Abstract
Proteases play causal roles in many aspects of the aggressive phenotype of tumors, yet many of the implicated proteases originate from tumor-associated cells or from responses of tumor cells to interactions with other cells. Therefore, to obtain a comprehensive view of tumor proteases, we need to be able to assess proteolysis in tumors that are interacting with their microenvironment. As this is difficult to do in vivo, we have developed functional live-cell optical imaging assays and 3D and 4D (i.e., 3D over time) coculture models. We present here a description of the probes used to measure proteolysis and protease activities, the methods used for imaging and analysis of proteolysis and the 3D and 4D models used in our laboratory. Of course, all assays have limitations; however, we suggest that the techniques discussed here will, with attention to their limitations, be useful as a screen for drugs to target the invasive pheno-type of tumors.
1. Introduction
Recent studies in our laboratory have focused on establishing live-cell assays to image the proteolysis that is associated with the progression of premalignant breast lesions to malignant carcinomas. This chapter will use our studies to illustrate how the interactions of tumor cells with their microenvironment contribute to this proteolysis and how functional imaging assays and 3D/4D coculture models might be used to identify druggable targets and screen therapeutic agents.
1.1. Why image proteolysis rather than protease activity?
Proteolysis or the hydrolytic degradation of proteins occurring as the result of interactions among proteases of more than one catalytic type, that is, proteolytic pathways or networks, has been shown to be critical to malignant progression (e.g., see DeClerck and Laug, 1996; Ellerbroek et al., 1998; Kim et al., 1998; Krol et al., 2003; Muehlenweg et al., 2000; Ramos-DeSimone et al., 1999). Prior research in our laboratory concentrated on one cysteine cathepsin, that is, cathepsin B (for reviews, see Cavallo-Medved and Sloane, 2003; Podgorski and Sloane, 2003; Roshy et al., 2003; Yan and Sloane, 2003). Analysis of any one protease or protease class, however, does not define the “tumor degradome” (Balbin et al., 2003). Therefore, we established an assay to study proteolysis of extracellular matrix substrates encountered by tumor cells as they invade into normal tissues surrounding tumors. This protein-based assay contrasts with those measuring degradation of synthetic substrates that are selective for one protease or one catalytic type of protease. Data generated in our laboratory (Cavallo-Medved et al., 2009; Jedeszko et al., 2009; Li et al., 2008; Podgorski et al., 2009; Sameni et al., 2000, 2001, 2003, 2008, 2009) and other laboratories (Kjoller et al., 2004; Madsen et al., 2011; Wolf et al., 2009) by means of functional proteolysis assays that employ protein substrates support our contention that meaningful analyses of tumor proteolysis require assessment of the roles played by multiple proteases as well as assessment of how those proteases interact to modulate the activities of other proteases.
1.2. Why image tumor proteolysis?
More than one catalytic type of protease has been implicated in the progression of human tumors (for reviews, see Choong and Nadesapillai, 2003; Fuchs, 2002; Hojilla et al., 2003; Podgorski and Sloane, 2003). Although the preclinical data implicating matrix metalloproteinases (MMPs) in malignant progression were particularly compelling, the clinical trials on MMP inhibitors (MMPIs) did not fulfill the promise of MMPs as therapeutic targets in cancer. There are several possible explanations for this apparent “disconnect” between the preclinical and clinical data; please see Chau et al. (2003), Coussens et al. (2002), Egeblad and Werb (2002) for insightful and thought-provoking reviews on this topic.
The failures of MMPIs in clinical trials have resulted in allegations that MMPs and, by extrapolation, other proteases are not appropriate therapeutic targets in cancer. Is this true or rather might the failure of the MMPI trials reflect problems in clinical trial design for cytostatic agents and in particular the need to use imaging (Adjei et al., 2009; Ang et al., 2010)? This is the case as among the critical questions is whether the MMPIs actually reached and reduced the activity of their target MMPs in vivo. The MMPI trials did not include surrogate endpoints so it is not known whether MMPs were actually inhibited in the patients enrolled in the trials (Chau et al., 2003; Li and Anderson, 2003; McIntyre and Matrisian, 2003). Clinical trials without surrogate endpoints to monitor and confirm the efficacy of the therapeutic strategies being tested should not be viewed as definitive (Adjei et al., 2009; Li and Anderson, 2003; McIntyre and Matrisian, 2003; Seymour et al., 2010). There are other concerns about the MMPI trials. Coussens et al. (2002) cite data revealing that the tumors studied in the clinical trials did not necessarily express the particular proteases targeted by the MMPIs. None of the clinical trials with BAY 12-9566 or other MMPIs included patients with breast cancers although this is a cancer for which there had been strong preclinical evidence that MMPs impact progression. For example, Sledge and colleagues (Nozaki et al., 2003) demonstrated efficacy for BAY 12-9566 in an orthotopic model in which human breast tumor cells were implanted in the mammary fat pads of mice. Furthermore, not all MMPs should be inhibited; this is clearly true for MMP-8 (collagenase-2), an MMP expressed by inflammatory neutrophils, which plays a protective role in skin cancer (Balbin et al., 2003). The data on MMP-8 and more recent data on a variety of other MMPs (for reviews, see Lopez-Otin and Matrisian, 2007; Lopez-Otin et al., 2009) support the concept that broad-spectrum MMPIs would have unanticipated side effects and indicate how essential it is “to define precisely the tumor degradome” (Balbin et al., 2003) before using MMPIs or for that matter other protease inhibitors for cancer therapy. In fact, unanticipated side effects did occur in the MMPI clinical trials leading to limitations in the amount of MMPIs that the patients could take and in some patients necessitating MMPI-free holidays (Coussens et al., 2002). How endogenous protease inhibitors factor into the equation is also of relevance. As just one example, TIMP-1 has been shown to promote carcinogenesis of squamous cell carcinoma of the skin, exerting “differential regulation on tissues in a stage-dependent manner” (Rhee et al., 2004). We are still far from having a thorough understanding of the roles of proteases in cancer: this includes understanding the roles of MMPs; the roles of other classes of proteases; the roles of endogenous protease inhibitors, activators and receptors, or binding proteins; that those roles are dynamic and may change during the course of malignant progression; whether the proteases playing critical roles in malignant progression come from tumor, stromal, or inflammatory cells; whether the critical proteases are affected by interactions of the tumor with its microenvironment; what the relevant substrates are for proteases that play causal roles in malignant progression; etc. Many of these issues are discussed in a large volume on what constitutes the cancer degradome that was published in 2008 (Edwards et al., 2008). Nonetheless, we do not yet know which protease(s) or proteolytic pathway is the most appropriate target for antiprotease therapies or when antiprotease therapies might prove most effective.
2. Assays for Functional Imaging of Proteolysis
The terminology “functional imaging” was originally used to describe imaging methods that assessed changes in physiological processes such as metabolism and blood flow, for example, positron emission tomography (PET) and magnetic resonance imaging. Here, we use this terminology because the live-cell assays that we have developed for imaging proteolysis can be used to quantify changes in proteolytic activity as well as to localize sites at which proteolysis is occurring (Jedeszko et al., 2008). Functional imaging often uses agents or probes; in PET, for example, a glucose analogue fluorodeoxyglucose is used as an F18 radioisotopic tracer to detect and localize the high levels of metabolic activity associated with tumors (Mankoff et al., 2007).
2.1. Probes
Probes for imaging proteases have been based on either substrates or inhibitors. A substrate probe can be a signal amplifier as, in the presence of an active target protease, the substrate will be cleaved continuously and thus the signal intensity increased. This property also makes substrate probes sensitive reagents for detecting reductions in protease activity due to protease inhibitors, etc. A potential negative with substrate probes is that the cleavage products may not remain at the site where cleavage occurred and thus would not accurately localize an active protease. This is especially true for extracellular or cell surface proteolysis where substrates and/or cleavage products might diffuse away from the site at which they are generated. Protease probes based on inhibitors that bind covalently to the active site of a protease are more effective in localizing protease activity. On the other hand, they cannot amplify the signal and therefore are less sensitive in detecting proteases that are not highly expressed or reductions in protease activity (Fonovic and Bogyo, 2007).
Whether protease probes are based on a substrate or an inhibitor, proteases are ideal targets for selective, activatable contrast agents. In this case, a fluorescent probe is synthesized in a quenched state in which a nonfluorescent quencher is attached to the probe in close proximity to the reporting fluorophore via a protease-selective peptide linker. The proximity of the fluorophore to the quencher causes transfer of energy from the former to the latter, thus preventing emission of fluorescence. When such a probe encounters an active target protease, the peptide will be cleaved and the quencher released, resulting in emission of a fluorescent signal. Alternatively, probes can be developed utilizing a synthetic (e.g., dendrimer or polylysine—see Marten et al., 2002; McIntyre et al., 2004, respectively) or protein (collagen) backbone to which a large number of reporters are attached via peptide linkers in close proximity to each other (for review see, Sloane et al., 2006). The overabundance of the reporter in close proximity causes self-quenching due to a FRET (Forster Resonance Energy Transfer) effect (Brzostowski et al., 2009). When an active target protease cleaves the reporter off the backbone, fluorescence is emitted. Such probes have been used for both in vitro and in vivo systems (Brzostowski et al., 2009; McIntyre et al., 2004).
2.1.1. Fluorescently tagged proteins
The primary probes that we use in our laboratory to image proteolysis are quenched fluorescent or dye quenched (DQ) extracellular matrix proteins that are commercially available from Invitrogen, that is, DQ-collagen IV and DQ-collagen I. Our use of these substrates was featured in their newsletter (Visualizing tumor metastasis: CellTracker™ dyes, DQ™ collagen, and Geltrex™ matrix. BioProbes 60, pp. 32–33, October 2009). We selected the two collagen substrates because type IV collagen is a major component of the basement membrane (Aumailley and Gayraud, 1998) and dissolution of type IV collagen has been shown to be integral to normal developmental processes and an early step in malignant progression (for review, see Liotta and Kohn, 2001). Fibrillar collagen I in the connective tissue through which tumor cells invade is an impediment to cell growth and invasion (Henriet et al., 2000; Sabeh et al., 2009).
By using quenched fluorescent derivatives of type IV and I collagen, we have been able to image and localize proteolysis, that is, a gain-of-function/fluorescence, by live cells (Sameni et al., 2000, 2001, 2003, 2009). This is in contrast to using nonquenched FITC-labeled proteins where one is imaging a loss of fluorescence and has to fix the cells and substrate before imaging (Demchik et al., 1999; Sloane, 1996).
A critical point is that, by virtue of their fluorescent labeling, the DQ-collagens are no longer native proteins. Therefore the ability to cleave these substrates may not be representative of an ability to cleave native forms of these proteins. This is likely more the case for DQ-collagen I as gelatin or denatured collagen I is readily degraded by many proteases. There are collagenase-resistant forms of collagen I in which the cleavage sites in the helical region have been mutated so that they cannot be degraded by true “collagenases” such as MMP-1 (Wu et al., 1990). Studies using collagenase-resistant collagen I have demonstrated that proteolysis of collagen I is required for motility of vascular smooth muscle cells on collagen I (Li et al., 2000) and for invasion of ovarian cancer cells into collagen I (Ellerbroek et al., 2001). Unfortunately, DQ-collagenase-resistant collagen I is not commercially available.
2.1.2. Activity-based probes (ABPs)
We also employ ABPs developed by Bogyo and colleagues for imaging cysteine proteases of the papain family (Greenbaum et al., 2002a,b). These probes are based on the broad-spectrum cysteine protease inhibitor E-64 and bind covalently to the active sites of the enzymes. Therefore, with these probes, one can image active cysteine cathepsins in situ in cells and also identify what active protease is being detected by visualizing the fluorescently tagged protease(s) in cell lysates/conditioned media on SDS-PAGE gels. These probes have been used to identify and image cysteine cathepsins in live cells in vitro (Blum et al., 2005) and in vivo in a transgenic mouse model of pancreatic cancer (Joyce et al., 2004) and a transgenic mouse model of mammary cancer (Vasiljeva et al., 2006). These ABPs are cell permeable and thus allow us to image the intracellular activity of cysteine cathepsins in the lysosomes (Blum et al., 2005). Furthermore, the ABPs are available in both unquenched and quenched versions albeit the selectivity of the ABPs for a given cysteine cathepsin seems to be reduced in the quenched versions (Blum et al., 2005).
2.1.3. Other protease probes
The two other types of protease probes that we are using in vitro are fluorogen-activating protein (FAP) protease biosensors developed by Berget and colleagues at Carnegie-Mellon and proteolytic beacons that Matrisian and colleagues have designed and developed for real-time analysis of MMP activity, including activity of individual MMPs like MMP-7 (McIntyre and Matrisian, 2009; McIntyre et al., 2004, 2010; Scherer et al., 2008). The FAPs are engineered proteins based on single chain antibodies (Falco et al., 2009). At present, FAPs to assess the activity on the surface of live cells of MMP-14, -2, -9, and cathepsin K are being tested. The proteolytic beacons are protease substrates, which are built on a nanodendron scaffold and use FRET between two fluorophores linked to a selective peptide substrate (see McIntyre and Matrisian, 2003; Scherer et al., 2008 for review). When in close proximity, the sensor fluorophore is quenched by the reference fluorophore, which also serves to monitor substrate concentration. The ability of these beacons to monitor how much of a probe is delivered and demonstrate that the probe is delivered is a crucial one. Proteolytic cleavage of the peptide linker results in increased fluorescence of the sensor so that the ratio of sensor to reference can be used as a quantitative measure of proteolytic activity. The design permits a great deal of flexibility: fluorophores can be in either the visible or near infrared range (NIR), protease selectivity can be modulated by alterations in the peptide sequence, clearance kinetics and route can be determined by the size and composition of the dendron backbone, and the multifunctionality of the dendrons allows for optimization of fluorophore concentration and solubility characteristics. Published proteolytic beacons include a visible one that is selective for MMP-7 (McIntyre and Matrisian, 2003; Wadsworth et al., 2010), NIR MMP-7- and MMP-9-selective beacons (McIntyre et al., 2010; Scherer et al., 2008), and a NIR beacon that detects general MMP activity (McIntyre et al., 2010).
2.2. Analysis
The development of new fluorogenic dyes and of confocal, multiphoton, and structured illumination microscopes has made optical imaging the method of choice for direct observations in living systems (for review, see Andrews et al., 2002; Swedlow and Platani, 2002). These state-of-the-art advanced imaging systems allow one to perform 3D and 4D analyses using multiple probes.
For live-cell imaging there is an absolute requirement to perform experiments under physiological conditions. For this reason all of our imaging systems are equipped with environmental chambers (temperature, CO2, and humidity controlled). This allows us to image live cultures over extended times without removing the cultures from the microscope stage. In addition, we can image the dynamics of proteolysis in real-time as impacted by tumor–stromal interactions. Another common feature of our systems is their motorized, fully automated stages. With this type of stage, we are able to select several areas of interest and program the system to image these areas sequentially at multiple time points and in 3D. The environmental chamber allows us to establish 3D models, place them on the microscope stage and program the microscope to acquire images as the cells attach, migrate, and invade into the surrounding extracellular matrices; we illustrate the association of proteolysis with cell migration and cell– cell interaction in Fig. 10.1. We now routinely label the various types of cells in our cocultures by prestaining with vital cytoplasmic dyes (Fig. 10.1) or transducing with fluorescent proteins (Fig. 10.2) so that they can be readily distinguished from one another when analyzing cell– cell interactions. We obtain optical sections through the entire volume of the fluorescently labeled specimen in real time at various time points. The data are then analyzed with the 3D and 4D image reconstruction software as described below and as illustrated in Fig. 10.2.
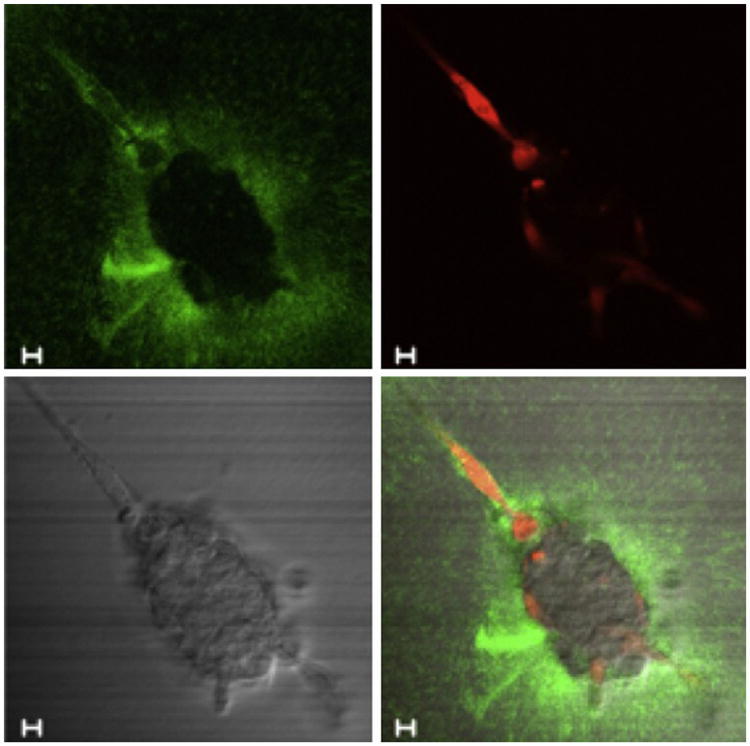
Figure 10.1.
Still images of time-lapse series (90 min total) to follow degradation of DQ-collagen IV (green fluorescence) by carcinoma cells in coculture with fibroblasts. CCD-112CoN colon fibroblasts (red due to prestaining with CellTracker Orange) were cocultured with a spheroid of HCT 116 colon carcinoma cells. In this image taken after an overnight period of coculture, the fibroblasts can be seen to be moving toward and entering the HCT 116 spheroid. Note the pericellular fluorescent cleavage products due to degradation of DQ-collagen IV by the fibroblasts as they infiltrate into the HCT 116 spheroid, which is more readily apparent in video format. The four panels of this figure represent degradation products of DQ-collagen IV (green), fibroblasts (red), spheroid of HCT 116 cells (DIC), and a composite of the other three images. Bar, 10 μm.
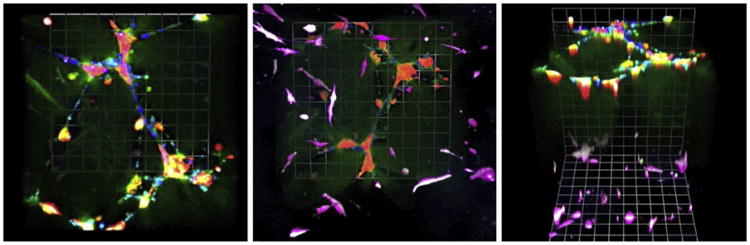
Figure 10.2.
MAME tripartite cocultures of human SUM102 breast carcinoma cells (red; transduced with Lenti-RFP) with human umbilicial vein endothelial cells (HUVEC; blue; stained with CellTrace Far Red) in reconstituted basement membrane above a bottom layer of WS-12Ti human breast tumor-associated fibroblasts (magenta; transduced with Lenti-YFP) in interstitial collagen I (see also Fig. 10.3A). At 2 days of coculture as illustrated here, the SUM102 cells cluster around the branching networks formed by the HUVECs. Degradation products of DQ-collagen IV (green) are apparent around the interacting cells. The fibroblasts, which are primarily in the layer of collagen I, are associated with pericellular degradation products of DQ-collagen I (white). A few fibroblasts, also associated with proteolysis, can be seen to have migrated into the upper layer. The three panels from left to right are 3D reconstructions of the MAME tripartite coculture from the top, the bottom and at a 45° angle. Magnification, 20×.
The operating software for the imaging systems performs basic quantitative analyses, but is limited in regard to the extensive 3D and 4D quantitation required by the studies described here. Therefore, we use advanced stand-alone image analysis software such as Metamorph™ 7.64 and Volocity™ 5.5 to analyze fluorescence intensities, volumetric areas, surface areas, etc. Volumetric measurements are critical for accurately assessing changes in cell–cell interactions over time in 3D, that is, in 4D. To assess interactions, we label samples for detection of the different cell types, specific target proteins, proteolytic degradation products, etc., as we have described (Jedeszko et al., 2008; Sameni et al., 2003, 2009). Volumetric measurements of the sample components are acquired in multiple channels with each channel representing a single target. Datasets from each time point are then loaded for 4D quantification, analysis, and rendering of the data in 3D for each time point. Importantly, the software allows one to mark specific regions of interests in the 3D image stacks. The software can then quantify each region in 3D as well as quantify changes over time, that is, in 4D. The compiled images can be presented in many formats, including movies of live events.
3. 3D/4D Models for Analysis of Biological Processes Linked to Proteolysis
The working hypothesis for ongoing studies in our laboratory is that 3D mammary cell-based models will recapitulate the proteolytic mechanisms integral to developmental and neoplastic processes. Using such models to image proteolysis over time (i.e., 4D imaging) should allow us to place proteolysis, including the proteases identified and their interactions, within the context of the signaling pathways and other functions already elucidated by Bissell and her many collaborators as essential to malignant progression of mammary cancer. Those studies have convincingly demonstrated the importance of context to developmental and neoplastic processes in the mammary gland (for review, see Lee et al., 2007; Schmeichel and Bissell, 2003). By growing mammary epithelial cells “within 3D basement membrane-like-matrices,” the Bissell laboratory has been able to reproduce signaling pathways and functions (e.g., milk protein production) in vitro that are not observed when the same cells are cultured in 2D monolayers. Furthermore, the in vitro 3D mammary models have shown the need to match cell types with appropriate extracellular matrices, in this case mammary epithelial cells with basement membrane-like-matrices. When Gudjonsson et al. (2002) substituted an interstitial connective tissue matrix protein, that is, collagen I, acini formed, but they were inside-out. Adding normal myoepithelial cells to the collagen I gels resulted in correctly polarized acini, an effect dependent on production by the myoepithelial cells of the α-1 chain of the basement membrane protein laminin. There is a wealth of studies by Bissell and her collaborators showing that 3D cell-based assays can be used to study mechanisms for morphogenesis and neoplasia of human breast in vitro (for review, see Gudjonsson et al., 2003; Schmeichel and Bissell, 2003). They also have demonstrated that proteases, in particular MMPs, are involved in morphogenesis and neoplasia. Studies by Weiss and his colleagues also implicate MMPs, in this case, MMP-14, or MT1-MMP, which they have shown to degrade collagen I and to be a prerequisite for proliferation of tumor cells in 3D collagen I gels (Hotary et al., 2003). The Brugge laboratory has used 3D monotypic cultures grown in reconstituted basement membrane (rBM) to demonstrate a role for caspase-family cysteine proteases and apoptosis in the formation of lumens in the mammary acini (Debnath et al., 2002; Shaw et al., 2004). They have shown that migration and invasion of the mammary epithelial cells can be induced by coexpression of activated ErbB2 and TGF-β (58), but have not analyzed the proteases responsible for the invasion. Further studies based on the Brugge 3D monotypic cultures by Debnath and colleagues have identified a role for lysosomal proteases. They found that autophagy and proteolysis within lysosomes plays both a suppressive and a promotion role that is context dependent (Chen and Debnath, 2010; Roy and Debnath, 2010).
Liotta and Kohn (2001) have suggested that cancer therapies should target the stroma or the tumor–stroma interface and hypothesized that stromal therapy could require lower doses than therapies that target the tumor. The need to target stroma would certainly appear to be true for protease inhibitors, as cells present in the tumor-associated stroma (e.g., fibroblasts, endothelial cells, inflammatory cells, myofibroblasts) are all important sources of proteases (for reviews, see Almholt and Johnsen, 2003; Bogenrieder and Herlyn, 2003; Coussens and Werb, 2001; DeClerck, 2000; Johnsen et al., 1998; van Kempen and Coussens, 2002). For example, MMP-8 is expressed by neutrophils (Balbin et al., 2003); urokinase plasminogen activator (uPA) by myofibroblasts, macrophages, and endothelial cells in ductal breast cancer (Nielsen et al., 2007); MMP-9 by neutrophils, macrophages, and mast cells in a mouse model of squamous cell carcinoma of the skin (Coussens et al., 2000); MMP-3 by subepithelial myofibroblasts in human colon (Bamba et al., 2003); the cysteine proteases cathepsins B, K, L, and S and MMP-7, -9, and -12 by macrophages (Filippov et al., 2003; Punturieri et al., 2000; Reddy et al., 1995); cathepsin B by endothelial cells from breast, glioma, and prostate (for review, see Keppler et al., 1996); and MMP-2 by endothelial cells (Han et al., 2003). Tumor-associated macrophages in human carcinomas also express high levels of cathepsin B (Campo et al., 1994; Fernandez et al., 2001; McKerrow et al., 2000) and this macrophage cathepsin B enhances malignant progression in mouse transgenic models for mammary carcinoma (Vasiljeva et al., 2006) and pancreatic carcinoma (Gocheva et al., 2010). Stromal cells can also affect tumor proteolysis through the expression of endogenous protease inhibitors. Myofibroblasts, for example, are suggested to be the primary source of plasminogen activator inhibitor-1 (PAI-1) in human breast carcinomas (Offersen et al., 2003). Overall such data indicate that we will not be able to define the “tumor degradome” (Balbin et al., 2003) unless we study proteolysis in the context of tumor cells interacting with their microenvironment, interactions that we contend can be modeled in vitro in organotypic cocultures.
Using cocultures, we have established that stromal cells significantly impact tumor proteolysis (Sameni et al., 2003). Degradation of DQ-collagen IV is increased as much as 17-fold in live 3D cocultures of stromal cells (fibroblasts or fibroblasts + macrophages) with breast or colon tumor cells (Sameni et al., 2003). Such findings are pertinent to whether protease inhibitors would be efficacious in vivo. Also relevant is that fibroblasts isolated from invasive ductal breast carcinomas, but not normal breast fibroblasts, can recruit infiltration of other stromal cells, in this case blood monocytes, into 3D spheroids (Silzle et al., 2003). Analyzing the contribution of inflammatory components to the “tumor degradome” is important as infiltration of macrophages in vivo potentiates malignant progression of tumors, for example, in a transgenic mouse model for mammary carcinoma (Gouon-Evans et al., 2002; Lin et al., 2001).
The composition and density of the extracellular matrix appears to be critical to tumor cell invasion as well as to fibril formation by collagen I. Brugge and colleagues (Seton-Rogers et al., 2004) have reported that breast epithelial cells do not invade in 3D cultures plated on undiluted rBM; however, when the basement membrane was diluted with collagen I (in this case pepsin-solubilized collagen I), the cells did invade. Such observations indicate the need to compare various matrix compositions if we are to reach any definitive conclusions about whether proteolysis is or is not required for tumor invasion in such models. Intriguing work by Weaver and colleagues suggests that increased cross-linking of collagen I promotes tumor invasion in vitro and in vivo (Levental et al., 2009). This finding seems counterintuitive to a role for proteolysis in tumor invasion, as cross-linked collagen is more resistant to proteolysis (Sabeh et al., 2009). It is the dynamics of collagen remodeling that appear to be critical (Egeblad et al., 2010), supporting a need for not only 3D models but also for studying 3D models over time, that is, in 4D.
4. Live-Cell Imaging of MAME Models: A Screening Tool for Drug Discovery
There is substantial evidence that 3D cultures are predictive of the resistance of tumor cells to cytotoxic therapy and can be used to identify targets and validate potential therapeutic agents (Li et al., 2008, 2010; Nam et al., 2010). We hypothesize that functional imaging of proteolysis by live cells in 3D and 4D can be used as an in vitro screen for testing alternative strategies to target the malignant phenotype of tumor cells, using as a readout proteolysis of extracellular matrix proteins. Cell-based 3D models have already been proposed for use in high-throughput screening of drugs (Schmeichel and Bissell, 2003) as well as their use for analyzing dynamic interactions between tumor cells and cellular and noncellular constituents of their microenvironment (Ng and Brugge, 2009). Therefore, we contend that the dimension of time needs to be part of high-throughput screening, including screening of therapeutic strategies to reduce protease activity, whether those strategies are ones that directly impact activity such as protease inhibitors or ones that target upstream effectors of protease activity.
We have developed a robust preclinical in vitro 3D/4D model to recapitulate paracrine interactions between tumor cells and other cells that comprise the tumor microenvironment. We have named these models MAME for mammary architecture and microenvironment engineering (Sameni et al., 2009). Our MAME models are designed to closely mimic the architecture of normal breast tissue, a need strongly advocated by Weigelt and Bissell (2008), as they provide a readily adaptable system through which to determine the contribution of individual cell types of the tumor microenvironment to the aggressive phenotype of breast cancers. In 4D (3D + time), MAME models in conjunction with live-cell imaging techniques are allowing us to determine the timing as well as the respective contributions of various cell types, proteolytic pathways, signaling pathways, etc., to progression from a preinvasive to an invasive phenotype.
Our MAME tripartite cocultures (Figs. 10.2 and 10.3A) consist of a bottom layer of interstitial type I collagen in which we embed breast fibroblasts, a second layer of rBM on which we plate normal breast epithelial cells, premalignant breast epithelial cells or breast carcinoma cells and a top layer of 2% rBM. Thus, the second and top layers are based on the rBM overlay cultures used by the Brugge laboratory (Debnath and Brugge, 2005; Debnath et al., 2003; Shaw et al., 2004). In order to study proteolysis, we incorporate DQ-collagen I in the collagen I layer and DQ-collagen IV in the second layer of rBM. Other iterations of the MAME models consist of only the second and top layers of rBM in which DQ-collagen IV is incorporated (Fig. 10.3B). The tripartite cocultures recapitulate tumor–tumor microenvironment interactions that occur in vivo in human breast tumors as a result of indirect interactions. With the tripartite model we have observed over time the migration of fibroblasts toward the tumor cells, eventually infiltrating into the tumor structures over a period of 7 days. The breast tumor cells also migrate towards the lower layer of fibroblasts, but do so more slowly over a period of 3 weeks. Proteolysis is associated with the migrating tumor cells and fibroblasts, in this case pericellular fluorescent degradation products. In contrast, there is extensive diffuse fluorescence associated with the bottom layer of fibroblasts, indicative of the high protease activity produced by fibroblasts. To date, we have maintained the tripartite MAME models for as long as 24 days and imaged live cultures at intervals over that period. If we use preinvasive breast epithelial cells in the MAME models, we are able to image the progression of those cells to an invasive phenotype, accompanied by an increase in proteolysis. Furthermore, we have demonstrated the ability of a variety of antagonists to reduce the invasive phenotype and the proteolysis (Jedeszko et al., 2009; Sameni et al., 2009). We have used comparable models for analysis of the invasive phenotype of a variety of cancers, including inflammatory breast cancer (Victor et al., in press), and prostate cancer (Hayward and Sloane, unpublished data). Recently, we have used MAME models to demonstrate that pericellular proteolysis is increased by incubating the cultures at a slightly acidic pH comparable to that found in tumors in vivo (Rothberg and Sloane, unpublished data).
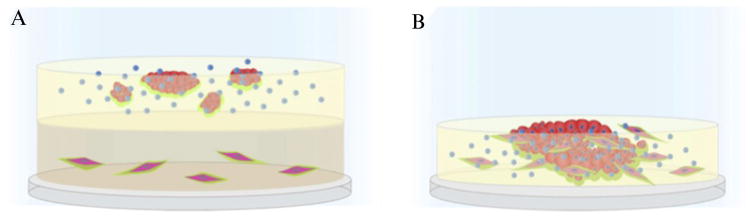
Figure 10.3.
Schematic of MAME tripartite and mixed cocultures of tumor cells, fibroblasts, and macrophages. (A) Coverslips are coated with collagen I containing DQ-collagen I™ and fibroblasts (elongated red cells). A second layer of rBM containing DQ-collagen IV™ is added and tumor cells (round red cells) plated on top along with macrophages (blue). The cultures are then overlaid with a third layer of 2% rBM, which also is included in subsequent changes of media. (B) Coverslips are coated with rBM containing DQ-collagen IV™ and a mixture of fibroblasts, tumor cells, and macrophages plated on top. Cocultures are imaged live to follow changes in morphogenesis and collagen degradation, depicted here in green.
Through live-cell imaging of MAME models, we are able to both image and quantify the cleavage products of the DQ-collagens (Jedeszko et al., 2008; Sameni et al., 2009). We are of course limited to localizing and quantifying cleavage of the labeled collagens and therefore our findings need to be considered in that context. Despite this limitation, however, we can visualize proteolysis associated with migration of individual cells and cellular structures and invasive protrusions from those structures (Jedeszko et al., 2009) and to do so over long time periods. In the case of individual endothelial cells migrating to form tube-like structures, we have imaged proteolysis over a 20-h period (Cavallo-Medved et al., 2009). That these labeled collagens may be more easily degraded than native collagens is another caveat that we must consider. Nonetheless, the labeled collagens do allow us to monitor protease activity and in combination with other protease probes such as those discussed above should allow us to identify proteases that participate in progression to an invasive phenotype and proteases that may be read-outs for therapeutic strategies to abrogate that progression.
We are now adapting a WaferGen SmartSlide Microincubation System for real-time monitoring of our MAME models. The system was designed for monolayer culture of cells in six-well plates on the stage of an inverted confocal microscope. Each well can be individually perfused and effluents individually collected for immunochemical and biochemical analysis of secreted proteins without disturbing the integrity of the cultures. Furthermore, we can acquire optical sections of the six wells sequentially at multiple time points. In addition, we can harvest media for immunochemical and biochemical analyses at times corresponding to changes in aggressive phenotype. We will be able to image and assay conditioned media from six cultures at once and at multiple time points, thus increasing the throughput of our live-cell imaging assays of MAME models. Although we are primarily interested in using MAME models for studying proteolysis, the tripartite and other modifications of the MAME models along with microincubation systems such as the Wafergen provide an experimental system in which one can test various cellular and noncellular aspects of the tumor microenvironment as it affects the progression of human breast cancer and other cancers.
Acknowledgments
The research described in this paper and the confocal facility in which the imaging was performed were supported by the National Cancer Institute, the National Center for Research Resources and the National Institute of Child Health and Human Development of the National Institutes of Health, a Department of Defense Breast Cancer Center of Excellence and the Avon Foundation.
References
- Adjei AA, Christian M, Ivy P. Novel designs and end points for phase II clinical trials. Clin Cancer Res. 2009;15:1866–1872. doi: 10.1158/1078-0432.CCR-08-2035. [DOI] [PubMed] [Google Scholar]
- Almholt K, Johnsen M. Stromal cell involvement in cancer. Recent Results Cancer Res. 2003;162:31–42. doi: 10.1007/978-3-642-59349-9_3. [DOI] [PubMed] [Google Scholar]
- Andrews PD, Harper IS, Swedlow JR. To 5D and beyond: Quantitative fluorescence microscopy in the postgenomic era. Traffic. 2002;3:29–36. doi: 10.1034/j.1600-0854.2002.30105.x. [DOI] [PubMed] [Google Scholar]
- Ang MK, Tan SB, Lim WT. Phase II clinical trials in oncology: Are we hitting the target? Expert Rev Anticancer Ther. 2010;10:427–438. doi: 10.1586/era.09.178. [DOI] [PubMed] [Google Scholar]
- Aumailley M, Gayraud B. Structure and biological activity of the extracellular matrix. J Mol Med (Berl) 1998;76:253–265. doi: 10.1007/s001090050215. [DOI] [PubMed] [Google Scholar]
- Balbin M, Fueyo A, Tester AM, Pendas AM, Pitiot AS, Astudillo A, Overall CM, Shapiro SD, Lopez-Otin C. Loss of collagenase-2 confers increased skin tumor susceptibility to male mice. Nat Genet. 2003;35:252–257. doi: 10.1038/ng1249. [DOI] [PubMed] [Google Scholar]
- Bamba S, Andoh A, Yasui H, Araki Y, Bamba T, Fujiyama Y. Matrix metalloproteinase-3 secretion from human colonic subepithelial myofibroblasts: Role of interleukin-17. J Gastroenterol. 2003;38:548–554. doi: 10.1007/s00535-002-1101-8. [DOI] [PubMed] [Google Scholar]
- Blum G, Mullins SR, Keren K, Fonovic M, Jedeszko C, Rice MJ, Sloane BF, Bogyo M. Dynamic imaging of protease activity with fluorescently quenched activity-based probes. Nat Chem Biol. 2005;1:203–209. doi: 10.1038/nchembio728. [DOI] [PubMed] [Google Scholar]
- Bogenrieder T, Herlyn M. Axis of evil: Molecular mechanisms of cancer metastasis. Oncogene. 2003;22:6524–6536. doi: 10.1038/sj.onc.1206757. [DOI] [PubMed] [Google Scholar]
- Brzostowski JA, Meckel T, Hong J, Chen A, Jin T. Imaging protein-protein interactions by Forster resonance energy transfer (FRET) microscopy in live cells. Curr Protoc Protein Sci. 2009;Chapter 19 doi: 10.1002/0471140864.ps1905s56. Unit19 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campo E, Munoz J, Miquel R, Palacin A, Cardesa A, Sloane BF, Emmert-Buck MR. Cathepsin B expression in colorectal carcinomas correlates with tumor progression and shortened patient survival. Am J Pathol. 1994;145:301–309. [PMC free article] [PubMed] [Google Scholar]
- Cavallo-Medved D, Sloane BF. Cell-surface cathepsin B: Understanding its functional significance. Curr Top Dev Biol. 2003;54:313–341. doi: 10.1016/s0070-2153(03)54013-3. [DOI] [PubMed] [Google Scholar]
- Cavallo-Medved D, Rudy D, Blum G, Bogyo M, Caglic D, Sloane BF. Live-cell imaging demonstrates extracellular matrix degradation in association with active cathepsin B in caveolae of endothelial cells during tube formation. Exp Cell Res. 2009;315:1234–1246. doi: 10.1016/j.yexcr.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau I, Rigg A, Cunningham D. Matrix metalloproteinase inhibitors—an emphasis on gastrointestinal malignancies. Crit Rev Oncol Hematol. 2003;45:151–176. doi: 10.1016/s1040-8428(02)00015-x. [DOI] [PubMed] [Google Scholar]
- Chen N, Debnath J. Autophagy and tumorigenesis. FEBS Lett. 2010;584:1427–1435. doi: 10.1016/j.febslet.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choong PF, Nadesapillai AP. Urokinase plasminogen activator system: A multifunctional role in tumor progression and metastasis. Clin Orthop Relat Res. 2003;(Suppl. 415):S46–S58. doi: 10.1097/01.blo.0000093845.72468.bd. [DOI] [PubMed] [Google Scholar]
- Coussens LM, Werb Z. Inflammatory cells and cancer: Think different! J Exp Med. 2001;193:F23–F26. doi: 10.1084/jem.193.6.f23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coussens LM, Tinkle CL, Hanahan D, Werb Z. MMP-9 supplied by bone marrow-derived cells contributes to skin carcinogenesis. Cell. 2000;103:481–490. doi: 10.1016/s0092-8674(00)00139-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coussens LM, Fingleton B, Matrisian LM. Matrix metalloproteinase inhibitors and cancer: Trials and tribulations. Science. 2002;295:2387–2392. doi: 10.1126/science.1067100. [DOI] [PubMed] [Google Scholar]
- Debnath J, Brugge JS. Modelling glandular epithelial cancers in three-dimensional cultures. Nat Rev Cancer. 2005;5:675–688. doi: 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- Debnath J, Mills KR, Collins NL, Reginato MJ, Muthuswamy SK, Brugge JS. The role of apoptosis in creating and maintaining luminal space within normal and oncogene-expressing mammary acini. Cell. 2002;111:29–40. doi: 10.1016/s0092-8674(02)01001-2. [DOI] [PubMed] [Google Scholar]
- Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- DeClerck YA. Interactions between tumour cells and stromal cells and proteolytic modification of the extracellular matrix by metalloproteinases in cancer. Eur J Cancer. 2000;36:1258–1268. doi: 10.1016/s0959-8049(00)00094-0. [DOI] [PubMed] [Google Scholar]
- DeClerck YA, Laug WE. Cooperation between matrix metalloproteinases and the plasminogen activator-plasmin system in tumor progression. Enzyme Protein. 1996;49:72–84. doi: 10.1159/000468617. [DOI] [PubMed] [Google Scholar]
- Demchik LL, Sameni M, Nelson K, Mikkelsen T, Sloane BF. Cathepsin B and glioma invasion. Int J Dev Neurosci. 1999;17:483–494. doi: 10.1016/s0736-5748(99)00011-8. [DOI] [PubMed] [Google Scholar]
- Edwards DR, Hoyer-Hansen G, Blasi F, Sloane BF. The Cancer Degradome—Proteases and Cancer Biology. Springer; New York: 2008. p. 896. [Google Scholar]
- Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer. 2002;2:161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- Egeblad M, Rasch MG, Weaver VM. Dynamic interplay between the collagen scaffold and tumor evolution. Curr Opin Cell Biol. 2010;22:697–706. doi: 10.1016/j.ceb.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellerbroek SM, Hudson LG, Stack MS. Proteinase requirements of epidermal growth factor-induced ovarian cancer cell invasion. Int J Cancer. 1998;78:331–337. doi: 10.1002/(SICI)1097-0215(19981029)78:3<331::AID-IJC13>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Ellerbroek SM, Wu YI, Overall CM, Stack MS. Functional interplay between type I collagen and cell surface matrix metalloproteinase activity. J Biol Chem. 2001;276:24833–24842. doi: 10.1074/jbc.M005631200. [DOI] [PubMed] [Google Scholar]
- Falco CN, Dykstra KM, Yates BP, Berget PB. scFv-based fluorogen activating proteins and variable domain inhibitors as fluorescent biosensor platforms. Biotechnol J. 2009;4:1328–1336. doi: 10.1002/biot.200900075. [DOI] [PubMed] [Google Scholar]
- Fernandez PL, Farre X, Nadal A, Fernandez E, Peiro N, Sloane BF, Shi GP, Chapman HA, Campo E, Cardesa A. Expression of cathepsins B and S in the progression of prostate carcinoma. Int J Cancer. 2001;95:51–55. doi: 10.1002/1097-0215(20010120)95:1<51::aid-ijc1009>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Filippov S, Caras I, Murray R, Matrisian LM, Chapman HA, Jr, Shapiro S, Weiss SJ. Matrilysin-dependent elastolysis by human macrophages. J Exp Med. 2003;198:925–935. doi: 10.1084/jem.20030626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonovic M, Bogyo M. Activity based probes for proteases: Applications to biomarker discovery, molecular imaging and drug screening. Curr Pharm Des. 2007;13:253–261. doi: 10.2174/138161207779313623. [DOI] [PubMed] [Google Scholar]
- Fuchs SY. The role of ubiquitin-proteasome pathway in oncogenic signaling. Cancer Biol Ther. 2002;1:337–341. [PubMed] [Google Scholar]
- Gocheva V, Wang HW, Gadea BB, Shree T, Hunter KE, Garfall AL, Berman T, Joyce JA. IL-4 induces cathepsin protease activity in tumor-associated macrophages to promote cancer growth and invasion. Genes Dev. 2010;24:241–255. doi: 10.1101/gad.1874010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouon-Evans V, Lin EY, Pollard JW. Requirement of macrophages and eosinophils and their cytokines/chemokines for mammary gland development. Breast Cancer Res. 2002;4:155–164. doi: 10.1186/bcr441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum D, Baruch A, Hayrapetian L, Darula Z, Burlingame A, Medzihradszky KF, Bogyo M. Chemical approaches for functionally probing the proteome. Mol Cell Proteomics. 2002a;1:60–68. doi: 10.1074/mcp.t100003-mcp200. [DOI] [PubMed] [Google Scholar]
- Greenbaum DC, Arnold WD, Lu F, Hayrapetian L, Baruch A, Krumrine J, Toba S, Chehade K, Bromme D, Kuntz ID, Bogyo M. Small molecule affinity fingerprinting. A tool for enzyme family subclassification, target identification, and inhibitor design. Chem Biol. 2002b;9:1085–1094. doi: 10.1016/s1074-5521(02)00238-7. [DOI] [PubMed] [Google Scholar]
- Gudjonsson T, Ronnov-Jessen L, Villadsen R, Rank F, Bissell MJ, Petersen OW. Normal and tumor-derived myoepithelial cells differ in their ability to interact with luminal breast epithelial cells for polarity and basement membrane deposition. J Cell Sci. 2002;115:39–50. doi: 10.1242/jcs.115.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudjonsson T, Ronnov-Jessen L, Villadsen R, Bissell MJ, Petersen OW. To create the correct microenvironment: Three-dimensional heterotypic collagen assays for human breast epithelial morphogenesis and neoplasia. Methods. 2003;30:247–255. doi: 10.1016/s1046-2023(03)00031-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Boyd PJ, Colgan S, Madri JA, Haas TL. Transcriptional up-regulation of endothelial cell matrix metalloproteinase-2 in response to extracellular cues involves GATA-2. J Biol Chem. 2003;278:47785–47791. doi: 10.1074/jbc.M309482200. [DOI] [PubMed] [Google Scholar]
- Henriet P, Zhong ZD, Brooks PC, Weinberg KI, DeClerck YA. Contact with fibrillar collagen inhibits melanoma cell proliferation by up-regulating p27KIP1. Proc Natl Acad Sci USA. 2000;97:10026–10031. doi: 10.1073/pnas.170290997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hojilla CV, Mohammed FF, Khokha R. Matrix metalloproteinases and their tissue inhibitors direct cell fate during cancer development. Br J Cancer. 2003;89:1817–1821. doi: 10.1038/sj.bjc.6601327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotary KB, Allen ED, Brooks PC, Datta NS, Long MW, Weiss SJ. Membrane type I matrix metalloproteinase usurps tumor growth control imposed by the three-dimensional extracellular matrix. Cell. 2003;114:33–45. doi: 10.1016/s0092-8674(03)00513-0. [DOI] [PubMed] [Google Scholar]
- Jedeszko C, Sameni M, Olive MB, Moin K, Sloane BF. Visualizing protease activity in living cells: From two dimensions to four dimensions. Curr Protoc Cell Biol. 2008;Chapter 4 doi: 10.1002/0471143030.cb0420s39. Unit 4 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedeszko C, Victor BC, Podgorski I, Sloane BF. Fibroblast hepatocyte growth factor promotes invasion of human mammary ductal carcinoma in situ. Cancer Res. 2009;69:9148–9155. doi: 10.1158/0008-5472.CAN-09-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsen M, Lund LR, Romer J, Almholt K, Dano K. Cancer invasion and tissue remodeling: Common themes in proteolytic matrix degradation. Curr Opin Cell Biol. 1998;10:667–671. doi: 10.1016/s0955-0674(98)80044-6. [DOI] [PubMed] [Google Scholar]
- Joyce JA, Baruch A, Chehade K, Meyer-Morse N, Giraudo E, Tsai FY, Greenbaum DC, Hager JH, Bogyo M, Hanahan D. Cathepsin cysteine proteases are effectors of invasive growth and angiogenesis during multistage tumorigenesis. Cancer Cell. 2004;5:443–453. doi: 10.1016/s1535-6108(04)00111-4. [DOI] [PubMed] [Google Scholar]
- Keppler D, Sameni M, Moin K, Mikkelsen T, Diglio CA, Sloane BF. Tumor progression and angiogenesis: Cathepsin B & Co. Biochem Cell Biol. 1996;74:799–810. doi: 10.1139/o96-086. [DOI] [PubMed] [Google Scholar]
- Kim J, Yu W, Kovalski K, Ossowski L. Requirement for specific proteases in cancer cell intravasation as revealed by a novel semiquantitative PCR-based assay. Cell. 1998;94:353–362. doi: 10.1016/s0092-8674(00)81478-6. [DOI] [PubMed] [Google Scholar]
- Kjoller L, Engelholm LH, Hoyer-Hansen M, Dano K, Bugge TH, Behrendt N. uPARAP/endo180 directs lysosomal delivery and degradation of collagen IV. Exp Cell Res. 2004;293:106–116. doi: 10.1016/j.yexcr.2003.10.008. [DOI] [PubMed] [Google Scholar]
- Krol J, Kopitz C, Kirschenhofer A, Schmitt M, Magdolen U, Kruger A, Magdolen V. Inhibition of intraperitoneal tumor growth of human ovarian cancer cells by bi- and trifunctional inhibitors of tumor-associated proteolytic systems. Biol Chem. 2003;384:1097–1102. doi: 10.1515/BC.2003.122. [DOI] [PubMed] [Google Scholar]
- Lee GY, Kenny PA, Lee EH, Bissell MJ. Three-dimensional culture models of normal and malignant breast epithelial cells. Nat Methods. 2007;4:359–365. doi: 10.1038/nmeth1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, Fong SF, Csiszar K, Giaccia A, Weninger W, Yamauchi M, Gasser DL, et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WP, Anderson CJ. Imaging matrix metalloproteinase expression in tumors. Q J Nucl Med. 2003;47:201–208. [PubMed] [Google Scholar]
- Li S, Chow LH, Pickering JG. Cell surface-bound collagenase-1 and focal substrate degradation stimulate the rear release of motile vascular smooth muscle cells. J Biol Chem. 2000;275:35384–35392. doi: 10.1074/jbc.M005139200. [DOI] [PubMed] [Google Scholar]
- Li Q, Mullins SR, Sloane BF, Mattingly RR. p21-Activated kinase 1 coordinates aberrant cell survival and pericellular proteolysis in a three-dimensional culture model for premalignant progression of human breast cancer. Neoplasia. 2008;10:314–329. doi: 10.1593/neo.07970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Chow AB, Mattingly RR. Three-dimensional overlay culture models of human breast cancer reveal a critical sensitivity to mitogen-activated protein kinase kinase inhibitors. J Pharmacol Exp Ther. 2010;332:821–828. doi: 10.1124/jpet.109.160390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin EY, Nguyen AV, Russell RG, Pollard JW. Colony-stimulating factor 1 promotes progression of mammary tumors to malignancy. J Exp Med. 2001;193:727–740. doi: 10.1084/jem.193.6.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liotta LA, Kohn EC. The microenvironment of the tumour-host interface. Nature. 2001;411:375–379. doi: 10.1038/35077241. [DOI] [PubMed] [Google Scholar]
- Lopez-Otin C, Matrisian LM. Emerging roles of proteases in tumour suppression. Nat Rev Cancer. 2007;7:800–808. doi: 10.1038/nrc2228. [DOI] [PubMed] [Google Scholar]
- Lopez-Otin C, Palavalli LH, Samuels Y. Protective roles of matrix metalloproteinases: From mouse models to human cancer. Cell Cycle. 2009;8:3657–3662. doi: 10.4161/cc.8.22.9956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen DH, Ingvarsen S, Jurgensen HJ, Melander MC, Kjoller L, Moyer A, Honore C, Madsen CA, Garred P, Burgdorf S, Bugge TH, Behrendt N, et al. The non-phagocytic route for collagen uptake: A distinct degradation pathway. J Biol Chem. 2011;286(30):26996–27010. doi: 10.1074/jbc.M110.208033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankoff DA, O'Sullivan F, Barlow WE, Krohn KA. Molecular imaging research in the outcomes era: Measuring outcomes for individualized cancer therapy. Acad Radiol. 2007;14:398–405. doi: 10.1016/j.acra.2007.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marten K, Bremer C, Khazaie K, Sameni M, Sloane B, Tung CH, Weissleder R. Detection of dysplastic intestinal adenomas using enzyme sensing molecular beacons. Gastroenterology. 2002;122(2):406–414. doi: 10.1053/gast.2002.30990. [DOI] [PubMed] [Google Scholar]
- McIntyre JO, Matrisian LM. Molecular imaging of proteolytic activity in cancer. J Cell Biochem. 2003;90:1087–1097. doi: 10.1002/jcb.10713. [DOI] [PubMed] [Google Scholar]
- McIntyre JO, Matrisian LM. Optical proteolytic beacons for in vivo detection of matrix metalloproteinase activity. Methods Mol Biol. 2009;539:155–174. doi: 10.1007/978-1-60327-003-8_9. [DOI] [PubMed] [Google Scholar]
- McIntyre JO, Fingleton B, Wells KS, Piston DW, Lynch CC, Gautam S, Matrisian LM. Development of a novel fluorogenic proteolytic beacon for in vivo detection and imaging of tumour-associated matrix metalloproteinase-7 activity. Biochem J. 2004;377:617–628. doi: 10.1042/BJ20030582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre JO, Scherer RL, Matrisian LM. Near-infrared optical proteo-lytic beacons for in vivo imaging of matrix metalloproteinase activity. Methods Mol Biol. 2010;622:279–304. doi: 10.1007/978-1-60327-299-5_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKerrow JH, Bhargava V, Hansell E, Huling S, Kuwahara T, Matley M, Coussens L, Warren R. A functional proteomics screen of proteases in colorectal carcinoma. Mol Med. 2000;6:450–460. [PMC free article] [PubMed] [Google Scholar]
- Muehlenweg B, Assfalg-Machleidt I, Parrado SG, Burgle M, Creutzburg S, Schmitt M, Auerswald EA, Machleidt W, Magdolen V. A novel type of bifunctional inhibitor directed against proteolytic activity and receptor/ligand interaction. Cystatin with a urokinase receptor binding site. J Biol Chem. 2000;275:33562–33566. doi: 10.1074/jbc.C000383200. [DOI] [PubMed] [Google Scholar]
- Nam JM, Onodera Y, Bissell MJ, Park CC. Breast cancer cells in three-dimensional culture display an enhanced radioresponse after coordinate targeting of integrin alpha5beta1 and fibronectin. Cancer Res. 2010;70:5238–5248. doi: 10.1158/0008-5472.CAN-09-2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng MR, Brugge JS. A stiff blow from the stroma: Collagen crosslinking drives tumor progression. Cancer Cell. 2009;16:455–457. doi: 10.1016/j.ccr.2009.11.013. [DOI] [PubMed] [Google Scholar]
- Nielsen BS, Rank F, Illemann M, Lund LR, Dano K. Stromal cells associated with early invasive foci in human mammary ductal carcinoma in situ coexpress urokinase and urokinase receptor. Int J Cancer. 2007;120:2086–2095. doi: 10.1002/ijc.22340. [DOI] [PubMed] [Google Scholar]
- Nozaki S, Sissons S, Chien DS, Sledge GW., Jr Activity of biphenyl matrix metalloproteinase inhibitor BAY 12–9566 in a human breast cancer orthotopic model. Clin Exp Metastasis. 2003;20:407–412. doi: 10.1023/a:1025473709656. [DOI] [PubMed] [Google Scholar]
- Offersen BV, Nielsen BS, Hoyer-Hansen G, Rank F, Hamilton-Dutoit S, Overgaard J, Andreasen PA. The myofibroblast is the predominant plasminogen activator inhibitor-1-expressing cell type in human breast carcinomas. Am J Pathol. 2003;163:1887–1899. doi: 10.1016/S0002-9440(10)63547-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podgorski I, Sloane BF. Cathepsin B and its role(s) in cancer progression. Biochem Soc Symp. 2003;70:263–276. doi: 10.1042/bss0700263. [DOI] [PubMed] [Google Scholar]
- Podgorski I, Linebaugh BE, Koblinski JE, Rudy DL, Herroon MK, Olive MB, Sloane BF. Bone marrow-derived cathepsin K cleaves SPARC in bone metastasis. Am J Pathol. 2009;175:1255–1269. doi: 10.2353/ajpath.2009.080906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punturieri A, Filippov S, Allen E, Caras I, Murray R, Reddy V, Weiss SJ. Regulation of elastinolytic cysteine proteinase activity in normal and cathepsin K-deficient human macrophages. J Exp Med. 2000;192:789–799. doi: 10.1084/jem.192.6.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-DeSimone N, Hahn-Dantona E, Sipley J, Nagase H, French DL, Quigley JP. Activation of matrix metalloproteinase-9 (MMP-9) via a converging plasmin/stromelysin-1 cascade enhances tumor cell invasion. J Biol Chem. 1999;274:13066–13076. doi: 10.1074/jbc.274.19.13066. [DOI] [PubMed] [Google Scholar]
- Reddy VY, Zhang QY, Weiss SJ. Pericellular mobilization of the tissue-destructive cysteine proteinases, cathepsins B, L, and S, by human monocyte-derived macrophages. Proc Natl Acad Sci USA. 1995;92:3849–3853. doi: 10.1073/pnas.92.9.3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee JS, Diaz R, Korets L, Hodgson JG, Coussens LM. TIMP-1 alters susceptibility to carcinogenesis. Cancer Res. 2004;64:952–961. doi: 10.1158/0008-5472.can-03-2445. [DOI] [PubMed] [Google Scholar]
- Roshy S, Sloane BF, Moin K. Pericellular cathepsin B and malignant progression. Cancer Metastasis Rev. 2003;22:271–286. doi: 10.1023/a:1023007717757. [DOI] [PubMed] [Google Scholar]
- Roy S, Debnath J. Autophagy and tumorigenesis. Semin Immunopathol. 2010;32:383–396. doi: 10.1007/s00281-010-0213-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeh F, Shimizu-Hirota R, Weiss SJ. Protease-dependent versus -independent cancer cell invasion programs: Three-dimensional amoeboid movement revisited. J Cell Biol. 2009;185:11–19. doi: 10.1083/jcb.200807195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sameni M, Moin K, Sloane BF. Imaging proteolysis by living human breast cancer cells. Neoplasia. 2000;2:496–504. doi: 10.1038/sj.neo.7900116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sameni M, Dosescu J, Sloane BF. Imaging proteolysis by living human glioma cells. Biol Chem. 2001;382:785–788. doi: 10.1515/BC.2001.094. [DOI] [PubMed] [Google Scholar]
- Sameni M, Dosescu J, Moin K, Sloane BF. Functional imaging of proteolysis: Stromal and inflammatory cells increase tumor proteolysis. Mol Imaging. 2003;2:159–175. doi: 10.1162/15353500200303136. [DOI] [PubMed] [Google Scholar]
- Sameni M, Dosescu J, Yamada KM, Sloane BF, Cavallo-Medved D. Functional live-cell imaging demonstrates that beta1-integrin promotes type IV collagen degradation by breast and prostate cancer cells. Mol Imaging. 2008;7:199–213. [PMC free article] [PubMed] [Google Scholar]
- Sameni M, Cavallo-Medved D, Dosescu J, Jedeszko C, Moin K, Mullins SR, Olive MB, Rudy D, Sloane BF. Imaging and quantifying the dynamics of tumor-associated proteolysis. Clin Exp Metastasis. 2009;26:299–309. doi: 10.1007/s10585-008-9218-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer RL, VanSaun MN, McIntyre JO, Matrisian LM. Optical imaging of matrix metalloproteinase-7 activity in vivo using a proteolytic nanobeacon. Mol Imaging. 2008;7:118–131. [PMC free article] [PubMed] [Google Scholar]
- Schmeichel KL, Bissell MJ. Modeling tissue-specific signaling and organ function in three dimensions. J Cell Sci. 2003;116:2377–2388. doi: 10.1242/jcs.00503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seton-Rogers SE, Lu Y, Hines LM, Koundinya M, LaBaer J, Muthuswamy SK, Brugge JS. Cooperation of the ErbB2 receptor and transforming growth factor beta in induction of migration and invasion in mammary epithelial cells. Proc Natl Acad Sci USA. 2004;101:1257–1262. doi: 10.1073/pnas.0308090100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour L, Ivy SP, Sargent D, Spriggs D, Baker L, Rubinstein L, Ratain MJ, Le Blanc M, Stewart D, Crowley J, Groshen S, Humphrey JS, et al. The design of phase II clinical trials testing cancer therapeutics: Consensus recommendations from the clinical trial design task force of the national cancer institute investigational drug steering committee. Clin Cancer Res. 2010;16:1764–1769. doi: 10.1158/1078-0432.CCR-09-3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw KR, Wrobel CN, Brugge JS. Use of three-dimensional basement membrane cultures to model oncogene-induced changes in mammary epithelial morphogenesis. J Mammary Gland Biol Neoplasia. 2004;9:297–310. doi: 10.1007/s10911-004-1402-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silzle T, Kreutz M, Dobler MA, Brockhoff G, Knuechel R, Kunz-Schughart LA. Tumor-associated fibroblasts recruit blood monocytes into tumor tissue. Eur J Immunol. 2003;33:1311–1320. doi: 10.1002/eji.200323057. [DOI] [PubMed] [Google Scholar]
- Sloane BF. Suicidal tumor proteases. Nat Biotechnol. 1996;14:826–827. doi: 10.1038/nbt0796-826b. [DOI] [PubMed] [Google Scholar]
- Sloane BF, Sameni M, Podgorski I, Cavallo-Medved D, Moin K. Functional imaging of tumor proteolysis. Annu Rev Pharmacol Toxicol. 2006;46:301–315. doi: 10.1146/annurev.pharmtox.45.120403.095853. [DOI] [PubMed] [Google Scholar]
- Swedlow JR, Platani M. Live cell imaging using wide-field microscopy and deconvolution. Cell Struct Funct. 2002;27:335–341. doi: 10.1247/csf.27.335. [DOI] [PubMed] [Google Scholar]
- van Kempen LC, Coussens LM. MMP9 potentiates pulmonary metastasis formation. Cancer Cell. 2002;2:251–252. doi: 10.1016/s1535-6108(02)00157-5. [DOI] [PubMed] [Google Scholar]
- Vasiljeva O, Papazoglou A, Kruger A, Brodoefel H, Korovin M, Deussing J, Augustin N, Nielsen BS, Almholt K, Bogyo M, Peters C, Reinheckel T. Tumor cell-derived and macrophage-derived cathepsin B promotes progression and lung metastasis of mammary cancer. Cancer Res. 2006;66:5242–5250. doi: 10.1158/0008-5472.CAN-05-4463. [DOI] [PubMed] [Google Scholar]
- Victor BC, Anbalagan A, Mohamed MM, Sloane BF, Cavallo-Medved D. Breast Cancer Res. doi: 10.1186/bcr3058. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadsworth SJ, Atsuta R, McIntyre JO, Hackett TL, Singhera GK, Dorscheid DR. IL-13 and TH2 cytokine exposure triggers matrix metalloproteinase 7-mediated Fas ligand cleavage from bronchial epithelial cells. J Allergy Clin Immunol. 2010;126(366–374):e361–e368. doi: 10.1016/j.jaci.2010.05.015. [DOI] [PubMed] [Google Scholar]
- Weigelt B, Bissell MJ. Unraveling the microenvironmental influences on the normal mammary gland and breast cancer. Semin Cancer Biol. 2008;18:311–321. doi: 10.1016/j.semcancer.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf K, Alexander S, Schacht V, Coussens LM, von Andrian UH, van Rheenen J, Deryugina E, Friedl P. Collagen-based cell migration models in vitro and in vivo. Semin Cell Dev Biol. 2009;20:931–941. doi: 10.1016/j.semcdb.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Byrne MH, Stacey A, Goldring MB, Birkhead JR, Jaenisch R, Krane SM. Generation of collagenase-resistant collagen by site-directed mutagenesis of murine pro alpha 1(I) collagen gene. Proc Natl Acad Sci USA. 1990;87:5888–5892. doi: 10.1073/pnas.87.15.5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S, Sloane BF. Molecular regulation of human cathepsin B: Implication in pathologies. Biol Chem. 2003;384:845–854. doi: 10.1515/BC.2003.095. [DOI] [PubMed] [Google Scholar]