Abstract
G protein coupled receptors (GPCR) constitute the largest group of cell surface receptors that transmit various signals across biological membranes through the binding and activation of heterotrimeric G proteins, which amplify the signal and activate downstream effectors leading to the biological responses. Thus, the first critical step in this signaling cascade is the interaction between receptor and its cognate G protein. Understanding this critical event at the molecular level is of high importance because abnormal function of GPCRs is associated with many diseases. Thus, these receptors are targets for drug development.
Keywords: G protein-coupled receptors, rhodopsin-transducin complex, binding interface, structural asymmetry
1. Structures of GPCRs and G proteins
GPCRs
G protein coupled receptors all share the same structural organization of seven transmembrane spanning domains connected by three short extracellular (ECL) and intracellular (ICL) loops. The GPCR family is divided into four subfamilies according to their sequence homology and pharmacological properties: rhodopsin-like family A, secretin-like family B, metabotropic/glutamate family C and frizzled receptors. The first crystal structure of any GPCR was solved from family A: inactive, dark state bovine rhodopsin in 2000. To date it is the only structure of GPCR in its native, genetically non-modified conformation (Palczewski et al. 2000). It took seven more years and a number of technological advances to solve a structure of another family A GPCR. In 2007 the first crystal structure of a GPCR activated by diffusible ligand, the β2 adrenergic receptor (β2AR), was solved (Cherezov et al. 2007; Rasmussen et al. 2007). Since then structures of many other family A GPCRs bound to different ligands, in different activation states have been determined. These revealed the overall structural architecture of these receptors is highly homologous despite differences in the sequence and ligand sensitivity. All of them feature general seven transmembrane architecture with cytoplasmic helix H8 laying parallel to the plasma membrane. They all contain conserved structural elements important in receptor activation: a) a disulfide bond linking ECL2 with TM III; b) the D[E]RY motif located on top of helix TM III that is a part of a hydrogen bonding network called the TM III - TM VI ionic lock; c) the WxP motif in helix VI; and d) the NPxxY motif in helix VII. The length of connecting loops differs among specific GPCRs which may be attributed to the binding specificity of G proteins. The largest diversity concerns the extracellular part of the receptor, specifically, the ligand binding region that usually is located in a cavity between TM helices. However, some GPCRs bind their diffusible ligands in their amino terminal region that form a distinct domain with a more open conformation.
Despite large diversity of the GPCR family, these membrane receptors interact with a relatively small number of G proteins.
Heterotrimeric G proteins
Heterotrimeric G proteins belong to a superfamily of regulatory GTP hydrolyses that are divided into main four classes based on the sequence similarity of Gα: namely Gαs, Gαi/Gαo, Gαq/Gα11, and Gα12/Gα13. Each G protein is composed of three subunits α, β and γ with a nucleotide binding pocket located in the α subunit. In the GDP-bound conformation, nucleotide release is inhibited by the βγ dimer, which stabilizes an inactive conformation of the αβγ heterotrimer. G proteins are activated by binding to activated GPCR which causes rapid GDP→GTP nucleotide exchange. Binding of GTP results in dissociation of the GPCR-G-protein complex as well as the G-protein into free, active subunits of Gα and Gβγ that subsequently activate different downstream effectors leading to specific physiological responses (Fig. 1). The crystal structures of the heterotrimeric G proteins Gt and Gi revealed the overall topology to be highly similar and likely typical for all heterotrimeric G proteins (Sondek et al. 1996; Lambright et al. 1996; Noel et al. 1993). The Gα subunit consists of two conserved domains: a GTP-ase domain and helical domain. The GTP-ase domain mediates hydrolysis of GTP and provides the binding surface for the βγ dimer, GPCR, and effector proteins. Three flexible loops known as switch I, II and III were identified in the crystal structure of the GTP-ase domain. These loops most likely play a significant role in GDP/GTP exchange during activation process because they display the most significant conformational differences in the active and inactive structures of Gα. The Gα helical domain is composed of six α-helices that cover up the nucleotide binding pocket located within GTP-ase domain, retaining the nucleotide in the core of protein. In all classes, the N-terminus of the Gα subunit is posttranslationally modified by fatty acid palmitate. The only exceptions are Gtα and Gαi which are modified by myristoyl group. The Gβ subunit features a seven-bladed β propeller composed of seven W40 repeats and it forms strong affinity functional unit with Gγ. The βγ complex is assembled from a repertoire of five β subunits and twelve γ subunits (Wettschureck and Offermanns 2005). All of the Gγ subunits are modified at their C-terminus by either a farnesyl or a gerenylgeranyl moiety. Although Gα and Gγ do not form direct contacts, their modified N- and C-termini are in close proximity and are responsible for membrane association.
Fig. 1.
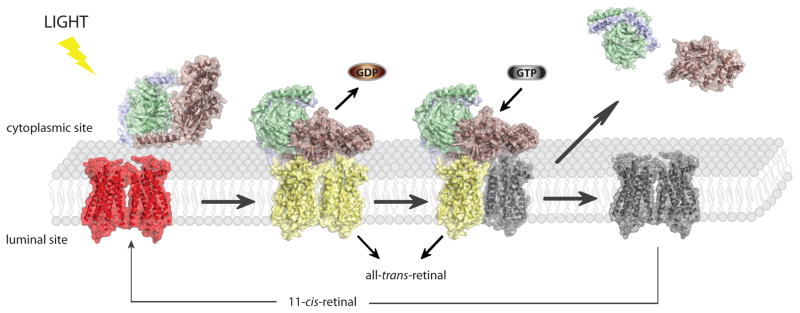
Rhodopsin signaling. Light activation of rhodopsin enables binding of its cognate G protein, transducin and triggers GDP→GTP exchange in its α subunit. Transiently, high affinity, nucleotide free rhodopsin-Gt complex is formed. Binding of GTP causes complex dissociation as well as dissociation of Gtα subunit from Gtβγ dimer and they activate different downstream pathways leading in effect to neuronal response in the brain. Dissociation of transducin from activated rhodopsin enables dissociation of is chromophore all-trans-retinal. Accumulated free opsin has to be regenerated with 11-cis-retinal in order to restore its function.
2. Mechanism of GPCR and G protein activation
The first insights into GPCR activation were gained after the crystal structures of the photoactivated rhodopsin intermediate (Salom et al. 2006), opsin (Standfuss et al. 2007), and the complex of opsin (Scheerer et al. 2008) with the C-terminal peptide of Gtα (Park et al. 2008) were obtained. The differences between dark state rhodopsin and its photoactivated intermediate were mainly associated with reorganization of the receptor’s cytoplasmic surface within ECL2, ECL3, and the C-terminus. However, the structure of opsin revealed both rearrangement of extracellular loops ECL2 and ECL3 and the movement of TM V and TM VI which broke the ionic lock, accompanied by a previously predicted ~ 6 Å outward movement of helix VI (Fig. 2). This reorganization at the cytoplasmic surface opens a groove enabling the binding of heterotimeric G protein, where the C-terminus of Gtα interacts with Arg135 from the D[E]RY motif. Smaller energy movements of TM VII lead to distortion of the NPxxY motif. Similar overall structural rearrangements have been observed in structures of other GPCRs crystallized in their activated states (Rasmussen et al. 2011b; Rasmussen et al. 2011a; Lebon et al. 2011; Xu et al. 2011; White et al. 2012), indicating a common activation mechanism for the whole family of GPCRs. Availability of GPCR crystal structures in different activation stages agrees with the existence of multiple conformations in the signaling cycle and structural dynamics of these receptors. Moreover, a multistage GPCR activation mechanism has been proposed by solid state H2 NMR studies of rhodopsin which found that retinal isomerization initiates fluctuation of the helices and cytoplasmic loops during transition to its active Meta II state (Struts et al. 2011). Changes in receptor flexibility upon activation were derived also from HDX exchange studies that suggested structural rigidity of dark state rhodopsin, its relaxation upon transition to Meta II and a following increase in structural rigidity upon binding of Gt (Orban et al. 2012). Conserved internal water molecules were identified in the core of the GPCR structures and their involvement in receptor activation and mediation of the signal across biological membranes has been proposed as well (Angel et al. 2009). Monitoring of dynamic changes and reorganization of these waters by radiolytic footprintng in combination with mass spectrometry revealed significant, water mediated modification of residues localized close to the chromophore binding pocket and cytoplasmic face of the receptor. Striking differences in the modification rates between dark state rhodopsin, rhodopsin activated by light, and rhodopsin in complex with Gt could be attributed to the differences in the arrangement of internal waters in all three rhodopsin states confirming their unique structural properties. Superposition of the dark state rhodopsin with its light activated conformer demonstrated a different arrangement of these internal water molecules. Moreover the importance of this internal water network was emphasized by the M257Y rhodopsin mutant that causes disruption of water mediating bonding network, affecting chromophore induced helix movement and resulting in receptor constitutive activity (Deupi et al. 2012). Therefore, along with changes in the protein structure stimulated by ligand, internal water molecules, involved in local changes can act as a prosthetic group by transmitting the signal from the extracellular part of the receptor to its intracellular, cytoplasmic surface that must be rearranged to allow G protein binding and activation.
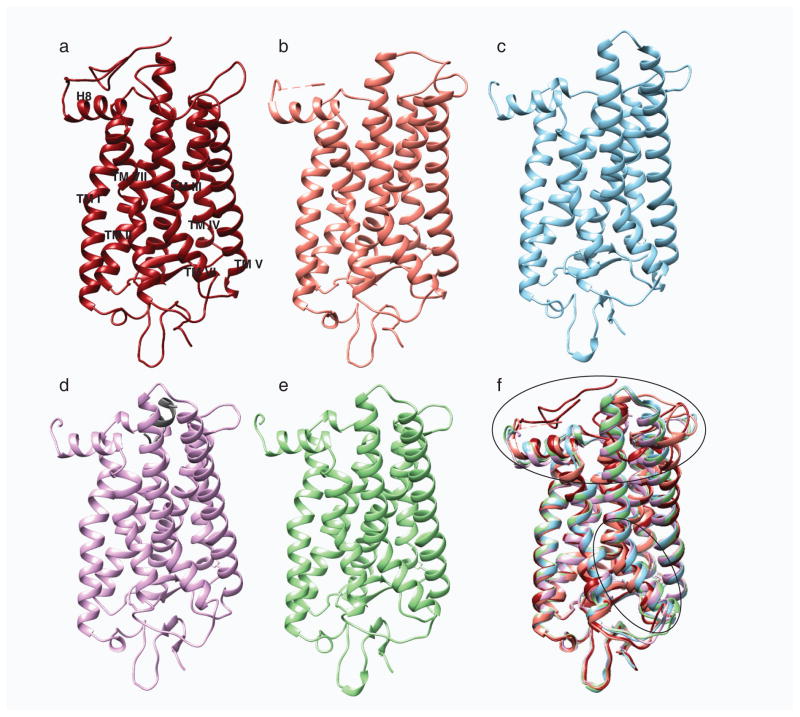
Fig. 2.
Structural comparison of different photo-states of rhodopsin. a, Structures of ground state rhodopsin (1u19) (Pescitelli et al. 2008); b, photoactivated rhodopsin (2i35) (Salom et al. 2006); c, opsin (3cap) (Park et al. 2008); d, opsin with bound C-terminal peptide of Gt (3dqb) (Scheerer et al. 2008) and e, Meta II (3pxo) (Choe et al. 2011) are shown in dark red, salmon, blue, purple and green, respectively. f, All activated rhodopsin states were superposed on its ground state structure. Superposition of these structures show rearrangement of the cytoplasmic loops and movement of helix TM VI depicted with black ovals.
Dating from the initial crystal structure of a G protein in 1993 (Noel et al. 1993), it was recognized that during activation release of nucleotide would probably require an opening between the GTP-ase and helical domain to form a nucleotide-free complex. But no experimental evidence supporting this idea was available. Flexibility of the Gα subunit upon binding of activated GPCR and possible opening of the interface between Gα and Gβγ required for GDP release was reported in 2006 by several groups (Gales et al. 2006; Van Eps et al. 2006; Oldham et al. 2006; Abdulaev et al. 2006). However, a true separation of the GTP-ase and helical domain was not demonstrated for the next several years. In 2011, Van Eps et al. by using site-directed spin labeling (SDSL) and double electron-electron resonance spectroscopy (DEER) demonstrated a significant increase ~ 20 Å in the distance between nitroxide probes positioned on the GTP-ase and helical domains during formation of the nucleotide free complex between Gαi and the activated GPCR, rhodopsin (Van Eps et al. 2011). Soon thereafter, a ~130 Å sideways rotation of the GTP-ase domain relative to the helical domain was found in the crystal structure of the nucleotide-free T4L-β2AR-Gs-nanobody complex, confirming the necessity of movement of these two domains to allow nucleotide exchange (Rasmussen et al. 2011b). However, the domain opening detected in the last crystal structure was unexpectedly large, possibly enforced by crystallization conditions and/or bound nanobodies. Therefore, this is rather unlikely to occur under physiological conditions where rapid G protein turnover is essential for fast biological responses. At the same time, at low resolution EM-derived 3D map of the photoactivated rhodopsin-Gt (Rho*-Gt) complex in the free-nucleotide state revealed its overall shape (Fig. 3). To achieve a ‘best fitting’ of the Gt structure into the obtained density map, an opening of Gα subunit at the nucleotide binding cleft by ~ 30 Å was applied, providing another demonstration of Gα flexibility and necessity of domain movements to allow GDP to GTP exchange (Fig. 3a) and (Jastrzebska et al. 2011). Moreover, HDX experiments confirmed that both nucleotide-free Gt and Gs in complexes with their respective receptors exhibited a significantly greater deuterium uptake (Chung et al. 2011; Orban et al. 2012)
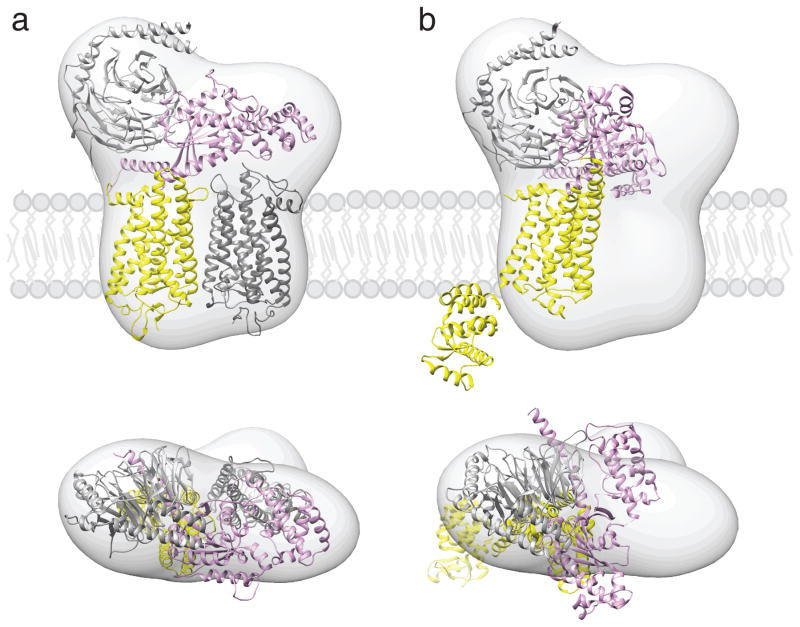
Fig. 3.
Proposed location of GPCR-G protein complexes in a phospholipid bilayer. Semi-empirical model of the complex formed between light-activated rhodopsin dimer and a Gt heterotrimer (a) (Jastrzebska et al. 2011) and a structure of T4L-β2AR-Gs complex (b) (Rasmussen et al. 2011b) were fitted into a 3D molecular map derived from EM-single particle reconstructions of negatively stained, crosslinked rhodopsin-Gt complexes purified in LMNG. Best fitting of the rhodopsin-Gt model was achieved for rhodopsin dimer and Gt hereotrimer (GOT1) after 30° hinge-like motion of the α-helical domain of Gt was applied. Fitting the T4L-β2AR-Gs-nanobody structure into our EM 3D map leaves enough unoccupied space to accommodate a second molecule of this receptor, conformation of Gsα helical domain is inconsistent with our EM-density. The photoactivated rhodopsin that binds C-terminus of Gtα and molecule of T4L-β2AR is depicted with yellow. The second rhodopsin molecule is shown in dark grey. Gα is colored pink; Gβ and Gγ are colored light grey.
3. Binding interface in GPCR-G protein complexes
A first glimpse of the contacting surfaces in the activated GPCR-G protein complex was delivered by using site directed cysteine mutagenesis in combination with crosslinking. These studies demonstrated that intracellular loops ICL2 and ICL3, and helix H8 in photoactivated rhodopsin, and the C-terminus of Gtα (residues 340–350) are important for the assembly of this complex (Itoh et al. 2001; Cai et al. 2001). Besides the α subunit, the C-terminus of Gtγ also interacts with the receptor’s helix H8 (Kisselev and Downs 2006). Because these two regions of Gt are located quite far apart, it is unlikely that a single receptor could provide a surface large enough to satisfy an interaction with both structural regions. Therefore, it is highly possible that a rhodopsin dimer provides an interacting platform to accommodate the Gt heterotrimer.
GPCR activation initiates structural rearrangements of ICL2 and ICL3 and opens the crevice at the cytoplasmic surface. This enables G protein binding and has been found in the crystal structures of both opsin and Meta II-like rhodopsin with bound C-terminal peptide of Gtα, confirming early predictions (Scheerer et al. 2008; Choe et al. 2011). However, the most detailed molecular determinants involved in the binding of GPCR and G protein were revealed by solving the crystal structure of the T4L-β2AR-Gs-nanobody complex (Rasmussen et al. 2011b). Interestingly, extensive interaction was found only between the receptor and GTP-ase domain of the α subunit. This interface is formed between ICL2, TM V and TM VI of the receptor and the C-terminal α5-helix, αN-b1 junction, the top of β3-strand and α4-helix of the Gs GTP-ase domain. Conformational changes in TM V and TM VI are smaller than in the Meta II-Gt peptide structure, and the position of the α5-helix of Gt is tilted ~ 30° relative to the position of the same region of Gs, which may be a consequence of more extensive contacts in the case of intact G protein. Surprisingly, no interaction between receptor and either the beta or gamma subunits have been detected raising the possibility that these interactions could be observed only in the dimerized receptor, whereas in the structure of the T4L-β2AR-Gs-nanobody complex just a single receptor is bound to the G protein. Nevertheless, the β2AR has a propensity to self-associate upon reconstitution into lipid vesicles, and it also forms oligomers in the native membranes (Fung et al. 2009; Vobornik et al. 2009). Therefore, its monomeric form found in the crystal structure of the complex with Gs could result from its selection during protein purification or due to crystallization conditions.
4. GPCR dimerization
All GPCRs including rhodopsin were initially assumed to be single receptors freely diffusing in the lipid bilayer that, upon activation, bind to heterotrimeric G proteins in a 1:1 stoichiometry. However, this view was challenged by growing evidence that GPCRs exist and function as dimers and/or higher oligomers. For most GPCRs evidence for oligomerization comes from indirect immunoprecipitation, crosslinking, fluorescence, or bioluminescence energy transfer experiments. However, the development of high resolution imaging techniques allowed direct visualization of some GPCRs in their native membranes. For example, existence of densely packed rows of rhodopsin dimers in native disk membranes (Fotiadis et al. 2004, 2003; Filipek et al. 2004; Liang et al. 2003) was revealed by atomic force microscopy (AFM). Interestingly, these oligomers could be extracted as separate dimers or entire rows of dimers in mild detergents (Jastrzebska et al. 2006). Clusters of β1AR and β2AR receptor molecules on the surface of cardiac myocytes were visualized by near field scanning optical microscopy (NSOM) (Ianoul et al. 2005; Vobornik et al. 2009). Dimers of CXCR4 were detected by using tetrametylrhodamine-labeled bivalent ligands conjugated with a short, rigid linker on the surface of cancerous cells expressing elevated levels of this receptor (Tanaka et al. 2010). Dimers of vasopressin receptors were found on the surface of mammary glands of lactating mice by using fluorescently labeled specific ligands and fluorescence resonance energy transfer (FRET) assays (Albizu et al. 2010). Therefore, the simplified view of GPCR as a single mobile signaling molecule must be revised.
Several crystal structures provide more evidence of GPCRs dimerization. The dimer of rhodopsin, although in an anti-parallel orientation, was detected in the first crystal structure of dark state bovine rhodopsin (Palczewski et al. 2000). Its dimeric organization was later observed in 2D crystals of rhodopsin Meta I (Ruprecht et al. 2004). These dimers were in a ‘head to head’ orientation, the same as in the membrane with the interface formed by TM I of each molecule. The short H8 helix located by the C-terminus formed a close interaction between subunits as well. Similar dimers were observed in 2D crystals of bovine and frog dark state rhodopsin (Schertler et al. 1993; Schertler and Hargrave 1995). The crystal structure of photoactivated rhodopsin also implicated a parallel dimer with contacts between TM I and H8 as well as TM II (Salom et al. 2006). Similar dimers in a ‘membrane-like’ orientation were also reported in more recently solved crystals of other family A GPCRs, including CXCR4, μ-opioid, κ-opioid receptors (Audet and Bouvier 2012) and β1-arenergic receptor (Huang et al. 2013). Significant protein-protein interfaces were found between TM V and TM VI in CXCR4 (Wu et al. 2010) and between TM II and H8 in κ-opioid receptor (Wu et al. 2012). In the μ-opioid receptor, contact surfaces were seen between TM V and TM VI and also between TM I, TM II and H8 (Manglik et al. 2012). Two contacting surfaces were identified also in β1AR where one employs TM I and H8 and the second involves TM IV, TM V, ICL2 and ECL2 (Huang et al. 2013). The presence of two distinct interfaces found on opposite sides of the μ-opioid and β1AR receptor raises the possibility of forming higher-ordered oligomers in a fashion similar to the packing observed by AFM for rhodopsin oligomers in native disk membranes (Fotiadis et al. 2003). Thus a ‘native-like’ dimeric organization observed in crystals of multiple family A GPCRs strongly suggests that dimerization of these receptors is the rule rather than an exception. Moreover, structures of GPCRs, G proteins and other GPCR-interacting proteins are highly homologous, implying conservation of their mechanisms. If oligomers serve as functional units for some class A GPCRs (Rivero-Muller et al. 2010), they likely do the same for most if not all GPCRs.
5. Binding stoichiometry of GPCR-G protein complexes
Cumulative evidence indicates that GPCRs can traffic and signal as multimeric complexes (Milligan 2010; Gurevich and Gurevich 2008a, b). The requirement of quaternary structure for cell surface delivery was initially noted for the family C GPCR, GABAB but further studies demonstrated that this also applies to family A GPCRs. For example, dimerization of β2AR was found to be obligatory for cell surface targeting and mutations at the dimeric interface led to receptor arrest in the ER (Salahpour et al. 2004; Kobayashi et al. 2009). GPCR dimerization also influences G protein binding. Even though activation of a single receptor suffices to trigger a physiological response in vitro (Baylor et al. 1979; Whorton et al. 2007; Ernst et al. 2007; Whorton et al. 2008; Bayburt et al. 2007), such experiments do not provide information about the actual stoichiometry of the receptor-G protein complex in the native membrane. In fact, binding of the receptor dimer to a single G protein and formation of pentameric complexes are supported by reconstitution experiments involving the leukotriene B4 receptor BLT1, serotonin 5HT4 receptor, and dopamine D2 receptor, and metabotropic receptor- each with their respective G proteins (Baneres and Parello 2003; Han et al. 2009; El Moustaine et al. 2012). Convincing evidence supporting the functional significance of a GPCR dimer in signal mediation can also be derived from in vivo studies on the luteinizing hormone receptor, which revealed intermolecular cooperation between a ligand-binding deficient GPCR and G protein activation deficient mutants (Rivero-Muller et al. 2010). Moreover, studies of rhodopsin lateral diffusion in photoreceptor membranes (Govardovskii et al. 2009) as well as Monte Carlo simulations of stochastic encounters between photoactivated rhodopsin and Gt in disk membranes (Dell’Orco and Schmidt 2008), also proposed the binding of one Gt to at least two receptors. These findings greatly support the concept of a GPCR dimer as the smallest functional unit, organized in higher ordered structures, to provide a kinetic advantage in rapidly responding to an appropriate stimulus (Dell’Orco and Schmidt 2008; Dell’Orco et al. 2009). Using transmission electron microscopy (TEM) and single particle reconstruction, we visualized and solved the three dimensional envelope of the rhodopsin-Gt complex which contained one rhodopsin dimer and one Gt heterotrimer (Jastrzebska et al. 2011) and (Fig. 3). The composition of this has been recently confirmed by labeling of rhodopsin molecules within the rhodopsin-Gt complex with succinylated concanavalin A (sConA), a lectin that binds mannose, one of two sugars decorating the rhodopsin structure (Jastrzebska et al. 2013b). In contrast, a crystal structure of T4L-β2AR complexed to Gs and nanobody provides the most detailed insight into GPCR-G protein interaction and was solved with a 1:1 binding stoichiometry (Rasmussen et al. 2011b). Interestingly, the crystal structure of T4L-β2AR-Gs-nanobody fits quite well into the EM density of the rhodopsin-Gt complex with the exception of the extremely opened conformation of helical domain and a significant unoccupied space corresponding to the position of the second rhodopsin (Fig. 3b). Despite substantial progress, the role of GPCR oligomerization in the molecular mechanism of activation is still not fully understood and these apparently conflicting observations could result from the dynamic nature of receptor oligomers that form and dissociate depending on the receptor cell cycle (Hern et al. 2010; Lambert 2010).
6. Functional and structural asymmetry of the GPCR dimer
Convincing evidence indicates that GPCR dimerization and activation are intricately associated. The functional consequences of GPCR dimerization are revealed by studies showing inter-communication between two protomers in the dimer. This communication can occur at the level of: i) agonist binding (Ilien et al. 2009), ii) ligand-induced allosteric modulation between receptor protomers (Vilardaga et al. 2008; Han et al. 2009; Albizu et al. 2010), and/or iii) binding of GPCR-associated proteins such as specific G proteins (Baneres and Parello 2003; Damian et al. 2006; Pellissier et al. 2011). Such cross-communication supports the asymmetric nature of GPCR dimers wherein each protomer performs a specific task. Analysis of the relationship between subunits of the leukotriene B4 receptor BLT1 revealed that ligand-induced activation of a single receptor can trigger G protein activation but that bound G protein restricts conformational changes in the second receptor protomer; thus only one subunit in the dimer could achieve the fully activated state (Damian et al. 2006). Moreover, dissociation of the dimer with TM VI peptide resulted in lower stability of the receptor-G protein complex. Similarly, activation of one protomer in the serotonin type 4 receptor (5-HT4R) dimer sufficed to stimulate G protein signaling but the binding affinity was two times greater when both protomers were activated (Pellissier et al. 2011). From this perspective, the dimeric assembly of GPCRs is required for more efficient coupling of different regions of the G protein to the receptor, thereby assuring rapid signal transduction (Jastrzebska et al. 2006). Dimer functional asymmetry agrees with studies of trans-complementation between a receptor defective in ligand binding and a receptor defective in G protein activation demonstrated for family A GPCRs, both in vitro (Pellissier et al. 2011) and, more importantly, in vivo (Rivero-Muller et al. 2010; Moepps et al. 2006; Struts et al. 2011). Recently structural asymmetry of the rhodopsin dimer was identified in the pentameric complex formed with heterotrimeric G protein (Jastrzebska et al. 2013a). Each rhodopsin molecule was found to be in a different activation state contributing unevenly to the rhodopsin-transducin complex (Fig. 4). Only one rhodopsin monomer in this complex, most likely this interacting with the C-terminus of Gtα, is stabilized in its Meta II conformation, whereas the second decays eventually to opsin and free retinal. Interestingly, this observation greatly supports the results obtained for the rhodopsin-arrestin complex, where again 50% of the receptor was stabilized in the Meta II state and the other 50% evolved toward an opsin conformation, suggesting a dimeric nature of the receptor bound to arrestin (Sommer et al. 2012). This opsin monomer within the rhodopsin-Gt complex can be regenerated in vitro with the iso-chromophore 9-cis-retinal. Subsequently, all-trans-retinal can be removed from the Meta II rhodopsin monomer by treatment with the strong nucleophile NH2OH, without influencing complex stability. Regeneration of such a derived complex with 11-cis-retinal results in formation of the rhodopsin-Gt complex containing one rhodopsin in a 9-cis-retinal bound conformation and the other surprisingly in an all-trans-retinal bound conformation (Fig. 4a). This result indicates that Gt stabilizes one rhodopsin in Meta II like state even after chromophore release and increases its structural rigidity. Thus, the receptor forces isomerization of unstable 11-cis-retinal to the all-trans configuration in the rigorously defined space of the chromophore binding pocket. This observation is in agreement with HDX exchange studies of rhodopsin-Gt that demonstrated tightening of the more relaxed Meta II structure upon Gt binding (Orban et al. 2012). Moreover treatment of rhodopsin-Gt with NH2OH can produce a chromophore depleted complex, which unlike free opsin (Heck et al. 2003) can take up all-trans-retinal and reform a Schiff base linkage (Jastrzebska et al. 2013a). Interestingly, only 50% of the complex can be regenerated with all-trans-retinal (Fig. 4b). Taking into account that precoupled rhodopsin-Gt complexes in the native membranes most likely exist in the dark and the fact that only one rhodopsin molecule provides an interface for Gt binding the possibility of asymmetric rhodopsin activation arises (Cangiano and Dell’Orco 2013). In such a scenario, photoexcitation of chromophore and its isomerization in the rhodopsin monomer, which does not physically interact with the G protein, induces conformational changes in the precoupled rhodopsin monomer leading to structural evolution toward the Meta II state (Neri et al. 2010). Thus, this tandem mechanism could be critical to prevent loss of energy, enhance the efficiency of receptor activation, and possibly its desensitization. Receptor inactivation is initiated by its phosphorylation with a specific kinase. Cross-phosphorylation between activated and non-activated receptors has been reported for rhodopsin expressed in mouse rod outer segments (Shi et al. 2005) and for chemokine CCR5 and C5a receptors as well (Huttenrauch et al. 2005). Following phosphorylation, the receptor is inactivated by coupling with visual arrestin. The bimodal structure of arrestin, which displays two clefts on its receptor binding surface with the size of individual rhodopsin molecules (Hirsch et al. 1999; Han et al. 2001; Sutton et al. 2005; Palczewski et al. 2000) suggests that a rhodopsin dimer could be accommodated by a single arrestin (Modzelewska et al. 2006).
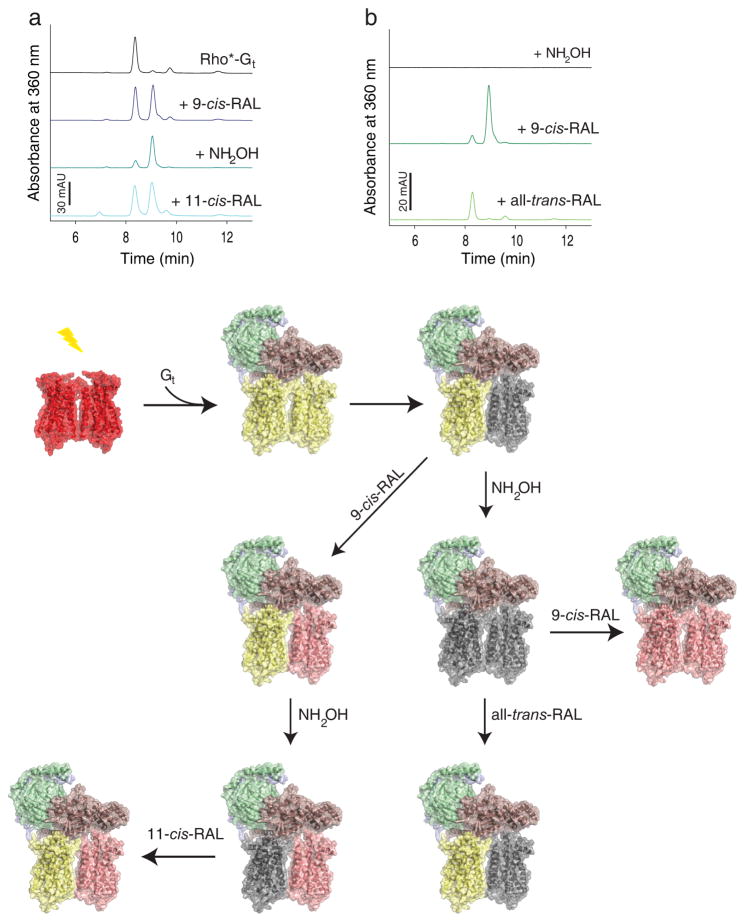
Fig. 4.
Asymmetry of activated rhodopsin (Rho*) dimer within rhodopsin-Gt (Rho*-Gt) complex. Gt binds to fully light activated rhodopsin (Rho) forming the Rho*-Gt complex. After light illumination, 11-cis-retinal (11-cis-RAL) of ground state rhodopsin (shown as red-red dimer) isomerizes to all-trans-retinal (all-trans-RAL) (yellow-yellow dimer) allowing binding of the Gt heterotrimer (Gtα is depicted in salmon, Gtβ in green and Gtγ in light blue). This binding prevents chromophore release from one rhodopsin in the dimer, introducing dimer asymmetry in the Rho*-Gt complex (shown as a yellow-grey dimer bound to Gt and black trace on the chromatogram). a, retinal-free rhodopsin molecule of the Rho*-Gt complex can be regenerated with 9-cis-retinal (9-cis-RAL) (shown as yellow-pink rhodopsin dimer bound to Gt and blue trays on the chromatogram). Release of chromophore from the Gt-protected rhodopsin molecule can be promoted by NH2OH, resulting then in formation of an asymmetric retinal free and 9-cis-retinal regenerated rhodopsin dimer complexed to Gt (shown as grey-pink rhodopsin dimer bound to Gt and dark cyan trace on the chromatogram). Regeneration of this complex with 11-cis-retinal surprisingly results in formation of all-trans-retinal bound and 9-cis-retinal bound asymmetric rhodopsin dimer in the complex with Gt (shown as yellow-pink rhodopsin dimer bound to Gt and cyan trace on the chromatogram). b, the chromophore-free Rho*-Gt complex (modeled as a grey-grey rhodopsin dimer bound to Gt and black lane on the chromatogram) can be generated by treatment of the Rho*-Gt complex with NH2OH. Then this chromophore-free Rho*-Gt complex can be regenerated with 9-cis-retinal which binds to both rhodopsin monomers within the dimer (shown as a pink-pink dimer bound to Gt and dark green trace on the chromatogram). Alternatively, it can be regenerated with all-trans-retinal but only one rhodopsin molecule, the one directly coupled to Gt binds all-trans-retinal (shown as a yellow-grey dimer bound to Gt and light green trace on the chromatogram).
7. Future directions
Constant improvements and great development and of crystallization technologies applicable to membrane binding proteins has already resulted in more than 75 structures of 18 different family A GPCRs in different activation states. Solving the crystal structure of β2AR in complex with its cognate G protein delivered an eagerly awaited “holy grail”, greatly furthering our understanding of the molecular mechanism of GPCRs/G protein activation. However, solving more high resolution structures of other GPCRs with their partner G proteins is required to evaluate the first one in adequate detail and learn about possible differences in the activation of specific G proteins and oligomeric organization of GPCRs within these complexes. Although the structure of T4L-β2AR-Gs-nanobody revealed 1:1 GPCR:G protein stoichiometry, an EM-derived, low resolution 3D structure of native rhodopsin-Gt complex indicated rhodopsin dimerization is important in binding with its cognate G protein, transducin. Moreover, the rhodopsin dimer within this complex displays asymmetric properties suggesting the possibility of a tandem asymmetric activation mechanism. Nevertheless, this concept needs to be tested experimentally by using electrophysiological approaches. Oligomerization of GPCRs is now a highly accepted phenomenon. If GPCR oligomerization is compromised, signal transduction could be impaired as well. Improper oligomerization therefore could be an underlying cause of some disease states. Thus, accurate identification of residues that interact at the dimer interface would further our understanding of GPCR signal transmission across the membranes and the mechanisms of GPCR-linked diseases. For example, missense and nonsense mutations of rhodopsin can cause at least three types of human visual disorders: autosomal dominant retinitis pigmentosa (adRP) (Farrar et al. 2011), congenital stationary night blindness (CSNB) (McAlear et al. 2010), and autosomal recessive retinitis pigmentosa (arRP) (Wen et al. 2011) and more than 60 rhodopsin mutations can cause RP. Interestingly, some of these mutations are located at the rhodopsin dimer interface, suggesting that improper rhodopsin oligomerization could contribute to this disease phenotype.
Acknowledgments
I thank Drs. Krzysztof Palczewski and Leslie T. Webster, Jr. (Case Western Reserve University) for valuable comments on the manuscript. This work was supported in part by NIH grants R01-EY0008061, and R01 GM079191.
Footnotes
Conflict of interest
The author declares that she has no conflict of interst.
References
- Abdulaev NG, Ngo T, Ramon E, Brabazon DM, Marino JP, Ridge KD. The receptor-bound “empty pocket” state of the heterotrimeric G-protein alpha-subunit is conformationally dynamic. Biochemistry. 2006;45 (43):12986–12997. doi: 10.1021/bi061088h. [DOI] [PubMed] [Google Scholar]
- Albizu L, Cottet M, Kralikova M, Stoev S, Seyer R, Brabet I, Roux T, Bazin H, Bourrier E, Lamarque L, Breton C, Rives ML, Newman A, Javitch J, Trinquet E, Manning M, Pin JP, Mouillac B, Durroux T. Time-resolved FRET between GPCR ligands reveals oligomers in native tissues. Nat Chem Biol. 2010;6 (8):587–594. doi: 10.1038/nchembio.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angel TE, Gupta S, Jastrzebska B, Palczewski K, Chance MR. Structural waters define a functional channel mediating activation of the GPCR, rhodopsin. Proc Natl Acad Sci U S A. 2009;106 (34):14367–14372. doi: 10.1073/pnas.0901074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audet M, Bouvier M. Restructuring G-protein-coupled receptor activation. Cell. 2012;151 (1):14–23. doi: 10.1016/j.cell.2012.09.003. [DOI] [PubMed] [Google Scholar]
- Baneres JL, Parello J. Structure-based analysis of GPCR function: evidence for a novel pentameric assembly between the dimeric leukotriene B4 receptor BLT1 and the G-protein. Journal of molecular biology. 2003;329 (4):815–829. doi: 10.1016/s0022-2836(03)00439-x. [DOI] [PubMed] [Google Scholar]
- Bayburt TH, Leitz AJ, Xie G, Oprian DD, Sligar SG. Transducin activation by nanoscale lipid bilayers containing one and two rhodopsins. The Journal of biological chemistry. 2007;282 (20):14875–14881. doi: 10.1074/jbc.M701433200. [DOI] [PubMed] [Google Scholar]
- Baylor DA, Lamb TD, Yau KW. Responses of retinal rods to single photons. The Journal of physiology. 1979;288:613–634. [PMC free article] [PubMed] [Google Scholar]
- Cai K, Itoh Y, Khorana HG. Mapping of contact sites in complex formation between transducin and light-activated rhodopsin by covalent crosslinking: use of a photoactivatable reagent. Proc Natl Acad Sci U S A. 2001;98 (9):4877–4882. doi: 10.1073/pnas.051632898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cangiano L, Dell’Orco D. Detecting single photons: a supramolecular matter? FEBS Lett. 2013;587 (1):1–4. doi: 10.1016/j.febslet.2012.11.015. [DOI] [PubMed] [Google Scholar]
- Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, Stevens RC. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318 (5854):1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe HW, Kim YJ, Park JH, Morizumi T, Pai EF, Krauss N, Hofmann KP, Scheerer P, Ernst OP. Crystal structure of metarhodopsin II. Nature. 2011;471 (7340):651–655. doi: 10.1038/nature09789. [DOI] [PubMed] [Google Scholar]
- Chung KY, Rasmussen SG, Liu T, Li S, DeVree BT, Chae PS, Calinski D, Kobilka BK, Woods VL, Jr, Sunahara RK. Conformational changes in the G protein Gs induced by the beta2 adrenergic receptor. Nature. 2011;477 (7366):611–615. doi: 10.1038/nature10488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damian M, Martin A, Mesnier D, Pin JP, Baneres JL. Asymmetric conformational changes in a GPCR dimer controlled by G-proteins. The EMBO journal. 2006;25 (24):5693–5702. doi: 10.1038/sj.emboj.7601449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell’Orco D, Schmidt H. Mesoscopic Monte Carlo simulations of stochastic encounters between photoactivated rhodopsin and transducin in disc membranes. J Phys Chem B. 2008;112 (14):4419–4426. doi: 10.1021/jp709963f. [DOI] [PubMed] [Google Scholar]
- Dell’Orco D, Schmidt H, Mariani S, Fanelli F. Network-level analysis of light adaptation in rod cells under normal and altered conditions. Molecular bioSystems. 2009;5 (10):1232–1246. doi: 10.1039/b908123b. [DOI] [PubMed] [Google Scholar]
- Deupi X, Edwards P, Singhal A, Nickle B, Oprian D, Schertler G, Standfuss J. Stabilized G protein binding site in the structure of constitutively active metarhodopsin-II. Proc Natl Acad Sci U S A. 2012;109 (1):119–124. doi: 10.1073/pnas.1114089108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Moustaine D, Granier S, Doumazane E, Scholler P, Rahmeh R, Bron P, Mouillac B, Baneres JL, Rondard P, Pin JP. Distinct roles of metabotropic glutamate receptor dimerization in agonist activation and G-protein coupling. Proc Natl Acad Sci U S A. 2012;109 (40):16342–16347. doi: 10.1073/pnas.1205838109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst OP, Gramse V, Kolbe M, Hofmann KP, Heck M. Monomeric G protein-coupled receptor rhodopsin in solution activates its G protein transducin at the diffusion limit. Proceedings of the National Academy of Sciences of the United States of America. 2007;104 (26):10859–10864. doi: 10.1073/pnas.0701967104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrar GJ, Millington-Ward S, Chadderton N, Humphries P, Kenna PF. Gene-based therapies for dominantly inherited retinopathies. Gene therapy. 2011 doi: 10.1038/gt.2011.172. [DOI] [PubMed] [Google Scholar]
- Filipek S, Krzysko KA, Fotiadis D, Liang Y, Saperstein DA, Engel A, Palczewski K. A concept for G protein activation by G protein-coupled receptor dimers: the transducin/rhodopsin interface. Photochem Photobiol Sci. 2004;3 (6):628–638. doi: 10.1039/b315661c. [DOI] [PubMed] [Google Scholar]
- Fotiadis D, Liang Y, Filipek S, Saperstein DA, Engel A, Palczewski K. Atomic-force microscopy: Rhodopsin dimers in native disc membranes. Nature. 2003;421 (6919):127–128. doi: 10.1038/421127a. [DOI] [PubMed] [Google Scholar]
- Fotiadis D, Liang Y, Filipek S, Saperstein DA, Engel A, Palczewski K. The G protein-coupled receptor rhodopsin in the native membrane. FEBS letters. 2004;564 (3):281–288. doi: 10.1016/S0014-5793(04)00194-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung JJ, Deupi X, Pardo L, Yao XJ, Velez-Ruiz GA, Devree BT, Sunahara RK, Kobilka BK. Ligand-regulated oligomerization of beta(2)-adrenoceptors in a model lipid bilayer. EMBO J. 2009;28 (21):3315–3328. doi: 10.1038/emboj.2009.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gales C, Van Durm JJ, Schaak S, Pontier S, Percherancier Y, Audet M, Paris H, Bouvier M. Probing the activation-promoted structural rearrangements in preassembled receptor-G protein complexes. Nature structural & molecular biology. 2006;13 (9):778–786. doi: 10.1038/nsmb1134. [DOI] [PubMed] [Google Scholar]
- Govardovskii VI, Korenyak DA, Shukolyukov SA, Zueva LV. Lateral diffusion of rhodopsin in photoreceptor membrane: a reappraisal. Molecular vision. 2009;15:1717–1729. [PMC free article] [PubMed] [Google Scholar]
- Gurevich VV, Gurevich EV. GPCR monomers and oligomers: it takes all kinds. Trends in neurosciences. 2008a;31 (2):74–81. doi: 10.1016/j.tins.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich VV, Gurevich EV. How and why do GPCRs dimerize? Trends in pharmacological sciences. 2008b;29 (5):234–240. doi: 10.1016/j.tips.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M, Gurevich VV, Vishnivetskiy SA, Sigler PB, Schubert C. Crystal structure of beta-arrestin at 1.9 A: possible mechanism of receptor binding and membrane Translocation. Structure. 2001;9 (9):869–880. doi: 10.1016/s0969-2126(01)00644-x. [DOI] [PubMed] [Google Scholar]
- Han Y, Moreira IS, Urizar E, Weinstein H, Javitch JA. Allosteric communication between protomers of dopamine class A GPCR dimers modulates activation. Nat Chem Biol. 2009;5 (9):688–695. doi: 10.1038/nchembio.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck M, Schadel SA, Maretzki D, Hofmann KP. Secondary binding sites of retinoids in opsin: characterization and role in regeneration. Vision Res. 2003;43 (28):3003–3010. doi: 10.1016/j.visres.2003.08.011. [DOI] [PubMed] [Google Scholar]
- Hern JA, Baig AH, Mashanov GI, Birdsall B, Corrie JE, Lazareno S, Molloy JE, Birdsall NJ. Formation and dissociation of M1 muscarinic receptor dimers seen by total internal reflection fluorescence imaging of single molecules. Proc Natl Acad Sci U S A. 2010;107 (6):2693–2698. doi: 10.1073/pnas.0907915107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch JA, Schubert C, Gurevich VV, Sigler PB. The 2.8 A crystal structure of visual arrestin: a model for arrestin’s regulation. Cell. 1999;97 (2):257–269. doi: 10.1016/s0092-8674(00)80735-7. [DOI] [PubMed] [Google Scholar]
- Huang J, Chen S, Zhang JJ, Huang XY. Crystal structure of oligomeric beta1-adrenergic G protein-coupled receptors in ligand-free basal state. Nature structural & molecular biology. 2013;20 (4):419–425. doi: 10.1038/nsmb.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenrauch F, Pollok-Kopp B, Oppermann M. G protein-coupled receptor kinases promote phosphorylation and beta-arrestin-mediated internalization of CCR5 homo- and hetero-oligomers. The Journal of biological chemistry. 2005;280 (45):37503–37515. doi: 10.1074/jbc.M500535200. [DOI] [PubMed] [Google Scholar]
- Ianoul A, Grant DD, Rouleau Y, Bani-Yaghoub M, Johnston LJ, Pezacki JP. Imaging nanometer domains of beta-adrenergic receptor complexes on the surface of cardiac myocytes. Nat Chem Biol. 2005;1 (4):196–202. doi: 10.1038/nchembio726. [DOI] [PubMed] [Google Scholar]
- Ilien B, Glasser N, Clamme JP, Didier P, Piemont E, Chinnappan R, Daval SB, Galzi JL, Mely Y. Pirenzepine promotes the dimerization of muscarinic M1 receptors through a three-step binding process. The Journal of biological chemistry. 2009;284 (29):19533–19543. doi: 10.1074/jbc.M109.017145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y, Cai K, Khorana HG. Mapping of contact sites in complex formation between light-activated rhodopsin and transducin by covalent crosslinking: use of a chemically preactivated reagent. Proc Natl Acad Sci U S A. 2001;98 (9):4883–4887. doi: 10.1073/pnas.051632998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jastrzebska B, Fotiadis D, Jang GF, Stenkamp RE, Engel A, Palczewski K. Functional and structural characterization of rhodopsin oligomers. J Biol Chem. 2006;281 (17):11917–11922. doi: 10.1074/jbc.M600422200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jastrzebska B, Orban T, Golczak M, Engel A, Palczewski K. Asymmetry of the rhodopsin dimer in complex with transducin. FASEB J. 2013a doi: 10.1096/fj.12-225383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jastrzebska B, Ringler P, Lodowski DT, Moiseenkova-Bell V, Golczak M, Muller SA, Palczewski K, Engel A. Rhodopsin-transducin heteropentamer: three-dimensional structure and biochemical characterization. J Struct Biol. 2011;176 (3):387–394. doi: 10.1016/j.jsb.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jastrzebska B, Ringler P, Palczewski K, Engel A. The Rhodopsin-Transducin Complex Houses Two Distinct Rhodopsin Molecules. J Struct Biol. 2013b doi: 10.1016/j.jsb.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisselev OG, Downs MA. Rhodopsin-interacting surface of the transducin gamma subunit. Biochemistry. 2006;45 (31):9386–9392. doi: 10.1021/bi060806x. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Ogawa K, Yao R, Lichtarge O, Bouvier M. Functional rescue of beta-adrenoceptor dimerization and trafficking by pharmacological chaperones. Traffic. 2009;10 (8):1019–1033. doi: 10.1111/j.1600-0854.2009.00932.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert NA. GPCR dimers fall apart. Science signaling. 2010;3(115):pe12. doi: 10.1126/scisignal.3115pe12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambright DG, Sondek J, Bohm A, Skiba NP, Hamm HE, Sigler PB. The 2.0 A crystal structure of a heterotrimeric G protein. Nature. 1996;379 (6563):311–319. doi: 10.1038/379311a0. [DOI] [PubMed] [Google Scholar]
- Lebon G, Warne T, Edwards PC, Bennett K, Langmead CJ, Leslie AG, Tate CG. Agonist-bound adenosine A2A receptor structures reveal common features of GPCR activation. Nature. 2011;474 (7352):521–525. doi: 10.1038/nature10136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Fotiadis D, Filipek S, Saperstein DA, Palczewski K, Engel A. Organization of the G protein-coupled receptors rhodopsin and opsin in native membranes. The Journal of biological chemistry. 2003;278 (24):21655–21662. doi: 10.1074/jbc.M302536200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manglik A, Kruse AC, Kobilka TS, Thian FS, Mathiesen JM, Sunahara RK, Pardo L, Weis WI, Kobilka BK, Granier S. Crystal structure of the micro-opioid receptor bound to a morphinan antagonist. Nature. 2012;485 (7398):321–326. doi: 10.1038/nature10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAlear SD, Kraft TW, Gross AK. 1 rhodopsin mutations in congenital night blindness. Advances in experimental medicine and biology. 2010;664:263–272. doi: 10.1007/978-1-4419-1399-9_30. [DOI] [PubMed] [Google Scholar]
- Milligan G. The role of dimerisation in the cellular trafficking of G-protein-coupled receptors. Current opinion in pharmacology. 2010;10 (1):23–29. doi: 10.1016/j.coph.2009.09.010. [DOI] [PubMed] [Google Scholar]
- Modzelewska A, Filipek S, Palczewski K, Park PS. Arrestin interaction with rhodopsin: conceptual models. Cell biochemistry and biophysics. 2006;46 (1):1–15. doi: 10.1385/CBB:46:1:1. [DOI] [PubMed] [Google Scholar]
- Moepps B, Nuesseler E, Braun M, Gierschik P. A homolog of the human chemokine receptor CXCR1 is expressed in the mouse. Molecular immunology. 2006;43 (7):897–914. doi: 10.1016/j.molimm.2005.06.043. [DOI] [PubMed] [Google Scholar]
- Neri M, Vanni S, Tavernelli I, Rothlisberger U. Role of aggregation in rhodopsin signal transduction. Biochemistry. 2010;49 (23):4827–4832. doi: 10.1021/bi100478j. [DOI] [PubMed] [Google Scholar]
- Noel JP, Hamm HE, Sigler PB. The 2.2 A crystal structure of transducin-alpha complexed with GTP gamma S. Nature. 1993;366 (6456):654–663. doi: 10.1038/366654a0. [DOI] [PubMed] [Google Scholar]
- Oldham WM, Van Eps N, Preininger AM, Hubbell WL, Hamm HE. Mechanism of the receptor-catalyzed activation of heterotrimeric G proteins. Nature structural & molecular biology. 2006;13 (9):772–777. doi: 10.1038/nsmb1129. [DOI] [PubMed] [Google Scholar]
- Orban T, Jastrzebska B, Gupta S, Wang B, Miyagi M, Chance MR, Palczewski K. Conformational dynamics of activation for the pentameric complex of dimeric G protein-coupled receptor and heterotrimeric G protein. Structure. 2012;20 (5):826–840. doi: 10.1016/j.str.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289 (5480):739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- Park JH, Scheerer P, Hofmann KP, Choe HW, Ernst OP. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature. 2008;454 (7201):183–187. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- Pellissier LP, Barthet G, Gaven F, Cassier E, Trinquet E, Pin JP, Marin P, Dumuis A, Bockaert J, Baneres JL, Claeysen S. G protein activation by serotonin type 4 receptor dimers: evidence that turning on two protomers is more efficient. J Biol Chem. 2011;286 (12):9985–9997. doi: 10.1074/jbc.M110.201939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pescitelli G, Sreerama N, Salvadori P, Nakanishi K, Berova N, Woody RW. Inherent chirality dominates the visible/near-ultraviolet CD spectrum of rhodopsin. J Am Chem Soc. 2008;130 (19):6170–6181. doi: 10.1021/ja711009y. [DOI] [PubMed] [Google Scholar]
- Rasmussen SG, Choi HJ, Fung JJ, Pardon E, Casarosa P, Chae PS, Devree BT, Rosenbaum DM, Thian FS, Kobilka TS, Schnapp A, Konetzki I, Sunahara RK, Gellman SH, Pautsch A, Steyaert J, Weis WI, Kobilka BK. Structure of a nanobody-stabilized active state of the beta(2) adrenoceptor. Nature. 2011a;469 (7329):175–180. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen SG, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VR, Sanishvili R, Fischetti RF, Schertler GF, Weis WI, Kobilka BK. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. 2007;450 (7168):383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- Rasmussen SG, DeVree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, Thian FS, Chae PS, Pardon E, Calinski D, Mathiesen JM, Shah ST, Lyons JA, Caffrey M, Gellman SH, Steyaert J, Skiniotis G, Weis WI, Sunahara RK, Kobilka BK. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature. 2011b;477 (7366):549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivero-Muller A, Chou YY, Ji I, Lajic S, Hanyaloglu AC, Jonas K, Rahman N, Ji TH, Huhtaniemi I. Rescue of defective G protein-coupled receptor function in vivo by intermolecular cooperation. Proc Natl Acad Sci U S A. 2010;107 (5):2319–2324. doi: 10.1073/pnas.0906695106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruprecht JJ, Mielke T, Vogel R, Villa C, Schertler GF. Electron crystallography reveals the structure of metarhodopsin I. The EMBO journal. 2004;23 (18):3609–3620. doi: 10.1038/sj.emboj.7600374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salahpour A, Angers S, Mercier JF, Lagace M, Marullo S, Bouvier M. Homodimerization of the beta2-adrenergic receptor as a prerequisite for cell surface targeting. The Journal of biological chemistry. 2004;279 (32):33390–33397. doi: 10.1074/jbc.M403363200. [DOI] [PubMed] [Google Scholar]
- Salom D, Lodowski DT, Stenkamp RE, Le Trong I, Golczak M, Jastrzebska B, Harris T, Ballesteros JA, Palczewski K. Crystal structure of a photoactivated deprotonated intermediate of rhodopsin. Proc Natl Acad Sci U S A. 2006;103 (44):16123–16128. doi: 10.1073/pnas.0608022103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheerer P, Park JH, Hildebrand PW, Kim YJ, Krauss N, Choe HW, Hofmann KP, Ernst OP. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455 (7212):497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- Schertler GF, Hargrave PA. Projection structure of frog rhodopsin in two crystal forms. Proceedings of the National Academy of Sciences of the United States of America. 1995;92 (25):11578–11582. doi: 10.1073/pnas.92.25.11578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schertler GF, Villa C, Henderson R. Projection structure of rhodopsin. Nature. 1993;362 (6422):770–772. doi: 10.1038/362770a0. [DOI] [PubMed] [Google Scholar]
- Shi GW, Chen J, Concepcion F, Motamedchaboki K, Marjoram P, Langen R. Light causes phosphorylation of nonactivated visual pigments in intact mouse rod photoreceptor cells. The Journal of biological chemistry. 2005;280 (50):41184–41191. doi: 10.1074/jbc.M506935200. [DOI] [PubMed] [Google Scholar]
- Sommer ME, Hofmann KP, Heck M. Distinct loops in arrestin differentially regulate ligand binding within the GPCR opsin. Nature communications. 2012;3:995. doi: 10.1038/ncomms2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondek J, Bohm A, Lambright DG, Hamm HE, Sigler PB. Crystal structure of a G-protein beta gamma dimer at 2.1A resolution. Nature. 1996;379 (6563):369–374. doi: 10.1038/379369a0. [DOI] [PubMed] [Google Scholar]
- Standfuss J, Xie G, Edwards PC, Burghammer M, Oprian DD, Schertler GF. Crystal structure of a thermally stable rhodopsin mutant. J Mol Biol. 2007;372 (5):1179–1188. doi: 10.1016/j.jmb.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struts AV, Salgado GF, Brown MF. Solid-state 2H NMR relaxation illuminates functional dynamics of retinal cofactor in membrane activation of rhodopsin. Proc Natl Acad Sci U S A. 2011;108 (20):8263–8268. doi: 10.1073/pnas.1014692108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton RB, Vishnivetskiy SA, Robert J, Hanson SM, Raman D, Knox BE, Kono M, Navarro J, Gurevich VV. Crystal structure of cone arrestin at 2.3A: evolution of receptor specificity. Journal of molecular biology. 2005;354 (5):1069–1080. doi: 10.1016/j.jmb.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Nomura W, Narumi T, Masuda A, Tamamura H. Bivalent ligands of CXCR4 with rigid linkers for elucidation of the dimerization state in cells. J Am Chem Soc. 2010;132 (45):15899–15901. doi: 10.1021/ja107447w. [DOI] [PubMed] [Google Scholar]
- Van Eps N, Oldham WM, Hamm HE, Hubbell WL. Structural and dynamical changes in an alpha-subunit of a heterotrimeric G protein along the activation pathway. Proc Natl Acad Sci U S A. 2006;103 (44):16194–16199. doi: 10.1073/pnas.0607972103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Eps N, Preininger AM, Alexander N, Kaya AI, Meier S, Meiler J, Hamm HE, Hubbell WL. Interaction of a G protein with an activated receptor opens the interdomain interface in the alpha subunit. Proc Natl Acad Sci U S A. 2011;108 (23):9420–9424. doi: 10.1073/pnas.1105810108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilardaga JP, Nikolaev VO, Lorenz K, Ferrandon S, Zhuang Z, Lohse MJ. Conformational cross-talk between alpha2A-adrenergic and mu-opioid receptors controls cell signaling. Nature chemical biology. 2008;4 (2):126–131. doi: 10.1038/nchembio.64. [DOI] [PubMed] [Google Scholar]
- Vobornik D, Rouleau Y, Haley J, Bani-Yaghoub M, Taylor R, Johnston LJ, Pezacki JP. Nanoscale organization of beta2-adrenergic receptor-Venus fusion protein domains on the surface of mammalian cells. Biochem Biophys Res Commun. 2009;382 (1):85–90. doi: 10.1016/j.bbrc.2009.02.144. [DOI] [PubMed] [Google Scholar]
- Wen Y, Locke KG, Hood DC, Birch DG. Rod photoreceptor temporal properties in retinitis pigmentosa. Experimental eye research. 2011;92 (3):202–208. doi: 10.1016/j.exer.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wettschureck N, Offermanns S. Mammalian G proteins and their cell type specific functions. Physiol Rev. 2005;85 (4):1159–1204. doi: 10.1152/physrev.00003.2005. [DOI] [PubMed] [Google Scholar]
- White JF, Noinaj N, Shibata Y, Love J, Kloss B, Xu F, Gvozdenovic-Jeremic J, Shah P, Shiloach J, Tate CG, Grisshammer R. Structure of the agonist-bound neurotensin receptor. Nature. 2012;490 (7421):508–513. doi: 10.1038/nature11558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whorton MR, Bokoch MP, Rasmussen SG, Huang B, Zare RN, Kobilka B, Sunahara RK. A monomeric G protein-coupled receptor isolated in a high-density lipoprotein particle efficiently activates its G protein. Proc Natl Acad Sci U S A. 2007;104 (18):7682–7687. doi: 10.1073/pnas.0611448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whorton MR, Jastrzebska B, Park PS, Fotiadis D, Engel A, Palczewski K, Sunahara RK. Efficient coupling of transducin to monomeric rhodopsin in a phospholipid bilayer. J Biol Chem. 2008;283 (7):4387–4394. doi: 10.1074/jbc.M703346200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Chien EY, Mol CD, Fenalti G, Liu W, Katritch V, Abagyan R, Brooun A, Wells P, Bi FC, Hamel DJ, Kuhn P, Handel TM, Cherezov V, Stevens RC. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science. 2010;330 (6007):1066–1071. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Wacker D, Mileni M, Katritch V, Han GW, Vardy E, Liu W, Thompson AA, Huang XP, Carroll FI, Mascarella SW, Westkaemper RB, Mosier PD, Roth BL, Cherezov V, Stevens RC. Structure of the human kappa-opioid receptor in complex with JDTic. Nature. 2012;485 (7398):327–332. doi: 10.1038/nature10939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Wu H, Katritch V, Han GW, Jacobson KA, Gao ZG, Cherezov V, Stevens RC. Structure of an agonist-bound human A2A adenosine receptor. Science. 2011;332 (6027):322–327. doi: 10.1126/science.1202793. [DOI] [PMC free article] [PubMed] [Google Scholar]