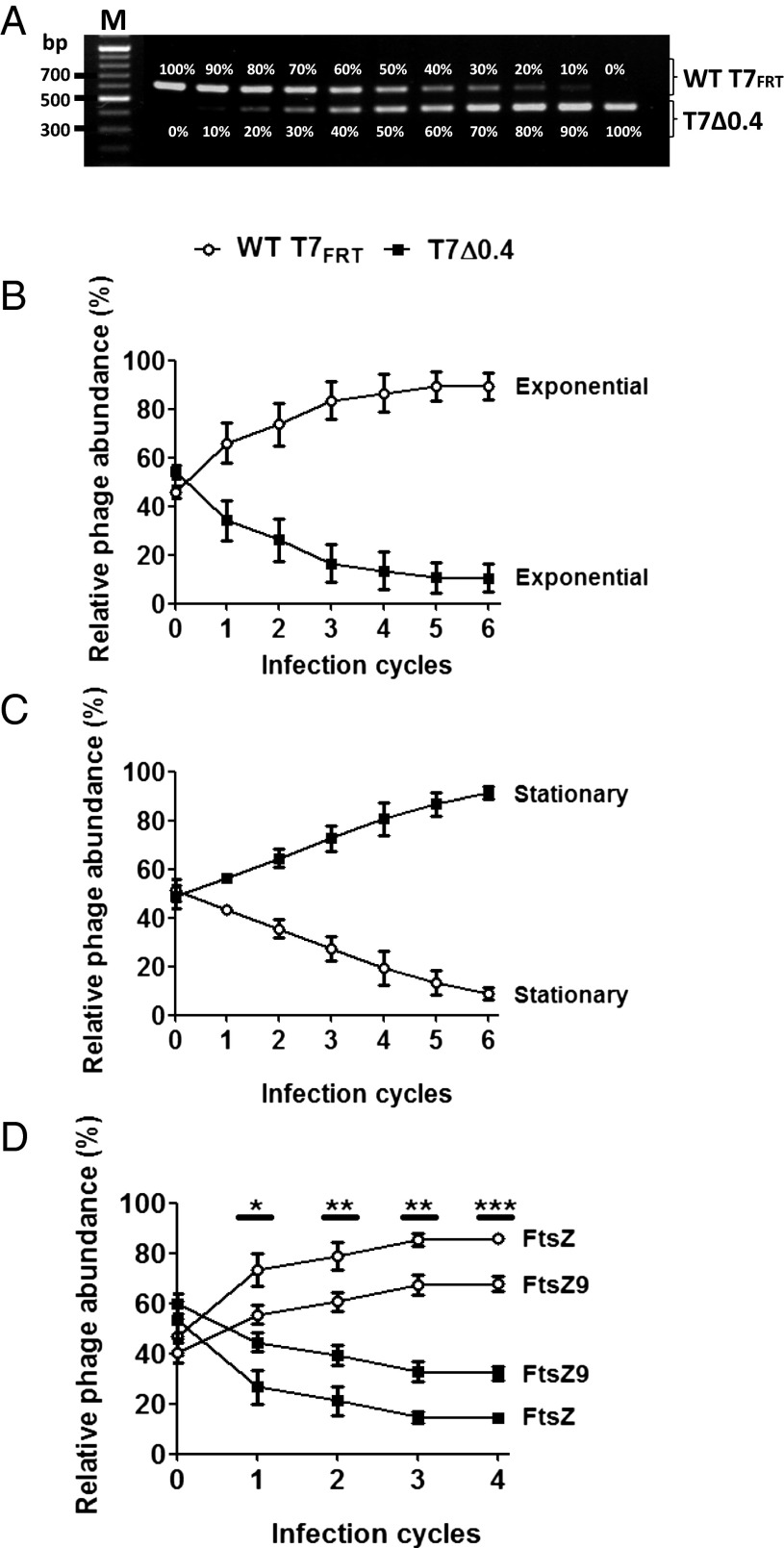
Finally, we speculated that the physiological role of Gp0.4 is to confer a competitive advantage to the phage infecting a dividing cell. If a cell divides early in infection, while there is only a single phage genome in the cell, one daughter cell will escape and phage propagation will be confined to only half of the cell resources. However, if the phage inhibits this daughter-cell escape using Gp0.4, then the entire cell resources are available for its progeny. To test whether this is indeed the case, we generated T7 phage lacking gene 0.4 (T7Δ0.4) and compared its competitive ability against the WT T7FRT (a phage having a similar “genetic scar” as does T7Δ0.4 but encodes gene 0.4; Table S1) in dividing or nondividing hosts. We expected that the WT T7FRT phage would have a significant advantage over T7Δ0.4 in dividing hosts, whereas this advantage would vanish in nondividing hosts. To determine the relative abundance of T7Δ0.4 compared with WT T7FRT in a phage mixture, we used PCR that amplifies the region flanking gene 0.4. This PCR discriminates between the two phages because amplification of the WT T7FRT results in a longer product than amplification of the deletion mutant. As can be seen in Fig. 5A, this assay is quantitative, enabling detection of different ratios of phage mixtures. To measure competition ability, we used a mixture containing an equal ratio of WT T7FRT to T7Δ0.4 to infect dividing E. coli hosts in the exponential-growth phase and nondividing E. coli hosts in the stationary-growth phase. Phage lysates were collected following each infection cycle, and PCR was carried out on these samples to measure the ratio of each phage. Obviously, a phage that is more competitive will produce more progeny in each infection cycle and therefore will eventually outcompete the other phage. As expected, WT T7FRT had a significant growth advantage on exponentially growing bacteria (Fig. 5B), indicating that Gp0.4 significantly increases the competitiveness of WT T7FRT phage on dividing bacteria. Remarkably, on nondividing hosts, the trend reversed showing that WT T7FRT was significantly less competitive than T7Δ0.4 (Fig. 5C). These results suggest that inhibition of cell division by Gp0.4 confers a competitive advantage to the phage. However, because stationary-phase bacteria also differ from exponential-phase bacteria in aspects other than cell division, we wanted to specifically test the effect on division inhibition. We therefore used E. coli encoding FtsZ9, the variant identified as resistant to Gp0.4 inhibition. We postulated that in this host, growing exponentially, the competitive advantage of WT T7FRT would be reduced because FtsZ9 is not inhibited. Indeed, on FtsZ9-encoding hosts WT T7FRT had a significantly lower competitive advantage compared with its advantage on FtsZ-encoding hosts. The fact that WT T7FRT still retains some competitive advantage is most likely because FtsZ9 is not completely refractory to Gp0.4 (Fig. 4). To verify that the measured DNA quantity in the above experiments corresponds to functional assembled phages, we plated dilutions of the lysates from the last cycle of each experiment. Individual plaque-forming units (pfu) were then picked and analyzed by PCR to determine whether they are WT T7FRT or T7Δ0.4. As expected, the percentage of the pfu in each lysate corroborated the results obtained by the quantitative PCR assay. The percentage of functional assembled phages of WT T7FRT recovered from the exponential phase culture was 78%, from the stationary phase 20%, from FtsZ-encoding cells 80%, and from FtsZ9-encoding cells 60%. These results indicate that the measured DNA levels correspond to functional phage particles. It should be noted that single-step burst size experiments showed that T7Δ0.4 produces less progeny per infected bacterium compared with WT-T7FRT (101 vs. 118 pfu per infected cell, respectively). However, because of technical limitations of the assay, the SDs were too high to demonstrate a statistical significance. Taken together, our results indicated that Gp0.4 has a physiological role in conferring a competitive advantage to the T7 phage through inhibition of FtsZ and, consequently, cell division.
Fig. 5.
PCR-quantified competition of phages. (A) PCR amplifying the region flanking gene 0.4 was carried out on T7Δ0.4 phage and WT T7FRT mixed at the indicated ratios. The upper bands are products obtained from amplifying the DNA of WT T7FRT, and the lower bands are from T7Δ0.4. (B) Graph showing the relative abundance of the indicated phages grown on an exponential-phase culture. (C) Graph showing the relative abundance of the indicated phages grown on a stationary-phase culture. (D) Graph showing the relative abundance of the indicated phages grown on an exponential-phase culture of E. coli encoding FtsZ9 or FtsZ. The PCR and relative abundance calculations were carried out as described in Materials and Methods. Graphs show averages ± SD of three independent experiments.*P = 0.01–0.05; **P = 0.001–0.01; ***P < 0.001.
Discussion
We showed that the T7-encoded protein Gp0.4 binds directly to, and inhibits, the division protein FtsZ. Bacteria overcoming this inhibition carry mutations in their ftsZ gene, resulting in a 2-aa insertion in the encoded protein. We further showed in vitro that Gp0.4 inhibits the assembly of FtsZ filaments. Accordingly, we demonstrated that in vivo, bacterial filamentation occurs when Gp0.4 is expressed, as expected from division inhibition. Importantly, the physiological benefit conferred on the T7 phage by expressing Gp0.4 is shown here. This inhibition of division seems to enable the phage to use the full resources of the cell, by preventing the escape of a daughter cell from infection. This function, which optimizes infection of T7 phage, concurs with its genetic location between gene 0.3 encoding a DNA-mimicking protein that prevents genomic cleavage by the host’s restriction enzymes, and gene 0.7 encoding a protein kinase that phosphorylates host proteins, both optimizing growth condition. The approach used in this study to identify protein–protein interactions biochemically demonstrate these interactions directly and show that their physiological importance may be applied to other phage proteins in this and other host–pathogen systems as well.
Gp0.4 can be used to study bacterial cell division, and it can also potentially serve as a tool in the fight against antibiotic-resistant bacteria. From a therapeutic viewpoint, FtsZ is a possible target for antibiotics because it is an essential bacterial protein, conserved across all known bacterial species and absent in eukaryotes. Indeed, several research groups have shown that small molecules inhibiting FtsZ can potentially serve as antibacterial drugs (e.g., refs. 19–21). Further studies on Gp0.4 to facilitate its use as an antimicrobial compound should determine the minimum effective peptide length for inhibition, its FtsZ inhibition capability across pathogenic bacterial species, its stability inside and outside mammalian tissues, and its penetration efficiency into the bacteria.
Why do phages require an inhibitor of division? T7 phage inhibits several host proteins, such as RNA polymerase (22), a dGTPase (23), the host restriction system (24), and presumably some RNases (25). In some cases, such as inhibition of a dGTPase that hydrolyzes dGTP into deoxyguanosine and triphosphate, the main reason is to preserve nucleotides for replication (23). RNA polymerase inhibition also secures more ribonucleotides for phage nucleic acid synthesis, in addition to preventing interference of the packaging step (26). We speculate that in the case of T7 inhibition of FtsZ, a major reason is to preserve cell resources. One aspect of achieving this goal is inhibition of FtsZ GTPase activity, which wastes energy at the expense of future phage progeny. Nevertheless, FtsZ GTPase activity wastes only ∼1/30,000th of the cell’s active metabolism (27), suggesting that this is not the major reason for FtsZ inhibition. A seemingly more important reason is that inhibiting division allows phage T7 to preserve half of the cell resources that would otherwise vanish with the dividing daughter cells. The advantage gained by the phage from inhibiting division of the host is substantial. To obtain a rough estimate of the beneficial role of such inhibition, one must calculate the percentage of dividing hosts infected by a single phage T7 genome out of the total infected hosts. Inhibiting division is beneficial only for a single genome of the phage in the cell. If there is more than one phage genome in the host, then division inhibition is less likely to be beneficial for the phage because these genomes will be distributed between both daughter cells regardless. The time taken for the T7 genome to produce its replication enzymes, and soon after have more than a single genome in the cell, under optimal conditions is ∼6 min, whereas the early T7 proteins, including Gp0.4, are expressed 2 min postinfection (1). This means that Gp0.4 is beneficial for a total period of 4 min. These 4 min constitute 20% of the generation time of the E. coli host, estimated to be ∼20 min under optimal conditions (28). In other words, 20% of the host cells infected with a single phage genome undergo division during this time. Thus, unless inhibited during this time, 20% of the infected bacterial hosts will divide and 50% of the daughter cells will not give rise to the phage progeny. This rough calculation estimates that a phage having a division inhibitor could increase its progeny by ∼10%, which is a significant benefit from such a small encoded molecule. These calculations assume that the daughter cells escape infection by the progeny of the infecting phage by diffusing away or actively swimming from the infection site or because of competition by other phages. We indeed show that a significant advantage is conferred to a phage encoding gene 0.4 compared with a phage lacking it (Fig. 5). Thus, it seems natural that a division inhibitor would be one of the earliest proteins encoded by phage T7, because earlier activity enhances the benefit. Taken together, our study highlights another aspect of phage–host interactions by experimentally demonstrating a strategy used by bacteriophages to markedly increase their progeny.
Materials and Methods
Reagents, Strains, and Plasmids.
Reagents, as well as the bacterial strains, phages, plasmids, and oligonucleotides, used in this study are listed in SI Materials and Methods and Table S1.
Strain and Plasmid Construction.
Strain construction by homologous recombination and plasmid construction are described in SI Materials and Methods.
TAP Assay.
E. coli strain K-12 was aerated overnight in LB at 37 °C. The overnight culture was diluted 1:100 in 750 mL of LB medium at 30 °C and aerated to an OD600 of 0.5. The cells were then infected with phage T7 having a tagged gene 0.4 with at a multiplicity of infection (MOI) of 4. The culture was aerated for 14 min at 30 °C and then cooled immediately to 0 °C. The bacteria were centrifuged for 10 min at 9,000 × g and 4 °C. The pellet was resuspended in 5 mL of lysis buffer [20 mM Tris⋅HCl (pH 8.0), 2 mM EDTA, 150 mM NaCl, 0.1% wt/vol Nonidet P-40 (Sigma), 200 µg/mL hen–egg lysozyme (Calbiochem), 2.5 U/mL benzonase (Novagen), and Complete Mini EDTA-free tablet (Roche)] and was frozen at −80 °C overnight. The sample was thawed in a room temperature (RT) water bath and then incubated for 30 min. It was then frozen again in liquid nitrogen and thawed in an RT water bath twice more. The lysate was injected through a 23-gauge needle five times and then centrifuged in a Sorvall SS34 rotor for 10 min at 9,000 × g and 4 °C. The samples were filtered through a 45-µL filter. IgG Sepharose beads (400 µL; Pharmacia) were added, and the suspension was incubated on a rotating platform at 4 °C for 1 h. The lysate and beads were poured onto a Bio-Rad Poly-Prep Chromatography Column (0.8 × 4 cm). The beads were washed three times with 10 mL of IPP150 buffer [10 mM Tris⋅HCl (pH 8.0), 150 mM NaCl, and 0.1% Nonidet P-40] and then with 10 mL TEV cleavage buffer [10 mM Tris⋅HCl (pH 8.0), 150 mM NaCl, 0.1% Nonidet P-40, 0.5 mM EDTA, 1.0 mM DTT (Calbiochem)]. The bottom of the column was sealed, and 500 µL of TEV cleavage buffer and 50 µL of TEV enzyme were added. The samples were incubated on a rotating platform at 16 °C for 2 h. TEV cleavage buffer (1 mL) was added to the columns. Three volumes of calmodulin binding buffer [10 mM Tris⋅HCl (pH 8.0), 150 mM NaCl, 1 mM Mg2+ acetate (Merck), 1 mM imidazole (Sigma), 2 mM CaCl2 (Merck), 10 mM β-mercaptoethanol (Sigma), and 0.1% Nonidet P-40], 3 µL 1 M CaCl2 per mL IgG eluate, and 300 µL calmodulin affinity resin (Stratagene) were added to the TEV supernatant. The sample was incubated on a rotating platform at 4 °C for 1 h. The lysate and beads were poured onto a new Bio-Rad Poly-Prep Chromatography Column. The beads were washed twice with 8 mL calmodulin binding buffer and then with 8 mL calmodulin binding buffer with a lower detergent concentration (0.02% Nonidet P-40 instead of 0.1% Nonidet P-40). The samples were eluted with 1 mL calmodulin elution buffer [10 mM Tris⋅HCl (pH 8.0), 150 mM NaCl, 0.02% Nonidet P-40, 1 mM Mg2+ acetate, 1 mM imidazole, 20 mM EGTA (Calbiochem) and 10 mM β-mercaptoethanol]. The elution was precipitated in trichloro-acetic acid (TCA) (Calbiochem) by adjusting it to 25% TCA (vol/vol), vortexing and placing it on ice overnight. The overnight elution was centrifuged at maximum speed at 4 °C for 5 min. The TCA pellet was first washed with 1 mL cold (−20 °C) acetone containing 0.05 N HCl (Bio-Lab) and centrifuged for 5 min at 13,000 × g and 4 °C. The second wash was carried out with 1 mL cold (−20 °C) acetone and centrifugation for 5 min at 13,000 × g and 4 °C. The supernatant was carefully removed. The pellet was dried for 5 min in a vacuum concentrator (Concentrator 5301; Eppendorf) with heating (45 °C). The pellet was used for mass spectrometry (Technion). Interactions of proteins identified by mass spectrometry with Gp0.4 were validated as described in SI Materials and Methods.
Gp0.4 Toxicity Assay.
Independent cultures of E. coli bacteria harboring pBAD-0.4 or pBAD18 as a control were aerated overnight in LB supplemented with 100 µg/mL ampicillin at 37 °C. Overnight cultures were diluted 1:100 in fresh LB supplemented with 100 µg/mL ampicillin at 37 °C and aerated to an OD600 of 0.1. The cultures were then centrifuged for 10 min at ∼4500g, 4 °C and resuspended to an OD600 of 1. Cultures were serially diluted and plated overnight at 37 °C on LB agar with 100 µg/mL ampicillin, with or without 0.2% (wt/vol) arabinose. The toxicity was calculated by dividing the number of cfu obtained in the presence of arabinose by that obtained in the absence of arabinose. E. coli NEB5α/pBAD-0.4 and E. coli NEB5α/pBAD18 colonies surviving on plates supplemented with arabinose were used as a template for PCR amplification using primers RK28F and RK28Rb (Table S1). The PCR product amplifying the ftsZ gene was DNA-sequenced using primers RK28F, RK28Ra and RK28Rb (Table S1). P1 transduction for transferring the obtained ftsZ mutations was carried out as described in SI Materials and Methods.
In Vitro FtsZ Assays.
FtsZ assembly was assayed by quenching of BODIPY fluorescence, as described previously (18, 29).
Phage Competition Assay.
To determine the effect of gene 0.4 in dividing and nondividing bacteria, BW25113ΔaceF (Table S1) bacteria were aerated overnight in LB supplemented with 25 µg/mL kanamycin at 37 °C. This culture was then kept on ice and used as the host culture in the stationary phase representing the nondividing bacteria. A sample of this overnight culture was diluted 1:100 in LB supplemented with 25 µg/mL kanamycin at 37 °C and aerated to an OD600 of ∼0.15. This culture was kept on ice and used as the culture in the exponential phase representing dividing bacteria. A mixture of T7Δ0.4 and WT T7FRT was used to infect the exponential- and stationary-phase cultures at a MOI of ∼0.01 at 37 °C. Each infection cycle was defined as coincubation of the phage mixture and the bacteria for 1 h at 37 °C with shaking. A diluted lysate was transferred from one cycle to the next, resulting in cumulative effect of the competition over time. The relative abundance of T7Δ0.4 compared with WT T7FRT in a phage mixture was determined for each cycle by PCR amplification of the region flanking gene 0.4 using primers 85F and SM24R1 (Table S1). The amplified DNA was 592 and 437 bp for WT T7FRT and T7Δ0.4, respectively. Band intensities were quantified by their digital densities using ImageJ software. Intensity values for the band representing WT T7FRT were multiplied by 0.74 to correct for size differences compared with T7Δ0.4. The relative abundance of each phage was calculated by dividing the density of the representative band by the total intensity of both bands. To verify that the intensities of the band correspond to functional phages, lysates were plated from the end point of four representative samples used to generate Fig. 5 B–D. Between 48 and 54 pfu from each of these plates were individually picked and analyzed by PCR for presence or absence of gene 0.4, and the percentage of each phage out of the total pfu was calculated.
To determine the effect of gene 0.4 on E. coli encoding WT-ftsZ compared with E. coli encoding ftsZ9, BW25113ΔaceF and RK6497 [BW25113ΔaceF, ftsZ9] (Table S1) bacteria were used as described above for the logarithmic-phase culture but grown at 32 °C because of growth deficiency of E. coli encoding ftsZ9 at higher temperature. An unpaired t test was used to analyze competition in each infection cycle. Statistical significance was determined using the GraphPad Prism software.
Supplementary Material
Acknowledgments
We thank Gabriel Mircus for technical help and Camille Vainstein for professional language editing. This research was supported by Binational Science Foundation Grant 2009218, a Sackler School of Medicine Research Grant, National Institutes of Health Grant GM66014, and Marie Curie International Reintegration Grant PIRG-GA-2009-256340.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1314096110/-/DCSupplemental.
References
- 1.Studier FW. Bacteriophage T7. Science. 1972;176(4033):367–376. doi: 10.1126/science.176.4033.367. [DOI] [PubMed] [Google Scholar]
- 2.Molineux IJ. In: The Bacteriophages. Abedon ST, Calendar RL, editors. Oxford: Oxford Univ Press; 2005. pp. 275–299. [Google Scholar]
- 3.Kitagawa M, et al. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): Unique resources for biological research. DNA Res. 2005;12(5):291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- 4.Dunn JJ, Studier FW. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983;166(4):477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- 5.Häuser R, et al. Bacteriophage protein-protein interactions. Adv Virus Res. 2012;83:219–298. doi: 10.1016/B978-0-12-394438-2.00006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vaughan S, Wickstead B, Gull K, Addinall SG. Molecular evolution of FtsZ protein sequences encoded within the genomes of archaea, bacteria, and eukaryota. J Mol Evol. 2004;58(1):19–29. doi: 10.1007/s00239-003-2523-5. [DOI] [PubMed] [Google Scholar]
- 7.Erickson HP, Osawa M. Cell division without FtsZ—a variety of redundant mechanisms. Mol Microbiol. 2010;78(2):267–270. doi: 10.1111/j.1365-2958.2010.07321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Boer PA, Crossley RE, Rothfield LI. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell. 1989;56(4):641–649. doi: 10.1016/0092-8674(89)90586-2. [DOI] [PubMed] [Google Scholar]
- 9.Johnson JE, Lackner LL, de Boer PA. Targeting of (D)MinC/MinD and (D)MinC/DicB complexes to septal rings in Escherichia coli suggests a multistep mechanism for MinC-mediated destruction of nascent FtsZ rings. J Bacteriol. 2002;184(11):2951–2962. doi: 10.1128/JB.184.11.2951-2962.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou H, Lutkenhaus J. MinC mutants deficient in MinD- and DicB-mediated cell division inhibition due to loss of interaction with MinD, DicB, or a septal component. J Bacteriol. 2005;187(8):2846–2857. doi: 10.1128/JB.187.8.2846-2857.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gottesman S, Halpern E, Trisler P. Role of sulA and sulB in filamentation by lon mutants of Escherichia coli K-12. J Bacteriol. 1981;148(1):265–273. doi: 10.1128/jb.148.1.265-273.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conter A, Bouché JP, Dassain M. Identification of a new inhibitor of essential division gene ftsZ as the kil gene of defective prophage Rac. J Bacteriol. 1996;178(17):5100–5104. doi: 10.1128/jb.178.17.5100-5104.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greer H. The kil gene of bacteriophage lambda. Virology. 1975;66(2):589–604. doi: 10.1016/0042-6822(75)90231-7. [DOI] [PubMed] [Google Scholar]
- 14.Ballesteros-Plaza D, Holguera I, Scheffers DJ, Salas M, Muñoz-Espín D. Phage 29 phi protein p1 promotes replication by associating with the FtsZ ring of the divisome in Bacillus subtilis. Proc Natl Acad Sci USA. 2013;110(30):12313–12318. doi: 10.1073/pnas.1311524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rigaut G, et al. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17(10):1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 16.Keseler IM, et al. EcoCyc: A comprehensive database of Escherichia coli biology. Nucleic Acids Res. 2011;39(Database issue):D583–D590. doi: 10.1093/nar/gkq1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dai K, Mukherjee A, Xu Y, Lutkenhaus J. Mutations in ftsZ that confer resistance to SulA affect the interaction of FtsZ with GTP. J Bacteriol. 1994;176(1):130–136. doi: 10.1128/jb.176.1.130-136.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Y, Erickson HP. Conformational changes of FtsZ reported by tryptophan mutants. Biochemistry. 2011;50(21):4675–4684. doi: 10.1021/bi200106d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J, et al. Discovery of a small molecule that inhibits cell division by blocking FtsZ, a novel therapeutic target of antibiotics. J Biol Chem. 2003;278(45):44424–44428. doi: 10.1074/jbc.M307625200. [DOI] [PubMed] [Google Scholar]
- 20.Margalit DN, et al. Targeting cell division: Small-molecule inhibitors of FtsZ GTPase perturb cytokinetic ring assembly and induce bacterial lethality. Proc Natl Acad Sci USA. 2004;101(32):11821–11826. doi: 10.1073/pnas.0404439101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haydon DJ, et al. An inhibitor of FtsZ with potent and selective anti-staphylococcal activity. Science. 2008;321(5896):1673–1675. doi: 10.1126/science.1159961. [DOI] [PubMed] [Google Scholar]
- 22.Hesselbach BA, Nakada D. “Host shutoff” function of bacteriophage T7: Involvement of T7 gene 2 and gene 0.7 in the inactivation of Escherichia coli RNA polymerase. J Virol. 1977;24(3):736–745. doi: 10.1128/jvi.24.3.736-745.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beauchamp BB, Richardson CC. A unique deoxyguanosine triphosphatase is responsible for the optA1 phenotype of Escherichia coli. Proc Natl Acad Sci USA. 1988;85(8):2563–2567. doi: 10.1073/pnas.85.8.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Studier FW. Gene 0.3 of bacteriophage T7 acts to overcome the DNA restriction system of the host. J Mol Biol. 1975;94(2):283–295. doi: 10.1016/0022-2836(75)90083-2. [DOI] [PubMed] [Google Scholar]
- 25.Marchand I, Nicholson AW, Dreyfus M. Bacteriophage T7 protein kinase phosphorylates RNase E and stabilizes mRNAs synthesized by T7 RNA polymerase. Mol Microbiol. 2001;42(3):767–776. doi: 10.1046/j.1365-2958.2001.02668.x. [DOI] [PubMed] [Google Scholar]
- 26.Qimron U, Kulczyk AW, Hamdan SM, Tabor S, Richardson CC. Inadequate inhibition of host RNA polymerase restricts T7 bacteriophage growth on hosts overexpressing udk. Mol Microbiol. 2008;67(2):448–457. doi: 10.1111/j.1365-2958.2007.06058.x. [DOI] [PubMed] [Google Scholar]
- 27.Stricker J, Maddox P, Salmon ED, Erickson HP. Rapid assembly dynamics of the Escherichia coli FtsZ-ring demonstrated by fluorescence recovery after photobleaching. Proc Natl Acad Sci USA. 2002;99(5):3171–3175. doi: 10.1073/pnas.052595099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cooper S, Helmstetter CE. Chromosome replication and the division cycle of Escherichia coli B/r. J Mol Biol. 1968;31(3):519–540. doi: 10.1016/0022-2836(68)90425-7. [DOI] [PubMed] [Google Scholar]
- 29.Chen Y, Milam SL, Erickson HP. SulA inhibits assembly of FtsZ by a simple sequestration mechanism. Biochemistry. 2012;51(14):3100–3109. doi: 10.1021/bi201669d. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.