Significance
Spinal muscular atrophy (SMA), a common genetic motor neuron (MN) degenerative disease and leading hereditary cause of infant mortality, results from survival of motor neuron (SMN) protein deficiency. However, SMN’s ubiquitous expression and housekeeping functions in biogenesis of snRNPs, the spliceosome’s subunits, seems difficult to reconcile with SMA’s MN selective pathology. Here, we sequenced transcriptomes of MNs and adjacent white matter microdissected from spinal cords of presymptomatic SMA mice. This process revealed selective and MN-specific splicing and expression-level perturbations of mRNAs, including those essential for establishing neuromuscular junctions, the first structures that degenerate in SMA. We suggest that SMN’s central role in transcriptome regulation explains the gene-expression perturbations that impair MN function and survival in SMA.
Keywords: transcriptome perturbations, Z+ (neuronal) agrin, C1q complex
Abstract
The motor neuron (MN) degenerative disease, spinal muscular atrophy (SMA) is caused by deficiency of SMN (survival motor neuron), a ubiquitous and indispensable protein essential for biogenesis of snRNPs, key components of pre-mRNA processing. However, SMA’s hallmark MN pathology, including neuromuscular junction (NMJ) disruption and sensory-motor circuitry impairment, remains unexplained. Toward this end, we used deep RNA sequencing (RNA-seq) to determine if there are any transcriptome changes in MNs and surrounding spinal cord glial cells (white matter, WM) microdissected from SMN-deficient SMA mouse model at presymptomatic postnatal day 1 (P1), before detectable MN pathology (P4–P5). The RNA-seq results, previously unavailable for SMA at any stage, revealed cell-specific selective mRNA dysregulations (∼300 of 11,000 expressed genes in each, MN and WM), many of which are known to impair neurons. Remarkably, these dysregulations include complete skipping of agrin’s Z exons, critical for NMJ maintenance, strong up-regulation of synapse pruning-promoting complement factor C1q, and down-regulation of Etv1/ER81, a transcription factor required for establishing sensory-motor circuitry. We propose that dysregulation of such specific MN synaptogenesis genes, compounded by many additional transcriptome abnormalities in MNs and WM, link SMN deficiency to SMA’s signature pathology.
Spinal muscular atrophy (SMA) is an autosomal recessive motor neuron (MN) degenerative disease and a leading genetic cause of infant mortality (1, 2). SMA is caused by deletions or point mutations in the survival of motor neurons 1 gene (SMN1) (3), exposing a splicing defect in a duplicated gene (SMN2), which produces predominantly exon 7-skipped mRNA (SMNΔ7) (4). This in turn creates a degron that destabilizes the SMNΔ7 protein, resulting in reduced SMN levels (SMN deficiency) (5). SMA pathology has been extensively characterized in patients as well as a widely used SMA mouse model (6). Major morphological and biochemical deficits have been found at neuromuscular junctions (NMJs) and sensory-motor synapses. NMJ defects are first detectable at postnatal day 5 (P5), including presynaptic defects of terminal arborization and intermediate neurofilament aggregation in MNs, poor postsynaptic organization of AChRs in muscle, as well as reduced synaptic vesicle density and release at the NMJ (7–9). Importantly, similar NMJ defects have been reported in type I (the most severe type) SMA human fetuses (10). SMA MNs have also been shown to be hyperexcitable and loss of proprioceptive synapses on the somata, and proximal dendrites of SMA MNs (MN deafferentation) occurs no later than P4 (11–13). These findings indicate that both peripheral (NMJs) and central (sensory-motor) synapses are affected in SMA mice at early stage of disease. Although SMA is clinically manifested primarily in MNs, recent studies have shown that other cell types and nonneuronal tissues are also involved (13, 14, 15).
SMN, as part of the SMN–Gemins complex (16), functions in the assembly of Sm protein cores on snRNAs (U1, U2, U4, U5, U11, U12, and U4atac), a key step in snRNP biogenesis (17–20). snRNPs are the major components of the spliceosome, the machinery that splices introns to produce mRNAs (21). In addition, U1 snRNP protects nascent transcripts from premature termination (a function termed telescripting) and regulates mRNA length (22, 23). Consistent with its general function, SMN is ubiquitously expressed and essential for viability of all eukaryotic cell types. Both snRNP assembly capacity and SMA severity correspond directly to the degree of SMN deficiency (24, 25). Interestingly, SMN decrease results in a nonuniform change of individual snRNP’s level, and thus an altered snRNP repertoire (abundance and stoichiometry), which is apparent in mouse and Drosophila SMA models (15, 26, 27). It is likely that this alteration contributes to the transcriptome changes observed in the spinal cord and other tissues of the SMA mouse model at P11 (15), a symptomatic stage for these mice that typically survive to P13 (6). The most striking of these changes are widespread tissue-specific differential expression and pre-mRNA splicing defects, consisting of both alternative splicing changes and noncanonical splice site use (15, 28). These findings indicate that mRNA perturbations are a feature of SMA and that they are not restricted to spinal cord or MNs, where the overt clinical phenotypes manifest (15).
Despite major advances in understanding the genetic basis of SMA and SMN’s function, the molecular basis of SMA pathogenesis has remained unclear, and a direct link between specific transcriptome abnormalities and SMA’s distinct synaptic pathogenesis in mammalian SMA models has not been established. Here, we performed RNA sequencing (RNA-seq) of laser-capture microdissected (LCM) MNs and neighboring white matter (WM) glia from the same SMA mouse model at P1, an early presymptomatic stage. Our studies identify expression-level changes and splicing abnormalities of specific mRNAs critical for MN function, revealing molecular events at early disease stage that could explain key aspects of SMA pathogenesis.
Results
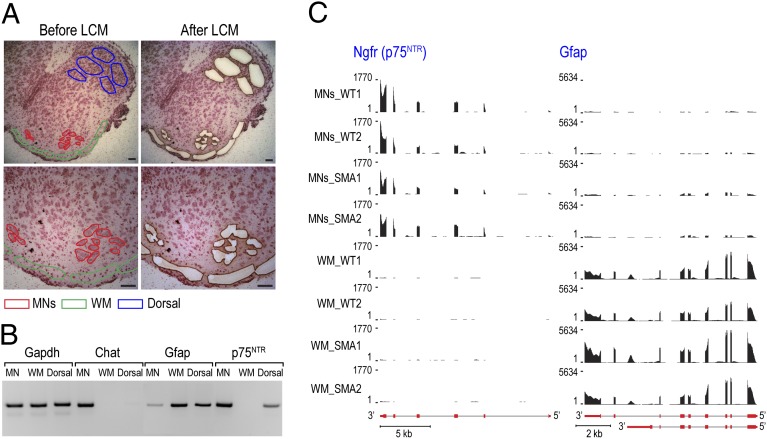
SMA primarily affects spinal cord MNs (1), and thus we probed directly for transcriptome changes in MNs of SMN-deficient mice (SMN2+/+;SMNΔ7+/+;Smn−/−) using LCM. We isolated 1,500–2,000 MN cell bodies from the lumbar level spinal cord of each mouse at age of P1. We also microdissected adjacent WM (including astrocytes, oligodendrocytes, and microglia) as well as neurons and glia of the dorsal horn region (referred to as Dorsal) from the same spinal cord sections (Fig. 1A), as nonneuronal and non-MN cell controls, respectively. RT-PCR of cell type-specific markers confirmed the effectiveness of the procedure, showing specific MN expression of choline acetyltransferase (Chat), and nerve growth factor receptor (p75NTR) preferentially expressed in spinal MNs of embryonic and neonatal mice (29). In contrast, Gapdh was ubiquitously expressed and glial fibrillary acidic protein (Gfap) expression was detected predominantly in WM and Dorsal (Fig. 1B).
Fig. 1.
Isolating MNs and surrounding WM from spinal cord of P1 SMA mice by LCM. (A) Frozen cross-sections of WT mouse lumbar spinal cord were prepared as described in Materials and Methods. Images were taken before and after LCM. (Scale bar, 50 μm.) (B) Purity of LCM samples was confirmed by RT-PCR detecting neuron (Chat and p75NTR) and astrocyte (Gfap) markers, using total RNA isolated from MNs, WM, and Dorsal of WT mice. (C) UCSC Genome Browser view of p75NTR and Gfap displaying RNA-seq mapped reads profile, showing four tracks for MNs and WM samples from each of the WT or SMA mouse, respectively. Gene structures (red boxes for exons and gray lines for introns) are depicted below the tracks. Strong p75NTR expression is observed in all four MNs samples and barely detectable in the WM, but Gfap expression is WM-specific.
For RNA-seq experiments, total RNA was isolated from MNs and WM of two pairs of SMA and WT littermates, derived from two unrelated litters, to control for litter-to-litter variations. Sequencing libraries were prepared from each mouse and profiled by Illumina high-throughput RNA sequencing. We obtained ∼100 million 100-bp paired-end reads from each sample, 50–63% of which were uniquely mapped to the mouse reference genome (University of California at Santa Cruz, UCSC, mm9) using TopHat (30) (Dataset S1). Consistent with the RT-PCR results (Fig. 1B) and confirming the quality of the LCM and RNA-seq procedures, sequencing profiles demonstrated specific expression of p75NTR in MNs, and Gfap in WM (Fig. 1C). As expected, no reads were aligned to the Smn1 locus in SMA MNs or WM.
We performed differential expression analysis using DESeq. (31), with a cutoff of at least twofold and P value < 0.05 to identify significant mRNA level changes in the replicate samples. This process revealed 248 affected genes in MNs (138 up-regulated and 110 down-regulated), and 212 affected genes in WM (125 up-regulated and 87 down-regulated) (Fig. 2A and Dataset S2). Although both cell populations expressed similar genes overall [∼93% overlap between 11,771 and 12,338 expressed genes in MNs and WM, respectively, based on reads per kilobase per million mapped reads > 2], only seven genes were affected in common (Fig. 2A and Dataset S2), indicating that SMN deficiency causes cell type-specific mRNA level changes. Examples of these mRNA level dysregulations are shown in Fig. S1, demonstrating MN-specific up-regulation (Fos) and down-regulation (Ociad2), and WM-specific up-regulation (Mt2) in SMA mice.
Fig. 2.
Summary of transcriptome changes identified by RNA-seq analysis in P1 SMA MNs and WM. (A) Venn diagrams showing up-regulated and down-regulated genes in SMA MNs and WM. (B) Pie charts showing splicing changes identified in SMA MNs and WM, classified by types of splicing events.
To identify alternative splicing changes in MNs and WM of SMA mice, we applied mixture of isoforms (MISO) analysis (32), with settings of Bayes factor > 10 and |ΔΨ| > 0.2. We also established two additional analyses to identify additional splicing changes: (i) Novel exons present only in SMA samples, including unannotated 5′ and 3′ splice sites according to common databases (RefSeq, UCSC, AceView or Ensembl), classified as “aberrant splicing” events; (ii) Events that skip two neighboring cassette exons, which we referred to as “two-exon skipped” (Materials and Methods). Overall, we identified 104 (100 genes) and 86 (84 genes) splicing events in MNs and WM, respectively in both replicates (Fig. 2B, and Datasets S3 and S4), most of which did not overlap with differentially expressed genes (only two and four genes in MNs and WM, respectively) (Dataset S5). This finding suggests that most of the splicing events affect the corresponding isoforms’ expression without changing the overall transcript level. Moreover, despite that the vast majority of genes affected by splicing are expressed in both MNs and WM, only eight genes showed similar splicing changes in both MNs and WM (Dataset S5), demonstrating that splicing changes caused by SMN deficiency are also cell-type specific.
Although the amount of RNA precluded measuring all of the snRNAs in the same samples, absolute quantitation at presymptomatic P3 revealed major changes in snRNP repertoire (Fig. S2), consistent with the findings that SMN deficiency results in individual snRNA-specific perturbations of snRNP levels (15, 26). Considering a recent report showing that inefficient splicing of the minor (U12) intron of CG8408 (Stasimon) contributes to NMJ defects in Drosophila smn mutants (27), we searched the U12 intron database (U12DB) (33) and found four and five genes that contain minor introns affected by splicing in P1 SMA MNs and WM, respectively (Dataset S6). However, none of those splicing changes occurs in regions where U12 introns reside (Dataset S6), and neither expression nor splicing was affected for Tmem41b (mouse homolog of Stasimon), indicating that the minor splicing pathway is not selectively perturbed in SMA at this early time point.
The total number of genes whose mRNAs are affected at P1 in MNs and WM are 348 and 296, about 3% and 2.4% of expressed genes in MNs and WM, respectively. Thus, SMN deficiency did not cause widespread transcriptome changes at P1. Consistently, Gene Ontology and pathway analysis by DAVID (34) did not reveal significant enrichment of any pathways or biological processes for genes up-regulated, down-regulated, or affected by splicing in MNs or WM. Nevertheless, a few splicing changes in mRNAs occurred that could explain the pathogenic features MNs exhibit at P4–P5, including defects in maturation and neurotransmission of NMJ and MN deafferentation.
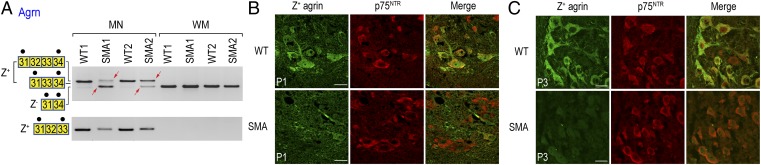
Considering SMA’s NMJ pathology, the heparin sulfate proteoglycan agrin (Agrn) is of particular interest. Agrin is an extracellular matrix protein that is expressed in spinal cord MNs as well as many other neuron types in the CNS, and is critical for the postsynaptic organization of the NMJ (35). Specifically, the neuronal agrin isoforms containing Z exons (exons 32, 33, or both) (Fig. 3A), Z+ agrin, are 1,000-fold more active than those lacking Z exons in inducing aggregation of AChR clusters at the NMJ (35, 36). Z exon splicing is developmentally regulated, with Z+ agrin mRNA expressed at its highest levels between embryonic day 18 and P6 (37). As expected, we observed that Z+ agrin mRNA isoforms were clearly neuron-specific, present in MNs but not in WM of WT mice (Fig. 3A). Strikingly, Z+ agrin mRNA was drastically reduced in MNs of both SMA mice. Instead of exclusively producing Z exon-containing isoforms as in WT MNs, SMA MNs expressed agrin mRNA that skipped Z exons (Fig. 3A). The loss of Z+ agrin mRNA in SMA MNs was further confirmed by RT-PCR with a second set of primers that specifically interrogated the Z+ isoform (Fig. 3A, Lower). Importantly, immunostaining of lumbar spinal cord cross sections using an antibody specific to agrin’s Z peptides (38) showed a corresponding loss of Z+ agrin protein in P1 SMA MNs (Fig. 3B). Although some SMA MNs retain residual Z+ agrin staining at P1 (consistent with RT-PCR results), Z+ agrin staining was undetectable at P3 (Fig. 3C).
Fig. 3.
Agrin Z exon skipping results in drastically decreased levels of Z+ agrin mRNA and protein in SMA MNs. (A) RT-PCR reactions confirmed agrin Z exon skipping in P1 SMA MNs. Similar results have been obtained from at least another P1 SMA mouse littermate. Spliced isoforms are shown as boxes labeled with the corresponding exon number. Black dots on top of the boxes indicate primer binding sites. The red arrows indicate splicing abnormalities observed in SMA mice. (B) Immunofluorescence staining was performed to detect Z+ agrin in lumbar MNs of P1 SMA mice. Dramatic loss of Z+ agrin was detected in P1 SMA MNs. Similar results have been obtained from at least three P1 SMA mice. Immunostaining of p75NTR was used to identify MNs in the anterior horn region of the lumbar spinal cord. (Scale bar, 25 μm.) (C) Immunofluorescence staining was performed to detect Z+ agrin in lumbar MNs of P3 SMA mice. Z+ agrin was completely undetectable in P3 SMA MNs. (Scale bar, 25 µm.) Similar results have been obtained from at least three P3 SMA mice.
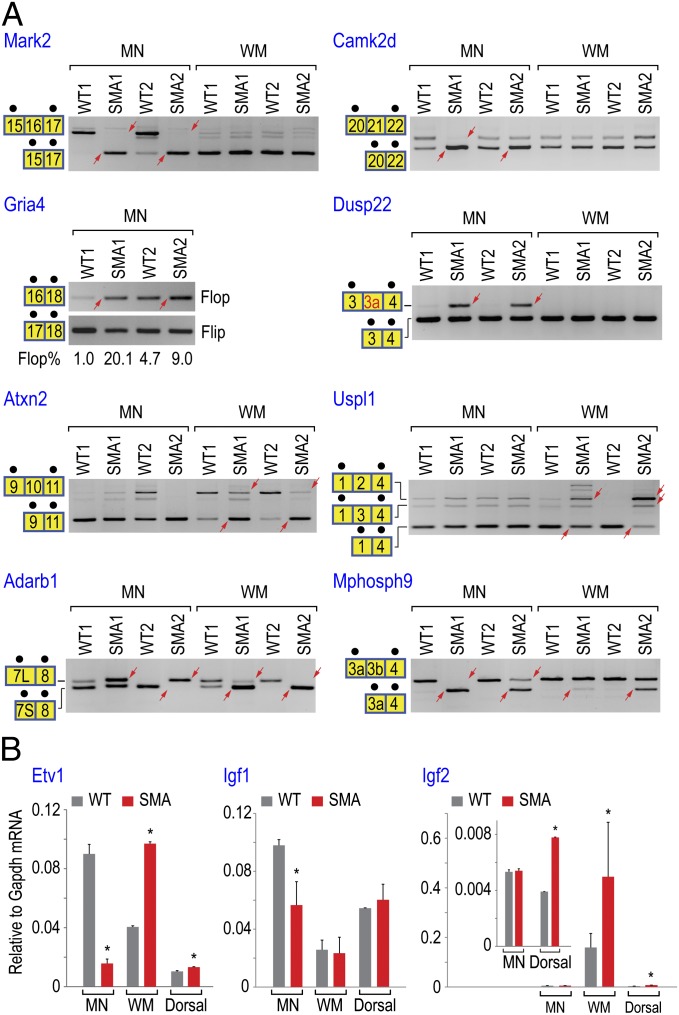
Focusing on genes critical for MN function, we performed additional RT-PCR and quantitative RT-PCR (RT-qPCR) experiments to test 29 splicing events and 6 differential expressed genes identified by RNA-seq, respectively, all of which were confirmed (Figs. 3A, 4, and 5A). These events include MN-specific splicing events in Mark2, Camk2d, Gria4, and Dusp22 (inclusion of a previously unannotated exon 3a), WM-specific events in Atxn2 and Uspl1, and splicing events that occurred in both MNs and WM (Adarb1 and Mphosph9) of SMA mice (Fig. 4A). Importantly, none of these transcripts spliced abnormally in the Dorsal region, except Mphosph9 and Uspl1 (Fig. S3), further supporting that splicing changes in SMA MNs and WMs are cell-type–specific.
Fig. 4.
RT-PCR and RT-qPCR experiments validated additional transcriptome changes occurring in genes important for MN functions. (A) RT-PCR reactions were performed as described in Fig. 3A. The abundance of Gria4 flop splice variant was quantitated by RT-qPCR. The unannotated exon 3a in Dusp22 is shown in red. Alternative 5′ splice sites created two exon 7 variants in Adarb1, indicated by 7L (long exon 7) and 7S (short exon 7). (B) RT-qPCR reactions confirmed differential expressed genes identified by RNA-seq. Each reaction used cDNA produced using MNs, WM, and Dorsal isolated from two WT or two SMA mice. Expression level of each gene was monitored by using validated Taqman gene-expression assay and normalized by Gapdh mRNA levels. Error bars indicate SD (*P < 0.05, Student t test).
Fig. 5.
Up-regulation of C1q mRNA in P1 SMA MNs results in dramatically increased level of C1q proteins at P2. (A) RT-qPCR reactions quantitating C1qb mRNA levels were performed as described in Fig. 4B, confirming MN-specific up-regulation. Error bars indicate SD (*P < 0.05, Student t test). (B) Immunofluorescence staining was performed to detect C1q complex in lumbar MNs of P2 SMA mice. Substantial increase of C1q was detected in P2 SMA MNs. Immunostaining of p75NTR was used as MN marker. (Scale bar, 25 µm.) Similar results have been obtained from at least three P2 SMA mice.
We have characterized the alternative splicing changes in Gria2 and Gria4, which encode core subunits of AMPA-type glutamate receptors that are critical for glutamatergic excitatory synapses (39). Alternative splicing via mutually exclusive exons generates flip or flop isoforms affecting the ligand-binding domain, which imparts different pharmacologic and kinetic properties on currents evoked by l-glutamate or AMPA (40). We observed that the abundance of Gria2 and Gria4 flop splice variants increased sharply in SMA MNs (2- to 20-fold increase) (Fig. 4A and Dataset S3). Because the flop isoform confers a faster desensitization rate (∼5- to 10-fold) than the flip (41), overabundance of flop isoforms in AMPA receptors is expected to cause a reduction of response to stimuli, consistent with the diminished synaptic transmission observed in SMA MNs (12). We then tested whether the splicing abnormalities in Agrn and Gria4 occur before P1, using MNs and WM isolated from SMA mice at P0. Similar splicing changes were found for both Agrn and Gria4 in P0 SMA MNs (Fig. S4), indicating that these splicing events take place at an even earlier time point.
Examples of differentially expressed genes are shown in Fig. 4B, including the transcription factor Etv1 (down-regulated in MNs and up-regulated in WM) and the neurotrophic factors Igf1 and Igf2 (down-regulated in MNs and up-regulated in WM, respectively). Etv1 controls a late step during establishment of functional sensory-motor circuitry in the developing spinal cord (42). Knockout of Etv1 causes deafferentation of MNs in the spinal cord and a diminished functional motor output in neonatal mice (42). The 77% decrease of Etv1 mRNA in MNs (Fig. 4B and Dataset S2) could explain impaired sensory-motor synapse development and MN deafferentation in SMA (11–13). Similarly, the down-regulation of Igf1 and one of its substrates Irs4 (48% and 60% decrease, respectively) in MNs and up-regulation of Igf2 (120% increase) in WM (Fig. 4B and Dataset S2) could contribute to the degeneration of SMA MNs, as both Igf1 and Igf2 neurotrophic factors are important for MN axonal outgrowth and NMJ formation (43).
An additional surprising finding from the transcript level analysis was the up-regulation of mRNAs for all three subunits of the C1q complex in SMA MNs (260%, 350%, and 200% increase for C1qa, C1qb, and C1qc, respectively) (Fig. 5A and Dataset S2). C1q is the initiating protein complex of the classic complement cascade. Besides its immune functions, C1q has recently been shown to play an essential role in synaptic pruning during embryonic and postnatal CNS development (44–46). We therefore interrogated the expression of C1q protein by immunostaining. C1q protein levels were low and unchanged at P1 in MNs of both WT and SMA mice (Fig. S5), but a much stronger staining is clearly observed at P2 in SMA MNs compared with WT MNs (Fig. 5B). The C1q mRNA up-regulation at P1 (Fig. 5A) immediately followed by its dramatic protein level increase at P2 (Fig. 5B) in SMA MNs could likely promote overpruning of synapses and impair neural connectivity, characteristic of the SMA phenotype.
Discussion
A lack of transcriptome profiles in SMA MNs, especially antecedent to overt MN pathology, has hampered understanding of the molecular basis of this degenerative disease. We have obtained RNA-seq from pure MN and WM populations of the lumbar sections, the spinal cord level that is particularly affected in SMA, and found that expression of only ∼300 genes were affected in each at P1. The lack of a significant effect on the vast majority of genes (> 11,000) indicates that the SMA MNs are not extensively or nonspecifically damaged at this early stage. The diversity of transcriptome perturbations, including both splicing changes and expression level changes, as well as of the functions of the affected genes, makes it difficult to ascribe the MNs’ dysfunction and demise to any one gene or mechanism. However, several of them are particularly striking as they have critical MN functions and impairment of each of them alone could explain a significant aspect of SMA’s synaptic pathology that manifest later, at P4–P5. Both the nature of these genes (discussed below) and the critical timing of their dysregulation suggest that they have a role in SMA pathogenesis. The transcriptome abnormalities at P1 suggest that some motor unit functions could be impaired even earlier than overt structural NMJ disruptions. Recent observations from a related SMA mouse model have indeed shown muscle weakness at P2 (7, 47).
Two transcriptome changes appear to be particularly relevant to SMA pathogenesis: the skipping of agrin Z exons and the up-regulation of C1q. Z+ agrin expression in mouse neuronal tissues correlates with developmental stage when it is required for organization of the postsynaptic AChR clusters into the complex pretzel-like pattern, and for stabilizing NMJ synaptic contact (35, 37). Our experiments revealed that expression of Z+ agrin mRNA is drastically reduced, and Z+ agrin protein is severely deficient in SMA MNs at P1 and completely absent at P3 (Fig. 3). Constitutive loss of Z+ agrin in all neurons has a much more severe phenotype than SMA, causing perinatal death in mouse (36). Z+ agrin is produced normally in dorsal region neurons of P1 SMA mice (Fig. S3), indicating that the SMA mice are capable of expressing Z+ agrin. However, the skipping of Z exons and the resulting Z+ agrin protein deficiency in SMA MNs between P1 and P3 would impair postsynaptic AChR clustering, as observed at P5 in this SMA model (7–9).
The up-regulation of complement factor C1q was striking and unexpected. Unlike amyotrophic lateral sclerosis (ALS), an adult onset MN degenerative disease, where neuroinflammation is well-documented (48), there is little evidence of inflammation involvement in SMA (49). Both neuron and microglia-derived C1q has an important role in the developing CNS, where it tags synapses for elimination through classic complement cascade (44–46). C1q-mediated synaptic pruning is essential for remodeling of immature synaptic circuits in the CNS of neonatal mice (46). Importantly, C1q up-regulation has been linked to neurodegeneration in the retina of a glaucoma mouse model (45). Up-regulation has also been observed in MNs of ALS mouse models (50, 51). We propose that aberrant C1q up-regulation could promote excessive loss of MN synapses in SMA (e.g., MN deafferentation observed in SMA mice) (11–13).
Additional abnormalities in several other genes, some of which have been associated with neurodegenerative diseases, likely contribute to the SMA phenotype and have a compounding effect. For example, a strong reduction in circulating Igf1 has also been reported in an SMA mouse model (14), as have alterations in AMPA receptors Gria2 and Gria4 splice isoforms, which would be expected to affect synaptic transmission at glutamatergic excitatory synapses (12, 41). AMPA receptor dysregulation has also been described in Alzheimer’s disease and clinical trials for Alzheimer’s disease patients are ongoing using AMPA receptor-positive modulators (41, 52), which we suggest could also be beneficial for SMA.
Consistent with previous observations (15), cell-type–specific mRNA abnormalities in P1 SMA mice are not limited to MNs and occur also in the surrounding glia. Many of these abnormalities would likely be deleterious to MNs. For example, mRNA expression of Ptgds, a key enzyme in prostaglandin D2 (PGD2) biogenesis, is up-regulated by ∼10-fold (Dataset S2). PGD2, an immune response mediator highly elevated in ALS glia, is toxic to MNs (53). In addition, expression of 12 genes encoding extracellular matrix and cell adhesion components (e.g., collagens and laminins), and 19 ion or solute transporters are dysregulated in WM (Dataset S2). Notably, collagen abnormalities have been described in spinal cord of ALS patients (54). These changes would disturb the MN’s microenvironment and spinal cord homeostasis, and likely contribute to SMA pathogenesis (13). A clear role for toxicity derived from nonneuronal supporting cells, such as glia, in MN dysfunction and death has been demonstrated in ALS and other neurodegenerative diseases (53, 55).
The intervening steps between SMN decrease and the mRNA abnormalities remain to be elucidated. SMN functions in snRNP biogenesis as part of the SMN–Gemins complex (16, 20) and SMN deficiency causes tissue-specific snRNA repertoire perturbations (15, 26), which likely elicit the mRNA expression and splicing abnormalities found in MNs and WM at this early presymptomatic stage. We have not detected changes in mRNAs encoding neuron-specific splicing factors, such as Nova1/2, which can regulate agrin’s Z exon splicing (56), or neural polypyrimidine tract-binding proteins (57) at P1. However, abnormalities detected in several other transcription and splicing factors (e.g., Etv1, Fos, Trp53, Tra2b and Celf3) (Datasets S2–S4) could potentially explain why the transcriptome defects in SMA exacerbate over time (15, 28). Our findings do not support a recent proposal of an especially important role for minor spliceosome disturbance in SMA pathology (27), as splicing of minor (U12) introns is not affected.
Previous investigations have left open the question of whether SMA’s synaptic pathogenesis results from a general transcriptome deterioration, specific mRNA abnormalities, or perhaps functions unrelated to mRNA biogenesis that SMN has been suggested to have (58). Our findings show that splicing and other mRNA expression abnormalities affect specific genes that are either required or would likely be deleterious to MN synapses (NMJ and motor-sensory) at a critical time window. By altering the mRNA processing machinery, SMN deficiency sets in motion a cascade of transcriptome and proteome changes in MNs and their supporting WM. The mRNA dysregulations explain the ensuing key features of SMA, linking SMN’s general function in RNA metabolism with SMA pathogenesis. In addition to the insights they provide into the mechanism of SMA pathogenesis, our findings reveal unique and potentially attractive targets for alleviating SMA severity, including correcting Z+ agrin splicing and inhibiting C1q. Importantly, our findings underscore SMN’s central role in transcriptome regulation.
Materials and Methods
Mouse Tissue Harvesting, Tissue Embedding, Sectioning, and LCM.
All mouse experiments were carried out in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee (IACUC Protocol #800714). Founders of the SMA mouse model (SMN2+/+;SMNΔ7+/+;Smn−/−) were purchased from Jackson laboratory (stock # 005025). For LCM, the lumbar spinal cord was dissected, embedded in NEG 50 (Richard-Allan Scientific) on dry ice/ethanol mixture, and stored at −80 °C. Cross-sections of 8-μm thickness were prepared in a cryostat and transferred to PEN membrane slides (Zeiss). Slides were stained by Mayer’s hematoxylin (Sigma), using an LCM staining kit (Ambion). LCM was performed on a PALM microbeam UV laser microdissection system (Zeiss). For this process, 45–50 sections from each mouse were used to isolate MNs, WM, and Dorsal samples.
RNA Purification, Amplification, and cDNA Processing for RNA-seq Analysis.
Total RNA from LCM samples was purified by using RNeasy Plus Micro kit (Qiagen). Average LCM dissected area for each mouse and yield of total RNA from each sample: MNs: ∼500,000 μm2 (∼1,500 cell bodies), 5–7 ng; WM: ∼2,000,000 μm2, 5–7 ng; Dorsal: ∼2,000,000 μm2, 17–20 ng. RNA quality (RIN 8–9) and quantity were analyzed on a 2100 Bioanalyzer (Agilent), using RNA 6000 Pico chips. ds-cDNA was produced by using Ovation RNA-seq System V2 (NuGEN) and fragmented by a Covaris S-series System (Covaris). DNA fragments in 150- to 300-bp size range were recovered to construct sequencing library, using Encore NGS Library System I (NuGEN), and submitted to Next-Generation Sequencing Core at University of Pennsylvania for 100 bp paired-end RNA-seq, using an Illumina HiSeq2000 sequencer.
RNA-seq Data Alignment, Differential Gene-Expression Analysis, and Alternative Splicing Events Analysis.
These analyses were performed using Tophat (30), DESEq. (31), and MISO (32), plus two custom analyses, respectively. See SI Materials and Methods for details.
Immunofluorescence Staining of Mouse Spinal Cord Cross-Sections.
Lumbar segment spinal cords were dissected after the mice were perfused with sterile PBS, fixed overnight in 4% (vol/vol) PFA at 4 °C. The segments were cryoprotected in 30% sucrose in PBS at 4 °C and mounted in NEG 50 (Richard-Allan Scientific), sectioned at 10-μm thickness in a cryostat and stored at −80 °C. Sections were postfixed in 4% PFA in PBS for 10 min, rinsed 2 × 5 min in PBS, and permeabilized with methanol for 5 min, then blocked for 1 h in 2% (wt/vol) BSA, 2% (vol/vol) NGS/PBST, and incubated overnight at 4 °C, using the following antibodies: mouse antiagrin (1:250; Chemicon, MAB5204) and rat anti-C1q (1:250; Abcam, ab11861). Sections were washed 4 × 5 min in PBST and incubated for 1 h with Alexa Fluor 488 goat anti-mouse IgG or goat anti-rat secondary antibody (Invitrogen), followed by 3 h incubation of rabbit anti-p75NTR (1:500; Abcam, ab38335) and Alexa Fluor 594 goat anti-rabbit IgG secondary antibody (Invitrogen). Slides were rinsed 4 × 5 min in PBST and mounted with ProLong Gold antifade mounting reagent (Invitrogen). Images were taken on a Zeiss LSM 510 NLO/META confocal microscope or a Leica TCS SP2 confocal microscope.
Supplementary Material
Acknowledgments
We thank members of our laboratory, especially Dr. Ihab Younis, for stimulating discussions and comments on the manuscript; Daniel Martinez (Children’s Hospital of Philadelphia) for technical support with laser-capture microdissection; and Xinyu Zhao and Dr. Andrea Stout (University of Pennsylvania) for expert help with confocal microscopy imaging. This work was supported by the Association Française Contre les Myopathies. G.D. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the NCBI GEO database (accession no. GSE51735).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1319280110/-/DCSupplemental.
References
- 1.Cifuentes-Diaz C, Frugier T, Melki J. Spinal muscular atrophy. Semin Pediatr Neurol. 2002;9(2):145–150. doi: 10.1053/spen.2002.33801. [DOI] [PubMed] [Google Scholar]
- 2.Wee CD, Kong L, Sumner CJ. The genetics of spinal muscular atrophies. Curr Opin Neurol. 2010;23(5):450–458. doi: 10.1097/WCO.0b013e32833e1765. [DOI] [PubMed] [Google Scholar]
- 3.Lefebvre S, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80(1):155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 4.Lorson CL, Hahnen E, Androphy EJ, Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc Natl Acad Sci USA. 1999;96(11):6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho S, Dreyfuss G. A degron created by SMN2 exon 7 skipping is a principal contributor to spinal muscular atrophy severity. Genes Dev. 2010;24(5):438–442. doi: 10.1101/gad.1884910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Le TT, et al. SMNDelta7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum Mol Genet. 2005;14(6):845–857. doi: 10.1093/hmg/ddi078. [DOI] [PubMed] [Google Scholar]
- 7.Kariya S, et al. Reduced SMN protein impairs maturation of the neuromuscular junctions in mouse models of spinal muscular atrophy. Hum Mol Genet. 2008;17(16):2552–2569. doi: 10.1093/hmg/ddn156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kong L, et al. Impaired synaptic vesicle release and immaturity of neuromuscular junctions in spinal muscular atrophy mice. J Neurosci. 2009;29(3):842–851. doi: 10.1523/JNEUROSCI.4434-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murray LM, et al. Selective vulnerability of motor neurons and dissociation of pre- and post-synaptic pathology at the neuromuscular junction in mouse models of spinal muscular atrophy. Hum Mol Genet. 2008;17(7):949–962. doi: 10.1093/hmg/ddm367. [DOI] [PubMed] [Google Scholar]
- 10.Martinez-Hernandez R, et al. Synaptic defects in type I spinal muscular atrophy in human development. J Pathol. 2013;229(1):49–61. doi: 10.1002/path.4080. [DOI] [PubMed] [Google Scholar]
- 11.Ling KK, Lin MY, Zingg B, Feng Z, Ko CP. Synaptic defects in the spinal and neuromuscular circuitry in a mouse model of spinal muscular atrophy. PLoS ONE. 2010;5(11):e15457. doi: 10.1371/journal.pone.0015457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mentis GZ, et al. Early functional impairment of sensory-motor connectivity in a mouse model of spinal muscular atrophy. Neuron. 2011;69(3):453–467. doi: 10.1016/j.neuron.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park GH, Maeno-Hikichi Y, Awano T, Landmesser LT, Monani UR. Reduced survival of motor neuron (SMN) protein in motor neuronal progenitors functions cell autonomously to cause spinal muscular atrophy in model mice expressing the human centromeric (SMN2) gene. J Neurosci. 2010;30(36):12005–12019. doi: 10.1523/JNEUROSCI.2208-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hua Y, et al. Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model. Nature. 2011;478(7367):123–126. doi: 10.1038/nature10485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Z, et al. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell. 2008;133(4):585–600. doi: 10.1016/j.cell.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cauchi RJ. SMN and Gemins: ‘We are family’ … or are we?: Insights into the partnership between Gemins and the spinal muscular atrophy disease protein SMN. Bioessays. 2010;32(12):1077–1089. doi: 10.1002/bies.201000088. [DOI] [PubMed] [Google Scholar]
- 17.Fischer U, Liu Q, Dreyfuss G. The SMN-SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell. 1997;90(6):1023–1029. doi: 10.1016/s0092-8674(00)80368-2. [DOI] [PubMed] [Google Scholar]
- 18.Pellizzoni L, Yong J, Dreyfuss G. Essential role for the SMN complex in the specificity of snRNP assembly. Science. 2002;298(5599):1775–1779. doi: 10.1126/science.1074962. [DOI] [PubMed] [Google Scholar]
- 19.Meister G, Bühler D, Pillai R, Lottspeich F, Fischer U. A multiprotein complex mediates the ATP-dependent assembly of spliceosomal U snRNPs. Nat Cell Biol. 2001;3(11):945–949. doi: 10.1038/ncb1101-945. [DOI] [PubMed] [Google Scholar]
- 20.Zhang R, et al. Structure of a key intermediate of the SMN complex reveals Gemin2’s crucial function in snRNP assembly. Cell. 2011;146(3):384–395. doi: 10.1016/j.cell.2011.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wahl MC, Will CL, Lührmann R. The spliceosome: Design principles of a dynamic RNP machine. Cell. 2009;136(4):701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 22.Berg MG, et al. U1 snRNP determines mRNA length and regulates isoform expression. Cell. 2012;150(1):53–64. doi: 10.1016/j.cell.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaida D, et al. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature. 2010;468(7324):664–668. doi: 10.1038/nature09479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lefebvre S, et al. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat Genet. 1997;16(3):265–269. doi: 10.1038/ng0797-265. [DOI] [PubMed] [Google Scholar]
- 25.Wan L, et al. The survival of motor neurons protein determines the capacity for snRNP assembly: Biochemical deficiency in spinal muscular atrophy. Mol Cell Biol. 2005;25(13):5543–5551. doi: 10.1128/MCB.25.13.5543-5551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gabanella F, et al. Ribonucleoprotein assembly defects correlate with spinal muscular atrophy severity and preferentially affect a subset of spliceosomal snRNPs. PLoS ONE. 2007;2(9):e921. doi: 10.1371/journal.pone.0000921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lotti F, et al. An SMN-dependent U12 splicing event essential for motor circuit function. Cell. 2012;151(2):440–454. doi: 10.1016/j.cell.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bäumer D, et al. Alternative splicing events are a late feature of pathology in a mouse model of spinal muscular atrophy. PLoS Genet. 2009;5(12):e1000773. doi: 10.1371/journal.pgen.1000773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anderson KN, et al. Isolation and culture of motor neurons from the newborn mouse spinal cord. Brain Res Brain Res Protoc. 2004;12(3):132–136. doi: 10.1016/j.brainresprot.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 30.Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katz Y, Wang ET, Airoldi EM, Burge CB. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods. 2010;7(12):1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alioto TS. U12DB: a database of orthologous U12-type spliceosomal introns. Nucleic Acids Res. 2007;35(Database issue):D110–D115. doi: 10.1093/nar/gkl796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 35.Bezakova G, Ruegg MA. New insights into the roles of agrin. Nat Rev Mol Cell Biol. 2003;4(4):295–308. doi: 10.1038/nrm1074. [DOI] [PubMed] [Google Scholar]
- 36.Burgess RW, Nguyen QT, Son YJ, Lichtman JW, Sanes JR. Alternatively spliced isoforms of nerve- and muscle-derived agrin: Their roles at the neuromuscular junction. Neuron. 1999;23(1):33–44. doi: 10.1016/s0896-6273(00)80751-5. [DOI] [PubMed] [Google Scholar]
- 37.Stephan A, et al. Neurotrypsin cleaves agrin locally at the synapse. FASEB J. 2008;22(6):1861–1873. doi: 10.1096/fj.07-100008. [DOI] [PubMed] [Google Scholar]
- 38.Hoch W, Campanelli JT, Harrison S, Scheller RH. Structural domains of agrin required for clustering of nicotinic acetylcholine receptors. EMBO J. 1994;13(12):2814–2821. doi: 10.1002/j.1460-2075.1994.tb06575.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Newpher TM, Ehlers MD. Glutamate receptor dynamics in dendritic microdomains. Neuron. 2008;58(4):472–497. doi: 10.1016/j.neuron.2008.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sommer B, et al. Flip and flop: A cell-specific functional switch in glutamate-operated channels of the CNS. Science. 1990;249(4976):1580–1585. doi: 10.1126/science.1699275. [DOI] [PubMed] [Google Scholar]
- 41.Traynelis SF, et al. Glutamate receptor ion channels: Structure, regulation, and function. Pharmacol Rev. 2010;62(3):405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arber S, Ladle DR, Lin JH, Frank E, Jessell TM. ETS gene Er81 controls the formation of functional connections between group Ia sensory afferents and motor neurons. Cell. 2000;101(5):485–498. doi: 10.1016/s0092-8674(00)80859-4. [DOI] [PubMed] [Google Scholar]
- 43.Sullivan KA, Kim B, Feldman EL. Insulin-like growth factors in the peripheral nervous system. Endocrinology. 2008;149(12):5963–5971. doi: 10.1210/en.2008-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stephan AH, Barres BA, Stevens B. The complement system: An unexpected role in synaptic pruning during development and disease. Annu Rev Neurosci. 2012;35:369–389. doi: 10.1146/annurev-neuro-061010-113810. [DOI] [PubMed] [Google Scholar]
- 45.Stevens B, et al. The classical complement cascade mediates CNS synapse elimination. Cell. 2007;131(6):1164–1178. doi: 10.1016/j.cell.2007.10.036. [DOI] [PubMed] [Google Scholar]
- 46.Schafer DP, et al. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron. 2012;74(4):691–705. doi: 10.1016/j.neuron.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.El-Khodor BF, et al. Identification of a battery of tests for drug candidate evaluation in the SMNDelta7 neonate model of spinal muscular atrophy. Exp Neurol. 2008;212(1):29–43. doi: 10.1016/j.expneurol.2008.02.025. [DOI] [PubMed] [Google Scholar]
- 48.Philips T, Robberecht W. Neuroinflammation in amyotrophic lateral sclerosis: Role of glial activation in motor neuron disease. Lancet Neurol. 2011;10(3):253–263. doi: 10.1016/S1474-4422(11)70015-1. [DOI] [PubMed] [Google Scholar]
- 49.Papadimitriou D, et al. Inflammation in ALS and SMA: Sorting out the good from the evil. Neurobiol Dis. 2010;37(3):493–502. doi: 10.1016/j.nbd.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saxena S, Cabuy E, Caroni P. A role for motoneuron subtype-selective ER stress in disease manifestations of FALS mice. Nat Neurosci. 2009;12(5):627–636. doi: 10.1038/nn.2297. [DOI] [PubMed] [Google Scholar]
- 51.Lobsiger CS, Boillée S, Cleveland DW. Toxicity from different SOD1 mutants dysregulates the complement system and the neuronal regenerative response in ALS motor neurons. Proc Natl Acad Sci USA. 2007;104(18):7319–7326. doi: 10.1073/pnas.0702230104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chappell AS, et al. AMPA potentiator treatment of cognitive deficits in Alzheimer disease. Neurology. 2007;68(13):1008–1012. doi: 10.1212/01.wnl.0000260240.46070.7c. [DOI] [PubMed] [Google Scholar]
- 53.Di Giorgio FP, Boulting GL, Bobrowicz S, Eggan KC. Human embryonic stem cell-derived motor neurons are sensitive to the toxic effect of glial cells carrying an ALS-causing mutation. Cell Stem Cell. 2008;3(6):637–648. doi: 10.1016/j.stem.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 54.Ono S, et al. Collagen abnormalities in the spinal cord from patients with amyotrophic lateral sclerosis. J Neurol Sci. 1998;160(2):140–147. doi: 10.1016/s0022-510x(98)00223-8. [DOI] [PubMed] [Google Scholar]
- 55.Ilieva H, Polymenidou M, Cleveland DW. Non-cell autonomous toxicity in neurodegenerative disorders: ALS and beyond. J Cell Biol. 2009;187(6):761–772. doi: 10.1083/jcb.200908164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ruggiu M, et al. Rescuing Z+ agrin splicing in Nova null mice restores synapse formation and unmasks a physiologic defect in motor neuron firing. Proc Natl Acad Sci USA. 2009;106(9):3513–3518. doi: 10.1073/pnas.0813112106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27(3):435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Burghes AH, Beattie CE. Spinal muscular atrophy: Why do low levels of survival motor neuron protein make motor neurons sick? Nat Rev Neurosci. 2009;10(8):597–609. doi: 10.1038/nrn2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.